Abstract
Under starvation conditions, the majority of intracellular degradation occurs at the lysosome or vacuole by the autophagy pathway. The cytoplasmic substrates destined for degradation are packaged inside unique double-membrane transport vesicles called autophagosomes and are targeted to the lysosome/vacuole for subsequent breakdown and recycling. Genetic analyses of yeast autophagy mutants, apg and aut, have begun to identify the molecular machinery as well as indicate a substantial overlap with the biosynthetic cytoplasm to vacuole targeting (Cvt) pathway. Transport vesicle formation is a key regulatory step of both pathways. In this study, we characterize the putative compartment from which both autophagosomes and the analogous Cvt vesicles may originate. Microscopy analyses identified a perivacuolar membrane as the resident compartment for both the Apg1-Cvt9 signaling complex, which mediates the switching between autophagic and Cvt transport, and the autophagy/Cvt-specific phosphatidylinositol 3-kinase complex. Furthermore, the perivacuolar compartment designates the initial site of membrane binding by the Apg/Cvt vesicle component Aut7, the Cvt cargo receptor Cvt19, and the Apg conjugation machinery, which functions in the de novo formation of vesicles. Biochemical isolation of the vesicle component Aut7 and density gradient analyses recapitulate the microscopy findings although also supporting the paradigm that components required for vesicle formation and packaging concentrate at subdomains within the donor membrane compartment.
Cellular homeostasis requires the regulated balance of biosynthetic and degradative activities. Autophagy functions as the major degradative pathway by which proteins and organelles are delivered to the lysosome or vacuole for breakdown and recycling. In addition to normal homeostatic function, autophagy is induced in response to certain environmental stress conditions such as serum deprivation, amino acid starvation, radiation damage (1), and in response to anti-estrogen agents (2). Autophagy-mediated degradation may be important during mammalian embryogenesis, differentiation, and aging (3, 4) and plays a critical role during type II non-apoptotic programmed cell death (reviewed in Ref. 5). Dysfunction of autophagy has been implicated in the etiology of an increasing number of genetic diseases, and cancer in particular (6). Autophagy also appears to be associated with neurodegenerative conditions such as Parkinson's and Alzheimer's diseases (7, 8). Mutations in lysosomal associated membrane protein 2 result in cardiomyopathy associated with Danon's disease, and lysosomal associated membrane protein 2-deficient mice show defects in the autophagy pathway (reviewed in Ref. 9). Therefore, understanding the basic mechanisms of autophagy may have wide therapeutic relevance.
The use of pharmacological agents in mammalian systems has identified the role of signaling components including various kinases, phosphatases, and heterotrimeric G proteins, in the regulation of autophagy (reviewed in Refs. 4 and 10). The morphology of mammalian autophagy has also been described in detail (reviewed in Ref. 11). In the past few years, a classical genetic approach in Saccharomyces cerevisiae has transformed our understanding of the autophagy pathway by identifying autophagy-specific protein machinery (reviewed in Refs. 12 and 13). Genetic screens based on starvation sensitivity or defects in the degradation of specific cytosolic proteins resulted in the isolation of the apg and aut mutants that are defective in autophagy (14, 15). In addition, these mutants overlap with mutants in the cytoplasm to vacuole targeting (Cvt)1 pathway (16, 17) that were isolated from a screen based on defects in the proteolytic processing of the resident vacuolar hydrolase aminopeptidase I (Ape1). Genetic, biochemical, and morphological studies all demonstrate that autophagy and the Cvt pathway share largely the same targeting machinery (17–20). Both pathways employ a double membrane transport vesicle (termed the autophagosome and Cvt vesicle, respectively) to sequester cytoplasmic cargo, followed by docking and fusion of the outer vesicle membrane with the vacuole to release the cargo-containing inner vesicle (the autophagic and Cvt bodies). The smaller Cvt vesicles (diameter 100–150 nm) appear to exclude bulk cytoplasm, specifically transporting precursor Ape1 (prApe1) and α-mannosidase. Starvation conditions induce the elevated expression of autophagic proteins (21, 22) to accommodate the formation of the much larger autophagosomes (diameter 300–900 nm) and the nonselective sequestration of cytoplasm in addition to resident vacuolar hydrolases (23).
The cloning and characterization of the gene products that complement the apg, aut, and cvt mutants have provided information about an increasing number of proteins required for the Cvt and autophagy pathways (reviewed in Refs. 10, 12, and 13). Mammalian orthologs for key autophagy genes have also been identified (reviewed in Refs. 12 and 13). Recent studies in yeast have begun to elucidate the molecular basis behind the mechanisms that mediate both the Cvt pathway and autophagy. However, the subcellular localization of many of the proteins that are involved in the complex membrane rearrangements that are the hallmark of these pathways has not been determined.
The least understood and most complex step of the Cvt and autophagy pathways is the formation of the sequestering vesicle. Vesicle formation in the secretory pathway has been relatively well characterized and includes a requisite cargo concentration step in which cargo receptors bind their substrates and associate with general cytosolic coat proteins at the site of vesicle budding (reviewed in Refs. 24 and 25). Similar to the formation of vesicles in the secretory pathway, many proteins appear to be required at the vesicle formation step of the Cvt and autophagy pathways. These include a cargo receptor, Cvt19, that is required for transport of resident hydrolases (26). In addition, a group of proteins involved in a novel conjugation reaction that covalently joins Apg12 to Apg5 are transiently associated with the forming vesicle (27–29). Although the exact function of Apg conjugation is not well understood, this process is required for the membrane recruitment of Aut7, a protein that plays a role in vesicle formation (22, 30–32) and expansion of the autophagosomal membrane (23). Aut7 is also unique in that it is the only characterized protein required for vesicle formation that remains associated with the completed vesicle. However, the initial site of Aut7 membrane binding, a location that may mark the origin of the membrane for the sequestering vesicle, has not been identified.
Although much progress has been made in the characterization of yeast autophagy gene products, either individually or as part of a protein complex, an overall comparative examination placing these multiple autophagy and Cvt components and complexes at their physical site(s) of function has remained an important goal. In this study, we investigate the site of function of many of the recently characterized Apg/Cvt proteins and complexes. We demonstrate that the majority of these components co-localize to a distinct perivacuolar compartment. Surprisingly, isopycnic centrifugation of the membrane fraction of the cell divided the proteins co-localizing by microscopy into two separate populations peaking at different densities, the lighter peak containing the conjugation components as well as the prApe1 receptor Cvt19 and the vesicle marker Aut7. These results are supported by biochemical studies based on immunoisolation of Aut7. Together, the data suggest that although most autophagy and Cvt proteins co-localize to a unique perivacuolar compartment, they may display a unique spatial distribution within this compartment that may be essential to their function and the formation of the autophagosome/Cvt vesicle.
EXPERIMENTAL PROCEDURES
Strains and Growth Media
Yeast cells were grown in rich medium (YPD; 1% yeast extract, 2% peptone, and 2% glucose) or synthetic minimal medium (SMD; 0.67% yeast nitrogen base without amino acids (YNB), 2% glucose, and auxotrophic amino acids and vitamins as needed). Starvation was carried out in the synthetic minimal medium lacking nitrogen (SD-N; 0.17% YNB without ammonium sulfate and amino acids, 2% glucose).
Strains used in this study are listed in Table I. The chromosomal APG1 locus was deleted from strain WPHYD2 by a PCR-based, one-step procedure to generate strain WPHYD102. The Schizosaccharomyces pombe HIS5 gene was amplified from the ME3 vector using the following oligonucleotides containing APG1 sequences at flanking regions: 5′-CAACACCAGACGAGAAATTAAGAAAATGGGAGACATTTAACCCGGGCTGCAGGAATTC-3′ and 5′-TTGCAATACTATCAATATACTTTCTTATGATTATTCTGTCGTCGACGGTATCGATAAG-3′. The PCR product was transformed into the WPHYD2 strain and grown on a plate lacking histidine and leucine. Correct transformants were then confirmed by complementation assay and PCR. A PCR-based, chromosomal disruption of the APG9 locus in the wild type background (SEY6210) has been described previously (33). We used the same method to disrupt the APG9 locus in the pep4Δ strain background (TVY1), resulting in the apg9Δ pep4Δ strain JKY009.
Table I.
Strains used in this study
Materials
OptiPrep™ was from Accurate Chemical and Scientific Corp. (Westbury, NY). The Dynabeads® M-500 Subcellular were from Dynal Biotech, Inc. (Lake Success, NY). Complete™ EDTA-free protease inhibitor mixture was from Roche Molecular Biochemicals. Oligonucleotides were synthesized by Operon and Invitrogen. All other reagents were from Sigma.
Antiserum
Antisera against Ape1 (34), Apg9 (33), Cvt9 (35), Cvt19 (26), and Aut7 (22) were described previously. Antibodies to alkaline phosphatase (Pho8), carboxypeptidase Y (Prc1), and dolichol-mannose phosphate synthase (Dpm1) were from Molecular Probes (Eugene, OR). Antibodies to the hemagglutinin and c-Myc epitopes were from Santa Cruz Biotechnology (Santa Cruz, CA). Antiserum against Anp1 and phosphoglycerate kinase 1 were generously provided by Dr. Sean Munro (MRC, Cambridge, UK) and Dr. Jeremy Thorner (University of California, Berkeley), respectively. Antiserum against Pep12 was generously provided by Dr. Scott Emr (University of California, San Diego, La Jolla, CA).
Plasmid Constructions
The plasmids used in this study are listed in Table II. The CVT19 gene with 534 bp of upstream and 246 bp of downstream sequences flanking the open reading frame (ORF) was PCR-amplified from genomic DNA by using the following 5′ and 3′ primers: 5′-GGTAAACTTAAATATAGGCGCGGCCGCTTAAAGCCGCCC-3′ and 5′-CTTGTTTCCCTGCTTTTCTCTGGGATCCGTGCGGAAGGCG-3′. The PCR product was ligated into pRS414 using the NotI and SalI sites engineered into the 5′ and 3′ primers, respectively. The resulting CVT19 clone, pCVT19(414)JK, was used as the template for all subsequent PCR-based gene manipulations.
Table II.
Plasmids used in this study
| Plasmid | Description | Source |
|---|---|---|
| pYK128; pHAAPG1(423)a | Apg1, endogenous promoter, N-terminal 3× HA tag | 41 |
| pPS101; pCuYFPAPG1(426) | Apg1, CUP1 promoter, N-terminal YFP fusion | This study |
| pPS104; pAPG5YFP(426) | Apg5, endogenous promoter, C-terminal YFP fusion | This study |
| pAPG5HA(424) | Apg5, endogenous promoter, C-terminal HA tag | 28 |
| pAPG9(424) | Apg9, endogenous promoter | 33 |
| pAPG9ts(414) | Apg9 temperature-sensitive, endogenous promoter | 33 |
| pPS100; pCuYFPAPG9(426) | Apg9, CUP1 promoter, N-terminal YFP fusion | This study |
| pPS106; pCuCFPAPG9(424) | Apg9, CUP1 promoter, N-terminal CFP fusion | This study |
| pHA-APG12(424) | Apg12, endogenous promoter, N-terminal 3× HA tag | 27 |
| pMyc-APG12(426) | Apg12, endogenous promoter, N-terminal 3× Myc tag | 27 |
| pPS102; pCuYFPAPG12(426) | Apg12, CUP1 promoter, N-terminal YFP fusion | This study |
| pPS103; pCuYFPAPG14(426) | Apg14, CUP1 promoter, N-terminal YFP fusion | This study |
| pPS99; pCuYFPAUT7(426) | Aut7, CUP1 promoter, N-terminal YFP fusion | This study |
| pCuGFPAUT7(416) | Aut7, CUP1 promoter, N-terminal GFP fusion | 32 |
| pRS416-CuProtA-AUT7ΔR | Aut7, CUP1 promoter, N-terminal protein A fusion | This study |
| pCuGFPAUT7ΔR(416) | Aut7, CUP1 promoter, N-terminal GFP fusion | This study |
| pCuCVT9(414, 415, 416) | Cvt9, CUP1 promoter | 35 |
| pCuGFPCVT9(416) | Cvt9, CUP1 promoter, N-terminal GFP fusion | 35 |
| pPS97; pCuYFPCvt9(426) | Cvt9, CUP1 promoter, N-terminal YFP fusion | This study |
| pPS98; pCuCFPCvt9(424) | Cvt9, CUP1 promoter, N-terminal CFP fusion | This study |
| pCVT19(414)JK | Cvt19, endogenous promoter | This study |
| pCVT19CFP(414, 416) | Cvt19, endogenous promoter, C-terminal CFP fusion | This study |
| pCVT19YFP(414, 416) | Cvt19, endogenous promoter, C-terminal YFP fusion | This study |
| pCuCFP(424) | Cloning vector, CUP1 promoter, N-terminal CFP | This study |
| pCuYFP(426) | Cloning vector, CUP1 promoter, N-terminal YFP | This study |
| pRS416-CuProtA | Cloning vector, CUP1 promoter, N-terminal protein A tag | This study |
The number in parentheses indicates the pRS plasmid vector.
To make C-terminal CFP and YFP fusion constructs, pRS-based vectors containing CFP and YFP were first constructed. The enhanced versions of CFP and YFP cassettes (ECFP and EYFP) were obtained from the CLONTECH (Palo Alto, CA) plasmids pECFP and pEYFP by XmaI/EcoRI digestions and subcloned into the pRS414, 424, 416, and 426 vectors. A CYC1 termination cassette obtained from the pCu(416) vector (36) was also added by XhoI/KpnI digestions, resulting in the final pC-ECFP and pC-EYFP plasmids. For the N-terminal CFP and YFP fusion constructs, plasmids with the CFP and YFP genes under CUP1 promoter control were first constructed. The CFP and YFP ORFs were PCR-amplified using the pECFP and pEYFP plasmids as templates. The 5′ primer, 5′-GAGGATCCCAGAGTACTAGTCGCCACCATGGTGAGCAAG-3′, destroys the XmaI and AgeI sites in the 5′ multicloning site of the pECFP and pEYFP plasmids and introduces an SpeI site directly upstream of the CFP and YFP start sites. The 3′ primer, 5′-CAGTTGGAATTCTAGAGTCGCGGCCGCCCGGGTTGTACAG-3′, replaces the penultimate Lys residue and the TAA stop codon with Asn and Pro residues, respectively. The PCR product was digested with SpeI and XmaI and cloned into the pCu(414, 424, 416, and 426) vectors (36), resulting in the plasmids pCuCFP and pCuYFP(414, 424, 416, and 426) plasmids.
For the Cvt19-CFP and -YFP C-terminal fusion constructs, the CVT19 ORF with 496-bp upstream sequences was cloned into pC-ECFP and pC-EYFP(414, 424, 416, 426) by PCR amplification of the pCVT19 (414)JK plasmid as the template with the following 5′ and 3′ primers, respectively: 5′-CACATCACATTGAGAACGAGCTCATTAGTAG-3 and 5′-GAGTTTGACCCGGGGTTCTTCCCAAGTCAG-3. By using the engineered SacI site in the 5′ primer and the 3′ primer XmaI site, which is in-frame with the ECFP and EYFP coding sequences, the CVT19 PCR product was digested and subcloned into pC-ECFP and pC-EYFP, resulting in the pCVT19CFP and pCVT19YFP fusion constructs.
The plasmid expressing an N-terminal fusion of protein A to Aut7 with the C-terminal arginine of Aut7 deleted, pRS416-CuProtA-AUT7ΔR, was generated as follows. The sequence encoding the IgG binding domain of protein A was released from plasmid pPHY1044 (provided by Dr. Paul Herman, Ohio State University, Columbus, OH) with BamHI and EcoRI enzyme treatment and ligated into the same cloning sites of the pCu416 plasmid, resulting in pRS416-CuProtA. The AUT7 gene was PCR-amplified from pRS416-AUT7 (22) to replace its C-terminal arginine with a new stop codon. The oligonucleotides incorporated an in-frame EcoRI site immediately preceding the AUT7 start codon in the 5′ primer (5′-GGAATTCGAAGTCTACATTTAAGTCTGAATATCC-3′) and a HindIII site engineered 9 bp downstream of the new stop codon in the 3′ primer (3′-CGCTTCATTTCTAAGCTTATAAAAGACTACTAGCCAAATG-3′). The PCR product was cloned into the EcoRI/HindIII sites of pRS416-CuProtA.
An N-terminal fusion of GFP to Apg14 (pRS416-CuGFP-APG14) was made by PCR amplification of the APG14 ORF and downstream sequences from genomic DNA. The 5′ and 3′ oligonucleotides used for the PCR amplification were 5′-CCTAGTACCCGGGGCATTGCCCAATTTGCCACCATAGAGCG-3′ and 5′-GCGAATTCTAGCCTACCACGTACC-3′, respectively. The 5′ oligonucleotide contains an in-frame XmaI site at the start codon, and the 3′ oligonucleotide contains an EcoRI site. The PCR product was cloned into the XmaI/EcoRI sites of the pCuGFP(416) plasmid (35). N-terminal YFP- and CFP-Cvt9 fusion plasmids pPS97 (pCuYFPCVT9(426)) and pPS98 (pCuCFPCvt9(424)) were made by excising CVT9 from pCuGFPCVT9(416) (35) with XmaI and XhoI and inserting into XmaI- and XhoI-cut pCuYFP(426) and pCuCFP(424), respectively. Plasmids pPS99 (pCuYFPAUT7(426)) and pPS103 (pCuYFPApg14(426)) were made similarly by excising AUT7 from pCuGFPAUT7(416) (32) and APG14 from pCuGFP-APG14(416) with XmaI and XhoI and inserting into XmaI- and XhoI-cut pCuYFP(426). Plasmids pPS100 (pCuYFPAPG9(426)) and pPS106 (pCuCFPAPG9(424)) encoding N-terminally YFP/CFP-tagged Apg9 were made by amplification of APG9 from genomic DNA with 5′- AAATCACGGAATTACCCGGGTATGGAGAGAGATGAATACCAGTTA-3′ and 5′-GAAAAGAACTACTCGAGTCTAAAACAGGGAAGGGATTCTCT-3′ and inserting into XmaI- and XhoI-cut pCuYFP(426) and pCuCFP(424). Plasmid pPS101 (pCuYFPAPG1(426)) containing N-terminally YFP-tagged Apg1 was made by amplification of APG1 from genomic DNA with 5′-AGACGAGAAATCCCGGGAATGGGAGACATTAAAAATAAAGAT-3′ and 5′-CTCGTAAAGCATCTCGAGAGTAGCATAACATAA-3′ and inserting into XmaI- and XhoI-cut pCuYFP(426). Plasmid pPS102 (pCuYFPAPG12(426)) containing N-terminally YFP-tagged Apg12 was made by amplification of APG12 from genomic DNA with 5′-GTAGAGTGAACCAACCCGGGTATGAGTAGGATCCTAGAGA-3′ and 5′-CATTCCAGCGCTCGAGTATTTAACCAAACGCTACGGA-3′ and inserting into XmaI- and XhoI-cut pCuYFP(426). Plasmid pCuGFPAUT7ΔR(416) encodes Aut7 lacking its ultimate arginine fused at the N terminus to GFP and under the CUP1-regulatory promoter. It was constructed by PCR-amplifying pAUT7(416) (22) DNA with 5′-CTAGAGACCCGGGGTCTACATTTAAGTCTGAATATCC-3′ and 5′-CGCTTCATTTCTTTTCATACTCGAGACTACTAGCCAAATG-3′ and cloning into the XmaI and XhoI sites of pCuGFP(416).
The pME3 vector, which contains an S. pombe HIS5 auxotrophic marker, was a gift from Dr. Neta Dean (State University of New York, Stony Brook, NY). The pMyc-APG12(426) (27) and pCuGFPAUT7(416) (32) plasmids have been described elsewhere. The pCu414, 415, and 416 plasmids were gifts from Dr. Dennis Thiele (University of Michigan, Ann Arbor, MI).
OptiPrep™ Density Gradients
OptiPrep™ density gradient analyses were performed using a modified method of one described previously (37). Strains harboring the Cvt9-expressing plasmid under the control of the CUP1 regulatory promoter (pCuCVT9(414, 415, or 416)) were grown to A600 = 0.3 and induced for 2 h with 50 μm CuSO4 prior to fractionation procedures. Strains containing both Cvt9 and GFPAut7 under CUP1 control were induced for 2 h with 10 μm CuSO4. Yeast cells were then converted into spheroplasts before being subjected to osmotic lysis in PS200 buffer containing 1 mm EDTA, 1 mm MgCl2, 1 mm dithiothreitol, and a protease inhibitor mixture (Complete™ EDTA-free protease inhibitor tablets, 1 μg/ml leupeptin, and 1 μg/ml pepstatin A). After a pre-clear centrifugation step at 800 × g for 5 min to remove the cell debris, the crude lysate (20 A600 units of cells) was centrifuged at 100,000 × g for 20 min at 4 °C. The resulting total membrane fraction was resuspended in the same osmotic lysis buffer and layered on top of a 10–55% OptiPrep™ linear density gradient in PS200 containing 1 mm EDTA, 1 mm DTT, 1 mm MgCl2, and a protease inhibitor mixture. The gradients were subjected to centrifugation at 100,000 × g for 12 h at 4 °C. Samples were collected from the top of the gradients into 14 fractions. The samples were analyzed by Western blot as described previously (16) using the SuperSignal® West Dura Extended Duration Substrate (Pierce) for chemiluminescent detection. The protein bands were quantitated with the Bio-Rad Fluor-S Max Imager using the Bio-Rad Quantity One® software.
Protein A-Aut7ΔR Pull-down Analysis
To identify potential autophagy components isolated from the Aut7-resident compartment, the pRS416-CuProtA-AUT7ΔR plasmid or the pRS416-CuProtA negative control plasmid was co-transformed into cells from the apg1Δ aut2Δ double knockout strain with one of the following plasmids: pCuCVT9(414) (35), pAPG9(424) (33), and pHA-APG12(424) (27). Strains containing CVT9 under CUP1 control were induced for 2.5 h with 20 μm CuSO4. Cells were grown to midlog, converted to spheroplasts, and lysed with a Dounce homogenizer at 30 A600/ml in PBS lysis buffer (1× PBS, 200 mm sorbitol, 5 mm MgCl2, and protease inhibitors) with or without 0.5% Nonidet P-40. Cell lysates were then incubated with 15 μl of Dynabeads® M-500 Subcellular cross-linked to human IgG for 1 h at 4 °C. The beads were then washed in PBS lysis buffer 4 times and eluted with Laemmli sample buffer. Proteins associated with Aut7ΔR were pulled down with Dynabeads-IgG, resolved by SDS-PAGE, and analyzed by immunoblot with corresponding antibodies essentially as described previously (38) with the above modifications.
Fluorescence Microscopy
For the time course analyses of GFP-Aut7 in the apg9Δ pep4Δ strain expressing the plasmid-based apg9ts protein, cells were grown at non-permissive temperature (37 °C) until midlog phase, labeled with FM 4–64 as described previously (32), and incubated for 2 h with 10 μm CuSO4 to induce GFPAUT7 expression under CUP1 control (pCuGFPAUT7(414)). The cells were then shifted to SD-N medium for 1 h and viewed directly at 37 °C or shifted to permissive temperature (25 °C) for the indicated times prior to microscopy analysis.
The plasmids expressing the various GFP, CFP, and YFP fusion proteins were transformed singly or in pairs for co-localization analysis into the indicated yeast strains, grown to midlog phase in SMD, induced with 10 μm CuSO4 where appropriate for 2 h (or 5 μm CuSO4, for 30 min for analysis of Apg5-YFP), and viewed on a Nikon E-800 fluorescent microscope. The following band pass filters from Chroma Technology Corp. (Brattleboro, VT) were used for the fluorescent microscopy analyses as follows: CFP filter (exciter 436/20, emitter 480/40), GFP filter (exciter 470/40, emitter 525/50), and YFP filter (exciter 500/20, emitter 535/30). Images were captured by a Hamamatsu Orca2 digital camera using Openlab 3 software (Improvision, Inc., Lexington, MA).
RESULTS
Machinery Required for Vesicle Formation Is Co-localized to the Perivacuolar Cvt9 Compartment
Most autophagy and Cvt components characterized thus far function as soluble proteins that are recruited to, and peripherally associate with, membrane compartments (21, 22, 26–28, 32, 35, 39–44). The products of an increasing number of autophagy and Cvt genes also display a perivacuolar punctate pattern. Therefore, in this study, we sought to determine whether these autophagy components co-localized to the same membrane compartment. Such a finding would imply a significant role for this perivacuolar compartment in the autophagy and Cvt pathways and would identify the physical residence at which various specialized autophagy and Cvt functions are coordinated.
We recently characterized Cvt9 as a protein showing a strong perivacuolar localization when fused to GFP (35). The cvt9 mutant displays a relatively minor defect in non-selective autophagy. In contrast, the Cvt9 protein is essential for the Cvt pathway as well as for selective autophagy of peroxisomes (pexophagy) and has been shown to interact physically with Apg1. Cvt9 also displays a unique distribution on isopycnic density gradients and does not overlap with either endoplasmic reticulum markers or Golgi markers (35). Furthermore, the localization of Cvt9 does not change significantly upon the growth stage or nutritional status of the cells, and Cvt9 does not travel to the vacuole with the Cvt vesicles or autophagosomes. We therefore chose Cvt9 as a reference to which we tried to co-localize other proteins specific to the Cvt and autophagy pathways.
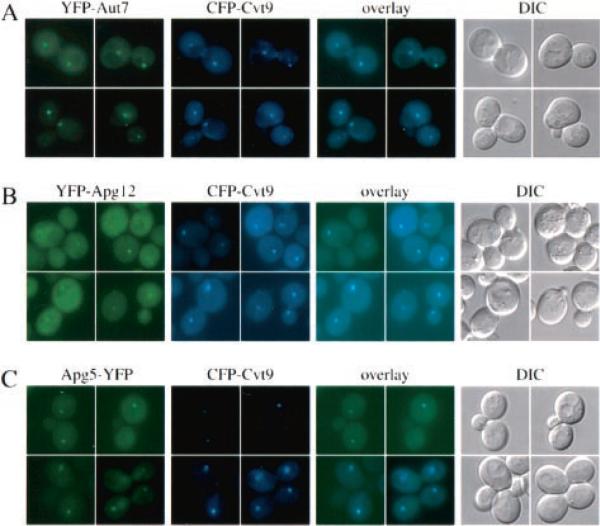
We first wanted to see if Aut7, a component that is involved in the formation of both the Cvt vesicle and autophagosome (30–32), co-localized to the same compartment as Cvt9. Aut7 is a small soluble protein that is conjugated to phosphatidylethanolamine (PE) through the action of Apg7 and Aut1 after the C-terminal arginine is removed by Aut2. The lipid-anchored Aut7 is membrane-associated, and steady-state localization of Aut7 appears to be predominantly at a membrane compartment in proximity to the vacuole in a wild type genetic background (32). YFP-tagged Aut7 showed a clear co-localization with CFP-tagged Cvt9 (Fig. 1A), and the identical result was obtained using the opposite fusion proteins (data not shown).
Fig. 1. Cvt9, the vesicle component Aut7, and the Apg12-Apg5 conjugation components co-localize by fluorescent microscopy.
Wild type (SEY6210) (A and B) or aut7Δ (WPHYD7) (C) cells were co-transformed with the following pairs of copper-inducible plasmids. A, YFP-Aut7 (pCuYFPAUT7(426)) and CFP-Cvt9 (pCuCFPCVT9(424)); B, YFP-Apg12 (pCuYFPAPG12(426)) and CFP-Cvt9; C, Apg5-YFP (pAPG5YFP(426)) and CFP-Cvt9. The transformed cells were grown to midlog stage in SMD, induced with 30 μm CuSO4 for 2 h (A and B) or with 5 μm CuSO4 for 30 min (C), and examined with a Nikon E-800 fluorescence microscope equipped with a Hamamatsu Orca 2 digital camera and processed with Openlab software. The YFP fusions to the vesicle component Aut7 and the conjugation components Apg12 and Apg5 co-localize with CFP-Cvt9. The DIC panels are images obtained with differential interference contrast optics.
Aut7 initially associates with both sides of the forming autophagosome/Cvt vesicle (21, 22). Prior to or upon vesicle completion, the PE is cleaved from Aut7 through the action of Aut2, removing Aut7 from the delimiting outer membrane. A portion of Aut7, however, remains sequestered within the completed vesicle and is transported along with the cargo to the vacuole lumen (22). To ensure that the Aut7 we found to co-localize with Cvt9 was associated with a structure prior to the completion of the Cvt vesicle or autophagosome, we also examined the co-localization of Aut7 with Cvt9 in the apg1Δ mutant that is completely blocked in both the Cvt pathway and autophagy. In rich medium, YFP-Aut7 did not show an obvious punctate pattern in this mutant.2 However, upon starvation, a strong perivacuolar localization of YFP-Aut7 that was completely overlapping with CFP-Cvt9 was seen in the apg1Δ strain (data not shown). These observations suggest that the Cvt9- and Aut7-localized perivacuolar compartment may represent the site of initial Aut7 membrane association prior to vesicle formation.
The lipid conjugation of Aut7 and concomitant membrane association have been shown to require the action of a novel protein conjugation system (reviewed in Ref. 45). Mutations in any one of the Apg conjugation components, Apg5, 7, 10, 12, and 16, prevent the formation or completion of both Cvt vesicles and autophagosomes as well as Aut7 conjugation to PE (27, 28, 32, 39, 42, 43). Recent studies in mouse embryonic stem cells demonstrated that Apg5, and by extension its conjugated partner Apg12, associated with a membrane structure at sites of vesicle formation (29). We therefore wanted to examine if some of the key conjugation components that are essential for Aut7 localization could be detected in the same perivacuolar compartment as Aut7 and Cvt9. YFP-tagged Apg12 showed a predominantly cytosolic localization when overexpressed, but a punctate signal was also seen when the protein was expressed at lower levels (Fig. 1B). Co-expression of YFP-Apg12 with CFP-Cvt9 in wild type cells revealed that the membrane-associated population of YFP-Apg12 co-localized with CFP-Cvt9 to the same perivacuolar compartment (Fig. 1B).
We also examined the localization of the Apg12 conjugation partner Apg5. Fluorescent fusion protein constructs of Apg5 generally display cytosolic staining patterns in wild type cells, but the occurrence of punctate structures has been reported for Apg5-GFP expressed in mutants defective in events leading up to Aut7 lipidation as well as in the aut7Δ strain (28, 46). Accordingly, we co-expressed Apg5-YFP and CFP-Cvt9 in an aut7Δ strain to simplify detection of the punctate Apg5-YFP signal. As with YFP-Apg12, both proteins co-localized to the perivacuolar structure (Fig. 1C).
Protein Kinase and Lipid Kinase Complexes Associate with the Apg/Cvt Perivacuolar Compartment
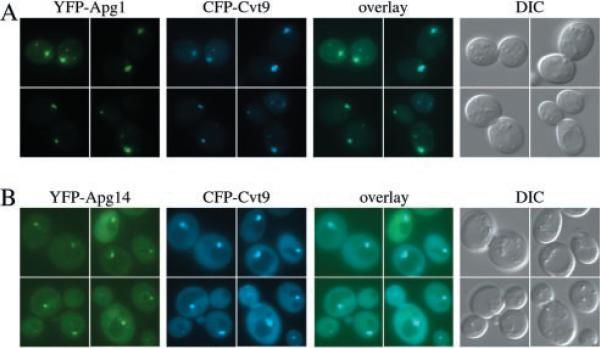
At present, Apg1 is the only putative protein kinase identified that may be dedicated to the Cvt pathway and autophagy. Complexed to other proteins, Apg1 regulates the transition between the two pathways upon nutrient limitation (40, 41). Apg1 is regulated upstream by Tor kinase, but downstream targets have not yet been identified. Two-hybrid and co-immunoprecipitation studies also indicate that Apg1 physically interacts with Cvt9 (35, 41), but previous studies have concluded that GFP-tagged Apg1 is cytosolic (47). In accordance with the GFP-tagged construct, YFP-Apg1 did not result in any discernible perivacuolar structures under nutrient-rich conditions.2 However, strong recruitment to the Cvt9 perivacuolar compartment was seen shortly after the cells were shifted to nitrogen starvation medium (Fig. 2A), the same conditions that both enhance the interaction between Apg1 and Apg13 as well as lead to increased kinase activity. The perivacuolar signal co-localized with CFP-Cvt9 (Fig. 2A) but was equally strong in the cvt9Δ strain (data not shown), suggesting that a factor other than Cvt9 may be crucial for the membrane recruitment of Apg1.
Fig. 2. The Apg1 protein kinase and the Apg14 component of the PI 3-kinase co-localize with Cvt9.
Wild type cells (SEY6210) were co-transformed with plasmids, under CUP1 copper-inducible control, expressing either YFP-Apg1 (pCuYFPAPG1(426)) and CFP-Cvt9 (pCuCFPCVT9(424)) (A) or YFP-Apg14 (pCuYFPAPG14(426)) and CFP-Cvt9 (B). Transformed cells were grown to midlog stage in SMD, induced for 1–2 h with 30 μm CuSO4, and viewed directly (B) or shifted to SD-N medium for 1 h prior to microscopy analysis (A). Images were captured and analyzed as in Fig. 1. The YFP fusion of the Apg1 kinase, and Apg14, a subunit of the Apg/Cvt-specific Vps15/34 PI-3 kinase complex, co-localize with the perivacuolar CFP-Cvt9 fusion protein. DIC, differential interference contrast.
In addition to the Apg1 protein kinase complex, a lipid kinase complex has recently been shown to function in Cvt and autophagy transport. Generation of phosphatidylinositol 3-phosphate by Vps34 plays key roles in membrane regulation and traffic between the late Golgi-to-PVC and PVC-to-vacuole (48–50). Activity of Vps34 requires binding to Vps15, a Ser/Thr kinase, as well as other proteins. The Vps34/Vps15 core complex associated with Vps30/Apg6 and Vps38 has been shown to be required for transport of Prc1 to the vacuole from the late Golgi, whereas substituting Vps38 with Apg14 results in PI 3-kinase activity required for Cvt and autophagy transport (51). The function of Apg14 and Vps38 is not known but could be to direct the PI 3-kinase holoenzyme to specific locations. To follow the location of the autophagy/Cvt-specific PI 3-kinase complex, we constructed YFP-tagged Apg14. Surprisingly, YFP-Apg14 showed a weak but distinct vacuolar rim staining (Fig. 2B). In addition, YFP-Apg14 also bound to a perivacuolar structure, and the YFP-Apg14 at this location showed complete co-localization with CFP-Cvt9. These results suggest that the perivacuolar compartment, to which Cvt9 and Aut7 are localized, may also be the site of autophagy- and Cvt-specific membrane traffic mediated by the PI 3-kinase complex.
The Precursor Aminopeptidase I Receptor Is Localized to the Cvt/Apg Compartment Prior to Sorting into the Cvt Vesicle or Autophagosome
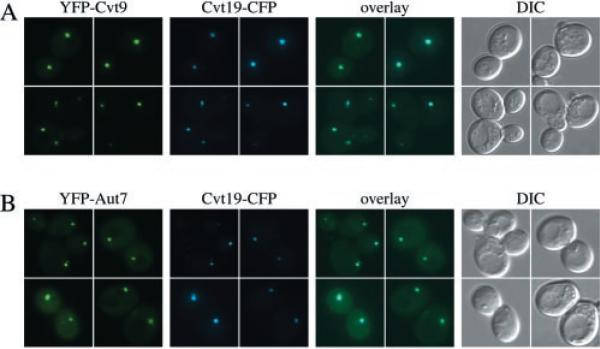
We recently characterized a protein receptor for the prApe1 complex (26). Cvt19 is not a conventional receptor in that it is not a transmembrane protein but still shows strong membrane association. The prApe1 complex is bound to the sequestering membrane mainly through interaction with the propeptide, and a fraction of Cvt19 is transported to the vacuole with the vesicle in a prApe1-dependent manner. Components including Apg1, Cvt9, and the conjugation machinery may transiently associate with the forming vesicle but have not been found to be part of the completed Cvt vesicle or autophagosome. Aut7, on the other hand, also functions at the formation step, but a fraction of it is transported to the vacuole with the vesicle. As we have shown, in wild type cells co-expressing functional CFP-Cvt9 and YFP-Aut7, both fusion proteins were localized to the same perivacuolar compartment (Fig. 1A). Co-localization of the Cvt19-CFP fusion protein with YFP-Cvt9 was also quite clear (Fig. 3A). Finally, as expected, YFP-Aut7 and Cvt19-CFP also appeared in the same perivacuolar structures (Fig. 3B). These observations suggest that the Cvt9- and Aut7-localized perivacuolar compartment, which is the initial site for Aut7 membrane association prior to vesicle formation, also is the compartment where the prApe1 cargo receptor Cvt19 is residing before being packaged into the nascent vesicle.
Fig. 3. Cvt9 co-localizes with the Cvt19 cargo receptor by fluorescence microscopy.
Wild type cells (SEY6210) were co-transformed with the following pairs of plasmids. A, copper-inducible YFP-Cvt9 (pCuYFP-CVT9(426)) and Cvt19-CFP under endogenous promoter control (pCVT19CFP(414)); B, copper-inducible YFP-Aut7 (pCuYFPAUT7(426)) and Cvt19-CFP under endogenous promoter control (pCVT19CFP(414)). The transformed cells were grown to midlog stage in SMD, induced with 10 μm CuSO4 for 2 h, and examined with a Nikon E-800 fluorescence microscope as described in Fig. 1 and under “Experimental Procedures.” A fluorescent protein fusion of Cvt19 co-localizes with both Cvt9 and Aut7 at the perivacuolar compartment. DIC, differential interference contrast.
Apg9 Is Present at the Apg/Cvt Compartment
Apg9 was the first identified integral membrane protein to function in both the autophagy and Cvt pathways (33). Immunoelectron microscopy studies revealed that Apg9 resided in a compartment separate from the vacuole. This was confirmed by density gradient analysis where Apg9 had a distinct density profile when compared with other vacuolar and endomembrane marker proteins. Although immunofluorescence suggested that Apg9 localizes to a single punctate, perivacuolar membrane structure, GFP-tagged Apg9 showed several membranous structures in proximity to the vacuole, and a similar result was obtained with YFP-tagged Apg9.2 Although Apg9 is an essential component for both the Cvt pathway and autophagy, it is not found on autophagosomes. Furthermore, Apg9 is not required for the localization of Cvt9 and Cvt19 to the perivacuolar compartment and hence does not appear to be the critical component that defines the compartment. The difference in the appearance of Apg9 by immunofluorescence and GFP/YFP fusion protein analysis suggests that the overexpression required to see the fluorescent fusion protein may lead to mis-localization of the protein. Alternatively, there may be multiple locations for Apg9 with the majority of the protein localized to a punctate perivacuolar structure.
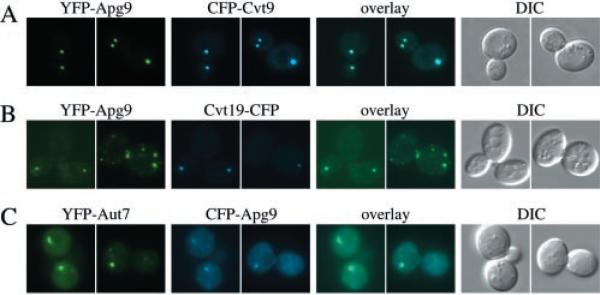
When expressed by itself in wild type cells, YFP-Apg9 showed multiple dots throughout the cell (data not shown), in agreement with previous results seen for GFP-Apg9 (33). In contrast, when YFP-Apg9 was co-expressed with CFP-Cvt9, both fusion proteins appeared as prominent punctate dots, generally with one or two dots per cell (Fig. 4A). Under these conditions, YFP-Apg9 showed complete co-localization with CFP-Cvt9. Identical results were obtained when co-expressing CFP-Apg9 with YFP-Cvt9 (data not shown). Shifting the cultures to starvation conditions did not alter this localization pattern.2 Because the localization of YFP-Apg9 expressed together with CFP-Cvt9 differed somewhat from that of YFP-Apg9 expressed alone, we also expressed YFP-Apg9 together with Cvt19-CFP (Fig. 4B) and CFP-Apg9 together with YFP-Aut7 (Fig. 4C). In both cases, Apg9 structures co-localized with Cvt19 and Aut7, but frequently additional Apg9 structures could be found that did not show any Cvt19 and Aut7 staining. Taken together, the data suggest that a population of Apg9 is present at the perivacuolar Apg/Cvt compartment and that co-expression of Cvt9 in particular may result in greater recruitment of Apg9 to the compartment.
Fig. 4. Cvt9 and Apg9 co-localize by fluorescent microscopy.
Wild type cells (SEY6210) were co-transformed with plasmids, under CUP1 copper-inducible control, expressing either YFP-Apg9 (pCuYFPAPG9(426)) and CFP-Cvt9 (pCuCFPCVT9(424)) (A), YFP-Apg9 and Cvt19-CFP (pCVT19CFP(414)) (B), or YFP-Aut7 (pCuYFPAUT7(426)) and CFP-Apg9 (pCuCFPAPG9(424)) (C). Transformed cells were grown to midlog stage in SMD and induced for 1–2 h with 30 μm CuSO4. Images were taken and examined with a Nikon E-800 fluorescence microscope as described in Fig. 1 and under “Experimental Procedures.” Apg9 displays multiple punctate dots when co-expressed with Cvt19 and Aut7 but only a single dot when co-expressed with Cvt9. The YFP-Apg9 and CFP-Cvt9 dots co-localize. DIC, differential interference contrast.
Subcellular Fractionation Divides Proteins at the Perivacuolar Apg/Cvt Compartment into Two Subpopulations
The microscopy data placed protein components of the conjugation machinery, the Apg1 signaling complex, the PI 3-kinase complex, the cargo receptor Cvt19, the vesicle component Aut7, and a population of the integral membrane protein Apg9 at the perivacuolar Apg/Cvt compartment along with Cvt9. Although the microscopy data suggested that all of these proteins are at the same subcellular location, the different components may be unevenly distributed spatially within this location, and they may even be on different membranous structures at this site. For example, Cvt9 is equally distributed between low and high speed pelletable fractions, whereas Apg5 and Cvt19 are recovered primarily in a low speed fraction following velocity sedimentation (26, 28, 35). Accordingly, we sought to recapitulate the in vivo co-localization results by linear density gradient analysis.
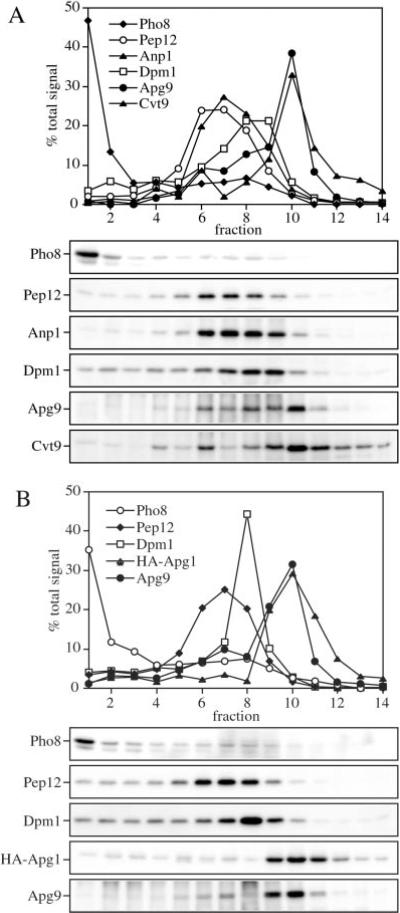
A total membrane fraction from wild type cells extragenically co-expressing combinations of Apg9, Cvt9, and/or an HA epitope-tagged Apg1 (HA-Apg1) was prepared from osmotically lysed spheroplasts and resolved on isopycnic OptiPrep gradients as described under “Experimental Procedures.” In agreement with previous findings, Apg9 and Cvt9 did not co-fractionate with any other resident organelle marker proteins including Pho8 (vacuole), Anp1 (Golgi), Pep12 (endosome), and Dpm1 (ER), (Fig. 5A; 33). Similarly, HA-Apg1 had a density profile that was distinct from these markers (Fig. 5B). All three proteins were recovered with a density peak in fraction 10 of the gradients, suggesting that they were co-localized to the same compartment. These results agree with previously published studies (35) showing that Cvt9 and Apg1 interact. Furthermore, the linear density gradient analysis agrees with the in vivo co-localization results in the present study demonstrating that both Apg1 and at least a population of Apg9 co-localize with Cvt9. Taken together, these results indicate that the Apg1 signaling complex and Cvt9 bind to a compartment with a density equal to the membranes spanned by Apg9.
Fig. 5. Cvt9, Apg9, and Apg1 co-localize with each other but not with other endomembrane markers by density gradient separation.
Subcellular co-localization of Apg9 and Cvt9 (A) and HA-Apg1 and Apg9 (B) by OptiPrep density gradients. The wild type strain (SEY6210) was co-transformed with the multicopy APG9 plasmid (pAPG9(426)) and the copper-inducible CVT9 plasmid (pCuCVT9(416)) (A) or a plasmid expressing an HA epitope-tagged Apg1 (pHAAPG1(423)) the multicopy APG9 plasmid (pAPG9(426)) (B). Cells were grown to midlog stage in SMD, and those in A incubated with 50 μm CuSO4 for 2 h to induce Cvt9 expression. The cells were converted to spheroplasts and lysed in PS200 buffer as described under “Experimental Procedures.” A total membrane fraction was isolated by centrifugation at 100,000 × g for 20 min and loaded to the top of a 10-ml OptiPrep linear gradient (10–55%). Following centrifugation at 100,000 × g for 12 h at 4 °C, 14 fractions were collected and analyzed by immunoblots with antiserum or antibodies to Pho8 (vacuole), Pep12 (endosome), Anp1 (cis-Golgi), Dpm1 (ER), Cvt9, Apg9, and the HA epitope (Apg1) as indicated. Indirect chemiluminescent detection and quantification of relative protein concentrations were performed with the Bio-Rad Fluor-S Max Imager. Cvt9, Apg1, and Apg9 co-localize to a dense part of the gradient and are separated from known endomembrane markers.
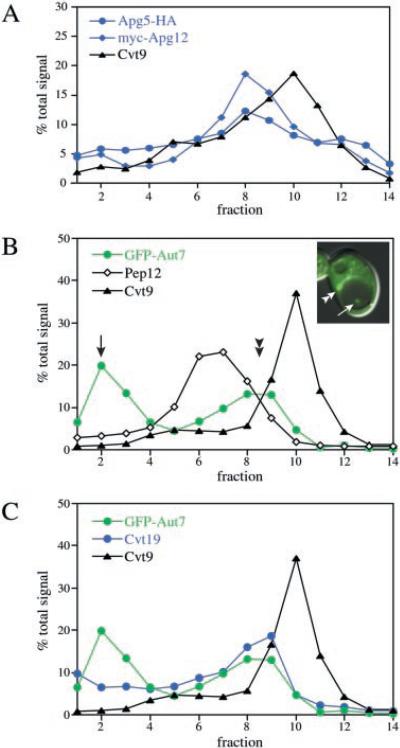
To examine further the perivacuolar Apg/Cvt compartment, an HA-tagged fusion of Apg5 was co-transformed with a plasmid expressing Myc-tagged Apg12. The density gradient resolution of the total membrane fraction indicated that both Apg5-HA and Myc-Apg12 co-migrated with peak concentrations appearing in fractions 8 and 9, offset from the peak levels of Cvt9, which remained in fraction 10 (Fig. 6A). Apg5 and Apg12 are both known to transiently associate with the forming autophagosome/Cvt vesicle. Accordingly, we extended our gradient analysis by examining Aut7, a component that remains associated with the completed vesicle. To correlate the microscopy observations with the gradient analysis, we decided to examine the GFP-Aut7 fusion protein in a proteinase A-deficient mutant (pep4Δ) co-transformed with the copper-inducible Cvt9 plasmid. The pep4Δ strain expressing GFP-Aut7 and Cvt9 was grown to midlog stage, induced with 10 μm CuSO4 for 2 h, and examined by microscopy just prior to preparing a total membrane fraction to be resolved by density gradients. In the pep4Δ strain, GFP-Aut7 displayed two localization patterns by microscopy (Fig. 6B, see inset). GFP-Aut7 was concentrated at the perivacuolar compartment (Fig. 6B, inset, double arrowhead), similar to its steady-state distribution in wild type cells (Fig. 1A). However, because the pep4Δ strain fails to break down subvacuolar vesicles, GFP-Aut7 could also be detected in accumulated Cvt bodies in the vacuolar lumen (Fig. 6B, inset, single arrow). A more pronounced accumulation of GFP-Aut7 could also be detected in autophagic bodies under nitrogen starvation conditions (data not shown) (32). Examination of the gradient density profile of GFP-Aut7 also indicated a bimodal distribution of the fusion protein. The first GFP-Aut7 peak appeared at the top of the gradient (Fig. 6B, single arrow), consistent with the localization of GFP-Aut7 in accumulated Cvt bodies inside the vacuole. The second GFP-Aut7 peak appeared in fractions 8 and 9 (Fig. 6B, double arrowhead), the same density peaks as Apg5 and Apg12 (Fig. 6A), but distinct from the Cvt9 peak collected in fraction 10.
Fig. 6. Co-localization analyses of Cvt9, the Apg conjugation proteins, and the vesicle components Aut7 and Cvt19.
A, linear OptiPrep density gradient profiles of Apg conjugation components. Wild type cells (SEY6210) were co-transformed with plasmids expressing an HA-epitope fusion to Apg5 (pAPG5HA(424)), Myc-epitope fusion to Apg12 (pMyc-APG12(426)), and copper-inducible Cvt9 (pCuCVT9(415)). Transformed cells were grown to midlog in SMD and induced with 50 mm CuSO4 for 2 h to induce Cvt9 expression. A total membrane fraction was resolved by linear OptiPrep gradients (10–55%), and the collected fractions were analyzed by quantitative immunoblots with antiserum to Cvt9 and the HA (Apg5) and Myc (Apg12) epitopes as described in Fig. 5 and under “Experimental Procedures.” The Apg5 and Apg12 conjugation components can be resolved from Cvt9 by density gradient separation. B and C, linear density gradient profiles of GFP-Aut7, Cvt9, and Cvt19. The pep4Δ strain (TVY1) was co-transformed with copper-inducible plasmids expressing GFP-Aut7 (pCuGFPAUT7(416) and Cvt9 (pCuCVT9(414)). The transformed strain was grown to midlog stage in SMD and incubated with 20 μm CuSO4 for 2 h to induce GFP-Aut7 and Cvt9 expression. Just prior to subjecting the cells to density gradient analysis, the distribution of GFP-Aut7 was examined with a fluorescence microscope (see inset) as described under “Experimental Procedures.” The induced cells were then converted to spheroplasts, osmotically lysed, and a total membrane fraction was obtained and resolved on a 10–55% OptiPrep density gradient as described above. The collected gradient fractions were analyzed and quantified after immunoblotting with antibodies or antiserum to Aut7, Pep12, and Cvt9 (B), and Aut7, Cvt19, and Cvt9 (C). The quantification graphs in B and C are from the same gradient but are presented separately for clarity. Aut7 displays a bimodal distribution and co-fractionates with Cvt19 in the higher density peak. Both Aut7 and Cvt19 peak in a fraction that is resolved from Cvt9.
Endogenous levels of Cvt19 were also examined in the same gradients and exhibited a co-fractionation pattern with GFP-Aut7 (Fig. 6C). A small fraction of the total cellular level of Cvt19 also accumulates in the vacuole, in agreement with our recent study (26) demonstrating that the receptor accumulates inside Cvt bodies in the protease-deficient pep4Δ strain (Fig. 6C). In conclusion, whereas the microscopy experiments placed these components at the same membrane compartment as Cvt9, the density gradient analysis indicated that the conjugation machinery, Aut7, and the prApe1 receptor may occupy a distinct density subdomain within this perivacuolar compartment. Alternatively, these components may occupy an entirely different membrane source within the same subcellular location.
Aut7 Interacts with Cvt19 and Apg12
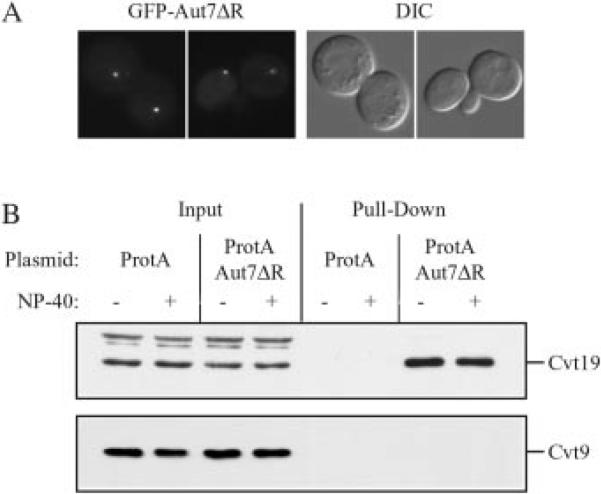
The gradient fractionation data suggested that the Apg/Cvt proteins localized to the punctate dot might reside within separate subdomains. We decided to extend our biochemical analysis by making a functional fusion of Aut7 to protein A. Aut7 undergoes a series of post-translational modifications. The ultimate arginine residue is proteolytically removed by Aut2, and the modified protein is subsequently attached to PE resulting in a tight attachment to the membrane (30, 31). The PE is subsequently cleaved from Aut7 in a reaction that is again dependent on Aut2. This processing event converts Aut7 back to a soluble form. To stabilize the interaction of Aut7 with the membrane, we constructed protein A-Aut7ΔR, lacking the C-terminal arginine residue. This construct was expressed in a strain having a deletion in the AUT2 gene to prevent the subsequent release of Aut7 from the membrane. The protein A-Aut7ΔR fusion was functionally based on complementation of the prApe1 accumulation phenotype of the aut7Δ strain (data not shown). Finally, to ensure that we were examining protein A-Aut7ΔR at the initial site of localization, we carried out our analysis in an aut2Δ apg1Δ double mutant strain that is also defective in autophagosome/Cvt vesicle formation. GFP-Aut7ΔR localized to a punctate dot in the aut2Δ apg1Δ strain, indicating that the modified protein was an appropriate marker for the perivacuolar compartment (Fig. 7A).
Fig. 7. Cvt19 is co-isolated with protein A-Aut7ΔR.
A, plasmid GFP-AUT7ΔR encoding GFP fused to Aut7 lacking the C-terminal arginine residue was transformed into the apg1Δ aut2Δ double knockout strain (WPHYD102). Cells were grown to midlog phase and examined by fluorescence microscopy as described under “Experimental Procedures.” GFP-Aut7ΔR localizes to a perivacuolar punctate dot. B, the protein A control vector (ProtA) or protein A-Aut7ΔR encoding plasmid (ProtA-AUT7ΔR) was co-transformed into the apg1Δ aut2Δ double knockout strain (WPHYD102) with one of the following plasmids: pCuCVT9(424), pAPG9(424), or pHA-APG12(424). Cells were collected either directly from midlog phase culture or after a 2.5-h induction with 20 μm CuSO4 to induce Cvt9 expression and converted to spheroplasts. Spheroplasts were then lysed in PBS lysis buffer in the absence or presence of 0.5% Nonidet P-40 (NP-40) as indicated. Lysates were incubated with Dynabeads M-500 cross-linked to human IgG. Protein A-Aut7ΔR and associated proteins were isolated by collecting Dynabeads from the crude cell lysates by magnetic isolation (Pull-Down). Proteins were resolved by SDS-PAGE and detected by immunoblot with antiserum against Cvt19, Cvt9, Apg9, or the HA epitope (Apg12). The positions of Cvt19 and Cvt9 are indicated. Input lanes corresponded to 2.5% of cell lysates used for each pull-down reaction. Protein A-Aut7ΔR pulls down Cvt19 but not Cvt9. The protein A-Aut7ΔR construct also pulled down HA-Apg12 but not Apg9 (data not shown, see text). DIC, differential interference contrast.
Protein A-Aut7ΔR, or a vector expressing only protein A, was co-expressed in combination with various Apg/Cvt proteins. Cells were grown to midlog phase, converted to spheroplasts, and lysed in the absence or presence of detergent. The protein A-Aut7ΔR fusion was then affinity-isolated by binding to Dynabeads that had been coupled to IgG (see “Experimental Procedures”). The recovered proteins were separated by SDS-PAGE and examined by Western blot. Protein A-Aut7ΔR was recovered along with endogenous Cvt19 in the absence or presence of detergent, suggesting that the two proteins interact directly (Fig. 7B). Cvt19 was not detected when the pull-down was performed with the control vector expressing protein A alone, indicating that the interaction was dependent on the Aut7ΔR domain. When protein A-Aut7ΔR was co-expressed with HA-Apg12, the Apg12 protein was also pulled down along with protein A-Aut7ΔR; however, the level was substantially lower than seen with Cvt19 (data not shown). In addition, very little Apg12 was recovered in the presence of detergent, suggesting that the interaction might not be direct. In contrast, neither Cvt9 (Fig. 7B) nor Apg9 (data not shown) was recovered along with protein A-Aut7ΔR, suggesting that these two proteins are not as close in proximity to Aut7 as Cvt19 and Apg12. These results further support the density fractionation data indicating that there are two populations of Apg/Cvt proteins localized to the perivacuolar structure.
The Perivacuolar Apg/Cvt Compartment Is a Physiological Intermediate in Vesicle Formation
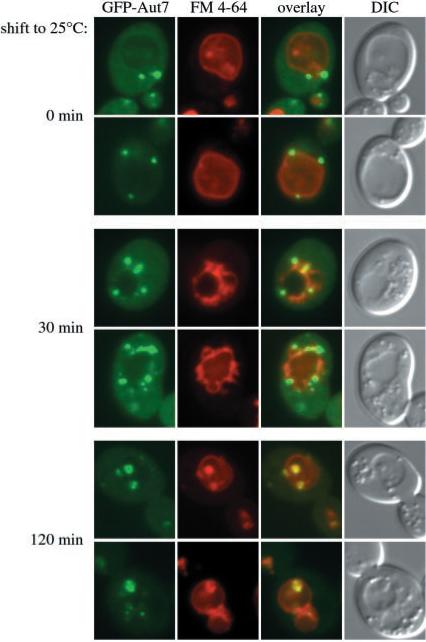
The data we have presented indicate that many of the Apg/Cvt components co-localize to a distinct perivacuolar membrane compartment. We next wanted to verify that this compartment was a physiologically relevant intermediate and did not represent a terminal destination. Accordingly, we followed the transit of the GFP-Aut7 vesicle marker to the vacuole in a temperature conditional mutant of Apg9. This allowed us to examine the physiological relevance of the perivacuolar compartment in the steps that lead to autophagosome or Cvt vesicle formation and subsequent delivery into the vacuole. We previously demonstrated that GFP-Aut7 may peripherally associate with a perivacuolar compartment in the apg9 null background (32). Because the apg9 mutation prevents vesicle formation, recruitment of Aut7 to this perivacuolar compartment may represent an early step leading to the generation of the transport vesicle. The association of GFP-Aut7 with the perivacuolar compartment in the apg9 mutant strain is weak under nutrient-rich conditions but becomes pronounced when the cells are shifted to starvation medium (32). The plasmid expressing the apg9ts allele was co-transformed with GFP-Aut7 into the apg9Δ pep4Δ strain and was grown to midlog phase at nonpermissive temperature. The strain was then stained with FM 4-64 to label the vacuole and shifted to nitrogen starvation medium for 1 h at nonpermissive temperature prior to microscopy analysis. By observing GFP-Aut7 in the temperature-sensitive background of apg9, we could arrest the transport of GFP-Aut7 at the stage of association with the perivacuolar compartment. Then, as the cells are shifted to permissive temperature, which results in the thermal reversal of the apg9ts block (33), we could kinetically follow the GFP-Aut7 itinerary from the perivacuolar compartment to the vacuolar lumen where it would potentially accumulate inside autophagic bodies due to the absence of the Pep4 protease.
In agreement with the perivacuolar distribution of GFP-Aut7 in the apg9 null strain (32), GFP-Aut7 appeared at the perivacuolar compartment in the apg9ts pep4Δ strain at the nonpermissive temperature (Fig. 8, 0-min shift). When the culture was then shifted to permissive temperature for 30 min, GFP-Aut7 was detected not only at the large perivacuolar structures but also in smaller punctate structures that were largely distributed around the perimeter of the vacuole (Fig. 8, 30-min shift). These structures may correspond to forming or completed autophagosomes. After 2 h at permissive temperature, GFP-Aut7 accumulated inside the vacuole; some perivacuolar labeling was also detected (Fig. 8, 120-min shift). These findings indicate three detectable steps in the transit itinerary of GFP-Aut7 as follows: initial recruitment to the perivacuolar compartment, followed by the emergence of smaller punctate membrane structures around the perimeter of the vacuole, and finally, transport into the vacuole and accumulation inside autophagic bodies in the pep4Δ background. Therefore, the perivacuolar compartment appears to serve a physiologically relevant role as the source membrane in the vacuolar targeting of Aut7 by the Cvt and autophagy pathways.
Fig. 8. The perivacuolar compartment is a physiological intermediate in the Cvt and autophagy pathways.
The apg9Δ pep4Δ strain was co-transformed with a plasmid expressing a temperature conditional allele of Apg9 (pAPG9ts(414)) and the copper-inducible GFP-Aut7 fusion protein (pCuGFPAUT7(416)). The transformed cells were grown at nonpermissive temperature (37 °C) to midlog stage, induced with 10 μm CuSO4 for 2 h, and labeled with FM 4–64 at 37 °C. The cultures were then incubated in SD-N medium for 1 h at 37 °C and viewed directly or shifted to permissive temperature (25 °C) for 30 and 120 min prior to fluorescence microscopy analysis as described in Fig. 1 and under “Experimental Procedures.” GFP-Aut7 fluorescence that accumulates at the PVC under nonpermissive conditions (0 min) can be seen to migrate to punctate structures (30 min) that ultimately end up in the vacuole lumen (120 min) at permissive temperature.
DISCUSSION
The transport of cargo to the vacuole by the Cvt and autophagy pathways differs significantly from classical modes of secretory traffic. Proteins within the secretory pathway that are destined for secretion or diverted to the vacuole are translocated into the endoplasmic reticulum and are packaged in a lumenal environment within single-membrane vesicles. In contrast, the Cvt and autophagy pathways capture cargo directly from the cytoplasm within double-membrane Cvt vesicles and autophagosomes. The key stage of regulation in this process occurs at the point of vesicle formation, as reflected by the finding that the majority of the genetically identified Cvt and Apg components function prior to, or at the step of, vesicle formation and completion (reviewed in Refs. 10, 12, and 13). Therefore, questions regarding the mechanisms of how Cvt vesicles and autophagosomes form remain a central focus of investigation in the field. In this study, we have demonstrated that a perivacuolar compartment provides residence for a remarkable array of specialized Cvt and Apg trafficking machinery that ultimately results in the formation of transport vesicles for the Cvt and autophagy pathways. These findings implicate the role of the Apg/Cvt perivacuolar compartment as the donor membrane for autophagosome and Cvt vesicle formation.
The Apg1 Signaling Complex
Both in vivo analyses using fluorescence microscopy and in vitro studies based on subcellular fractionation and density gradient separation demonstrate the co-localization of Apg1 and Cvt9 (Figs. 2 and 4). These results agree with previous studies that demonstrated an interaction between these two proteins based on two-hybrid interactions and co-immunoprecipitation (35) and validate the approach taken in the current analysis. Furthermore, the present study clearly places the Apg1 kinase complex at the perivacuolar compartment and assigns an essential role for this membrane structure in the regulation of Cvt and autophagy transport. The kinase activity of Apg1 depends on upstream nutrient signals that are initially relayed through the Tor kinase (41, 52). Tor kinase repression leads to the induction of autophagy and increased Apg1 kinase activity by enhancing the binding of Apg13, an Apg1 activator, to Apg1 (41). Growing evidence suggests that the Apg1 kinase complex plays a key role in switching between Cvt transport and autophagy. Apg1 physically interacts with Cvt9, a protein required for the Cvt pathway and selective peroxisome degradation but not autophagy (35, 41). In addition, an autophagy-specific component, Apg17, also physically interacts with Apg1, suggesting that Apg1, through its interactions with pathway-specific components, may dictate the initiation of either the Cvt or autophagy programs based on upstream nutrient signals (41).
Components Required for Cargo Packaging and Vesicle Formation
Aut7 is the only characterized protein required for the autophagy/Cvt pathways that is substantially induced under starvation conditions (21, 22). Aut7 remains associated with the completed autophagosome/Cvt vesicle (21, 22), and the increase in the level of Aut7 is critical for the expansion of the autophagosomal membrane (23). Aut7 and a set of proteins involved in the conjugation of Apg12 to Apg5 function at the step of vesicle formation. The E1-like conjugation component, Apg7, activates Aut7 for subsequent reactions that lead to Aut7 lipidation (31) and demonstrates that Aut7 and the Apg conjugation system function at the same stage in vesicle formation. Cvt19 is another component that remains associated with the completed transport vesicles, but unlike Aut7, Cvt19 does not appear to play a role in vesicle formation. Instead, Cvt19 appears to function as a specific receptor for concentrating biosynthetic cargo, including prApe1, that transits inside of autophagosomes/Cvt vesicles (26). Perhaps concomitant with the Aut7 membrane recruitment events, the cargo-bound Cvt19 receptor presumably concentrates at the membrane sites of nascent vesicle formation so that efficient cargo packaging can occur. Our microscopy results place the Apg conjugation proteins, the Cvt19 cargo receptor, and the Aut7 vesicle component in the same perivacuolar compartment as Cvt9 and Apg1 (Figs. 1–3). The co-localization of the cargo packaging and vesicle formation machinery to the same compartment as the signaling complex suggests that the transduction of the Tor kinase-dependent signals to the Apg1 kinase complex and then to downstream effectors all occurs at the perivacuolar Apg/Cvt compartment.
The density gradient analyses revealed that while Aut7, Cvt19, and the Apg conjugation components co-fractionated with each other, they did not share the same density profile with Cvt9 and the Apg1 signaling complex (Figs. 5 and 6). Machinery involved in or associated with vesicle formation may be recruited to subdomains within the perivacuolar compartment. The segregation of Aut7, Cvt19, and Apg12/Apg5 into a separate subdomain is further suggested by biochemical immunoisolation experiments using protein A-Aut7ΔR (Fig. 7). Cvt19, and to a lesser extent Apg12, could be recovered along with protein A-Aut7ΔR, whereas Cvt9 and Apg9 were not co-purified with this fusion protein. The biochemical isolation and manipulation of organelles tend to vesiculate membranes, resulting in a heterogeneous vesicle population with some fractions containing protein complexes that are concentrated to localized regions within the organelle. In that regard, the density gradients may be able to provide increased resolution of a given membrane compartment. There is increasing evidence for subdomains in the endomembrane system. For example, the transitional ER is a specialized region containing COPII components that are involved in packaging proteins for transport to the Golgi complex (53, 54). The Golgi complex has long been divided into separate cisternae based on biochemical and morphological criteria. However, recent data (reviewed in Ref. 55) have indicated that this is a highly dynamic organelle composed of constantly changing subdomains that are associated with specific protein components. Similarly, in mammalian cells, different endosomal populations are associated with distinct populations of Rab proteins (reviewed in Ref. 56).
Alternatively, the peak membrane fractions to which Aut7, Cvt19, and the Apg conjugation components all co-localize by density gradients may constitute a distinct compartment that substantially overlaps, both in density and physical proximity, with the Apg1/Cvt9-localized membrane. According to this model, the close proximity of the two perivacuolar compartments would make resolving them by conventional fluorescence microscopy unlikely but instead would require immunoelectron microscopy analysis.
Canonical cargo receptors recycle between the donor and the acceptor compartments to ensure continued rounds of cargo binding at the donor and release at the acceptor membranes. For example, the cargo receptor Vps10 transports Prc1 from the late Golgi to the PVC. The steady-state, subcellular distribution of Vps10 appears predominantly at the late Golgi donor compartment, where Prc1 binding and packaging into transport vesicles occurs (57, 58). Analogous to Vps10, Cvt19 appeared distributed to two subcellular locations (26) (Fig. 6B). The predominant pool of Cvt19 resides at the perivacuolar Apg/Cvt compartment, whereas a much smaller level can be detected accumulated in the vacuole of pep4Δ cells. Extrapolating from the donor/acceptor compartment distribution of Vps10, the localization pattern for the Cvt19 cargo receptor suggests that the perivacuolar Apg/Cvt compartment may serve as the donor membrane compartment for Cvt vesicles and autophagosomes.
A recent study (29) of the Apg5 conjugation component in mouse embryonic stem cells indicated that Apg5 is localized to forming vesicles but dissociates at a step just prior to vesicle completion. Importantly, the major species of Apg5 was detected in the Apg12-conjugated form. We also detected the Apg12-Apg5 conjugate by density gradients (data not shown). However, unlike the free monomers of Apg5 and Apg12, the Apg12-Apg5 conjugate did not co-fractionate with other components that function during vesicle formation (e.g. Aut7 and Cvt19) but instead peaked in fraction 2 of the density gradients (data not shown), in agreement with previously published density reports of the conjugate (27). The study in mouse embryonic stem cells localized the Apg12-Apg5 conjugate to a membrane structure that may represent an intermediate, incomplete transport vesicle, termed the isolation membrane. If an equivalent mechanism exists in yeast, then the free forms of Apg5 and Apg12 that reside at the perivacuolar compartment would exit the donor membrane in the conjugated form and associate with the pre-vesicular, isolation membranes. Subsequent vesicle completion and dissociation of the Apg12-Apg5 conjugate would then be followed by vesicle targeting to the vacuole. Further studies will be needed to clarify this issue.
A PI 3-Kinase Complex Required for the Apg/Cvt Pathways Is Localized to the Perivacuolar Compartment
In this study, we co-localized Apg14, a putative specificity factor for the Cvt and autophagy-specific PI 3-kinase complex, to the Cvt9-associated perivacuolar compartment. The core PI 3-kinase complex (Vps15, -30, and -34) may function at different membrane sites as has been suggested recently (51). The PI 3-kinase complex containing Vps38 may function primarily at the Golgi to mediate Prc1 targeting to the PVC, whereas the Apg14-associated PI 3-kinase complex may function at the perivacuolar, endosomal compartment to mediate Cvt and autophagy membrane traffic to the vacuole.
The localization of Apg14, and by extension, the Apg/Cvt-specific PI 3-kinase complex, to the perivacuolar compartment (Fig. 2) has significant mechanistic consequences for the role of this compartment in membrane movement and vesicle formation. Phosphoinositide modifications by their kinases can be concentrated to membrane microdomains that serve to spatially restrict signaling to a localized region of a membrane compartment. A null mutation in APG14 prevents vesicle formation in both the autophagy and Cvt pathways, suggesting a key role for the Apg/Cvt PI 3-kinase complex in the formation of these transport vesicles and implicating the Apg/Cvt perivacuolar compartment as the potential donor compartment for vesicle formation.
The Perivacuolar Compartment Is the Donor Site for Vesicle Formation
To illustrate the physiological relevance of the Apg/Cvt perivacuolar compartment, we followed the steps in GFP-Aut7 transport to the vacuole by real time microscopy in the apg9ts pep4Δ background (Fig. 8). At nonpermissive temperature, a significant population of GFP-Aut7 was arrested at its initial site of recruitment to the perivacuolar compartment, prior to vesicle formation. A shift to permissive temperature resulted in increased membrane binding and the emergence of smaller GFP-Aut7 localized membrane structures. These structures may represent completed transport vesicles; alternatively, they may be equivalent to the pre-vesicular isolation membranes observed in mouse embryonic stem cells. Finally, GFP-Aut7 was detected in the vacuolar lumen, indicating the completion of the GFP-Aut7 transport itinerary. Future research will focus on the mechanisms that mediate membrane exit from the perivacuolar compartment as well as in vitro reconstitution of the vesicle formation process.
Acknowledgments
We thank Dan Nice and Drs. Sarah Teter and Hagai Abeliovich for helpful discussions.
Footnotes
This work was supported by United States Public Health Service Grant GM53396 from the National Institutes of Health (to D. J. K.) and by a grant from The Research Council of Norway (to P. E. S.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The abbreviations used are: Cvt, cytoplasm to vacuole targeting; Apg, autophagy; CFP, cyan fluorescent protein; Dpm1, dolichol-phosphate mannose synthase; GFP, green fluorescent protein; ORF, open reading frame; PE, phosphatidylethanolamine; Pho8, alkaline phosphatase; PI, phosphatidylinositol; Prc1, carboxypeptidase Y; YFP, yellow fluorescent protein; ECFP, enhanced CFP; EYFP, enhanced YFP; ER, endoplasmic reticulum; Ape1, aminopeptidase I; HA, hemagglutinin; prApe1, precursor Ape1; PVC, prevacuolar compartment.
J. Kim, W.-P. Huang, P. E. Stromhaug, and D. J. Klionsky, unpublished results.
REFERENCES
- 1.Paglin S, Hollister T, Delohery T, Hackett N, McMahill M, Sphicas E, Domingo D, Yahalom J. Cancer Res. 2001;61:439–444. [PubMed] [Google Scholar]
- 2.Bursch W, Ellinger A, Kienzl H, Török L, Pandey S, Sikorska M, Walker R, Hermann RS. Carcinogenesis. 1996;17:1595–1607. doi: 10.1093/carcin/17.8.1595. [DOI] [PubMed] [Google Scholar]
- 3.Cavallini G, Donati A, Gori Z, Pollera M, Bergamini E. Exp. Gerontol. 2001;36:497–506. doi: 10.1016/s0531-5565(00)00224-2. [DOI] [PubMed] [Google Scholar]
- 4.Klionsky DJ, Emr SD. Science. 2000;290:1717–1721. doi: 10.1126/science.290.5497.1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bursch W. Cell Death Differ. 2001;8:569–581. doi: 10.1038/sj.cdd.4400852. [DOI] [PubMed] [Google Scholar]
- 6.Liang XH, Jackson S, Seaman M, Brown K, Kempkes B, Hibshoosh H, Levine B. Nature. 1999;402:672–676. doi: 10.1038/45257. [DOI] [PubMed] [Google Scholar]
- 7.Anglade P, Vyas S, Javoy-Agid F, Herrero MT, Michel PP, Marquez J, Mouatt-Prigent A, Ruberg M, Hirsch EC, Agid Y. Histol. Histopathol. 1997;12:25–31. [PubMed] [Google Scholar]
- 8.Cataldo AM, Barnett JL, Berman SA, Li J, Quarless S, Bursztajn S, Lippa C, Nixon RA. Neuron. 1995;14:671–680. doi: 10.1016/0896-6273(95)90324-0. [DOI] [PubMed] [Google Scholar]
- 9.Saftig P, Tanaka Y, Lullmann-Rauch R, von Figura K. Trends Mol. Med. 2001;7:37–39. doi: 10.1016/s1471-4914(00)01868-2. [DOI] [PubMed] [Google Scholar]
- 10.Kim J, Klionsky DJ. Annu. Rev. Biochem. 2000;69:303–342. doi: 10.1146/annurev.biochem.69.1.303. [DOI] [PubMed] [Google Scholar]
- 11.Dunn WA., Jr. Trends Cell Biol. 1994;4:139–143. doi: 10.1016/0962-8924(94)90069-8. [DOI] [PubMed] [Google Scholar]
- 12.Stromhaug PE, Klionsky DJ. Traffic. 2001;2:524–531. doi: 10.1034/j.1600-0854.2001.20802.x. [DOI] [PubMed] [Google Scholar]
- 13.Abeliovich H, Klionsky DJ. Microbiol. Mol. Biol. Rev. 2001;65:463–479. doi: 10.1128/MMBR.65.3.463-479.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tsukada M, Ohsumi Y. FEBS Lett. 1993;333:169–174. doi: 10.1016/0014-5793(93)80398-e. [DOI] [PubMed] [Google Scholar]
- 15.Thumm M, Egner R, Koch M, Schlumpberger M, Straub M, Veenhuis M, Wolf DH. FEBS Lett. 1994;349:275–280. doi: 10.1016/0014-5793(94)00672-5. [DOI] [PubMed] [Google Scholar]
- 16.Harding TM, Morano KA, Scott SV, Klionsky DJ. J. Cell Biol. 1995;131:591–602. doi: 10.1083/jcb.131.3.591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Harding TM, Hefner-Gravink A, Thumm M, Klionsky DJ. J. Biol. Chem. 1996;271:17621–17624. doi: 10.1074/jbc.271.30.17621. [DOI] [PubMed] [Google Scholar]
- 18.Baba M, Osumi M, Scott SV, Klionsky DJ, Ohsumi Y. J. Cell Biol. 1997;139:1687–1695. doi: 10.1083/jcb.139.7.1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Scott SV, Baba M, Ohsumi Y, Klionsky DJ. J. Cell Biol. 1997;138:37–44. doi: 10.1083/jcb.138.1.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Scott SV, Hefner-Gravink A, Morano KA, Noda T, Ohsumi Y, Klionsky DJ. Proc. Natl. Acad. Sci. U. S. A. 1996;93:12304–12308. doi: 10.1073/pnas.93.22.12304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kirisako T, Baba M, Ishihara N, Miyazawa K, Ohsumi M, Yoshimori T, Noda T, Ohsumi Y. J. Cell Biol. 1999;147:435–446. doi: 10.1083/jcb.147.2.435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huang W-P, Scott SV, Kim J, Klionsky DJ. J. Biol. Chem. 2000;275:5845–5851. doi: 10.1074/jbc.275.8.5845. [DOI] [PubMed] [Google Scholar]
- 23.Abeliovich H, Dunn WA, Jr., Kim J, Klionsky DJ. J. Cell Biol. 2000;151:1025–1034. doi: 10.1083/jcb.151.5.1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Springer S, Spang A, Schekman R. Cell. 1999;97:145–148. doi: 10.1016/s0092-8674(00)80722-9. [DOI] [PubMed] [Google Scholar]
- 25.Kirchhausen T. Nat. Rev. Mol. Cell. Biol. 2000;1:187–198. doi: 10.1038/35043117. [DOI] [PubMed] [Google Scholar]
- 26.Scott SV, Guan J, Hutchins MU, Kim J, Klionsky DJ. Mol. Cell. 2001;7:1131–1141. doi: 10.1016/s1097-2765(01)00263-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mizushima N, Noda T, Yoshimori T, Tanaka Y, Ishii T, George MD, Klionsky DJ, Ohsumi M, Ohsumi Y. Nature. 1998;395:395–398. doi: 10.1038/26506. [DOI] [PubMed] [Google Scholar]
- 28.George MD, Baba M, Scott SV, Mizushima N, Garrison BS, Ohsumi Y, Klionsky DJ. Mol. Biol. Cell. 2000;11:969–982. doi: 10.1091/mbc.11.3.969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mizushima N, Yamamoto A, Hatano M, Kobayashi Y, Kabeya Y, Suzuki K, Tokuhisa T, Ohsumi Y, Yoshimori T. J. Cell Biol. 2001;152:657–667. doi: 10.1083/jcb.152.4.657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kirisako T, Ichimura Y, Okada H, Kabeya Y, Mizushima N, Yoshimori T, Ohsumi M, Takao T, Noda T, Ohsumi Y. J. Cell Biol. 2000;151:263–276. doi: 10.1083/jcb.151.2.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ichimura Y, Kirisako T, Takao T, Satomi Y, Shimonishi Y, Ishihara N, Mizushima N, Tanida I, Kominami E, Ohsumi M, Noda T, Ohsumi Y. Nature. 2000;408:488–492. doi: 10.1038/35044114. [DOI] [PubMed] [Google Scholar]
- 32.Kim J, Huang W-P, Klionsky DJ. J. Cell Biol. 2001;152:51–64. doi: 10.1083/jcb.152.1.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Noda T, Kim J, Huang W-P, Baba M, Tokunaga C, Ohsumi Y, Klionsky DJ. J. Cell Biol. 2000;148:465–480. doi: 10.1083/jcb.148.3.465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Klionsky DJ, Cueva R, Yaver DS. J. Cell Biol. 1992;119:287–299. doi: 10.1083/jcb.119.2.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kim J, Kamada Y, Stromhaug PE, Guan J, Hefner-Gravink A, Baba M, Scott SV, Ohsumi Y, Dunn WA, Jr., Klionsky DJ. J. Cell Biol. 2001;153:381–396. doi: 10.1083/jcb.153.2.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Labbé S, Thiele DJ. Methods Enzymol. 1999;306:145–153. doi: 10.1016/s0076-6879(99)06010-3. [DOI] [PubMed] [Google Scholar]
- 37.Wang C-W, Kim J, Huang W-P, Abeliovich H, Stromhaug PE, Dunn WA, Jr., Klionsky DJ. J. Biol. Chem. 2001;276:30442–30451. doi: 10.1074/jbc.M102342200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wurmser AE, Sato TK, Emr SD. J. Cell Biol. 2000;151:551–562. doi: 10.1083/jcb.151.3.551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kim J, Dalton VM, Eggerton KP, Scott SV, Klionsky DJ. Mol. Biol. Cell. 1999;10:1337–1351. doi: 10.1091/mbc.10.5.1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Scott SV, Nice DC, III, Nau JJ, Weisman LS, Kamada Y, Keizer-Gunnink I, Funakoshi T, Veenhuis M, Ohsumi Y, Klionsky DJ. J. Biol. Chem. 2000;275:25840–25849. doi: 10.1074/jbc.M002813200. [DOI] [PubMed] [Google Scholar]
- 41.Kamada Y, Funakoshi T, Shintani T, Nagano K, Ohsumi M, Ohsumi Y. J. Cell Biol. 2000;150:1507–1513. doi: 10.1083/jcb.150.6.1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mizushima N, Noda T, Ohsumi Y. EMBO J. 1999;18:3888–3896. doi: 10.1093/emboj/18.14.3888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shintani T, Mizushima N, Ogawa Y, Matsuura A, Noda T, Ohsumi Y. EMBO J. 1999;18:5234–5241. doi: 10.1093/emboj/18.19.5234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kametaka S, Okano T, Ohsumi M, Ohsumi Y. J. Biol. Chem. 1998;273:22284–22291. doi: 10.1074/jbc.273.35.22284. [DOI] [PubMed] [Google Scholar]
- 45.Ohsumi Y. Nat. Rev. Mol. Cell. Biol. 2001;2:211–216. doi: 10.1038/35056522. [DOI] [PubMed] [Google Scholar]
- 46.Guan J, Stromhaug PE, George MD, Habibzadegah-Tari P, Bevan A, Dunn WA, Jr., Klionsky DJ. Mol. Biol. Cell. 2001;12:3821–3838. doi: 10.1091/mbc.12.12.3821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Straub M, Bredschneider M, Thumm M. J. Bacteriol. 1997;179:3875–3883. doi: 10.1128/jb.179.12.3875-3883.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Simonsen A, Wurmser AE, Emr SD, Stenmark H. Curr. Opin. Cell Biol. 2001;13:485–492. doi: 10.1016/s0955-0674(00)00240-4. [DOI] [PubMed] [Google Scholar]
- 49.Stack JH, Dewald DB, Takegawa K, Emr SD. J. Cell Biol. 1995;129:321–334. doi: 10.1083/jcb.129.2.321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Herman PK, Emr SD. Mol. Cell. Biol. 1990;10:6742–6754. doi: 10.1128/mcb.10.12.6742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kihara A, Noda T, Ishihara N, Ohsumi Y. J. Cell Biol. 2001;152:519–530. doi: 10.1083/jcb.152.3.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Noda T, Ohsumi Y. J. Biol. Chem. 1998;273:3963–3966. doi: 10.1074/jbc.273.7.3963. [DOI] [PubMed] [Google Scholar]
- 53.Kuehn MJ, Schekman R. Curr. Opin. Cell Biol. 1997;9:477–483. doi: 10.1016/s0955-0674(97)80022-1. [DOI] [PubMed] [Google Scholar]
- 54.Orci L, Ravazzola M, Meda P, Holcomb C, Moore HP, Hicke L, Schekman R. Proc. Natl. Acad. Sci. U. S. A. 1991;88:8611–8615. doi: 10.1073/pnas.88.19.8611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Glick BS. Curr. Opin. Cell Biol. 2000;12:450–456. doi: 10.1016/s0955-0674(00)00116-2. [DOI] [PubMed] [Google Scholar]
- 56.Zerial M, McBride H. Nat. Rev. Mol. Cell. Biol. 2001;2:107–117. doi: 10.1038/35052055. [DOI] [PubMed] [Google Scholar]
- 57.Marcusson EG, Horazdovsky BF, Cereghino JL, Gharakhanian E, Emr SD. Cell. 1994;77:579–586. doi: 10.1016/0092-8674(94)90219-4. [DOI] [PubMed] [Google Scholar]
- 58.Cooper AA, Stevens TH. J. Cell Biol. 1996;133:529–541. doi: 10.1083/jcb.133.3.529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Robinson JS, Klionsky DJ, Banta LM, Emr SD. Mol. Cell. Biol. 1988;8:4936–4948. doi: 10.1128/mcb.8.11.4936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gerhardt B, Kordas TJ, Thompson CM, Patel P, Vida T. J. Biol. Chem. 1998;273:15818–15829. doi: 10.1074/jbc.273.25.15818. [DOI] [PubMed] [Google Scholar]