Abstract
There is increasing interest in endocytosis that occurs independently of clathrin coats and the fates of membrane proteins internalized by this mechanism. The appearance of clathrin-independent endocytic and membrane recycling pathways seems to vary with different cell types and cargo molecules. In this review we focus on studies that have been performed using Hela and COS cells as model systems for understanding this membrane trafficking system. These endosomal membranes contain signaling molecules including H-Ras, Rac1, Arf6 and Rab proteins, and a lipid environment rich in cholesterol and PIP2 providing a unique platform for cell signaling. Furthermore, activation of some of these signaling molecules (H-Ras, Rac and Arf6) can switch the constitutive form of clathrin-independent endocytosis into a stimulated one, associated with PM ruffling and macropinocytosis.
Keywords: Arf6, clathrin-independent, endocytosis, macropinocytosis, phosphoinositides, signaling, src, ras
1. Introduction
Endocytosis is a mechanism for cells to remove ligands, nutrients, and plasma membrane (PM) proteins and lipids from the cell surface, bringing them into the cell interior. Once internalized, the membrane and content of the resulting endosome can meet different fates such as movement to late endosomes and lysosomes for degradation, or recycling back out to the PM. Most of the current research on endocytosis has focused on clathrin-dependent endocytosis (CDE).
The hallmark of CDE is the selective sorting of PM proteins by cytosolic adaptor proteins (APs) into clathrin-coated depressions at the surface prior to formation of the clathrin-coated vesicle. Trans-membrane proteins entering through CDE have sequences in their cytoplasmic domains that bind to the APs and enable their rapid removal from the PM [1]. In addition to APs and clathrin, there are numerous accessory proteins that are involved in CDE [2] including dynamin, a GTPase involved in vesicle scission. Immediately after endocytosis, the clathrin/AP coat is released and the vesicle then fuses with the “classical” early endosomal compartment that is defined by the presence of Rab5 and phosphatidylinositol 3-phosphate (PI3P) (see Fig. 1). Once in the early endosome, membrane proteins, lipids and the fluid content of the endosome are sorted and transported either to the trans-Golgi network, to late endosomes and lysosomes for degradation, or into membrane carriers that recycle back to the PM. The CDE pathway is important not only to facilitate the uptake of nutrients, such as iron-loaded transferrin and LDL, into the cell but also for the rapid internalization of most signaling receptors after ligand binding.
Figure 1.
Schematic illustration of CIE and CDE pathways showing lipid content and regulatory molecules. Constitutive CIE vesicles contain cargo such as MHCI (blue bars) while CDE vesicles contain cargo such as transferrin receptor (green bars). CIE vesicles lose PIP2 and may acquire Rab5 prior to fusion with Rab5, EEA1-positive, “classical” early endosome (EE) containing CDE cargo. From here, CIE cargo may go on to late endosomes (LE) and lysosomes for degradation. CIE cargo destined for recycling reaches the endosomal recycling compartment (ERC) either directly or after fusion with “classical” EE and is recycled back out to the PM via tubular membranes that do not contain CDE cargo. Recycling back out to the PM requires Rab22 and probably Rab8 for tubule formation and Rab11, Arf6, Par 3, Par6, Cdc42 and actin for fusion back to the PM. Activation of Arf6, Ras and Src leads to PM ruffling and macropinocytosis that alters the architecture of the CIE. Macropinosomes mature from PIP2/PIP3-containing by loss of PIP2, acquisition of Rab5 and loss of PIP3. The bulk of the macropinosome membrane is recycled back out to the PM using CIE pathway recycling components.
There are alterative mechanisms to bring membrane into the cell independently of clathrin. Endocytosis can occur through structures coated with the caveolin protein, and this caveolae-mediate endocytosis, like CDE, is dependent upon dynamin and is responsible for endocytosis of some proteins that partition into cholesterol-rich membrane domains, especially in endothelial cells. There are also two mechanisms of specialized endocytosis that are strictly dependent on cortical actin: phagocytosis, carried out by specialized cells for internalization of large particles such as bacteria, and macropinocytosis, associated with membrane ruffling and involving internalization of large volumes of extracellular fluid and the associated membrane.
Finally, investigators have been studying endocytosis that occurs independently of both clathrin and dynamin. There is some evidence that there might be distinct mechanisms of clathrin-independent endocytosis (CIE) depending upon the cargo and the cell type. The reader is referred to a thorough discussion of this in reviews by Mayor and Pagano [3] and Sandvig et al. [4]. Here we will confine our discussion to the CIE pathway that we and others have been studying in HeLa and COS cells that we believe can serve as a model system for understanding these alternative endosomal systems. By “CIE”, we are referring to the mechanism of endocytosis and when we refer to “CIE pathways” we are referring not only to the mode of endocytosis but also the itinerary and fates of proteins and lipids that are associated with these membranes.
2. CIE in HeLa cells - unique sets of cargo, regulatory GTPases and membrane lipids
For more than ten years, we have been studying a CIE pathway in HeLa cells that is clearly distinct from, yet intersects with, the CDE pathway (Fig. 1). A variety of endogenous proteins thought to reside only at the PM have been identified that travel into the cell along this pathway including the major histocompatibility Class I protein (MHCI) [5, 6], integrins [7, 8], K channels [9], E-cadherin [10], Syndecan 1 [11] and CD59, a protein anchored to the membrane by a glycosylphosphatidyl inositol (GPI) moiety [12]. Additionally, other cargo proteins have been shown to traffic through this pathway in transfection experiments including the metabotropic glutamate receptor 7 [13], a major myelin-associated protein PMP22 [14], the cation channel protein mucolipin- 2 [15], peptide loaded MHC Class II [16] and SNAP25, a SNARE protein [17]. The cargo proteins that travel along the CIE pathway include both detergent-resistant, “raft” proteins (CD59) and non-raft proteins, and range in function from regulating cell-cell and cell-matrix interactions to cell signaling. The list of PM proteins that follow this CIE pathway is likely to grow in the future.
The itinerary of two of these proteins, MHCI and CD59, has been studied in detail in HeLa cells (Fig. 1). Both MHCI and CD59 can be observed in the same endosome shortly (3–5 minutes) after internalization. These endosomes are distinct from those containing cargo, such as transferrin receptor, that enters by CDE. At later times (10–15 min), MHCI and CD59 can be observed in classical early endosomes containing the transferrin receptor, Rab5, and the early endosomal antigen 1 (EEA1). EEA1 is a tethering protein that is recruited to membranes by phosphatidylinositol 3-phosphate (PI3P) and Rab5-GTP, and together they facilitate early endosome fusion. From here, MHCI and CD59 can be either routed to late endosomes for degradation [5] or recycled back to the PM via distinctive tubular membranes, lacking tranferrin receptor, that emanate from the juxtanuclear endocytic recycling compartment (ERC) [5, 12, 18].
Recycling of MHCI and β1-integrin back to the PM requires the functioning of Rab and Arf6 GTPases (Fig. 1). Rab22 is specifically required for the formation of the recycling membranous tubules carrying MHCI back to the PM [18]. Additionally, Rab11 is necessary for recycling of MHCI and integrin back to the PM [8, 18]. The recycling of transferrin receptor back to the PM in separate carriers also requires Rab11, but not Rab22 [18]. Rab8 may act similarly to Rab22 [19], and Rab35 has also been observed on the recycling membranes [16]. Arf6 activity is required for MHCI [6], integrin [8] and syndecan 1 [11] recycling, and part of this Arf6 requirement is for the activation of phospholipase D [20]. The final step of recycling back to the PM is dependent on actin. Treatment of Hela cells with inhibitors of actin polymerization blocks recycling [8, 18] and causes a build up of tubular endosomes that are characteristic of the CIE pathway in HeLa cells [6]. These tubules are aligned along microtubules and we have found that recycling back to the PM is also microtubule dependent (unpublished observations).
Other accessory proteins have been identified that are involved in the sorting and routing of membrane through this pathway. The epsin homology domain (EHD) proteins 1 and 3 have been implicated in recycling of MHCI and also transferrin receptor back to the PM [21–23]. The cation channel mucolipin-2 is a cargo protein that travels alont the CIE pathway. Its overexpression activates Arf6 and its depletion inhibits recycling of CIE cargo back to the PM [15] suggesting a role in recycling. Furthermore, there are Rab11 interacting proteins, FIP2 and 3, that appear to act at particular sites in the pathway [22, 23]. Rab11-FIP3 also can associate with active Arf6, suggesting some Rab11/Arf6 coordination [24]. There is also evidence that a myosin might function during recycling, either in conjunction with Rab11 or Rab8 [25]. Interestingly, a recent study in C. elegans has identified anterior partitioning (PAR) proteins including Par3, Par6, and Cdc42 as being important for endosomal recycling in both coelomocytes and for MHCI recycling in Hela cells [26].
Why do cells have an endocytic pathway parallel to that of CDE? One explanation is that this allows those PM proteins that lack AP2 sorting sequences to be endocytosed into cells so that they may be routed to late endosomes for degradation. In addition, the CIE internalization and recycling pathway would allow cells to recycle membrane to particular sites at the PM. This may be why in many cellular contexts, Arf6 activation and subsequent membrane recycling is required for changes in the cortical actin cytoskeleton at the PM. Arf6 activation is necessary for a wide variety of such PM restructuring including PM ruffling, cell migration, wound healing and cancer cell metastasis [27]. In addition to providing membrane for structuring such events, the recycling membrane includes proteins that may participate in these alterations of membrane structure. The presence of integrins, cadherins, syndecans, Rac and Arf6 on these recycled membranes supports this idea. Furthermore, the trafficking of these signaling molecules into and out of cells via the CIE pathway may allow them to avoid the degradative compartments encountered in the CDE pathway. Indeed, this pathway may be the route of entry for a number of viruses and bacteria, and could provide a suitable intracellular niche for bacteria.
The CIE and CDE pathways in HeLa cells run in parallel in some respects but may differ in the step that is most regulated. The requirement for so many components for recycling in the CIE pathway (see Fig. 1) contrasts with relatively few for recycling from CDE pathways. On the other hand, very few requirements for internalization have been identified for CIE whereas CDE involves numerous accessory proteins [2]. It is intriguing that some of the proteins required for CDE such as Par3, Par6 and CDC42, turn up as requirements for recycling in the CIE pathway [28]. Additionally, actin is required for recycling in the CIE pathway [6] but may be involved in the CDE internalization step [29]. The use of the same regulators for endocytosis in one pathway and recycling in the other might facilitate the coordination of these two pathways and maintenance of PM homeostasis.
A distinguishing feature of the CIE pathway is that the lipid composition of the endocytic structures and the resultant endosomal compartments resemble that of the PM. Incoming CIE endosomes contain cholesterol and PIP2 [12]. Cholesterol appears to be important for CIE since the cholesterol binding drug filipin inhibits internalization of both MHCI and CD59, but not transferrin receptor [12]. Indeed, as noted earlier, both detergent-resistant proteins that reside in cholesterol and glycosphinoglipid-enriched membrane domains and proteins that are fully detergent soluble move into and out of cells via this pathway. The presence of phosphatidylinositol 4,5-bisphosphate (PIP2) on substantial parts of the CIE pathway is due to the presence of phosphatidylinositol 4-phosphate 5-kinase (PIP5-kinase) and Arf6 which activates PIP5-kinase on these membranes [7, 30]. However, Arf6 needs to be inactivated and PIP2 lost shortly after internalization since overexpression of either the constitutively active mutant of Arf6, Q67L, or PIP5-kinase leads to accumulation of enlarged vacuolar membranes that form from the fusion of newly internalized membranes [7]. These vacuolar structures are enriched in PIP2 and cholesterol, are actin coated, and contain cargo proteins that are internalized via CIE [5, 7, 9, 11, 12]. As a consequence, further transport of membrane and cargo from this compartment to classical early endosomes, late endosomes or to recycling is blocked. There is presumably an Arf6 GAP that acts to inactivate Arf6 at this step. The loss of PIP2 comes about either by a PIP5-phosphatase or by the action of a phospholipase C (PLC). Arf6 appears to remain inactive until the PM recycling step where Arf6 must again become activated to stimulate PLD [20] and probably PIP5-kinase[7, 20] to generate PA and PIP2.
When considering the possible interaction and cross talk between the two pathways, the CDE pathway is in some respects dependent upon normal functioning of the CIE pathway. Arf6 stimulation of PIP5-kinase maintains the PIP2 levels at the PM required for AP2 and clathrin assembly. Although there is some evidence for Arf6 binding to AP2 in vitro that would suggest a direct role in clathrin assembly [31], it is likely that perturbation of CDE by expression of Arf6Q67L or Arf6 T27N leads to diminished PM PIP2 by sequestering the PIP5-kinase in internal vacuoles and tubules, respectively [7]. More generally, Arf6-GTP at the PM may be critically important for endocytosis of a number of cargo proteins into different endocytic pathways. For example, siRNA depletion of Arf6 inhibits ligand-stimulated internalization of a variety of G protein coupled receptors regardless of their mode of endocytosis [32]. Arf6 is also required for the PM internalization and recycling back to the Golgi of the carboxypeptide E cargo receptor [33]. It is notable that CIE appears to be responsible for the PM cholesterol that enters the cell and is the source of surface cholesterol that reaches later endosomal compartments [12]. On the other hand, it is also likely that inhibition of CDE may affect the itinerary and fate of proteins following the CIE pathway since the composition of the classical early endosomal compartment would be altered. Understanding when and where these pathways converge and the effect of their convergence is an area of research that warrants further study.
Another feature of the CIE membranes is that many proteins associated with PM signaling are present on these membranes. In addition to Arf6, H-Ras, Rac, Erk and Src are found on these membranes. Intriguingly, these regulatory proteins appear to move into and out of the cell along this pathway without crossing over to the classical early endosomes [34] as cargo proteins like MHCI and CD59 do [5, 12]. This raises the question as to why they move into and out of the cell in this manner. The recycling of membrane back to the PM from the CIE pathway appears to be required in instances where changes in the structure of the PM are induced and these signaling molecules reappearing at the PM may be important for initiating these changes.
3. Macropinocytosis is a stimulated form of CIE
An intriguing feature of CIE that sets it apart from CDE is that it can operate constitutively in “resting” cells but, upon stimulation, the architecture of the pathway can change to internalization by macropinocytosis during PM ruffling (Fig. 1). In HeLa and COS cells, this switch to macropinocytosis involves the same cargo proteins and membranes involved in CIE. We have found that this transformation can be initiated through the activation of a number of those signaling molecules that travel along the CIE pathway. Activation of Arf6, Rac, Ras, and Src all lead to PM ruffling and corresponding macropinocytosis. Here we will consider how Arf6, Ras and Src in particular can shift this endocytic pathway into a stimulated one.
We initially encountered macropinocytosis years ago when we were examining the effect of “activation” of Arf6 by treatment of Arf6 overexpressing cells with aluminum fluoride [35]. Such cells appear similar to untransfected cells in the absence of stimulation but upon aluminum fluoride treatment exhibit actin-driven protrusive structures that internalize fluid phase tracers [35] and CIE cargo proteins [7] into large macropinosomes. Most of the fluid and membrane internalized is returned back to the PM. The same phenotype of protrusions and macropinocytosis could be observed by expressing a guanine nucleotide exchange factor (GEF), EFA6, in cells that activates endogenous Arf6 [7]. Furthermore, expression of the downstream effector of Arf6, PIP5-kinase, and resultant generation of PIP2 contributes to these changes in PM dynamics. Further examination of the changes in phosphoinositide composition during EFA6 induced macropinocytosis revealed that in addition to PIP2, phosphatidylinositol 3,4,5 trisphosphate (PIP3) was also present at the PM and on the incoming macropinosome [34]. During maturation of the macropinosome, PIP2 is lost from the macropinosome prior to the loss of PIP3 [34]. For the cell to continue PM ruffling, Arf6 needs to be inactivated and the PIP2 lost for the membrane to move further into the cell and then recycle back out to the PM. Expression of either Arf6Q67L or overexpression of PIP5-kinase leads to a block in recycling and accumulation of vacuolar membranes. These vacuoles, induced by Arf6Q67L, represent trapped macropinosomes.
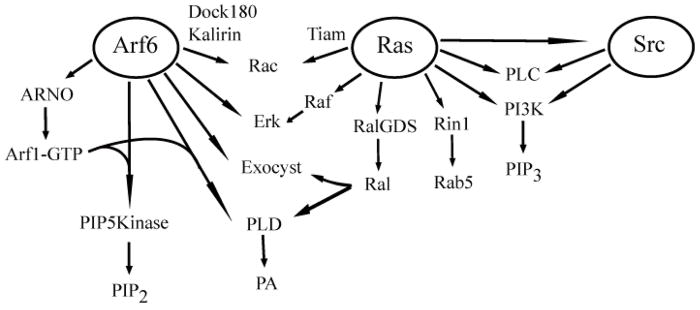
Another way that Arf6 can lead macropinocytosis is through the activation of Rac (Fig. 2), which has itself been linked to ruffling and macropinocytosis [36]. Expression of the Arf GEFs EFA6 [37] and ARNO [38], which activate Arf6, subsequently lead to the activation of Rac. Arf6 may promote Rac activation through endosomal trafficking [39] or through interaction with Rac GEFs Dock180/ELMO [40] and Kalirin [41]. Some TBC (Tre-2, Bu2, Cdc16) domain containing proteins have also been shown to affect Arf6 activation [42] and lead to macropinocytosis [43].
Figure 2.
Downstream effectors of Arf6, Ras and Src that contribute to PM ruffling, macropinocytosis, and membrane recycling. A partial list of effector proteins involved in macropinocytosis emphasizing overlapping targets. See text for details.
In addition to activating Rac, we recently found that Arf6-GTP can amplify its signal at the PM by activating “Golgi” Arfs, such as Arf1 at the PM. Arf6-GTP binds directly to the pleckstrin homology (PH) domains of ARNO/Cytohesin GEFs and thereby recruits these GEFs to the PM where they can activate Arf1. Although ARNO GEFs can activate Arf6 to some extent, in cells and in in vitro biochemical assays Arf1 is the preferred substrate [44, 45]. The binding of the PH domains to Arf6 is also dependent upon the presence of phosphosphoinositides, either PIP2 or PIP3, at the PM and thus recruitment of ARNO is sensitive to both Arf6-GTP and the presence of PIPs [44]. Recently the structure of Grp1, an ARNO/Cytohesin family member, was solved and revealed that the PH domain acts to autoinhibit the Sec7 catalytic domain and that the inhibition is relieved by Arf6-GTP binding to the PH domain [46].
The sequential activation of Arf6 and then Arf1 at the PM amplifies Arf signaling at the PM and possibly on macropinosomes. Since Arf6 is of low abundance in most cell types, the reserve of Arfs that cycle on and off of Golgi membranes are available to function transiently at the PM. Indeed, the activation of Arf1 can itself lead to generation of PM ruffling and macropinocytosis [44]. Arf6 had previously been associated with Fc-mediated phagocytosis, a related, actin-dependent process. A recent study showed that during phagocytosis there is a sequential activation of Arf6 to initiate phagocytic cup formation followed by Arf1 to induce closure [47]. The use of tandem activation provides acute control of signal amplification since Arf6 remains associated with membranes regardless of nucleotide status, whereas ARNO and Arf1 recruitment are dependent upon the presence of Arf6-GTP and PIP2 [44]. Following Arf6 inactivation, ARNO and Arf1 will return to the cytoplasm.
The switch from constitutive to stimulated macropinocytosis observed with activation of Arf6 can also be initiated by activation of H-Ras[48]. Ras proteins regulate diverse cellular functions that range from cell proliferation and differentiation to apoptosis and survival. To carry out these functions activated Ras interacts with a large array of effector proteins. There are three major Ras isoforms, H-, N- and K-Ras, which differ in their carboxyl terminal amino acid sequences and lipid modification required for membrane attachment. It is speculated that Ras protein involvement in many major signaling pathways is achieved through localization to different intracellular compartments. This allows accessibility to different effectors and hence a unique signaling output [49]. Besides the PM, Ras proteins localize and signal from the endoplasmic reticulum (ER) and Golgi membranes [50–52], the mitochondria [53], and “rasosomes” [54]. Finally, Ras proteins were shown to signal from endosomes [55–57] however, the nature of these endosomes was not clearly defined.
We found that wild type H-Ras traffics along the CIE pathway in HeLa and COS cells. Like activation of Arf6, activation of H-Ras either by EGF stimulation or expression of the constitutively active mutant of H-Ras, G12V, leads to PM ruffling and macropinocytosis. The H-Ras-induced macropinocytosis is a stimulated form of CIE that requires proper functioning of Arf6. Expression of Arf6Q67L or Arf6T27N traps RasG12V in vacuolar membranes and tubular recycling endosomes, respectively [34].
During macropinocytosis stimulated by activation of either H-Ras or Arf6, there are changes to the phosphoinositide and protein composition as the macropinosome is brought into the cell (Fig. 1). Both PIP2 and PIP3 are present at the PM and on the incoming macropinosome, but as the macropinosome matures, PIP2 is lost from the membrane first, followed by the loss of PIP3 [34]. In COS cells where this has been examined, we believe that the PIP2 is lost due to the activity of a PIP-phosphatase or phospholipase (PLC). Following the loss of PIP2, Rab5 is recruited onto the macropinosome whether macropinocytosis was induced by H-Ras or Arf6 activation. Rab5 is initially recruited onto the macropinosome while PIP3 is present. However, PI3P and EEA1 do not appear to be associated with these membranes. This Rab5 compartment is thus distinctly different from the Rab5-PI3P-EEA1-positive, “classical” early endosome. It resembles the APPL compartment described by Zerial and colleagues [58] that contains EGF receptor, Rab5 and not EEA1. Interestingly, Rab5 can also recruit the PIP5-phosphatase OCRL [59, 60] onto this compartment which might keep PIP2levels reduced. Following the recruitment of Rab5, the macropinosome becomes more dynamic and can extrude tubular membranes, suggesting that some type of cargo sorting event is occurring (our unpublished observations). At this point it seems that the bulk of the macropinosomal membrane is then recycled back out to the PM along the same pathway that occurs during constitutive endocytosis and recycling (see Fig. 1). Consistent with this, treatment of cells expressing activated H-Ras [34] or activated Arf6 [7] with cytochalsin D (or other actin inhibitors) leads to accumulation of membrane in the tubular recycling endosomes.
Although the same changes in phosphoinositide content and Rab5 recruitment are observed whether macropinosomes were induced through H-Ras or Arf6, the macropinosomes were not identical. Namely, the H-Ras effector Akt is recruited onto Ras-induced macropinosomes but not onto those induced by Arf6 [34]. In contrast, Arf1 is associated with Arf6 but not H-Ras generated macropinosomes (unpublished observations). This raises the possibility that the macropinosome could provide a platform for distinctive signaling by Ras, Arf6 and other signaling molecules.
Src activation also stimulates PM ruffling and macropinosome formation in various cell types [61]. The Src family of tyrosine kinases is comprised of seven highly related members involved in cell division, reorganization of the actin cytoskeleton and other functions in specialized cells [62]. In MDCK cells, macropinocytosis is dependent on PI3K, PLC, PLD and actin remodeling, all of which are Src effectors (Fig 2). Furthermore, these macropinosomes are associated with a unique set of Rab5 effectors, but not with EEA1 [63, 64]. Similarly, the Arf6 or Ras induced macropinosomes were Rab5 positive, but mostly were not associated with EEA1 [34].
What is the relationship between macropinocytosis induced by Ras, Arf6 and Src? First, it is important to note that these three important signaling molecules share some common biological outputs through the activation of common effectors (Fig 2). Furthermore, all are activated in response to different growth factor and have major roles in cell transformation and metastasis. This raises the possibility that in some respects Ras, Arf6 and Src have redundant biological activities, including the induction of membrane ruffling and macropinocytosis. However, as we previously demonstrated for Arf6 and Ras, despite these common features, each can recruit unique effectors to macropinosomes [34, 44]. This might also be the case for Src signaling. Although we have not directly demonstrated that Src macropinocytosis utilizes the CIE membranes, we have observed that phosphorylated forms of endogenous Src are associated with CIE membranes (unpublished observations). Further studies are needed to identify these unique downstream effectors of Arf6, Ras and Src.
4. Endosomal recycling is required for PM remodeling and related to regulated exocytosis
Regardless of whether membrane enters via the constitutive or stimulated macropinocytic pathway, endosomal membranes are recycled back out to the PM in distinctive carriers and exocytosis is dependent upon the factors listed in Fig. 1. In the case of macropinocytosis, it appears as though the recycling occurs rapidly such that the tubular nature of the recycling endosomes is not observed unless recycling is blocked, for instance by inhibitors of actin polymerization or dominant negative Arf6.
The recycling of membrane back to the PM is the step that is most regulated in the CIE pathway and is required for changes in PM structure and architecture. This is most clearly demonstrated by over-expression of proteins that block Arf6 activation, including Arf6T27N and the Arf6 GAPs ACAP1 and 2, which lead to blocks in membrane recycling, cell spreading, and PM ruffling [6, 65, 66]. It can be argued that many of Arf6’s effects such as activation of PIP5kinase, PLD and Rac, might take place at the PM to bring about these changes in cortical actin cytoskeleton and that membrane recycling may not be necessary. However, a recent study in fibroblasts revealed that the recycling of raft-like membrane requires Arf6 [67] and it is this recycling of raft-like membranes that is the requirement for Arf6 in cell spreading. Another study has demonstrated that the requirement for Arf6 in Fc-mediated phagocytosis is for the delivery of endosomal membrane to the forming phagosome [68]. Finally, since signaling molecules, such as Arf6, Rac, Ras, and Src, PIP5kinase and PLD2 are present on these endosomal membranes and may be critical for generating changes in the cell surface structure, inhibition of membrane recycling will diminish their presence at the PM.
Another interesting aspect of the many proteins that control CIE recycling is that the same proteins are utilized during regulated exocytosis. Chromaffin granule exocytosis requires Arf6, PLD, Cdc42, actin [69] and Ral [70]. Insulin-stimulation of Glut4 translocation in adipocytes requires Rab11, actin [71], Arf6 and ACAP1, an Arf6 GAP [72]. Additionally, Arf6 was shown to be required for generation of PIP2 to sustain stimulated insulin secretion from pancreatic beta cells [73].
5. Concluding remarks
In this review we have focused on studies that examined how a CIE membrane system that exists in HeLa and COS cells is regulated and impacts cell surface architecture. There are many questions still to answer in these model systems such as: To what extent are the trafficking pathways altered when the pathway is shifted from constitutive to stimulated macropinocytosis? What is the relationship of the CIE to the CDE pathway? It is important to emphasize that the model depicted in Fig. 1 is meant to provide a general framework for how these noncanonical endosomal membrane systems function. The involvement of particular regulators in any one step of the pathway might be different in different cell types. Hence, it will be important to extend these findings to different cell types to understand how these types of endosomal membranes and their trafficking might be similar or different to that described here.
A key and unifying feature of these CIE membrane systems might be the types of cargo and lipids that move into and out of cells by this pathway and how their directed recycling could control cell architecture and behavior. In addition to the cargo proteins identified that are associated with cell adhesion, the signaling proteins and their effectors present on these endosomal membranes could contribute to cell surface changes associated with a range of cellular activities from cytokinesis and wound healing to cell invasion and migration.
Acknowledgments
We thank Ed Korn and Craig Eyster for comments. This work was supported by the Intramural Research Program of the National Heart, Lung and Blood Institute, NIH.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Conner SD, Schmid SL. Nature. 2003;422(6927):37–44. doi: 10.1038/nature01451. [DOI] [PubMed] [Google Scholar]
- 2.Slepnev VI, De Camilli P. Nature Reviews Neuroscience. 2000;1(3):161–172. doi: 10.1038/35044540. [DOI] [PubMed] [Google Scholar]
- 3.Mayor S, Pagano RE. Nat Rev Mol Cell Biol. 2007;8(8):603–612. doi: 10.1038/nrm2216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sandvig K, Torgersen ML, Raa HA, van Deurs B. Histochem Cell Biol. 2008;129(3):267–276. doi: 10.1007/s00418-007-0376-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Naslavsky N, Weigert R, Donaldson JG. Mol Biol Cell. 2003;14(2):417–431. doi: 10.1091/mbc.02-04-0053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Radhakrishna H, Donaldson JG. J Cell Biol. 1997;139(1):49–61. doi: 10.1083/jcb.139.1.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brown FD, Rozelle AL, Yin HL, Balla T, Donaldson JG. J Cell Biol. 2001;154(5):1007–1017. doi: 10.1083/jcb.200103107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Powelka AM, Sun J, Li J, Gao M, Shaw LM, Sonnenberg A, Hsu VW. Traffic. 2004;5(1):20–36. doi: 10.1111/j.1600-0854.2004.00150.x. [DOI] [PubMed] [Google Scholar]
- 9.Gong Q, Weide M, Huntsman C, Xu Z, Jan LY, Ma D. J Biol Chem. 2007;282(17):13087–13097. doi: 10.1074/jbc.M700767200. [DOI] [PubMed] [Google Scholar]
- 10.Paterson AD, Parton RG, Ferguson C, Stow JL, Yap AS. J Biol Chem. 2003;278(23):21050–21057. doi: 10.1074/jbc.M300082200. [DOI] [PubMed] [Google Scholar]
- 11.Zimmermann P, Zhang Z, Degeest G, Mortier E, Leenaerts I, Coomans C, Schulz J, N’Kuli F, Courtoy PJ, David G. Dev Cell. 2005;9(3):377–388. doi: 10.1016/j.devcel.2005.07.011. [DOI] [PubMed] [Google Scholar]
- 12.Naslavsky N, Weigert R, Donaldson JG. Mol Biol Cell. 2004;15(8):3542–3552. doi: 10.1091/mbc.E04-02-0151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lavezzari G, Roche KW. Neuropharmacology. 2007;52(1):100–107. doi: 10.1016/j.neuropharm.2006.07.011. [DOI] [PubMed] [Google Scholar]
- 14.Chies R, Nobbio L, Edomi P, Schenone A, Schneider C, Brancolini C. J Cell Sci. 2003;116(Pt 6):987–999. doi: 10.1242/jcs.00326. [DOI] [PubMed] [Google Scholar]
- 15.Karacsonyi C, Miguel AS, Puertollano R. Traffic. 2007;8(10):1404–1414. doi: 10.1111/j.1600-0854.2007.00619.x. [DOI] [PubMed] [Google Scholar]
- 16.Walseng E, Bakke O, Roche PA. J Biol Chem. 2008 doi: 10.1074/jbc.M801070200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Aikawa Y, Xia X, Martin TF. Mol Biol Cell. 2006;17(2):711–722. doi: 10.1091/mbc.E05-05-0382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Weigert R, Yeung AC, Li J, Donaldson JG. Mol Biol Cell. 2004;15(8):3758–3770. doi: 10.1091/mbc.E04-04-0342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hattula K, Furuhjelm J, Tikkanen J, Tanhuanpaa K, Laakkonen P, Peranen J. J Cell Sci. 2006;119(Pt 23):4866–4877. doi: 10.1242/jcs.03275. [DOI] [PubMed] [Google Scholar]
- 20.Jovanovic OA, Brown FD, Donaldson JG. Mol Biol Cell. 2006;17(1):327–335. doi: 10.1091/mbc.E05-06-0523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Caplan S, Naslavsky N, Hartnell LM, Lodge R, Polishchuk RS, Donaldson JG, Bonifacino JS. Embo J. 2002;21(11):2557–2567. doi: 10.1093/emboj/21.11.2557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Naslavsky N, Rahajeng J, Sharma M, Jovic M, Caplan S. Mol Biol Cell. 2006;17(1):163–177. doi: 10.1091/mbc.E05-05-0466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wilson GM, Fielding AB, Simon GC, Yu X, Andrews PD, Hames RS, Frey AM, Peden AA, Gould GW, Prekeris R. Mol Biol Cell. 2005;16(2):849–860. doi: 10.1091/mbc.E04-10-0927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fielding AB, Schonteich E, Matheson J, Wilson G, Yu X, Hickson GR, Srivastava S, Baldwin SA, Prekeris R, Gould GW. Embo J. 2005 doi: 10.1038/sj.emboj.7600803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Roland JT, Kenworthy AK, Peranen J, Caplan S, Goldenring JR. Mol Biol Cell. 2007;18(8):2828–2837. doi: 10.1091/mbc.E07-02-0169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Balklava Z, Pant S, Fares H, Grant BD. Nat Cell Biol. 2007;9(9):1066–1073. doi: 10.1038/ncb1627. [DOI] [PubMed] [Google Scholar]
- 27.D’Souza-Schorey C, Chavrier P. Nat Rev Mol Cell Biol. 2006;7(5):347–358. doi: 10.1038/nrm1910. [DOI] [PubMed] [Google Scholar]
- 28.Wissler F, Labouesse M. Nat Cell Biol. 2007;9(9):1027–1029. doi: 10.1038/ncb0907-1027. [DOI] [PubMed] [Google Scholar]
- 29.Merrifield CJ, Feldman ME, Wan L, Almers W. Nat Cell Biol. 2002;4(9):691–698. doi: 10.1038/ncb837. [DOI] [PubMed] [Google Scholar]
- 30.Honda A, Nogami M, Yokozeki T, Yamazaki M, Nakamura H, Watanabe H, Kawamoto K, Nakayama K, Morris AJ, Frohman MA, Kanaho Y. Cell. 1999;99(5):521–532. doi: 10.1016/s0092-8674(00)81540-8. [DOI] [PubMed] [Google Scholar]
- 31.Paleotti O, Macia E, Luton F, Klein S, Partisani M, Chardin P, Kirchhausen T, Franco M. J Biol Chem. 2005;280(22):21661–21666. doi: 10.1074/jbc.M503099200. [DOI] [PubMed] [Google Scholar]
- 32.Houndolo T, Boulay PL, Claing A. J Biol Chem. 2005;280(7):5598–5604. doi: 10.1074/jbc.M411456200. [DOI] [PubMed] [Google Scholar]
- 33.Arnaoutova I, Jackson CL, Al-Awar OS, Donaldson JG, Loh YP. Mol Biol Cell. 2003;14(11):4448–4457. doi: 10.1091/mbc.E02-11-0758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Porat-Shliom N, Kloog Y, Donaldson JG. Mol Biol Cell. 2008;19(3):765–775. doi: 10.1091/mbc.E07-08-0841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Radhakrishna H, Klausner RD, Donaldson JG. J Cell Biol. 1996;134(4):935–947. doi: 10.1083/jcb.134.4.935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dharmawardhane S, Schurmann A, Sells MA, Chernoff J, Schmid SL, Bokoch GM. Mol Biol Cell. 2000;11(10):3341–3352. doi: 10.1091/mbc.11.10.3341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Franco M, Peters PJ, Boretto J, van Donselaar E, Neri A, D’Souza-Schorey C, Chavrier P. Embo J. 1999;18(6):1480–1491. doi: 10.1093/emboj/18.6.1480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Santy LC, Casanova JE. J Cell Biol. 2001;154(3):599–610. doi: 10.1083/jcb.200104019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Radhakrishna H, Al-Awar O, Khachikian Z, Donaldson JG. J Cell Sci. 1999;112( Pt 6):855–866. doi: 10.1242/jcs.112.6.855. [DOI] [PubMed] [Google Scholar]
- 40.Santy LC, Ravichandran KS, Casanova JE. Curr Biol. 2005;15(19):1749–1754. doi: 10.1016/j.cub.2005.08.052. [DOI] [PubMed] [Google Scholar]
- 41.Koo TH, Eipper BA, Donaldson JG. BMC Cell Biol. 2007;8:29. doi: 10.1186/1471-2121-8-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Martinu L, Masuda-Robens JM, Robertson SE, Santy LC, Casanova JE, Chou MM. Mol Cell Biol. 2004;24(22):9752–9762. doi: 10.1128/MCB.24.22.9752-9762.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Frittoli E, Palamidessi A, Pizzigoni A, Lanzetti L, Garre M, Troglio F, Troilo A, Fukuda M, Di Fiore PP, Scita G, Confalonieri S. Mol Biol Cell. 2008;19(4):1304–1316. doi: 10.1091/mbc.E07-06-0594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cohen LA, Honda A, Varnai P, Brown FD, Balla T, Donaldson JG. Mol Biol Cell. 2007;18(6):2244–2253. doi: 10.1091/mbc.E06-11-0998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Macia E, Chabre M, Franco M. J Biol Chem. 2001;276(27):24925–24930. doi: 10.1074/jbc.M103284200. [DOI] [PubMed] [Google Scholar]
- 46.DiNitto JP, Delprato A, Gabe Lee MT, Cronin TC, Huang S, Guilherme A, Czech MP, Lambright DG. Mol Cell. 2007;28(4):569–583. doi: 10.1016/j.molcel.2007.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Beemiller P, Hoppe AD, Swanson JA. PLoS Biol. 2006;4(6):e162. doi: 10.1371/journal.pbio.0040162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bar-Sagi D, Feramisco JR. Science. 1986;233(4768):1061–1068. doi: 10.1126/science.3090687. [DOI] [PubMed] [Google Scholar]
- 49.Hancock JF. Nat Rev Mol Cell Biol. 2003;4(5):373–384. doi: 10.1038/nrm1105. [DOI] [PubMed] [Google Scholar]
- 50.Chiu VK, Bivona T, Hach A, Sajous JB, Silletti J, Wiener H, Johnson RL, 2nd, Cox AD, Philips MR. Nat Cell Biol. 2002;4(5):343–350. doi: 10.1038/ncb783. [DOI] [PubMed] [Google Scholar]
- 51.Goodwin JS, Drake KR, Rogers C, Wright L, Lippincott-Schwartz J, Philips MR, Kenworthy AK. J Cell Biol. 2005;170(2):261–272. doi: 10.1083/jcb.200502063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rocks O, Peyker A, Kahms M, Verveer PJ, Koerner C, Lumbierres M, Kuhlmann J, Waldmann H, Wittinghofer A, Bastiaens PI. Science. 2005;307(5716):1746–1752. doi: 10.1126/science.1105654. [DOI] [PubMed] [Google Scholar]
- 53.Bivona TG, Quatela SE, Bodemann BO, Ahearn IM, Soskis MJ, Mor A, Miura J, Wiener HH, Wright L, Saba SG, Yim D, Fein A, Perez de Castro I, Li C, Thompson CB, Cox AD, Philips MR. Mol Cell. 2006;21(4):481–493. doi: 10.1016/j.molcel.2006.01.012. [DOI] [PubMed] [Google Scholar]
- 54.Rotblat B, Yizhar O, Haklai R, Ashery U, Kloog Y. Cancer Res. 2006;66(4):1974–1981. doi: 10.1158/0008-5472.CAN-05-3791. [DOI] [PubMed] [Google Scholar]
- 55.Rizzo MA, Kraft CA, Watkins SC, Levitan ES, Romero G. J Biol Chem. 2001;276(37):34928–34933. doi: 10.1074/jbc.M105918200. [DOI] [PubMed] [Google Scholar]
- 56.Jiang X, Sorkin A. Mol Biol Cell. 2002;13(5):1522–1535. doi: 10.1091/mbc.01-11-0552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Roy S, Wyse B, Hancock JF. Mol Cell Biol. 2002;22(14):5128–5140. doi: 10.1128/MCB.22.14.5128-5140.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Miaczynska M, Christoforidis S, Giner A, Shevchenko A, Uttenweiler-Joseph S, Habermann B, Wilm M, Parton RG, Zerial M. Cell. 2004;116(3):445–456. doi: 10.1016/s0092-8674(04)00117-5. [DOI] [PubMed] [Google Scholar]
- 59.Erdmann KS, Mao Y, McCrea HJ, Zoncu R, Lee S, Paradise S, Modregger J, Biemesderfer D, Toomre D, De Camilli P. Dev Cell. 2007;13(3):377–390. doi: 10.1016/j.devcel.2007.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hyvola N, Diao A, McKenzie E, Skippen A, Cockcroft S, Lowe M. Embo J. 2006;25(16):3750–3761. doi: 10.1038/sj.emboj.7601274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Veithen A, Cupers P, Baudhuin P, Courtoy PJ. J Cell Sci. 1996;109( Pt 8):2005–2012. doi: 10.1242/jcs.109.8.2005. [DOI] [PubMed] [Google Scholar]
- 62.Courtneidge SA. Biochem Soc Trans. 2002;30(2):11–17. doi: 10.1042/. [DOI] [PubMed] [Google Scholar]
- 63.Amyere M, Payrastre B, Krause U, Van Der Smissen P, Veithen A, Courtoy PJ. Mol Biol Cell. 2000;11(10):3453–3467. doi: 10.1091/mbc.11.10.3453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Mettlen M, Platek A, Van Der Smissen P, Carpentier S, Amyere M, Lanzetti L, de Diesbach P, Tyteca D, Courtoy PJ. Traffic. 2006;7(5):589–603. doi: 10.1111/j.1600-0854.2006.00412.x. [DOI] [PubMed] [Google Scholar]
- 65.Jackson TR, Brown FD, Nie Z, Miura K, Foroni L, Sun J, Hsu VW, Donaldson JG, Randazzo PA. J Cell Biol. 2000;151(3):627–638. doi: 10.1083/jcb.151.3.627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Song J, Khachikian Z, Radhakrishna H, Donaldson JG. J Cell Sci. 1998;111( Pt 15):2257–2267. doi: 10.1242/jcs.111.15.2257. [DOI] [PubMed] [Google Scholar]
- 67.Balasubramanian N, Scott DW, Castle JD, Casanova JE, Schwartz MA. Nat Cell Biol. 2007;9(12):1381–1391. doi: 10.1038/ncb1657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Niedergang F, Colucci-Guyon E, Dubois T, Raposo G, Chavrier P. J Cell Biol. 2003;161:1143–1150. doi: 10.1083/jcb.200210069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Malacombe M, Bader MF, Gasman S. Biochim Biophys Acta. 2006;1763(11):1175–1183. doi: 10.1016/j.bbamcr.2006.09.004. [DOI] [PubMed] [Google Scholar]
- 70.Vitale N, Mawet J, Camonis J, Regazzi R, Bader MF, Chasserot-Golaz S. J Biol Chem. 2005;280(33):29921–29928. doi: 10.1074/jbc.M413748200. [DOI] [PubMed] [Google Scholar]
- 71.Pilch PF. Acta Physiol (Oxf) 2008;192(1):89–101. doi: 10.1111/j.1748-1716.2007.01788.x. [DOI] [PubMed] [Google Scholar]
- 72.Li J, Peters PJ, Bai M, Dai J, Bos E, Kirchhausen T, Kandror KV, Hsu VW. J Cell Biol. 2007;178(3):453–464. doi: 10.1083/jcb.200608033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lawrence JT, Birnbaum MJ. Proc Natl Acad Sci U S A. 2003;100(23):13320–13325. doi: 10.1073/pnas.2232129100. [DOI] [PMC free article] [PubMed] [Google Scholar]