Abstract
Exegesis is a procedure to refine the gene predictions that are produced for complex genomes, e.g. those of humans and mice. It uses the program Genewise, sequences determined by experiment, experimental maps of gene segment libraries and a new browser that allows the user to rapidly inspect and compare multiple gene maps to regions of genomic sequences. The procedure should be of general use. Here, we use the procedure to find members of the immunoglobulin superfamily in the human and mouse genomes. To do this, Exegesis was used to process the original gene predictions from the automated Ensembl annotation pipeline. Exegesis produced (i) many more complete genes and new transcripts and (ii) a mapping of the immunoglobulin and T cell receptor gene libraries to the genome, which are largely absent in the Ensembl set.
INTRODUCTION
We are interested in protein families that have played a major role in evolution. One of these is the immunoglobulin superfamily (IgSF), which is defined as those proteins containing at least one IgSF domain. Some members of this family play a major role in the development and function of the nervous system and of the immune system; other members have important roles in the structure and function of muscle. We began an investigation of the members of the IgSF in humans and mice. To do this we used the predicted protein sequences produced by the analysis of the DNA sequence of the human and mouse genomes. The predicted protein sequences are made available through the Ensembl database (1). Inconsistencies between the IgSF sequence sets in different releases of the Ensembl database, and also discrepancies between Ensembl sequences and those determined by experiment, indicated that there are problems with some of these predictions.
We have developed a procedure we call ‘Exegesis’ that, using gene predictions from the Ensembl annotation method as a starting point, identifies problems and produces solutions to some of them. Here we describe this procedure and show that it makes significant improvements to the predictions of human and mouse IgSF proteins. This procedure is likely to be of general use in improving the prediction of other proteins in the genomes of higher organisms, particularly those that have long sequences.
The predicted human and mouse protein sequences provided by the Ensembl database
A genomic assembly for mouse or human each creates a three billion base sequence space in which to look for a comparatively minute subset of coding regions. As improvements in the quality of the human genome sequence have progressed over the past 3 years, a snapshot of all available valid raw DNA sequence ‘reads’ in the central database has been processed at various intervals to produce a new assembly. These assemblies are known as ‘Freeze Sets’ and each of them is given a sequential number. From each new Freeze Set, new predictions are made for coding regions and hence protein sequences. The protein predictions released by the Ensembl group for the first 11 human Freeze Sets are the basis of much of the work described here. Before going on to describe the use we make of these predictions it is useful to briefly describe how they are derived.
The Ensembl automatic annotation system (V.Curwen, D.Andrews, L.Clarke, E.Eyras, E.Mongin, S.Searle and M.Clamp, submitted for publication) proceeds as follows. For a given DNA Freeze Set, the procedure starts off by masking unwanted repeat regions. The masked DNA is then scanned, using Genscan (2), for ab initio exons (i.e. exon features deduced from their sequence composition without any homology reference whatsoever). Then, using BLAST (3), the resultant Genscan peptide sequences are matched against experimental sequences in SPTREMBL (4), the vertebrate mRNA EMBL subset (5) and sequences from Unigene clusters (6). In the subsequent ‘genebuild’ stage, novel Genscan peptide matches are used to direct Genewise (7) calculations for novel paralogues and orthologues using known human and non-human SPTREMBL sequences, respectively. A parallel source of gene maps in the pipeline involves the use of large-scale mRNA/cDNA/EST matching against the genome using Exonerate (G.Slater, unpublished). Transcriptional splice alternatives are then extracted from contigs of overlapping maps using Est2Genome (8). Genewise maps from these procedures are combined to create Ensembl protein predictions.
PROBLEMS WITH ENSEMBL PROTEIN PREDICTIONS
There are three main issues of concern with Ensembl gene predictions: first, the consistency of the protein predictions from the different human Freeze Sets; secondly, the relation between predictions and known experimental sequences, and thirdly, very few accurate predictions are available for the immunoglobulin and T cell receptor variable (V), joining (J) and diversity (D) gene segment libraries.
Loss of information in successive Freeze Sets
We would expect a comparison of sequence predictions made from the successive human Freeze Sets to show some variation. However, the extent and nature of the variation found when different Freeze Sets were compared was much greater than we might expect from a simple improvement in quality and quantity of the human DNA assembly. It is expected that modifications to the prediction procedure between Freeze Sets would account for some proportion of this discrepancy. To determine the nature of these variations we carried out the following procedure.
(i) In this paper we are concerned with proteins that are members of the IgSF. To identify these, each of the 11 available Ensembl human prediction sets, up to and including the version from the NCBI version 29 assembly (v29), were scanned using SAM software (9) with the same set of SUPERFAMILY IgSF HMMs (10) and an E-value inclusion threshold of 10–2. The same procedure was used to identify mouse IgSF proteins using the predicted sequences from MGSC v3 (v3).
(ii) Those predictions that were found to contain an IgSF domain by the above procedure, which we will call Ensembl IgSF proteins, were matched to the Non-Redundant DataBase (NRDB) protein sequence set from NCBI. The sources for NRDB are GenBank translations (11), NBRF PIR (12), SWISS-PROT (4), RCSB Protein Data Bank (13) and the NCBI Reference Sequence resource (14).
(iii) Different hits to NRDB sequences may arise from particular sequences with minor improvements in successive human Ensembl sets. To eliminate such differences from our comparisons for the human sets the MCD-HIT program (15) was used to cluster the NRDB sequences into groups whose members have sequence identities of 60% or more. Examination of matches made to these clusters gave us the ability to focus on significant match differences made by successive Ensembl releases. An examination of the remaining matches indicated that between successive Freeze Sets there is some loss of what appear to be reasonable predictions.
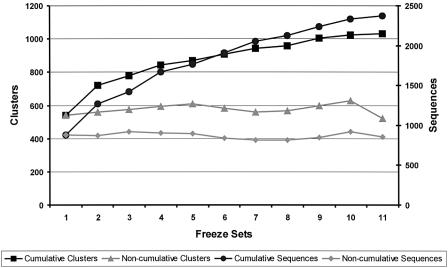
The extent of this loss between subsequent sets of human predictions is shown in Figure 1. In the lower part of the figure we indicate the number of NRDB sequences and sequence clusters individually matched by the chronologically ordered sets of human Ensembl IgSF proteins. This number of distinct matches is very similar for the different sets. However, for each new release, there are new matches to NRDB sequences and sequence clusters, i.e. matches not found by previous releases, and also the absence of some matches that were made by predictions in previous releases. In the upper part of the graph we show the cumulative number of distinct sequences, and sequence clusters, hit by the 11 human Ensembl sets. The cumulative number of sequences is circa three times greater than that which the last set, or any other Ensembl set, could provide. These results indicate that there may be a significant information loss between the prediction calculations of successive Freeze Set releases. The extent of the probable loss is discussed below.
Figure 1.
NRDB WU-BLAST hits from IgSF-bearing Ensembl sequence sets from 11 consecutive Freeze Sets up to and including NCBI build 29. The same set of SUPERFAMILY IgSF HMMs was used for every Ensembl release. These sets are lined up along the x-axis in chronological order of release. Set 11 is the last release. For every Ensembl sequence matched using WU-BLAST, the best NRDB hit was recorded. The same NRDB sequence set was used as target for all 11 sets in this analysis. The graph depicts four approaches to counting best hits. The number of sequences hit is recorded against the right y-axis. (i) Non-cumulative sequence matches: for every Ensembl IgSF set, the absolute number of NRDB sequences matched is depicted. (ii) Cumulative sequence matches: considering the sets on the x-axis from 1 to 11, this plot illustrates the number of distinct NRDB sequences hit at least once by the particular set or any set before it. The number of clusters hit is recorded against the left y-axis. (iii) Non-cumulative cluster hits: NRDB is now considered as a set of sequence clusters over a sequence identity cut-off of 60%. For every Ensembl IgSF set, the absolute number of NRDB clusters hit is shown. (iv) Cumulative cluster hits: considering the sets on the x-axis from 1 to 11, depicted is the number of distinct NRDB clusters hit at least once by the particular set or any set before it.
Length discrepancy between Ensembl and experimental sequences
An important indicator of whether or not a protein prediction is complete can be found from the comparison of prediction lengths with those of any corresponding experimental version of the sequence, as well as the determination of whether or not a stop codon is found at the end of the predicted sequence.
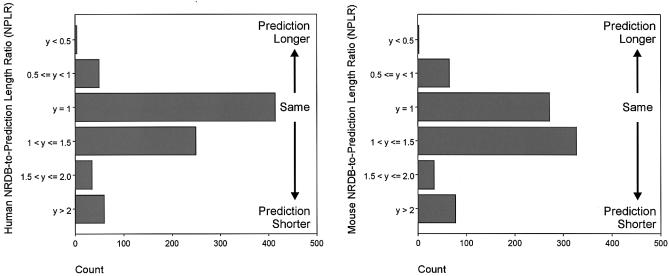
The WU-BLAST mapping to NRDB sequences of the human IgSF predicted proteins detected in the Ensembl v29 was used to ascertain the length relationship between the two. Of the 812 human IgSF proteins, 414 (51%) do have a length identical to the closest NRDB protein but the average length of all the predicted proteins is 70% of the length of the NRDB sequences that they match best (Fig. 2). Note, that in a small number of cases, predominantly involving predictions using fully formed antibody sequences, the predictions are longer than the experimental sequence.
Figure 2.
The distribution of the NRDB-to-prediction length ratio for human (Ensembl v29) and mouse (Ensembl v3) sequences matched against NRDB. The mean ratio for human was 1.4 and that for mouse was 1.6.
The presence of an in-frame stop codon is a complementary index of completeness in the assessment of genes that are not part of an immune system library of gene segments. Testing for the presence of an in-frame stop codon on the 812 human Ensembl v29 gene maps showed that 129 (16%) have a terminator immediately adjacent to the 3′ end, although 342 (45%) have a stop codon within 10 in-frame codons of the 3′ end.
The immunoglobulin and T cell receptor gene segment libraries
Immunoglobulin (antibody) and T cell receptor V, D and J genes rearrange in the developing B and T lymphocytes, respectively, and form mature chains with the constant (C) genes. From previous experimental work, there are known to be over 200 V and C segments in the human genome (Table 1). Ensembl has reported predictions at just over half the sites known from experimental work. However, most of the predictions come from fragments of mature sequences and are usually some 50% greater in length than those of the known gene segments. A similar situation is found for current predictions in the mouse genome.
Table 1. Reference list for experimental work and collected data tables about immune system library genes.
| Overviews and data sets | |
|---|---|
| 1. General reviews | LeFranc and LeFranc (21); LeFranc and LeFranc (22) |
| 2. Databases | IMGT (http://imgt.cines.fr), VBASE (http://www.mrc-cpe.cam.ac.uk) |
| 3. Original experimental papers | |
| (i) Immunoglobulin antibodies | |
| Heavy chain sequences | Tomlinson et al. (23); Matsuda et al. (24) |
| Kappa chain sequences | Zachau (25); Kirschbaum et al. (26); Kirschbaum et al. (27); Roschenthaler et al. (28) |
| Lambda chain sequences | Frippiat et al. (29); Williams et al. (30) |
| (ii) T cell receptors | |
| Alpha/delta chain sequences | Koop et al. (31); Bosc and LeFranc (32) |
| Beta chain sequences | Rowen et al. (33) |
| Gamma chain sequences | LeFranc et al. (34) |
A PROCEDURE FOR PREDICTION REFINEMENT
In view of the issues outlined in the previous section, a procedure was developed to improve the human and mouse IgSF predictions, and at the same time curate a more complete set of gene annotations. In the case of human annotation, full use was made of the information obtained from the predictions made in the present and previous Freeze Set assemblies.
New gene maps based on Ensembl predictions
Many of the improvements in predictions produced by the work described here comes from (i) the mapping, on to the appropriate chromosomal region, of those experimentally derived protein sequences from NRDB that are similar to but not the same as the Ensembl predictions and (ii) the visual inspection of these maps using a specially created browser. The gene mapping was carried out using Genewise calculations targeted, in the majority of cases, over the same DNA region as that of the original Ensembl predictions. The top NRDB WU-BLAST match for every prediction is the protein reference for the Genewise scan. When older human predictions from Freeze Sets prior to NCBI v29 were utilized, the Genewise calculation was performed on the entire region of the prediction’s clone of origin within the NCBI v29 Golden Path assembly.
All gene maps generated by the Genewise calculations were visualized using the Exegesis browser. This browser, together with information from the Genewise calculations, allows the gene maps to be edited according to a specific protocol. First, those Genewise maps showing a poor fit to the underlying DNA sequence were eliminated. Subsequently redundant maps were also removed. In this case, redundancy takes the form of a transcript map (i) being a fragment of another transcript, or (ii) showing the absence of experimental sequence evidence for it being a likely splice alternative. If two overlapping transcripts were deemed to be largely equivalent, the lower scoring Genewise map was eliminated. In the case of human gene map equivalence (i.e. two or more maps giving rise to the same product), the one generated by the older prediction was removed in preference to the one derived using the latest Ensembl.
Any overlapping gene maps that remained after this were treated as splice alternatives of one gene. If a map generated a translation shorter than the NRDB search sequence, a new Genewise run with an extended DNA target range was carried out in an attempt to achieve greater coverage.
Of the maps generated by this procedure, 180 human and 52 mouse transcripts were initially derived from NRDB entries that were not experimental sequences at the time, but genome predictions carried out by non-Ensembl groups. Since then, 89 of the human sequences, and 44 of the mouse sequences, have been verified by experimental work.
Gene maps for the immunoglobulin and T cell receptor genes
Immune system library genes are usually much shorter than most complete IgSF sequences—some consist of only a few nucleotides. The immunoglobulin and T cell receptor V, D and J genes also differ in not having stop codons at their 3′ ends. V and D segments terminate with a specific type of recombination signal. These features make it difficult to accurately predict gene segments using the Ensembl procedure (E. Birney, personal communication). We located the genes for the human immune gene segments and for the mouse T cell receptors using information from the earlier experimentally determined gene maps, as well as apposite sequence databases. The list of the resources used in this work can be found in Table 1.
Visual comparison with experimentally derived gene locus maps was carried out, and appropriate Genewise annotation generated, for immunoglobulin VH, VK and Vλ, as well as T cell receptor Vα, Vβ, Vγ and Vδ genes. Details of this work will be published elsewhere.
DETECTION OF NOVEL GENES AND EXONS USING HMMS
So far our discussion has been based on previous Ensembl predictions and experimental sequences. As part of the procedure, six-frame translations (6FT) of the complete human and mouse chromosomal sequences were also matched by Martin Madera to SUPERFAMILY HMMs for IgSF domains (Table 2). Good matches were found in 6FT regions that have no Ensembl annotation. These translated regions were matched to NRDB sequences and those with good scores were collected (325 for human, 468 for mouse). Then, using Genewise, the collected sequences were matched back onto the relevant regions of the genomes. For the human genome this work resulted in the prediction of 12 new genes, which gave predictions for 13 new transcripts. For the mouse genome, new IgSF-bearing exons in 123 genes were detected, of which 89 are new genes and 34 extended the description of genes already in Ensembl. Analysis of these two sets produced 144 and 43 new mouse transcripts, respectively. A detailed description of this work will be published elsewhere by Martin Madera. These new resultant gene and transcript predictions are included in the comparisons described below.
Table 2. A comparison of the Exegesis and Ensembl gene maps for members of the IgSF in the human and mouse genome.
| Human | Mouse | |||
|---|---|---|---|---|
| Exegesis | Ensembl | Exegesis | Ensembl | |
| Total number of genes | 893 | 644 | 811 | 556 |
| Total number of transcripts | 1266 | 812 | 934 | 774 |
| Coverage of potential IgSF coding regions (covered/total) | 2168/2807 | 1938/2807 | 1651/2217 | 1298/2217 |
| a) Whole IgSF sequences | ||||
| Genes | 532 | 508 | 525 | 437 |
| Total transcripts | 901 | 664 | 646 | 622 |
| Genes in common | 475 | 475 + 16 | 421 | 421 + 13 |
| Transcripts from genes in common | 837 | 646 | 532 | 618 |
| Transcripts of the same length from genes in common | 389 | 247 | ||
| Terminator immediately at the transcript 3′ end | 656 | 129 | 420 | 141 |
| Terminator within 10 in-frame codons of 3′ end | 720 | 342 | 497 | 358 |
| No terminator within 50 in-frame codons | 79 | 222 | 126 | 140 |
| Probable pseudogenes | 81 | 78 | 51 | 51 |
| b) Immune system gene segments | ||||
| V and C gene segments | 222 | 148 | 286 | 152 |
| J and D gene segments | 139 | 0 | 0 | 0 |
| Segments in common of same length | 17 | 22 | ||
DETECTION OF PSEUDOGENES
As will be described below, more than half the protein sequences predicted by this work are either significantly modified versions of Ensembl predictions or new transcript maps. During the post-processing stage of the procedure it is important to establish whether or not any of these might be pseudogenes.
Functional expressed proteins are under selective pressure. This means that in most cases synonymous mutations are usually more acceptable than non-synonymous mutations. Pseudogenes, on the other hand, are not under selective pressure and both types of mutations occur at similar rates (16). One test for whether a predicted gene is a pseudogene is to compare its DNA to that of a homologue known to be functional and to calculate the rate at which non-synonymous (KA) and synonymous (KS) mutations have occurred.
This assessment can be carried out using the program yn00, a component of the PAML suite (17) of phylogenetic analysis tools. This procedure calculates the rates of synonymous and non-synonymous substitutions between two aligned DNA sequences. If one of the two sequences is known to be expressed and the second is a predicted transcript, this calculation can give an indication of the likelihood that the second sequence is a pseudogene. The rate of non-synonymous (KA) substitutions divided by the synonymous rate (KS) is known as the omega (ω) score.
The ω scores of the 12 845 human–mouse orthologous pairs, which have a mean sequence identity of 78%, were determined by Mouse Genome Sequencing Consortium (18). For whole sequences, they found that the mean ω value is 0.115 and that most values are between 0.036 and 0.275. In an extensive analysis of over 200 pairs of known functional proteins from seven different families, Rajkumar Sasidharan found that when the absolute number of mutations is greater than 30, ω scores that exceed 0.3 are rare but, below the cut-off of 30 mutations, the values of ω can be higher (personal communication). The results described above suggest that a reasonable criteria to tag an EXEGESIS gene map as a likely pseudogene is an ω score of >0.3 when the number of DNA mutations is >30.
We determined the ω scores for all EXEGESIS sequences except for those that are immune system library genes. Using the program yn00 the ω values were calculated for the Genewise-derived DNA sequence of the EXEGESIS map aligned to the GenBank cDNA of the NRDB protein used to produce the map: the mean ω value is 0.14.
In terms of the criteria described above, 81 of the human EXEGESIS sequences are likely to be pseudogenes. For mouse sequences this number is 52. Of these, 78 of the human sequences and all 52 of the mouse sequences are also present in human Ensembl v29 and mouse Ensembl v3 predictions, respectively. The characteristics of these sequences are illustrated in Figure 3.
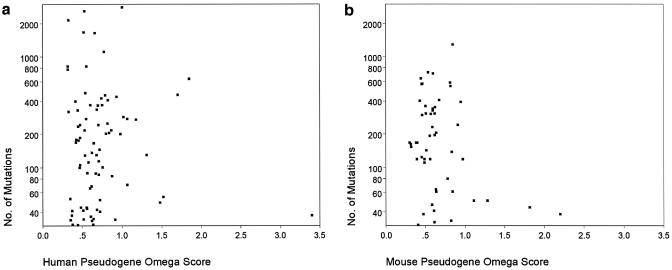
Figure 3.
The distribution of the number of mutations between the aligned Exegesis and NRDB Genbank cDNAs with respect to the ω score, for the likely pseudogenes in the human (a) (mean ω score 0.71) and mouse (b) (mean ω score 0.67) IgSF gene maps.
COMPARISON OF GENE MAPS MADE BY EXEGESIS AND ENSEMBL
In comparing these two sets of annotations it is useful to distinguish between (i) gene maps and protein transcripts and (ii) immune system library genes and whole IgSF sequences. The distinction between the terms in (i) is reasonably clear. As mentioned above, the distinction between the second two (ii) arises from the V, D and J immunoglobulin and T cell receptor genes being shorter than the other IgSF and not having stop codons at their 3′ ends.
A summary of the results obtained from the predictions available for human and mouse genomes from human Ensembl v29 and mouse Ensembl v3, as well as those produced by the Exegesis procedure are given in Table 2. The following paragraphs comment on the data in this table and add some additional information.
Whole IgSF genes and their transcripts in Exegesis and Ensembl v29
In this category, Ensembl v29 predicts for the human genome 508 genes and 664 transcripts, whilst Exegesis predicts 532 genes and 901 transcripts. For the mouse genome, Ensembl v3 predicts 437 genes and 622 transcripts whilst Exegesis predicts 525 genes and 646 transcripts. An important indicator of improved genome coverage involves the number of potential IgSF coding regions, detected by Madera’s SUPERFAMILY IgSF HMMs, covered by Exegesis vis-à-vis those claimed by Ensembl. The Exegesis mappings cover 10 and 27% more of these regions for human and mouse genomes, respectively.
Underlying these overall figures and those in Table 2, there are a number of important differences in the Ensembl and the Exegesis repertoires.
(i) Human genes: of the 532 genes in Exegesis, 459 are the same as those in Ensembl v29 (with respect to genome location and sequence identity) and 16 Exegesis maps cover 16 pairs of adjacent Ensembl predictions, which were subsequently fused by the Exegesis process (475 = 16 + 459). The other 57 [532 – (459 + 16)] Exegesis genes are derived Ensembl predictions prior to v29 (45 genes) or from Madera’s 6FT HMM ORFs (12 genes). There are 17 Ensembl v29 genes not included in Exegesis because our calculations indicate they are pseudogene V orphons.
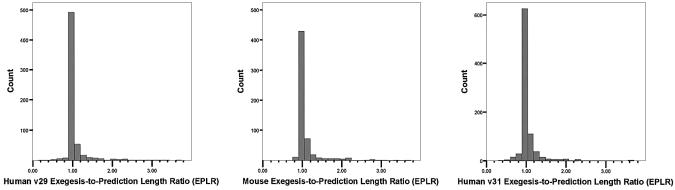
(ii) Human transcripts: Exegesis has 901 transcripts and Ensembl has 664. There are 389 transcripts in the two sets that have identical sequences and are of the same length. Another 256 transcripts have the same sequence but different lengths: in Exegesis 220 of these are longer than in Ensembl and 36 are shorter. On average, the Exegesis sequences were 1.4 times longer than the corresponding prediction (Fig. 4).
Figure 4.
The distribution of the Exegesis-to-prediction length ratio for human (Ensembl v29), mouse (Ensembl v3) and human (Ensembl v31) sequences matched against non-immune segment Exegesis sequences. The mean ratio for human (v29 and v31) was 1.4 in both cases and that for mouse was 1.5.
The remaining 256 transcripts in Exegesis are not in Ensembl: 188 come from genes common to both sets and 68 are derived from new genes. Of the 664 Ensembl transcripts, 424 have a stop codon either at the 3′ end of coding region of within 50 codons of the end. For the 901 Exegesis predictions this number is 828.
(iii) Mouse genes: there are 525 genes in Exegesis of which 408 are the same as those in Ensembl v3 and 13 that fuse 13 pairs of adjacent Ensembl predictions (421 = 13 + 408). There are three Ensembl v3 genes not included in Exegesis because our calculations indicate they are orphon V segment pseudogenes.
Fifteen genes were re-located to a genomic region in the vicinity of the original Ensembl genes that triggered the Genewise calculation. The 6FT HMM calculations gave rise to 89 new genes for Exegesis and extended the description of 34 genes that are common to the two mouse sets.
(iv) Mouse transcripts: Exegesis has in all 646 transcripts, and Ensembl has 622. The two sets of predictions have 247 transcripts that are common to the two mouse gene sets and have identical sequences as well as the same length. A further 285 transcripts are also derived from the common gene set and have the same sequence but differ in length: in Exegesis, 207 of these are longer than in Ensembl and 78 are shorter. On average, the Exegesis sequences were 1.5 times longer than the corresponding Ensembl prediction (Fig. 4).
The remaining 87 transcripts that Ensembl has for these common genes do not make a distinct match to any NRDB sequence or to any of 34 000 sequences in the Riken library of mouse cDNAs (19) that would confirm their existence as distinct splice alternatives. Of the 87, 36 are probably pseudogenes: they come from genes that have high ω values, and 26 do not have a stop codon either at the 3′ end of the coding region or within 50 codons of the end. Three sequences have both these characteristics.
There are 153 Exegesis transcripts that are not in Ensembl. Of these, 39 come from genes common to both sets and 114 come from new genes.
Of the 622 Ensembl transcripts, 478 have a stop codon at the 3′ end of coding region or within 50 codons of the end. For the 646 Exegesis predictions this number is 508.
Immune system library genes in Exegesis and Ensembl v29
Of the 250 V and C segments that are known to form human antibodies and T cell receptors, we mapped 222 onto the sequence of the human genome. The complete set of mouse immune library gene repertoire is not fully determined and 286 known functional segments were mapped on to the sequence of the mouse genome.
The number of Ensembl predictions that have the same sequence, length and positions as the segments determined by experiment is small (see Table 2).
The segments that form mouse heavy antibody chains have not been systematically mapped and sequenced; however, a list of putative V and C segments is given in the IMGT database (20). This list is believed to include numerous alleles. We matched these putative segments against the appropriate regions of the mouse genome. Of the 201 putative sequences 62 made good matches. These matches are currently being examined to determine which are likely to be functional and which are likely to be pseudogenes.
Comparison of Exegesis with Ensembl v31 and Vega
Subsequent to the work described above two additional human Ensembl versions have been released: v30 and v31. Their protein predictions are based on improved assemblies of the human genome. So far, the predictions in these releases have not been put through the Exegesis procedure. However, we have made a preliminary comparison of the Exegesis results with those in Ensembl v31. This version has in all 614 IgSF genes and 1091 transcripts. Compared with version 29 these numbers involve a small reduction in the total number of genes and a large increase in the number of transcripts. Of these, 495 families and 943 transcripts belong to what we have defined as the ‘Whole IgSF gene’ group. For the ‘Immune system library genes group’ Ensembl 31 has 119 genes and 148 transcripts: numbers close to those in version 29.
A transcript-by-transcript examination of the human ‘Whole IgSF gene’ group shows 439 Exegesis genes (with 780 transcripts) to map to 417 genes in Ensembl v31 (with 826 transcripts) on the basis of sequence identity. Three pairs of Exegesis genes were shown to have merged in the v31 assembly. The gene products of the remaining 19 [439 – (3 + 417)] outstanding pairs of Exegesis genes matched the transcripts of 19 individual Ensembl genes, but were not fragments of the latter (unlike the previous three pairs). It could be shown that these 38 Exegesis transcripts had a better or equal level of NRDB sequence length coverage when compared with the corresponding Ensembl v31 gene products to which these pairs converge.
There is also a non-overlapping gene subset from both Exegesis (93 genes, 121 transcripts) and Ensembl v31 (78 genes, 117 transcripts) that cannot be matched to each other on the basis of sequence similarity alone. Some of these Exegesis cases consist of the results of 6FT HMM searches and the use of older predictions as described above. The unique Ensembl v31 genes probably come from improvements in the DNA assembly, and more experimentally determined protein sequences than were otherwise available when assembly v29 was released. A more complete picture of these similarities and differences will become apparent from using the Exegesis procedure on later DNA assemblies.
The similarities of the genes and transcripts in v29 and v31 of Ensembl for the immune system library genes group mean that, for this group, the results of the comparisons made above between Exegesis and Ensembl v29 also apply to Ensembl v31.
The newly available Vega annotations (http://vega.sanger.ac.uk/), manually carried out on finished clone sequences (currently available for human Chr 6, 13, 14, 20 and 22) were also compared with the Exegesis results. At the time of writing, Vega had covered 58 IgSF genes (92 transcripts) over the five chromosomes for annotation. The Exegesis set for the same region suggests the presence of 197 IgSF genes (255 transcripts). The Vega transcripts matched Exegesis with a mean percentage identity of 98%. Sixty-two of the 92 transcripts were 100% identical. On average Exegesis sequences were 10% longer than Vega, 30 of the 92 transcripts having exactly the same length.
DISCUSSION
The genes of IgSF proteins tend to be difficult to predict by automatic procedures. Regions containing immune library genes are particularly problematic for automatic prediction because of the high density of IgSF coding regions that are very similar to each other (M.Clamp, personal communication). The results described above show that the Exegesis refinement of the Ensembl gene predictions provides a significant improvement and this enhancement appears to be maintained when map lengths are compared with IgSF sets from later assemblies (Fig. 4). The two main Exegesis achievements are (i) improved IgSF coding region coverage as is indicated by its producing more sequences with experimental support and a stop codon at the 3′ end or close to it and (ii) a greatly improved coverage of the immune system library genes.
We are aware, however, that the current results have certain limitations. A number of gene maps still lack a legitimate stop codon. It may well be the case that IgSF genes still lie undiscovered due to the lack of a close homologue to confirm them or because of the incomplete nature of the genome sequences. We have identified and ‘tagged’ potential pseudogenes in our sets on the basis of their ω values but we have not identified those pseudogenes that arise from mutations in key residues or in the regions that regulate them. There are certainly more splice variants than those described here.
The improvements produced by Exegesis come from two sources. First, the browser that allows both the rapid treatment of every map on an individual basis and the comparison of overlapping splice alternatives. When the length of a transcript map was significantly shorter than the NRDB sequence used for the mapping, the segment of DNA scanned by Genewise was extended in the appropriate direction for a new calculation. Where possible, more detailed gene information was sought from the literature during the refinement stage. The second source is the use of experimentally determined immunoglobulin and T cell receptor locus maps, and those derived from the detailed inspection and curation of non-segment genes (Table 2).
The Exegesis procedure can be used to improve or check any set of gene predictions over any DNA assembly. It is expected that the improvements will be greater for those proteins that, like IgSF molecules, have long sequences with alternative splice forms.
Details of the Exegesis IgSF gene maps for human (v29) and mouse (v3) genomes, as well as their annotations, are available over the internet at the following web address at http://www.genomesapiens.org/igsf/index.html.
Acknowledgments
ACKNOWLEDGEMENTS
We are indebted to Martin Madera for providing the 6-frame translation chromosomal annotations using IgSF SUPERFAMILY HMMs, as well as for comments on the manuscript. We are grateful to the Ensembl ‘pipeline’ team for fielding questions and promptly responding to numerous requests for further clarifications, during meetings and over e-mail, as to the specific details of their automatic gene annotation methods. Our special thanks go to Michele Clamp, Val Curwen, Laura Clarke, Eduardo Eyras and Simon Potter. We would like to thank Matthew Bashton, Michele Clamp and Sarah Teichmann for comments on the manuscript.
REFERENCES
- 1.Hubbard T., Barker,D., Birney,E., Cameron,G., Chen,Y., Clark,L., Cox,T., Cuff,J., Curwen,V., Down,T. et al. (2002) The Ensembl genome database project. Nucleic Acids Res., 30, 38–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Burge C. and Karlin,S. (1997) Prediction of complete gene structures in human genomic DNA. J. Mol. Biol., 268, 78–94. [DOI] [PubMed] [Google Scholar]
- 3.Altschul S.F., Gish,W., Miller,W., Myers,E.W. and Lipman,D.J. (1990) Basic local alignment search tool. J. Mol. Biol., 215, 403–410. [DOI] [PubMed] [Google Scholar]
- 4.Bairoch A. and Apweiler,R. (2000) The SWISS-PROT protein sequence database and its supplement TrEMBL in 2000. Nucleic Acids Res., 28, 45–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stoesser G., Baker,W., van den Broek,A., Camon,E., Garcia-Pastor,M., Kanz,C., Kulikova,T., Leinonen,R., Lin,Q., Lombard,V. et al. (2002) The EMBL Nucleotide Sequence Database. Nucleic Acids Res., 30, 21–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schuler G.D. (1997) Pieces of the puzzle: expressed sequence tags and the catalog of human genes. J. Mol. Med., 75, 694–698. [DOI] [PubMed] [Google Scholar]
- 7.Birney E. and Durbin,R. (1997) Dynamite: a flexible code generating language for dynamic programming methods used in sequence comparison. Proc. Int. Conf. Intell. Syst. Mol. Biol., 5, 56–64. [PubMed] [Google Scholar]
- 8.Mott R. (1997) EST_GENOME: a program to align spliced DNA sequences to unspliced genomic DNA. Comput. Appl. Biosci., 13, 477–478. [DOI] [PubMed] [Google Scholar]
- 9.Hughey R. and Krogh,A. (1996) Hidden Markov models for sequence analysis: extension and analysis of the basic method. Comput. Appl. Biosci., 12, 95–107. [DOI] [PubMed] [Google Scholar]
- 10.Gough J. and Chothia,C. (2002) SUPERFAMILY: HMMs representing all proteins of known structure. SCOP sequence searches, alignments and genome assignments. Nucleic Acids Res., 30, 268–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Benson D.A., Karsch-Mizrachi,I., Lipman,D.J., Ostell,J. and Wheeler,D.L. (2003) GenBank. Nucleic Acids Res., 31, 23–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wu C.H., Huang,H., Arminski,L., Castro-Alvear,J., Chen,Y., Hu,Z.Z., Ledley,R.S., Lewis,K.C., Mewes,H.W., Orcutt,B.C. et al. (2002) The Protein Information Resource: an integrated public resource of functional annotation of proteins. Nucleic Acids Res., 30, 35–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Berman H.M., Battistuz,T., Bhat,T.N., Bluhm,W.F., Bourne,P.E., Burkhardt,K., Feng,Z., Gilliland,G.L., Iype,L., Jain,S. et al. (2002) The Protein Data Bank. Acta Crystallogr. D Biol. Crystallogr., 58, 899–907. [DOI] [PubMed] [Google Scholar]
- 14.Pruitt K.D. and Maglott,D.R. (2001) RefSeq and LocusLink: NCBI gene-centered resources. Nucleic Acids Res., 29, 137–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li W., Jaroszewski,L. and Godzik,A. (2001) Clustering of highly homologous sequences to reduce the size of large protein databases. Bioinformatics, 17, 282–283. [DOI] [PubMed] [Google Scholar]
- 16.Bustamante C.D., Nielsen,R. and Hartl,D.L. (2002) A maximum likelihood method for analyzing pseudogene evolution: implications for silent site evolution in humans and rodents. Mol. Biol. Evol., 19, 110–117. [DOI] [PubMed] [Google Scholar]
- 17.Yang Z. (1997) PAML: a program package for phylogenetic analysis by maximum likelihood. Comput. Appl. Biosci., 13, 555–556. [DOI] [PubMed] [Google Scholar]
- 18.Waterston R.H., Lindblad-Toh,K., Birney,E., Rogers,J., Abril,J.F., Agarwal,P., Agarwala,R., Ainscough,R., Alexandersson,M., An,P. et al. (2002) Initial sequencing and comparative analysis of the mouse genome. Nature, 420, 520–562. [DOI] [PubMed] [Google Scholar]
- 19.Okazaki Y., Furuno,M., Kasukawa,T., Adachi,J., Bono,H., Kondo,S., Nikaido,I., Osato,N., Saito,R., Suzuki,H. et al. (2002) Analysis of the mouse transcriptome based on functional annotation of 60,770 full-length cDNAs. Nature, 420, 563–573. [DOI] [PubMed] [Google Scholar]
- 20.Lefranc M.P. (2001) IMGT, the international ImMunoGeneTics database. Nucleic Acids Res., 29, 207–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lefranc M.P. and LeFranc,G. (2001) The Immunoglobulin FactsBook. Academic Press, London, UK. [Google Scholar]
- 22.Lefranc M.P. and LeFranc,G. (2001) The T Cell Receptor FactsBook. Academic Press, London, UK. [Google Scholar]
- 23.Tomlinson I.M., Walter,G., Marks,J.D., Llewelyn,M.B. and Winter,G. (1992) The repertoire of human germline VH sequences reveals about fifty groups of VH segments with different hypervariable loops. J. Mol. Biol., 227, 776–798. [DOI] [PubMed] [Google Scholar]
- 24.Matsuda F., Ishii,K., Bourvagnet,P., Kuma,K., Hayashida,H., Miyata,T. and Honjo,T. (1998) The complete nucleotide sequence of the human immunoglobulin heavy chain variable region locus. J. Exp. Med., 188, 2151–2162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zachau H.G. (1990) The human immunoglobulin kappa locus and some of its acrobatics. A review presented on the occasion of receiving the ‘Otto-Warburg-Medaille’ of Gesellschaft fur Biologische Chemie at Osnabruck on September 13, 1989. Biol. Chem. Hoppe Seyler, 371, 1–6. [DOI] [PubMed] [Google Scholar]
- 26.Kirschbaum T., Pourrajabi,S., Zocher,I., Schwendinger,J., Heim,V., Roschenthaler,F., Kirschbaum,V. and Zachau,H.G. (1998) The 3′ part of the immunoglobulin kappa locus of the mouse. Eur. J. Immunol., 28, 1458–1466. [DOI] [PubMed] [Google Scholar]
- 27.Kirschbaum T., Roschenthaler,F., Bensch,A., Holscher,B., Lautner-Rieske,A., Ohnrich,M., Pourrajabi,S., Schwendinger,J., Zocher,I. and Zachau,H.G. (1999) The central part of the mouse immunoglobulin kappa locus. Eur. J. Immunol., 29, 2057–2064. [DOI] [PubMed] [Google Scholar]
- 28.Roschenthaler F., Kirschbaum,T., Heim,V., Kirschbaum,V., Schable,K.F., Schwendinger,J., Zocher,I. and Zachau,H.G. (1999) The 5′ part of the mouse immunoglobulin kappa locus. Eur. J. Immunol., 29, 2065–2071. [DOI] [PubMed] [Google Scholar]
- 29.Frippiat J.P., Williams,S.C., Tomlinson,I.M., Cook,G.P., Cherif,D., Le Paslier,D., Collins,J.E., Dunham,I., Winter,G. and Lefranc,M.P. (1995) Organization of the human immunoglobulin lambda light-chain locus on chromosome 22q11.2. Hum. Mol. Genet., 4, 983–991. [DOI] [PubMed] [Google Scholar]
- 30.Williams S.C., Frippiat,J.P., Tomlinson,I.M., Ignatovich,O., Lefranc,M.P. and Winter,G. (1996) Sequence and evolution of the human germline V lambda repertoire. J. Mol. Biol., 264, 220–232. [DOI] [PubMed] [Google Scholar]
- 31.Koop B.F., Rowen,L., Wang,K., Kuo,C.L., Seto,D., Lenstra,J.A., Howard,S., Shan,W., Deshpande,P. and Hood,L. (1994) The human T-cell receptor TCRAC/TCRDC (C α/C δ) region: organization, sequence and evolution of 97.6 kb of DNA. Genomics, 19, 478–493. [DOI] [PubMed] [Google Scholar]
- 32.Bosc N. and Lefranc,M.P. (2003) The mouse (Mus musculus) T cell receptor alpha (TRA) and delta (TRD) variable genes. Dev. Comp. Immunol., 27, 465–497. [DOI] [PubMed] [Google Scholar]
- 33.Rowen L., Koop,B.F. and Hood,L. (1996) The complete 685-kilobase DNA sequence of the human beta T cell receptor locus. Science, 272, 1755–1762. [DOI] [PubMed] [Google Scholar]
- 34.Lefranc M.P., Chuchana,P., Dariavach,P., Nguyen,C., Huck,S., Brockly,F., Jordan,B. and Lefranc,G. (1989) Molecular mapping of the human T cell receptor gamma (TRG) genes and linkage of the variable and constant regions. Eur. J. Immunol., 19, 989–994. [DOI] [PubMed] [Google Scholar]