Abstract
Autophagy is a catabolic membrane-trafficking mechanism involved in cell maintenance and development. Most components of autophagy also function in the cytoplasm to vacuole targeting (Cvt) pathway, a constitutive biosynthetic pathway required for the transport of aminopeptidase I (Ape1). The protein components of autophagy and the Cvt pathway include a putative complex composed of Apg1 kinase and several interacting proteins that are specific for either the Cvt pathway or autophagy. A second required complex includes a phosphatidylinositol (PtdIns) 3-kinase and associated proteins that are involved in its activation and localization. The majority of proteins required for the Cvt and autophagy pathways localize to a perivacuolar pre-autophagosomal structure. We show that the Cvt13 and Cvt20 proteins are required for transport of precursor Ape1 through the Cvt pathway. Both proteins contain phox homology domains that bind PtdIns(3)P and are necessary for membrane localization to the pre-autophagosomal structure. Functional phox homology domains are required for Cvt pathway function. Cvt13 and Cvt20 interact with each other and with an autophagy-specific protein, Apg17, that interacts with Apg1 kinase. These results provide the first functional connection between the Apg1 and PtdIns 3-kinase complexes. The data suggest a role for PtdIns(3)P in the Cvt pathway and demonstrate that this lipid is required at the pre-autophagosomal structure.
Autophagy (Apg)1 and the cytoplasm to vacuole targeting (Cvt) pathway are distinct membrane trafficking processes that share overlapping mechanistic components. During vegetative conditions, these components are constitutively employed in a biosynthetic capacity by the Cvt pathway. However, under nutrient starvation conditions, intracellular signaling events induce the autophagic pathway, enabling the degradative recycling of proteins and organelles in the lysosome-like vacuole of yeast (1–4). Efficient regulatory mechanisms are necessary to balance these biosynthetic and degradative activities and maintain cellular homeostasis.
The majority of cvt mutants is allelic with apg and aut mutants that are defective in autophagy (1, 2). The characterization of their gene products has begun to delineate the molecular events of the Cvt and autophagy pathways (reviewed in Refs. 5 and 6). There is significant overlap between the targeting and transport mechanisms of these pathways; however, there also exist proteins specific for each. Interestingly, the specific components appear to interact directly or indirectly with a central component, Apg1. Apg1 is a Ser/Thr protein kinase, which is required for both the Apg and Cvt pathways (7–9). The substrate of Apg1 kinase has not been identified, although Apg1 is known to be involved in autophosphorylation (7). Apg1 catalytic activity is presumably regulated through the association of specific proteins such as Apg13, another component required for both the Cvt pathway and autophagy (8–10). Apg13 interacts with Vac8, a Cvt-specific protein initially characterized as functioning in vacuolar inheritance (9, 11). The Apg1 kinase also physically interacts with Cvt9 and Apg17, proteins that are specific for the Cvt and Apg pathways, respectively (8, 12).
A second complex that is required for both the Cvt and autophagy pathways includes the yeast PtdIns 3-kinase Vps34. This lipid kinase functions as part of a core complex, consisting of Vps34, Vps15, and Vps30/Apg6. Either Vps38, which is required for Prc1 transport, or Apg14, which is required for the autophagy and Cvt pathways, are also part of the complex (13). The association of Vps38 or Apg14 appears to confer specificity, perhaps by targeting the PtdIns 3-kinase to its functional location. Both the Apg1 kinase and the Apg14-containing lipid kinase complex localize to perivacuolar punctate structures. At present, a direct connection between these complexes has not been established.
To gain a better understanding of the molecular mechanisms involved in the Cvt and autophagy pathways, we carried out a systematic analysis of a yeast ORF deletion library. Our screen for strains defective for import of precursor Ape1 (prApe1) identified several previously uncharacterized mutants. Two of these mutants, yjl036w/snx4 and ydl113c, renamed cvt13 and cvt20, respectively, correspond to genes that encode proteins reported to interact with Apg17 by the two-hybrid assay (14). In addition, both proteins contain phox homology (PX) domains. The PX domain is an ~120 amino acid domain that functions as a phosphoinositide-binding module. Protein-lipid interactions enable the PX domain specifically to target proteins to organelle membranes. Through regulation of intracellular localization, the PX domain has been implicated in a wide variety of cellular processes including cell growth, intracellular signaling, cytoskeletal organization, neutrophil defense, protein transport, and vesicular trafficking (reviewed in Refs. 15 and 16). A recent analysis extended phosphoinositide binding to all 15 PX domains encoded by the Saccharomyces cerevisiae genome (17). In most cases, however, a connection has not yet been made between the PX domain and physiological function.
In this paper we show that Cvt13 and Cvt20 interact with a component of the Apg1 kinase complex and are specifically required for the Cvt pathway. The PX domains within Cvt13 and Cvt20 drive PtdIns(3)P-specific recruitment to a perivacuolar structure. Mutations in the PX domain block function. Finally, we demonstrate that correct localization of Cvt13 and Cvt20 requires the synthesis of PtdIns(3)P by the Apg14-containing lipid kinase complex at the pre-autophagosomal structure.
EXPERIMENTAL PROCEDURES
Strains and Media
The S. cerevisiae knockout library in strains BY4742 and BY4743 was purchased from ResGen™ (Invitrogen). The strains used in this study are listed in Table I. The cvt13Δ, cvt20Δ, and apg17Δ strains were generated by PCR-mediated disruption of the YJL036w, YDL113c, and YLR423c loci, respectively, using amplified sequences from either the S. cerevisiae TRP1 gene, the Saccharomyces kluyveri HIS3 gene, or the Escherichia coli kanr gene (18) flanked by sequences homologous to the CVT13, CVT20, or APG17 coding sequences. All primer sequences will be made available upon request. PCR-based integration of YFP at the 3′ end of CVT13 and CVT20 was used to generate strains expressing fusion protein under control of the native CVT13 or CVT20 promoter. The template for integration of YFP was pDH5 (19).
Table I.
S. cerevisiae strains used in this study
| Strain | Genotype | Ref. |
|---|---|---|
| SEY6210 | MATα leu2–3,112 ura3–52 his3-Δ200 trp1-Δ901 lys2–801 suc2-Δ9 GAL | 45 |
| TN124 | MATa leu2–3,112 trp1 ura3–52 pho8::pho8Δ60 pho13Δ::LEU2 | 35 |
| BY4742 | MATα his3Δ leu2Δ lys2Δ ura3Δ | ResGen |
| cvtl3Δ | BY4742 cvt13Δ::KAN | ResGen |
| cvt20Δ | BY4742 cvt20Δ::KAN | ResGen |
| apg17Δ | BY4742 apg17Δ::KAN | ResGen |
| apg9Δ | BY4742 apg9Δ::KAN | ResGen |
| fab1Δ | BY4742 fab1Δ::KAN | ResGen |
| vps34Δ | SEY6210 vps34Δ::TRP1 | 41 |
| NNY20 | MATa leu2–3,112 trp1 ura3–52 apg1Δ::LEU2 | 7 |
| D3Y108 | SEY6210 cvt13Δ::HIS3 S.k. | This study |
| D3Y109 | SEY6210 cvt20Δ::HIS3 S.k. | This study |
| D3Y111 | TN124 cvt13Δ::TRP1 | This study |
| D3Y112 | TN124 cvt20Δ::TRP1 | This study |
| D3Y113 | TN124 apg17Δ::TRP1 | This study |
| PSY10 | D3Y108 cvt20Δ::TRP1 | This study |
| PSY13 | D3Y112 cvt13Δ::KAN | This study |
| PSY14 | SEY6210 cvt13-YFP::HIS5 S.p. | This study |
| PSY15 | SEY6210 cvt20-YFP::HIS5 S.p. | This study |
| WPHYD7 | SEY6210 aut7Δ::LEU2 | 25 |
| PSY24 | WPHYD7 cvt13-YFP::HIS5 S.p. | This study |
| PSY25 | WPHYD7 cvt20-YFP::HIS5 S.p. | This study |
| KTY1003 | SEY6210 apg14Δ::LEU2 | This study |
| PSY26 | KTY1003 cvt13-YFP::HIS5 S.p. | This study |
| PSY27 | KTY1003 cvt20-YFP::HIS5 S.p. | This study |
Media for growth of yeast strains, starvation, and induction of peroxisomes were described previously (20).
Materials and Antiserum
The preparation of antisera to Ape1 (21), Fox3 (22), Prc1, and Pho8 (23) were described previously. Antibodies to GST and the hemagglutinin (HA) and c-Myc epitopes were from Santa Cruz Biotechnology (Santa Cruz, CA). Anti-mouse horseradish peroxidase conjugate was from Zymed Laboratories Inc. (South San Francisco, CA). Antiserum to Pgk1 and Apg13 was generously provided by Dr. Jeremy Thorner (University of California, Berkeley) and Dr. Yoshinori Ohsumi (National Institute for Basic Biology, Okazaki, Japan), respectively. Other reagents were identical to those described previously (20, 24, 25).
Plasmid Constructions
A carboxyl-terminal fusion of HA to Cvt13 (pCVT13-HA(426)) was made by PCR amplification of the CVT13 ORF and upstream sequence from genomic DNA and ligation into pRS426–3xHA. Additional details of the plasmid constructions will be provided upon request. To construct a carboxyl-terminal fusion of GFP to Cvt13, GFP was excised from pRS416GFP (26) using EagI and XhoI. After EagI, but prior to XhoI digestion, the 5′ overhang was filled in, creating a blunt end on GFP. Plasmid pCVT13-HA was digested with SmaI and XhoI to remove the DNA encoding the HA epitope and subsequently ligated with GFP, yielding pCVT13-GFP(426).
Amino-terminal fusions of protein A to Cvt13 (pProtA-CVT13(424)), Cvt20 (pProtA-CVT20(424)), and Apg17 (pProtA-APG17(424)) were made by PCR amplification of the corresponding ORFs and ligation into the ClaI/SalI sites of pRS424-CuProtA (27).
The amino-terminal GFP fusion plasmids pGFP-CVT20(416), pGFP-CVT20(415) (pPS108), and pGFP-APG17(426) were made by excising CVT20 or APG17 from pProtA-CVT20 or pProtA-APG17 with ClaI and SalI and inserting into the ClaI/SalI sites of pCuGFP(416) or pCuGFP(426), respectively (25).
To construct amino-terminal HA fusion plasmids, the 3xHA epitope was PCR-amplified from pRS416–3xHA. The PCR product was digested with SpeI and ClaI. Plasmid pProtA-CVT20 was digested with SpeI and ClaI to remove the DNA encoding protein A and subsequently ligated with the above PCR product, yielding pHA-Cvt20(416,424).
DNA encoding the PX domain of Cvt20 was amplified by PCR with primers containing 5′ restriction enzyme sites that allow for in-frame gene fusions with the carboxyl terminus of GST. Amplification included the DNA encoding the PX domain of Cvt13 (amino acids 156–295) and Cvt20 (amino acids 173–301). PCR product and plasmid pGEX-KG (Amersham Biosciences) were digested with the appropriate restriction enzymes and ligated to generate plasmid pGEX-KG-CVT13PX and pGEX-KG-CVT20PX.
Plasmids pCvt13Y79A-GFP(416), pCvt13Y79A-HA(416), pGFP-Cvt20Y193A(416), and pGFP-Cvt20Y193A(415) (pPS109) were constructed using the QuikChange™ Site-directed Mutagenesis Kit (Stratagene). Plasmids pCu-APG13(426) (9), pCVT19-CFP(414), pCuCFP-CVT9(424), and pCuCFP-AUT7(424) (27) were described previously.
Pulse/Chase, Nitrogen Starvation, and Pexophagy
Pulse/chase immunoprecipitation, nitrogen starvation, and pexophagy experiments were conducted as described previously (2, 22, 28).
Alkaline Phosphatase Enzyme Assay
Induction of autophagy was estimated by measuring the activity of alkaline phosphatase from Pho8Δ60 with p-nitrophenyl phosphate as substrate essentially according to methods published previously (29). Protein concentrations were determined using the BCA protein assay (Pierce).
Subcellular Fractionation
For subcellular fractionation experiments cvt13Δ (D3Y108) cells transformed with the pCVT13-HA(416) plasmid or cvt20Δ (D3Y109) cells transformed with the pHA-CVT20(426) plasmid were grown to midlog phase in SMD and then converted to spheroplasts as described previously (30). Spheroplasts were subjected to differential lysis in PS200 lysis buffer (20 mm PIPES, pH 6.8, 200 mm sorbitol) containing 5 mm MgCl2 essentially as described previously (25). Fractionated samples were subjected to immunoblot analysis.
Protein A Affinity Isolation Analyses
For isolation of protein A fusion proteins and associated proteins, 30 A600 equivalents of yeast cells were converted to spheroplasts as described previously (30). Spheroplasts were washed in phosphate-buffered saline, pH 7.4, 1 m sorbitol, and 1 mm phenylmethylsulfonyl fluoride. Spheroplasts were lysed on ice in phosphate-buffered saline lysis buffer, and protein A was recovered on Dynabeads® as described previously (27). Bound protein was eluted with sample buffer, resolved by SDS-PAGE, and visualized by immunoblotting.
Fluorescence Microscopy
Cells were grown to midlog phase in SMD, induced with 10 μm CuSO4 where appropriate for 1–2 h, and viewed on a Nikon E-800 fluorescent microscope as described previously (27). FM 4-64 labeling was as described previously (12).
Protein Purification and Protein-Lipid Overlay Assays
GST-Cvt20 PX domain fusion protein was purified from glutathione-Sepharose beads by elution with reduced glutathione according to manufacturer's specifications. Purified protein was then desalted with PD-10 columns and concentrated to 5 μg/ml with Centriprep YM-30 centrifugal filters. Protein-lipid overlay assays were conducted as described previously (24, 31). Membranes were incubated with purified GST-Cvt13 and GST-Cvt20 PX domains at 0.05 μg/ml overnight at 4 °C. Binding of GST fusions to phosphoinositides was detected by enhanced chemiluminescence. No binding of GST alone to phosphoinositides was detected.
RESULTS
cvt13Δ and cvt20Δ Cells Are Specifically Defective in the Cvt Pathway
In order to develop a comprehensive understanding of the protein components involved in cytoplasm to vacuole protein transport pathways, we undertook a systematic analysis of the yeast ORF deletion library. Each strain was initially screened for the accumulation of precursor aminopeptidase I (prApe1). The vacuolar hydrolase, aminopeptidase I (Ape1), is synthesized in the cytosol as a precursor protein and is transported to the vacuole through the Cvt or Apg pathway (reviewed in Refs. 5 and 6).
Two of the mutants that we identified were cvt13 (YJL036w/snx4) and cvt20 (YDL113c). Both mutants accumulated the precursor form of Ape1 in rich medium, indicating a block in the Cvt pathway (Fig. 1A). The cvt13 mutant was initially isolated in a random screen for mutants defective in prApe1 maturation (1), but the corresponding gene had not been identified. A yeast genome database search for sequences with similarity to human sorting nexins (SNXs) identified a number of genes, including the open reading frame YJL036w (32). The sequence similarity of YJL036w with human SNX4 grouped it as the yeast ortholog SceSNX4. However, due to its functional placement within the Cvt transport pathway, YJL036w was renamed CVT13. Sequencing of the YJL036w gene from the cvt13 mutant revealed that deletion of the guanine base at position 113 of the ORF changed amino acid 38 and induced a frameshift.2 No function for the CVT20 gene product has been shown previously in prApe1 transport.
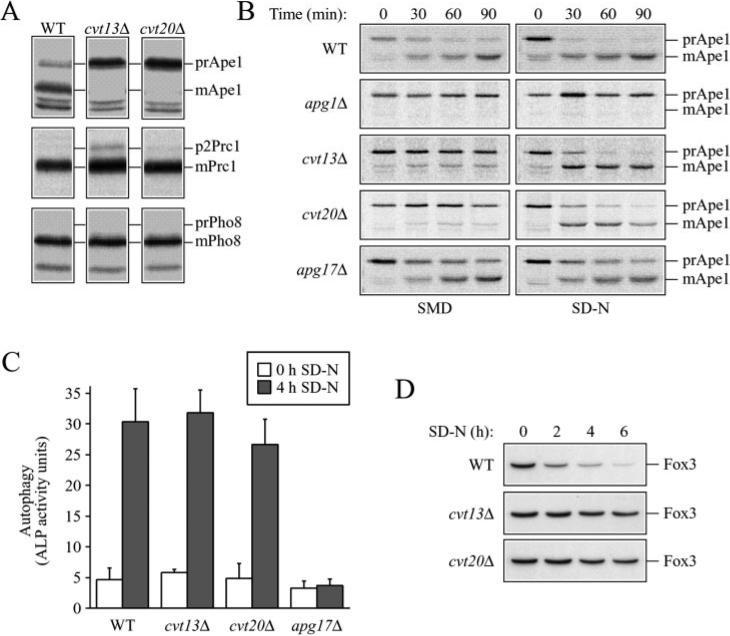
Fig. 1. The cvt13 and cvt20 mutants are specifically defective in the Cvt pathway.
A, processing of vacuolar hydrolases. Cells from wild type (WT) (SEY6210), cvt13Δ, and cvt20Δ strains were pulse-labeled for 10 min and chased for either 90 (Ape1) or 30 min (Prc1 and Pho8). Ape1, Prc1, and Pho8 were immunoprecipitated from cell lysates and resolved by SDS-PAGE. Prc1 and Pho8 processing was essentially normal in both mutant strains. B, kinetics of transport by the Cvt pathway. Cells from wild type (SEY6210), apg1Δ (NNY20), cvt13Δ (D3Y108), cvt20Δ (D3Y109), and apg17Δ strains were pulse-labeled for 10 min in SMD, washed, and resuspended in either SMD or SD-N medium, and chased for the indicated times. Ape1 was immunoprecipitated from cell lysates and resolved by SDS-PAGE. Note that there is a background band that migrates just below the position of mApe1. C, Pho8 activity assay for autophagy. Wild type (TN124), cvt13Δ (D3Y111), cvt20Δ (D3Y112), and apg17Δ (D3Y113) cells expressing Pho8Δ60 were shifted from nutrient-rich medium (white bars) to SD-N medium (black bars) for 4 h. Autophagy induction was determined by a Pho8 activity assay as described under “Experimental Procedures.” Error bars represent the standard deviation from three separate experiments. The cvt13 and cvt20 mutants are not defective in autophagic induction. D, pexophagy analysis. Cells from wild type (BY4742) and the cvt13Δ and cvt20Δ mutant strains in the BY4742 background were grown under conditions that induce peroxisomes (see “Experimental Procedures”), washed, and resuspended in SD-N for the time indicated. The presence of the peroxisomal thiolase enzyme, Fox3, was detected by immunoblotting. The S. cerevisiae strains cvt13Δ and cvt20Δ are defective in pexophagy.
To determine whether these proteins functioned in additional vacuolar transport pathways, we examined the processing of resident vacuolar hydrolases carboxypeptidase Y (Prc1) and alkaline phosphatase (Pho8). Prc1 and Pho8 transit to the vacuole through the CPY and ALP pathways, respectively, and are useful marker proteins to monitor the state of these transport processes (33). Both Prc1 and Pho8 undergo glycosylation in the endoplasmic reticulum and Golgi complex and are proteolytically processed in the vacuole. The molecular mass shifts that correspond to these modifications provide a convenient means to monitor the transport process. Yeast cells were subjected to a radioactive pulse/chase analysis to determine the kinetics of protein delivery to the vacuole. Wild type, cvt13Δ, and cvt20Δ cells efficiently processed both proteins, although the cvt13Δ strain displayed a weak defect in processing of Prc1 and accumulated ~5% of the protein in the p2 Golgi-modified form (Fig. 1A). These results suggest that the Cvt13 and Cvt20 proteins do not have major functions in Prc1 and Pho8 vacuolar delivery pathways.
Precursor Ape1 is transported to the vacuole through both the Cvt pathway and autophagy (2). Most of the characterized apg/cvt mutants are defective in delivery through both pathways and accumulate prApe1 under both rich and starvation conditions (reviewed in Ref. 34). Some mutants, however, are specific to each pathway. For example, cvt9Δ and vac8Δ strains are blocked in the Cvt pathway but are not completely blocked for autophagic import. These mutants are able to “reverse” their prApe1 accumulation defect through starvation-induced autophagy (9, 12). In wild type cells, prApe1 is matured with a half-time of ~30 min in synthetic minimal medium containing nitrogen (SMD). In contrast, very little mature Ape1 was detected in cvt13Δ and cvt20Δ cells under these conditions (Fig. 1B). The cvt13 and cvt20 mutants reversed the accumulation defect and processed prApe1 when shifted to SD-N, nitrogen starvation conditions (Fig. 1B). As a control, we examined the apg1Δ mutant. Apg1 is a Ser/Thr kinase essential for both Cvt transport and autophagy (7–9). As expected, apg1Δ cells did not import prApe1 under either condition.
High throughput two-hybrid analysis indicated a possible three-way interaction between Cvt13, Cvt20, and Apg17 (8, 14). In contrast to cvt9Δ and vac8Δ strains, the apg17Δ mutant is not defective for prApe1 import through the Cvt pathway but is defective for autophagy (8, 9, 12). We found that apg17Δ cells processed prApe1, albeit with a slightly lower efficiency (Fig. 1B). It is thought that starvation conditions result in a signal transduction event that induces autophagy and causes the Apg/Cvt machinery to switch from the production of Cvt vesicles to the formation of autophagosomes. As a result, precursor Ape1 is selectively transported to the vacuole by autophagosomes under starvation conditions (3). Even though autophagy is defective in the apg17Δ mutant (8), we found that prApe1 processing occurred under starvation conditions in apg17Δ cells (Fig. 1B). This result suggests either that transport through the Cvt pathway normally continues even when the autophagic pathway is induced or that Apg17 is necessary for the negative regulation of the Cvt pathway.
The data for the apg17Δ strain indicate that the ability to process prApe1 under starvation conditions is not a valid measure of autophagic function. Accordingly, we utilized an alternative approach to assess further the autophagic capacity of the cvt13Δ and cvt20Δ mutant strains. Pho8Δ60 is a soluble derivative of the integral membrane vacuolar protein Pho8 (alkaline phosphatase) that lacks the transmembrane domain. Pho8Δ60 is only delivered to the vacuole through the autophagy pathway (35). Upon vacuole delivery it is matured into its active form. Cells from wild type (TN124) and the cvt13Δ, cvt20Δ, and apg17Δ mutant strains expressing the Pho8Δ60 protein were measured for alkaline phosphatase activity following a shift to starvation conditions. Comparable with wild type cells, alkaline phosphatase enzymatic activity in cvt13Δ and cvt20Δ cells increased significantly after shifting to SD-N (Fig. 1C), indicating that Pho8Δ60 delivery and subsequent processing to its enzymatically active form occurred in the vacuole. By contrast, no increase in enzyme activity was detected upon shift to SD-N in the autophagy mutant apg17Δ, in agreement with previous studies (8). We also examined a cvt13Δ cvt20Δ double mutant strain to determine whether the autophagic capacity of the cvt13Δ and cvt20Δ strains was due to functional overlap between these two proteins. Alkaline phosphatase enzymatic activity in the double mutant strain was comparable with wild type (data not shown), suggesting that Cvt13 and Cvt20 are not essential for autophagy. Yeast mutants defective for autophagy are unable to recycle cytosolic material and thus die rapidly upon shifting to starvation conditions. When cvt13Δ and cvt20Δ cells were shifted to SD-N, their viability was comparable with wild type cells (data not shown), further indicating that autophagy is not defective in these strains.
Peroxisomes are induced under specific nutritional conditions such as growth in oleic acid as the sole carbon source. When cells grown on oleic acid are shifted to glucose-containing medium, peroxisomes are specifically degraded by a process termed pexophagy. The molecular machinery involved in pexophagy has been shown to overlap with that of the autophagy and Cvt pathways (reviewed in Ref. 34). We examined the degradation of the peroxisomal enzyme Fox3 (thiolase) to determine the requirement of Cvt13 and Cvt20 for the specific uptake of peroxisomes by pexophagy. Cells were grown in an oleic acid-containing medium to induce peroxisomes, switched to SD-N, and the level of Fox3 monitored by immunoblotting (22). In wild type cells, Fox3 was rapidly degraded. However, Fox3 was stable in cvt13Δ and cvt20Δ cells, indicating they are defective in the uptake of peroxisomes by pexophagy (Fig. 1D). We did detect a very slight decrease in Fox3 over the time course examined in the cvt13Δ and cvt20Δ strains, but the level was clearly stabilized relative to the wild type strain. With the exception of Cvt19, all cvt and apg mutants characterized thus far display defects in degradation of peroxisomes by pexophagy (9, 22).
Both Cvt13 and Cvt20 Distribute between the Cytosol and Punctate Structures near the Vacuole
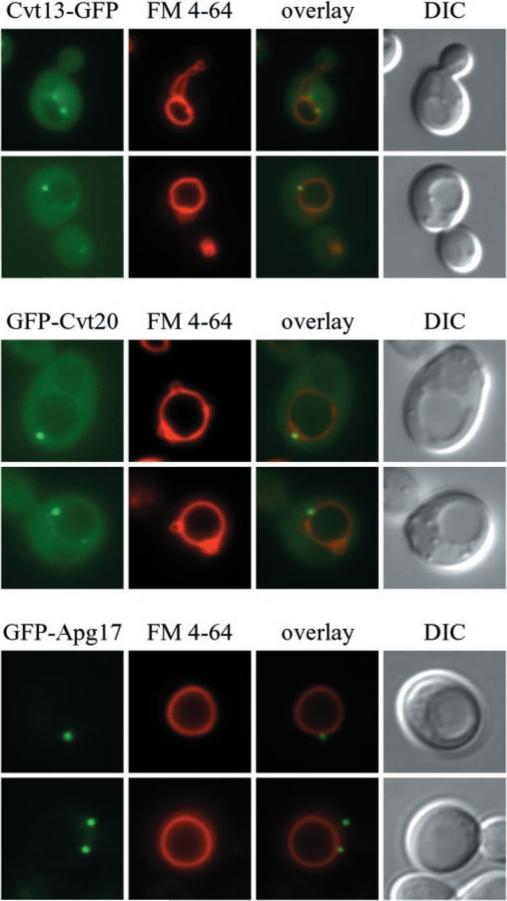
To gain insight into the function of Cvt13 and Cvt20, we examined the localization of these proteins in vivo through the use of GFP fusion proteins. Expression of Cvt13-GFP under the endogenous CVT13 promoter fully complemented the prApe1 transport defect in a cvt13Δ mutant, indicating that it was a functional chimera (data not shown). GFP-Cvt20 and GFP-Apg17, both under control of the CUP1 promoter, were induced with a relatively low concentration of CuSO4 (10 μm) for 1–2 h. GFP-Cvt20 expression fully complemented the prApe1 defect in a cvt20Δ mutant (data not shown). After labeling the vacuoles with the dye FM 4-64, cells expressing the various GFP constructs were examined by fluorescence microscopy. Cvt13-GFP and GFP-Cvt20 displayed both a diffuse cytosolic distribution and punctate structures near or on the surface of FM 4-64-labeled vacuoles (Fig. 2). Additional faint perivacuolar and vacuolar rim staining was also detectable. GFP-Apg17 displayed punctate structures similar to those seen with Cvt13-GFP and GFP-Cvt20, but no cytosolic staining was evident. GFP-Apg17 punctate structures were found adjacent to the vacuole, however, in contrast to Cvt13-GFP and GFP-Cvt20, they were also seen in other areas of the cell (Fig. 2).
Fig. 2. Localization of Cvt13-GFP, GFP-Cvt20, and GFP-Apg17.
The cvt13Δ (D3Y108), cvt20Δ (D3Y109), and apg17Δ strain were transformed with plasmids encoding GFP fused to the carboxyl or amino termini of the corresponding ORFs (pCVT13-GFP, pGFP-CVT20 and pGFP-APG17, respectively). Cells were grown in SMD, treated with FM 4-64 to label vacuoles, and visualized by fluorescence microscopy as described under “Experimental Procedures.” Cvt13-GFP and GFP-Cvt20 display punctate perivacuolar dots and diffuse cytosolic staining. DIC, differential interference contrast.
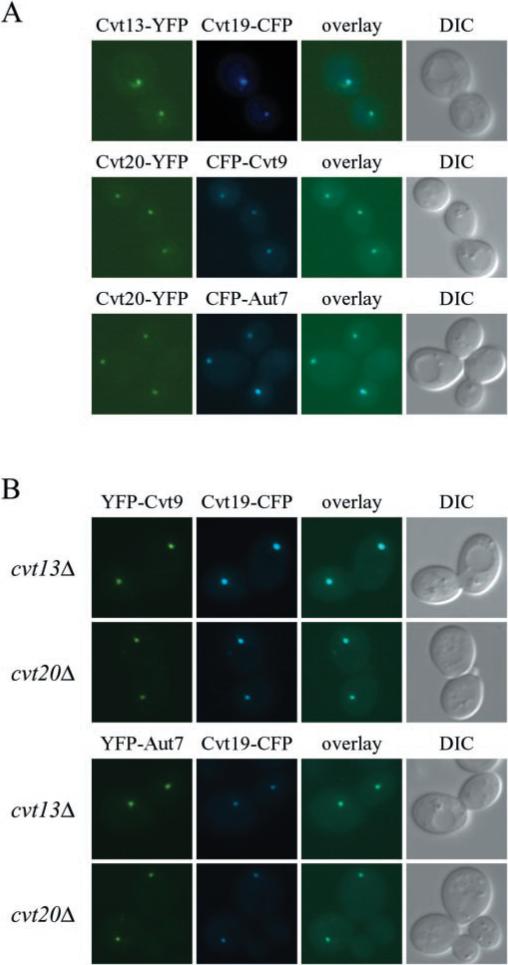
The perivacuolar staining seen with both GFP-Cvt20 and Cvt13-GFP is reminiscent of the staining pattern seen with many of the proteins that are involved in the Apg and Cvt pathways (12, 20, 25, 27). Although the nature of this perivacuolar structure is still unknown, it appears to play a physiological role in Cvt vesicle and autophagosome formation (27, 36). We co-localized Cvt13 and Cvt20 with Cvt9, Cvt19, and Aut7 to determine whether these components localize to the same perivacuolar structure. Cvt19 is a cargo receptor or adaptor required for transport of the prApe1 complex (37), and Aut7 plays a role in vesicle formation and size regulation (25, 38–40). Co-localization of Aut7, Cvt19, and Cvt9 has been demonstrated recently (27). Chromosomally expressed Cvt13-YFP or Cvt20-YFP displayed punctate structures that co-localized with Cvt19-CFP, CFP-Cvt9, and CFP-Aut7 (Fig. 3A). A number of cells expressing Cvt13-YFP and Cvt20-YFP also displayed diffuse cytosolic staining. These results indicate that a population of Cvt13 and Cvt20 localizes to the same perivacuolar structure as Cvt9, Cvt19, and Aut7.
Fig. 3. Cvt13, Cvt20, Cvt9, the cargo specificity factor Cvt19, and the vesicle component Aut7 co-localize on a punctate, perivacuolar structure.
A, cells of strains PSY14 (CVT13-YFP) and PSY15 (CVT20-YFP) were transformed with pCVT19-CFP, pCuCFP-CVT9, or pCuCFP-AUT7. Transformed cells were grown to midlog stage in SMD and shifted to YPD for 2–3 h prior to microscopy analysis. Images were captured and analyzed as described under “Experimental Procedures.” B, co-localization of Cvt19 with Cvt9 and Aut7 occurs independent of Cvt13 and Cvt20. D3Y108 (cvt13Δ) and D3Y109 (cvt20Δ) cells expressing YFP-Cvt9 or YFP-Aut7 along with Cvt19-CFP were prepared and examined as in A. DIC, differential interference contrast.
The localization of some Apg/Cvt proteins has been shown to depend on other proteins within these pathways. For example, membrane recruitment of Aut7 requires the function of the Apg12-Apg5 and the Aut1 conjugation systems (25, 40). We examined the localization of Cvt19-CFP and its co-localization with YFP-Cvt9 and YFP-Aut7 in the cvt13Δ and cvt20Δ strains. Tagged proteins were co-expressed in all combinations, and the localization of each protein appeared normal (Fig. 3B and data not shown). These data suggest that Cvt19, Cvt9, and Aut7 localize to the perivacuolar structure independent of Cvt13 and Cvt20. Furthermore, these results imply that Cvt13 and Cvt20 are not required for the functions associated with these proteins, such as Cvt19-prApe1 complex binding (37), and Aut7-dependent membrane expansion (38).
Cvt13 and Cvt20 Fractionate with Membranes
To extend the localization analysis in vitro, we examined the subcellular localization of Cvt13 and Cvt20 by differential centrifugation. The cvt13Δ and cvt20Δ strains were transformed with plasmids pCVT13-HA and pHA-CVT20, respectively, and grown to midlog phase in SMD. The expression of the respective HA epitope-tagged proteins completely complemented the prApe1 accumulation defect in the cvt13Δ and cvt20Δ strains (data not shown), indicating that the addition of the HA epitope did not alter Cvt13 and Cvt20 function. Spheroplasts were lysed and centrifuged to separate the lysates into low speed supernatant (S13) and pellet (P13) fractions (see “Experimental Procedures”). The S13 fraction was then further separated into high speed supernatant (S100) and pellet (P100) fractions. Immunoblot analysis revealed that HA-tagged Cvt13 and Cvt20 were distributed essentially equally throughout all pellet and supernatant fractions (data not shown), suggesting the presence of both cytosolic and membrane-associated protein. The distribution of the cytosolic protein, phosphoglycerate kinase (Pgk1), confirmed separation of membrane and soluble proteins.
The nature of the Cvt13 and Cvt20 pellet association was examined by treating the pellet fraction with various reagents. Treating the pellet fractions with 1 m salt, 3 m urea, or 1% Triton X-100 resulted in partial stripping of HA-tagged Cvt13 and Cvt20 into the supernatant fraction. However, both proteins were completely removed from the pellet fraction by extraction with 0.1 m Na2CO3, pH 10.5 (data not shown). These results indicate that Cvt13 and Cvt20 exist as both cytosolic and peripheral membrane proteins, and are in agreement with the in vivo fluorescent analysis that revealed both a punctate and diffuse cytosolic pool of both proteins (Fig. 2).
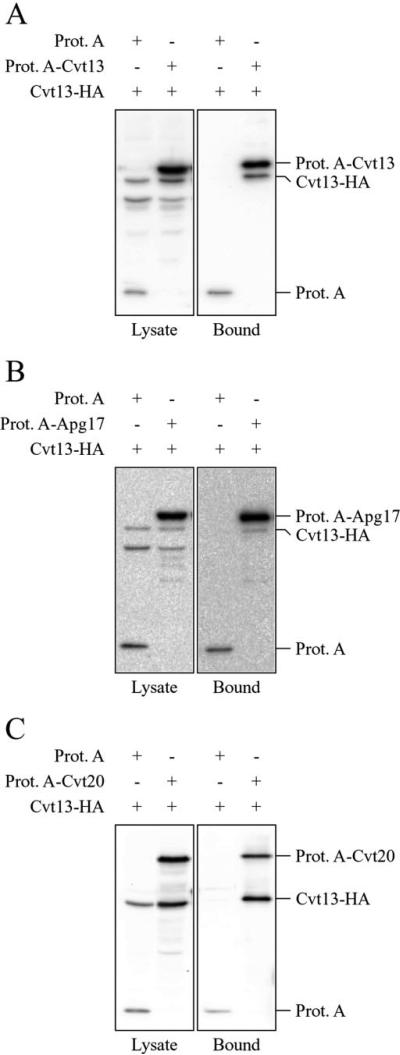
Cvt13, Cvt20, and Apg17 Interact
The analyses of GFP localization patterns along with two-hybrid studies suggested that Cvt13, Cvt20, and Apg17 may compose a protein complex (14). Previously published data have demonstrated that Apg1 physically interacts with Apg17, an autophagy-specific component, and Cvt9, a Cvt-specific component (8, 12). To investigate potential physical associations among Cvt13, Cvt20, and Apg17, a series of affinity purification experiments was undertaken with fusions to protein A. Either protein A alone or a protein A fusion was co-expressed in combination with Cvt13-HA. The expression of protein A-tagged Cvt13 or Cvt20 complemented the prApe1 accumulation defect in cvt13Δ and cvt20Δ strains, respectively (data not shown). Cells were grown to midlog phase, converted to spheroplasts, and lysed in the presence of detergent. Protein A and associated proteins were then affinity-isolated by binding to IgG-coupled Dynabeads as described under “Experimental Procedures.” The recovered proteins were separated by SDS-PAGE and examined by immunoblot. Cvt13-HA bound to protein A-Cvt13, protein A-Cvt20, and protein A-Apg17 (Fig. 4). Cvt13-HA did not bind to protein A alone, indicating that the interaction with Cvt13-HA was dependent on Cvt13, Cvt20, or Apg17 fused to protein A. These data support the existence of a protein complex with a three-way physical interaction between Cvt13, Cvt20, and Apg17. This complex may also include multiple copies of Cvt13.
Fig. 4. Cvt13 associates with Cvt20 and Apg17.
Detergent-solubilized extracts from wild type (SEY6210) cells expressing Cvt13-HA and protein A (Prot. A) and Cvt13-HA and either protein A-Cvt13 (A), protein A-Apg17 (B), or protein A-Cvt20 (C) were prepared as described under “Experimental Procedures.” Associated proteins were isolated with IgG-coated Dynabeads and visualized by immunoblotting with antiserum to HA. For each experiment ~2% of the total lysate or 10% of the total eluate was loaded per lane.
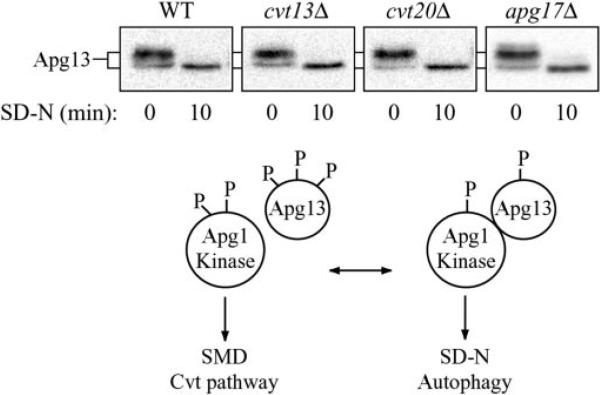
At least two proteins that interact with Apg1 modulate its kinase activity. One of these proteins, Apg13, is hyperphosphorylated in a Tor-dependent manner under rich media conditions. This form of Apg13 has a low affinity for Apg1, and under these conditions in vitro Apg1 kinase activity is reduced (8). Starvation results in dephosphorylation of Apg13, a greater affinity for Apg1, and an increase in Apg1 kinase activity in vitro. It is possible that the phosphorylation state of Apg13 is the signal that regulates the conversion between the Cvt and Apg pathways. We determined whether the phosphorylation state of Apg13 was altered in the cvt13Δ, cvt20Δ, or apg17Δ mutants. Wild type or mutant cells were grown in SMD and shifted to starvation conditions for 10 min, and Apg13 was examined by immunoblot (Fig. 5). In both wild type and mutant cells, Apg13 was seen primarily as the hyperphosphorylated form in SMD prior to shifting to starvation conditions. A smaller population of the protein was also detected as the hypophosphorylated form. Within 10 min in SD-N medium, the protein was efficiently dephosphorylated and migrated at a single position on the gel in all of the strains examined. Shifting the culture back to SMD resulted in rapid hyperphosphorylation (data not shown). These results suggest that Cvt13, Cvt20, and Apg17 do not affect the Cvt and Apg pathways by regulating the phosphorylation of Apg13.
Fig. 5. Phosphorylation state of Apg13.
Cells from wild type (WT) (SEY6210), cvt13Δ (D3Y108), cvt20Δ (D3Y109), or apg17Δ strains over-expressing Apg13 were grown in SMD and then shifted to SD-N at time 0. Samples were collected after 10 min, and Apg13 was detected by immunoblotting. Nutrient-dependent dephosphorylation of Apg13 is normal in strains cvt13Δ, cvt20Δ, and apg17Δ. A schematic representation of the Apg13 dephosphorylation and interaction with Apg1 is depicted below the blots.
The PX Domains of Cvt13 and Cvt20 Bind the Phosphoinositide PtdIns(3)P and Are Necessary for Cvt Transport
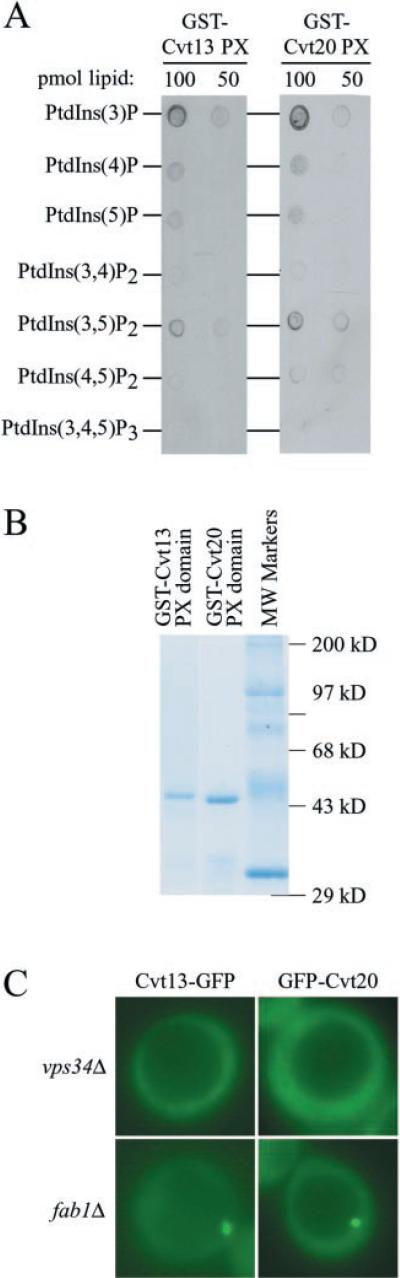
The PX domain has been identified as a novel ~120-residue domain that functions as a phosphoinositide-binding module (24). Sequence database analyses identified a number of proteins in yeast, including Cvt13 and Cvt20, that contain a PX domain (17). However, the in vivo role of the PX domain for Cvt13 and Cvt20 has not been addressed. The PX domain binding specificity of Cvt13 and Cvt20 was examined using a protein-lipid binding assay (31). Nitrocellulose membranes were spotted with decreasing amounts of various phosphoinositides and incubated with affinity-purified PX domains of Cvt13 or Cvt20 fused to glutathione S-transferase (GST) (Fig. 6, A and B). Both GST-PX domain fusions displayed a preferential interaction with PtdIns(3)P and a weaker interaction with PtdIns(3,5)P2.
Fig. 6. Specific phosphatidylinositol 3-phosphate (PtdIns(3)P) recognition by the Cvt20 PX domain.
A, protein-lipid overlay assay. Serial dilutions of phosphoinositides were spotted onto nitrocellulose membrane and incubated with purified GST-Cvt13 PX or GST-Cvt20 PX domain fusion protein. The membrane was washed and analyzed by immunoblot with antibodies to GST. B, Coomassie-stained gel of purified GST-Cvt13 PX and GST-Cvt20 PX domain fusion protein. C, PtdIns(3)P-dependent localization of Cvt13-GFP and GFP-Cvt20 by fluorescence microscopy. Cells from vps34Δ and fab1Δ strains expressing either Cvt13-GFP or GFP-Cvt20 were grown in SMD and visualized by fluorescence microscopy.
The VPS34 gene encodes the sole yeast PtdIns 3-kinase (41). Fab1 is a PtdIns(3)P 5-kinase, which phosphorylates PtdIns(3)P to produce PtdIns(3,5)P2 (42). Localization of Cvt13-GFP and GFP-Cvt20 was examined in both vps34Δ and fab1Δ strains to confirm the PtdIns(3)P specificity and determine whether membrane binding was dependent upon phosphoinositide synthesis. Cvt13-GFP and GFP-Cvt20 punctate staining completely redistributed into the cytosol in the vps34Δ strain (Fig. 6C). In contrast, fluorescence staining in the fab1Δ strain remained similar to that seen in wild type cells, with distribution between the cytosol and punctate structures. These data indicate that PtdIns(3)P binding is necessary for Cvt13 and Cvt20 membrane localization to perivacuolar punctate structures.
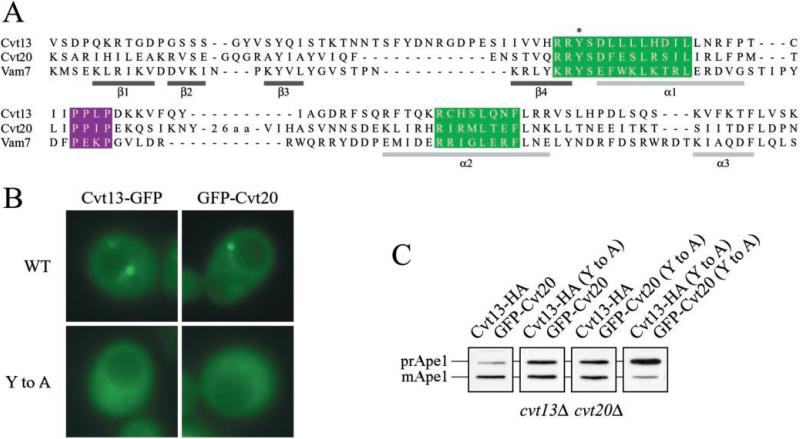
Analysis of the PX domain by NMR spectroscopy identified a basic binding pocket and membrane attachment loop as the structural basis for PtdIns(3)P binding (24). Altering a key tyrosine residue in one of two basic motifs, RR(Y/F) (where R is arginine, Y is tyrosine, and F is phenylalanine) within the binding pocket (Fig. 7A), has been shown to block phosphoinositide-binding (24, 43). A Tyr to Ala mutation of the highly conserved RRY motif was generated within the PX domain of Cvt13 and Cvt20. Cvt13Y79A-GFP and GFP-Cvt20Y193A punctate staining completely redistributed to the cytosol (Fig. 7B). Immunoblot analysis indicated that Cvt13Y79A-GFP and GFP-Cvt20Y193A were stable and corresponded to predicted molecular weights (data not shown). Thus, depleting the cell of PtdIns(3)P or mutating the PX domain abolished intracellular punctate localization of Cvt13 and Cvt20.
Fig. 7. Mutations in the PX domain of Cvt13 and Cvt20 abolish punctate localization and prApe1 trafficking.
A, alignment of PX domain sequences (17, 24). Elements of Vam7 secondary structure are shown below the sequences (24). Two highly conserved PX domain sequence motifs, (R/K)(R/K)(Y/F)XXFXXLXXXL and R(R/K)XXLXX(Y/F), are highlighted in green and the proline-rich motif, PXXP, is highlighted in purple (reviewed in Ref. 15). The Cvt13 and Cvt20 point mutations are indicated by an asterisk. B, Cvt13 and Cvt20 punctate localization requires a functional PX domain. D3Y108 (cvt13Δ) cells expressing Cvt13-GFP (WT) or Cvt13Y79A-GFP (Tyr to Ala (Y to A)) and D3Y109 (cvt20Δ) cells expressing GFP-Cvt20 (WT) or GFP-Cvt20Y193A (Tyr to Ala (Y to A)) were grown in SMD and visualized by fluorescence microscopy. C, prApe1 trafficking requires functional Cvt13 and Cvt20 PX domains. PSY10 (cvt13Δ cvt20Δ) cells expressing combinations of Cvt13-HA, GFP-Cvt20, Cvt13Y79A-HA, and GFP-Cvt20Y193A, as indicated, were grown to midlog phase in SMD. Protein extracts were prepared and analyzed by immunoblot with antiserum to Ape1.
Next, we investigated the role of Cvt13 and Cvt20 PX domains in Cvt transport. Expression of Cvt13Y79A-GFP in the cvt13Δ strain showed an incomplete block in prApe1 processing (data not shown). A similar result was seen with GFP-Cvt20Y193A expressed in the cvt20Δ background. Because Cvt13 and Cvt20 physically interact, partial complementation of prApe1 transport in these single PX domain mutants may have occurred through membrane targeting by the wild type PX domain partner. To address this possibility, we generated a cvt13Δ cvt20Δ double mutant strain expressing Cvt13-HA, Cvt13Y79A-HA, GFP-Cvt20, and GFP-Cvt20Y193A in varying combinations. When the PX domain of either Cvt13 or Cvt20 contained the Tyr to Ala mutation, prApe1 processing was reduced to ~50%. However, when both PX domains contained the Tyr to Ala mutation, prApe1 processing was almost completely blocked (Fig. 7C). Thus, Cvt13 and Cvt20 PtdIns(3)P-binding capacity is necessary for Cvt transport.
Apg14, a Component of the PtdIns 3-Kinase Complex, Is Necessary for Cvt13 and Cvt20 Punctate, Perivacuolar Localization
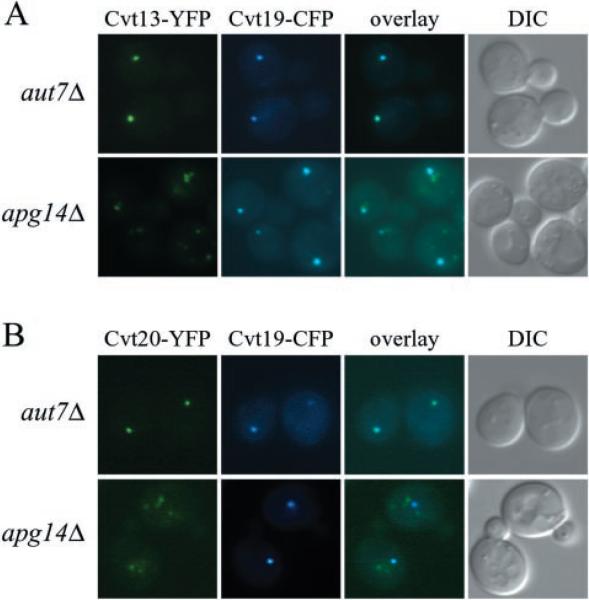
To investigate further possible Cvt13 and Cvt20 connections with the PtdIns 3-kinase complex, co-localization with Cvt19 was examined in the apg14Δ mutant strain. Apg14 is a component of the PtdIns 3-kinase complex required for the Cvt pathway and autophagy but not for the CPY pathway. As shown in Fig. 3, Cvt13-YFP and Cvt20-YFP co-localize with Cvt19-CFP in wild type cells. Similarly, Cvt19-CFP co-localization with Cvt13-YFP and Cvt20-YFP was normal in aut7Δ cells that are defective in the Cvt pathway and autophagy (Fig. 8). In contrast, punctate structures labeled with Cvt13-YFP or Cvt20-YFP were less frequent and of lower intensity in the apg14Δ strain (Fig. 8). Furthermore, in those cells that displayed punctate staining for Cvt13-YFP or Cvt20-YFP, the labeled structures seldom co-localized with Cvt19-CFP (Fig. 8). Loss of Cvt13 and Cvt20 punctate perivacuolar localization in this mutant strain occurred concomitant with an increased staining of more diffuse punctate structures near the vacuole and in the cytosol. These results suggest Apg14 may be dispensable for Cvt13 and Cvt20 PtdIns(3)P binding at other membranes that contain this lipid, such as the endosome, but necessary for recruitment to the pre-autophagosomal structure where they participate in Cvt transport. This result is in contrast to the complete loss of punctate staining seen with the vps34Δ strain that completely lacks PtdIns 3-kinase activity (Fig. 6). These data represent the first evidence of the requirement of PtdIns(3)P at the presumed site of pre-autophagosome/Cvt vesicle formation.
Fig. 8. Deletion of APG14 disrupts Cvt13 and Cvt20 co-localization with Cvt19.
The aut7Δ and apg14Δ strains expressing chromosomal Cvt13-YFP or Cvt20-YFP, PSY24, PSY25, PSY26 and PSY27, respectively, were transformed with pCVT19-CFP. Cells were prepared and examined as in Fig. 3. Contrast was enhanced for YFP in the apg14Δ strain relative to the aut7Δ strain to allow detection of the Cvt13-YFP and Cvt20-YFP signals. DIC, differential interference contrast.
DISCUSSION
Cvt13 and Cvt20 Are PX Domain Proteins That Associate with Components of the Apg1 Kinase Signaling Complex
The studies presented in this paper have defined two new proteins in the Cvt pathway, Cvt13 and Cvt20. These proteins contain PX domains that bind to PtdIns(3)P. Both Cvt13 and Cvt20 are localized to a punctate perivacuolar structure, and binding to PtdIns(3)P is critical for this localization. Finally, correct localization of both proteins is necessary for import of prApe1 through the Cvt pathway. Cvt13 is the first Cvt pathway-specific protein that has been characterized that has a mammalian homolog, Snx4 (32). This is an interesting finding because there are no data at present demonstrating that the Cvt pathway exists in any organism other than S. cerevisiae.
Autophagy and the Cvt pathway are distinct membrane trafficking processes that share overlapping mechanistic components. These components include the Apg1 kinase and its associated proteins, Apg13, Apg17, and Cvt9 (8, 12). A second set of proteins required for both pathways is the PtdIns 3-kinase complex composed of Vps34, Vps15, Apg6, and Apg14 (13). A separate PtdIns 3-kinase complex containing Vps38 instead of Apg14 is needed for protein transport through the CPY pathway. This latter complex is responsible for the synthesis of the majority of PtdIns(3)P in the cell and at present it has not been possible to assess directly the site of action of the Apg14-dependent PtdIns 3-kinase.
Cvt13 and Cvt20 were shown to interact with Apg17 by a two-hybrid screen (14). These proteins were also identified as belonging to a family containing the PX domain (17). However, neither of these analyses provided information on the physiological roles of Cvt13 or Cvt20. We identified the cvt13Δ and cvt20Δ strains in a screen of the S. cerevisiae deletion library as being defective in the processing of the vacuolar hydrolase Ape1. Both proteins are necessary for vacuole delivery of prApe1, and the mutant strains accumulated the precursor form of Ape1 in rich medium, indicating a defect in the Cvt pathway (Fig. 1). In contrast, the cvt13Δ and cvt20Δ strains were not defective in autophagy. Other vacuolar transport systems including the CPY and ALP pathways are essentially normal in cvt13Δ and cvt20Δ cells. However, both mutants are defective in the uptake of peroxisomes by pexophagy.
Consistent with two-hybrid results (14), immunoisolation experiments demonstrate that Cvt13, Cvt20, and Apg17 directly interact with each other (Fig. 4). In addition, Cvt13 forms homodimers or higher order oligomers. These data support a model in which Cvt20, Apg17, and one or more Cvt13 subunits form a protein complex. This complex may include additional components, such as Apg1, Apg13, and Vac8. Apg1 has recently been ascribed a central role in the events signaling a switch between the Cvt and autophagy pathways (8). We were unable to detect an interaction between Apg1 and either Cvt13 or Cvt20 in our affinity isolation experiments (Fig. 4). This may simply mean that Apg1 does not interact directly with either protein, and our experimental conditions did not preserve ternary or higher order complexes. Alternatively, this result may indicate the presence of multiple complexes containing discrete subsets of proteins.
Cvt13, Cvt20, Cvt9, Cvt19, and Aut7 Co-Localize on a Punctate, Perivacuolar Structure
Biochemical analyses indicate that Cvt13 and Cvt20 are membrane-associating proteins present in both a soluble and membrane fraction. In vivo analyses of GFP-tagged proteins suggest that Cvt13 and Cvt20 distribute between the cytosol and punctate structures near the vacuole (Fig. 2). Recent studies have placed Apg1 and Cvt9 at a perivacuolar compartment (12, 27). Various other components involved in the Cvt and Apg pathways including the autophagy/Cvt-specific PtdIns 3-kinase complex have also been localized to this pre-autophagosomal structure (27, 36). Fluorescence microscopy showed Cvt13 and Cvt20 co-localization with Cvt9, Cvt19, and Aut7 (Fig. 3A). This perivacuolar compartment may mark the site of vesicle nucleation and cargo sequestration and has been termed the pre-autophagosomal structure (36).
Cvt13 and Cvt20 PX Domains Are PtdIns(3)P-specific Binding Modules That Direct Recruitment to a Perivacuolar Structure and Are Necessary for Cvt Transport
PX domains are involved in the targeting of various proteins to membranes that contain PtdIns(3)P. Proteins with PX domains have been implicated in a range of cellular processes (reviewed in Ref. 15). Until now, PX domain-containing proteins have not been implicated in the Cvt pathway or autophagy. A mutation in the RR(Y/F) basic motif demonstrates that Cvt13 and Cvt20 membrane association requires a functional PX domain (Fig. 7). The presence of a PX domain in both associating proteins, Cvt13 and Cvt20, may allow for increased affinity and PtdIns(3)P-binding capacity. Similarly, the selective targeting of Cvt13 and Cvt20 to perivacuolar structures may be directed through interaction with additional proteins. The PX domains in Cvt13 and Cvt20 bind PtdIns(3)P with relatively low affinity (Fig. 6; 17) so that they likely require interaction with other components to direct membrane binding and membrane specificity. We are currently investigating the interaction of Cvt13 and Cvt20 with other Apg and Cvt proteins. Vps34, the yeast PtdIns 3-kinase, interacts with Vps15, a membrane-associated Ser/Thr kinase that regulates Vps34 activity (44). Vps15 and Vps34 are subunits of a core PtdIns 3-kinase complex that may function at different membrane sites, regulating different protein trafficking events (13). Vps15/Vps34 complexed with accessory proteins Vps30 and Vps38 may concentrate primarily at the Golgi, functioning in Prc1 targeting to the endosome/prevacuolar compartment. Vps15/Vps34 complexed with accessory proteins Vps30 and Apg14 may concentrate primarily at the pre-autophagosomal structure, functioning in Cvt and autophagy membrane traffic to the vacuole. Processing of prApe1 is completely blocked in vps15, vps30, vps34, and apg14, but not vps38 mutant strains (13). Alkaline phosphatase activity assays employing Pho8Δ60 indicate severely impaired autophagic capacity in vps15, vps30, vps34, and apg14 mutant cells in response to starvation. Thus, a functional autophagy/Cvt-specific PtdIns 3-kinase complex and PtdIns(3)P production is necessary for autophagy and Cvt transport, although the site of action of the Apg14-dependent PtdIns 3-kinase has not been demonstrated. PtdIns(3)P concentration by specific PtdIns 3-kinase complexes may target PX domain proteins to their site of function on discrete membrane domains. Indeed, Cvt13 and Cvt20 punctate perivacuolar localization was lost in apg14Δ cells (Fig. 8), suggesting that the Apg14-dependent PtdIns 3-kinase must function at the pre-autophagosomal structure.
Cvt13 and Cvt20 PtdIns(3)P binding and targeted membrane association is required for prApe1 transport through the Cvt pathway. Mutating tyrosine in the PX domain RR(Y/F) basic motif of either Cvt13 or Cvt20 resulted in diminished prApe1 processing (Fig. 7). With only one functional PX domain, the interaction of Cvt13 and Cvt20 may enable partial, reduced capacity Cvt transport. The PX domain mutation appears to abolish localization of the protein to the punctate perivacuolar structure; however, the partial function suggests that a low level of the protein may still be binding membrane. If so, this level is below detection. Alternatively, Cvt13 and Cvt20 retain sufficient activity in the absence of efficient membrane binding of either individual protein to permit partial import of prApe1. When the PX domain of both Cvt13 and Cvt20 are mutated, Cvt transport is blocked (Fig. 7). These data indicate a direct physiological function for the yeast PX domain. Furthermore, we know that a PtdIns 3-kinase complex is required for the autophagy/Cvt pathways (13). The PX domains of Cvt13 and Cvt20 function as PtdIns(3)P-binding modules; thus these data provide the first molecular connection between the autophagy/Cvt pathways and the autophagy/Cvt specific-PtdIns 3-kinase complex.
Acknowledgments
We thank Wei-Pang Huang and Dr. John Kim for valuable discussions.
Footnotes
This work was supported in part by United States Public Health Service Grants GM53396 (to D. J. K.) and CA58689 (to S. D. E.) from the National Institutes of Health. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The abbreviations used are: Apg, autophagy; Ape1, aminopeptidase I; CFP, cyan fluorescent protein; Cvt, cytoplasm to vacuole targeting; FM 4-64, N-(triethylammoniumpropyl)-4-(p-diethylaminophenylhexatrienyl) pyridinium dibromide; GFP, green fluorescent protein; ORF, open reading frame; prApe1, precursor Ape1; Pgk1, phosphoglycerate kinase; Pho8, alkaline phosphatase; Prc1, carboxypeptidase Y; PtdIns(3)P, phosphatidylinositol 3-phosphate; PX, phox homology; YFP, yellow fluorescent protein; PIPES, 1,4-piperazinediethanesulfonic acid; GST, glutathione S-transferase; HA, hemagglutinin; SNXs, sorting nexins.
Z. Xie and H. Abeliovich, unpublished results.
REFERENCES
- 1.Harding TM, Hefner-Gravink A, Thumm M, Klionsky DJ. J. Biol. Chem. 1996;271:17621–17624. doi: 10.1074/jbc.271.30.17621. [DOI] [PubMed] [Google Scholar]
- 2.Scott SV, Hefner-Gravink A, Morano KA, Noda T, Ohsumi Y, Klionsky DJ. Proc. Natl. Acad. Sci. U. S. A. 1996;93:12304–12308. doi: 10.1073/pnas.93.22.12304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baba M, Osumi M, Scott SV, Klionsky DJ, Ohsumi Y. J. Cell Biol. 1997;139:1687–1695. doi: 10.1083/jcb.139.7.1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Scott SV, Baba M, Ohsumi Y, Klionsky DJ. J. Cell Biol. 1997;138:37–44. doi: 10.1083/jcb.138.1.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stromhaug PE, Klionsky DJ. Traffic. 2001;2:524–531. doi: 10.1034/j.1600-0854.2001.20802.x. [DOI] [PubMed] [Google Scholar]
- 6.Reggiori F, Klionsky DJ. Euk. Cell. 2002;1:11–21. doi: 10.1128/EC.01.1.11-21.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Matsuura A, Tsukada M, Wada Y, Ohsumi Y. Gene (Amst.) 1997;192:245–250. doi: 10.1016/s0378-1119(97)00084-x. [DOI] [PubMed] [Google Scholar]
- 8.Kamada Y, Funakoshi T, Shintani T, Nagano K, Ohsumi M, Ohsumi Y. J. Cell Biol. 2000;150:1507–1513. doi: 10.1083/jcb.150.6.1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Scott SV, Nice DC, III, Nau JJ, Weisman LS, Kamada Y, Keizer-Gunnink I, Funakoshi T, Veenhuis M, Ohsumi Y, Klionsky DJ. J. Biol. Chem. 2000;275:25840–25849. doi: 10.1074/jbc.M002813200. [DOI] [PubMed] [Google Scholar]
- 10.Funakoshi T, Matsuura A, Noda T, Ohsumi Y. Gene (Amst.) 1997;192:207–213. doi: 10.1016/s0378-1119(97)00031-0. [DOI] [PubMed] [Google Scholar]
- 11.Wang YX, Zhao H, Harding TM, Gomes de Mesquita DS, Woldringh CL, Klionsky DJ, Munn AL, Weisman LS. Mol. Biol. Cell. 1996;7:1375–1389. doi: 10.1091/mbc.7.9.1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim J, Kamada Y, Stromhaug PE, Guan J, Hefner-Gravink A, Baba M, Scott SV, Ohsumi Y, Dunn WA, Jr., Klionsky DJ. J. Cell Biol. 2001;153:381–396. doi: 10.1083/jcb.153.2.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kihara A, Noda T, Ishihara N, Ohsumi Y. J. Cell Biol. 2001;152:519–530. doi: 10.1083/jcb.152.3.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Uetz P, Giot L, Cagney G, Mansfield TA, Judson RS, Knight JR, Lockshon D, Narayan V, Srinivasan M, Pochart P, Qureshi-Emili A, Li Y, Godwin B, Conover D, Kalbfleisch T, Vijayadamodar G, Yang M, Johnston M, Fields S, Rothberg JM. Nature. 2000;403:623–627. doi: 10.1038/35001009. [DOI] [PubMed] [Google Scholar]
- 15.Sato TK, Overduin M, Emr SD. Science. 2001;294:1881–1885. doi: 10.1126/science.1065763. [DOI] [PubMed] [Google Scholar]
- 16.Wishart MJ, Taylor GS, Dixon JE. Cell. 2001;105:817–820. doi: 10.1016/s0092-8674(01)00414-7. [DOI] [PubMed] [Google Scholar]
- 17.Yu JW, Lemmon MA. J. Biol. Chem. 2001;276:44179–44184. doi: 10.1074/jbc.M108811200. [DOI] [PubMed] [Google Scholar]
- 18.Longtine MS, McKenzie A, III, Demarini DJ, Shah NG, Wach A, Brachat A, Philippsen P, Pringle JR. Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 19.Drees BL, Sundin B, Brazeau E, Caviston JP, Chen GC, Guo W, Kozminski KG, Lau MW, Moskow JJ, Tong A, Schenkman LR, McKenzie A, III, Brennwald P, Longtine M, Bi E, Chan C, Novick P, Boone C, Pringle JR, Davis TN, Fields S, Drubin DG. J. Cell Biol. 2001;154:549–571. doi: 10.1083/jcb.200104057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang C-W, Kim J, Huang W-P, Abeliovich H, Stromhaug PE, Dunn WA, Jr., Klionsky DJ. J. Biol. Chem. 2001;276:30442–30451. doi: 10.1074/jbc.M102342200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Klionsky DJ, Cueva R, Yaver DS. J. Cell Biol. 1992;119:287–299. doi: 10.1083/jcb.119.2.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hutchins MU, Veenhuis M, Klionsky DJ. J. Cell Sci. 1999;112:4079–4087. doi: 10.1242/jcs.112.22.4079. [DOI] [PubMed] [Google Scholar]
- 23.Klionsky DJ, Emr SD. EMBO J. 1989;8:2241–2250. doi: 10.1002/j.1460-2075.1989.tb08348.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cheever ML, Sato TK, de Beer T, Kutateladze TG, Emr SD, Overduin M. Nat. Cell Biol. 2001;3:613–618. doi: 10.1038/35083000. [DOI] [PubMed] [Google Scholar]
- 25.Kim J, Huang W-P, Klionsky DJ. J. Cell Biol. 2001;152:51–64. doi: 10.1083/jcb.152.1.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim J, Dalton VM, Eggerton KP, Scott SV, Klionsky DJ. Mol. Biol. Cell. 1999;10:1337–1351. doi: 10.1091/mbc.10.5.1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim J, Huang W-P, Stromhaug PE, Klionsky DJ. J. Biol. Chem. 2002;277:763–773. doi: 10.1074/jbc.M109134200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sato TK, Darsow T, Emr SD. Mol. Cell. Biol. 1998;18:5308–5319. doi: 10.1128/mcb.18.9.5308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mitchell JK, Fonzi WA, Wilkerson J, Opheim DJ. Biochim. Biophys. Acta. 1981;657:482–494. doi: 10.1016/0005-2744(81)90333-8. [DOI] [PubMed] [Google Scholar]
- 30.Scott SV, Klionsky DJ. J. Cell Biol. 1995;131:1727–1735. doi: 10.1083/jcb.131.6.1727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dowler S, Currie RA, Downes CP, Alessi DR. Biochem. J. 1999;342:7–12. [PMC free article] [PubMed] [Google Scholar]
- 32.Haft CR, de la Luz Sierra M, Barr VA, Haft DH, Taylor SI. Mol. Cell. Biol. 1998;18:7278–7287. doi: 10.1128/mcb.18.12.7278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kim J, Scott SV, Klionsky DJ. Int. Rev. Cytol. 2000;198:153–201. doi: 10.1016/s0074-7696(00)98005-7. [DOI] [PubMed] [Google Scholar]
- 34.Kim J, Klionsky DJ. Annu. Rev. Biochem. 2000;69:303–342. doi: 10.1146/annurev.biochem.69.1.303. [DOI] [PubMed] [Google Scholar]
- 35.Noda T, Matsuura A, Wada Y, Ohsumi Y. Biochem. Biophys. Res. Commun. 1995;210:126–132. doi: 10.1006/bbrc.1995.1636. [DOI] [PubMed] [Google Scholar]
- 36.Suzuki K, Kirisako T, Kamada Y, Mizushima N, Noda T, Ohsumi Y. EMBO J. 2001;20:5971–5981. doi: 10.1093/emboj/20.21.5971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Scott SV, Guan J, Hutchins MU, Kim J, Klionsky DJ. Mol. Cell. 2001;7:1131–1141. doi: 10.1016/s1097-2765(01)00263-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Abeliovich H, Dunn WA, Jr., Kim J, Klionsky DJ. J. Cell Biol. 2000;151:1025–1034. doi: 10.1083/jcb.151.5.1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ichimura Y, Kirisako T, Takao T, Satomi Y, Shimonishi Y, Ishihara N, Mizushima N, Tanida I, Kominami E, Ohsumi M, Noda T, Ohsumi Y. Nature. 2000;408:488–492. doi: 10.1038/35044114. [DOI] [PubMed] [Google Scholar]
- 40.Kirisako T, Ichimura Y, Okada H, Kabeya Y, Mizushima N, Yoshimori T, Ohsumi M, Takao T, Noda T, Ohsumi Y. J. Cell Biol. 2000;151:263–276. doi: 10.1083/jcb.151.2.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schu PV, Takegawa K, Fry MJ, Stack JH, Waterfield MD, Emr SD. Science. 1993;260:88–91. doi: 10.1126/science.8385367. [DOI] [PubMed] [Google Scholar]
- 42.Gary JD, Wurmser AE, Bonangelino CJ, Weisman LS, Emr SD. J. Cell Biol. 1998;143:65–79. doi: 10.1083/jcb.143.1.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kanai F, Liu H, Field SJ, Akbary H, Matsuo T, Brown GE, Cantley LC, Yaffe MB. Nat. Cell Biol. 2001;3:675–678. doi: 10.1038/35083070. [DOI] [PubMed] [Google Scholar]
- 44.Stack JH, Herman PK, Schu PV, Emr SD. EMBO J. 1993;12:2195–2204. doi: 10.1002/j.1460-2075.1993.tb05867.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Robinson JS, Klionsky DJ, Banta LM, Emr SD. Mol. Cell. Biol. 1988;8:4936–4948. doi: 10.1128/mcb.8.11.4936. [DOI] [PMC free article] [PubMed] [Google Scholar]