Abstract
Mutations in particular codons of c-Ha-ras have a strong activating potential, and an activated ras oncogene has been found in a number of human cancers. Using fragments of the human c-Ha-ras gene containing 8-hydroxyguanine (8-OH-G) in codon 12, we provide evidence for highly complex biochemical events leading to activation of the oncogene. Replication with DNA polymerases α (Polα) and β (Polβ) led to misincorporation of dAMP, while DNA polymerase η (Polη) caused additional insertion of dGMP. For the first time we report an ‘action-at-a-distance’ mutagenic effect for Polη. Replication catalyzed by this enzyme resulted in misincorporating dAMP, dTMP and dGMP opposite non-oxidized guanine 3′-flanked by 8-OH-G. Interestingly, two adjacent 8-OH-G residues greatly relaxed the specificity of Polη, which in this system was able to incorporate all four nucleotides. Moreover, two adjacent 8-OH-G residues completely blocked Polα and strongly inhibited Polβ, whereas Polη was entirely resistant to this inhibition. These results suggest an important role for Polη in inducing hypermutability in codon 12. Our observations are important for understanding the consequences of 8-OH-G being positioned within the mutational hot spots of oncogenes, the outcome of which appears to be relatively complex even in minimal in vitro systems.
INTRODUCTION
Activated c-Ha-ras genes containing single substitutions in particular positions in the gene locus were found in ∼10% of human cancers examined (1). The mutational hotspots responsible for the gene activation were shown to be located in codons 12, 13, 61 or 117 (2). High frequencies of G→T transversions in codons 12 and 61 were observed in some types of cancer, e.g. in squamous cell carcinomas and basal cell carcinomas (3–5). Much evidence supports the hypothesis that reactive oxygen species take part in this mutagenic event. Du et al. (6) observed transforming activity of plasmids containing human c-Ha-ras fragments pretreated with an oxygen-free radicals-generating system transfected into NIH3T3 cells. The mutations responsible for the gene activation in transformed foci were found to be G→T transversions in codons 12 and 61, and A→T transitions in codon 61. The G→T transversion is the most frequent mutation caused by 8-hydroxyguanine [also known as 7,8-dihydro-8-oxoguanine (8-OH-G)] (7,8), a useful marker of DNA oxidation (9). Since 8-OH-G is believed to play an important role in carcinogenesis, the biological effects of its occurrence in DNA have been extensively studied under both in vitro and in vivo conditions (9,10). Systems containing a single 8-OH-G in codons 12 or 61 of synthetic fragments of the c-Ha-ras gene have been established and widely used to study activating mutagenic events. Using these models it has been directly shown that 8-OH-G can cause all possible substitutions, and predominately G→T transversions (11–15). Moreover, a ras gene containing 8-OH-G in codon 12 had remarkable transforming activity in NIH3T3 cells (16).
Further studies revealed that the frequencies of mutations caused by 8-OH-G depend on the DNA polymerase catalyzing the translesion replication and the sequence context. Two eukaryotic DNA polymerases have been used so far to replicate past 8-OH-G in codon 12 of the c-Ha-ras gene fragment under in vitro conditions. DNA polymerase α (Polα) incorporated dAMP and dCMP opposite 8-OH-G in the first position of the codon (12), whereas dAMP was inserted exclusively when 8-OH-G was in the second position (11,12). DNA polymerase β (Polβ) incorporated dCMP and dAMP opposite 8-OH-G in the second position of codon 12 (11).
Kuchino et al. (17) were the first to report the ability of 8-OH-G to stimulate mispairing at a neighboring template site in a system using bacterial DNA polymerase I (Klenow fragment). Kamiya et al. (13,16) extended the knowledge on this interesting phenomenon to a eukaryotic system, using c-Ha-ras vectors containing 8-OH-G and replicated in NIH3T3 cells. In this experimental system, a misreading at a base adjacent to the lesion was observed in codons 12 and 61. Later, this effect was referred to as ‘action-at-a-distance’ mutagenesis, and was also shown for eukaryotic Polβ (18).
Miscoding is not the only effect 8-OH-G exerts on DNA replication. When Klenow fragments of Escherichia coli pol I and Polα were used to insert dCMP opposite 8-OH-G, higher Km and lower Vmax values were found than with the insertion of dCMP opposite unmodified guanine, indicating that this lesion may decrease the efficiency of replication (7). However, the recently characterized DNA polymerase η (Polη) encoded by the Xeroderma pigmentosum variant (XPV) gene (19,20), was reported to have unique translesion replication abilities, being able efficiently to insert dCMP opposite 8-OH-G with Km and Kmax values almost identical with those obtained for unmodified templates (21). Polη was also found to be able to carry out an efficient and accurate translesion replication past thymine dimers (22), and to play an important role in the error-free bypass of UV lesions in an in vivo yeast system (23). On the other hand, Polη is known to be the least accurate among wild-type DNA polymerases (24). Moreover, Polη catalyzes efficient, but inaccurate, translesion replication past platinum complexes–DNA adducts (25) and 1,N6-ethenoadenine (26). Zhang et al. (27) provided further evidence for inaccurate translesion DNA synthesis by Polη using templates containing several types of lesions, including an abasic site, benzo[a]pyrene adduct and 8-OH-G. Therefore, the accuracy of the DNA synthesis catalyzed by Polη seems to depend on the kind of DNA lesion and the experimental system used. In particular, the results obtained for 8-OH-G are ambiguous. In some in vitro studies, the high accuracy of Polη was associated with its inability to continue the DNA synthesis from incorrect base pairs (28) or a low processivity of extension (29). This mechanism requires extrinsic proof-reading exonuclease activity in in vivo systems. However, it is noteworthy that all previously reported results concerning in vitro translesion DNA replication by Polη were obtained using oligonucleotide templates not corresponding to any known gene sequence.
We used a well-established experimental system consisting of fragments of the human c-Ha-ras gene containing 8-OH-G in codon 12, for systematic study of the effects of this lesion on DNA synthesis carried out by polymerases involved in DNA replication (Polα) and repair (Polβ). The unique properties of Polη reported previously using unspecific oligonucleotide sequences were thoroughly analyzed in the in vitro c-Ha-ras gene system. We also attempted to address the question of whether the presence of two adjacent 8-OH-G residues can modulate (enhance) the mutagenicity observed in codon 12 of ras genes. Kinetic parameters of the insertions of a single nucleotide by Polα, Polβ and Polη were measured using templates containing 8-OH-G in the first and second position of codon 12. Since the insertion of a nucleotide opposite the lesion may block further replication, we also studied the ability of each DNA polymerase used to extend from nucleotides incorporated in the insertion assay.
MATERIALS AND METHODS
Oligonucleotides were purchased from the Hokkaido System Science, Japan. Sequences of templates and primers are shown in Figure 1. Calf thymus Polα and human Polβ were from Chimerx, Milwaukee, WI. Human recombinant Polη was purified as described previously (20).
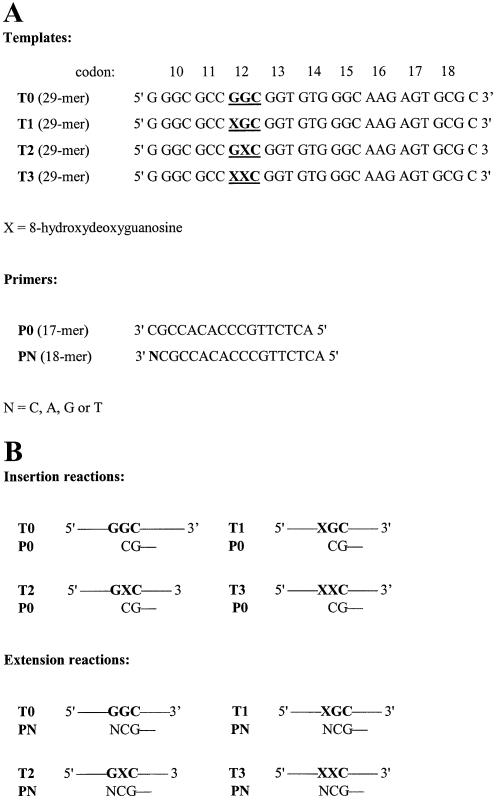
Figure 1.
Oligonucleotides corresponding to the region of human c-Ha-ras gene used in the study. (A) Templates and primers, the position of codon 12 is indicated. (B) Experimental systems used in the insertion and extension assay, only codon 12 of the templates is shown. Codon 12 is highlighted.
Insertion assay
Procedures from Haracska et al. (21) were adopted with minor modifications. Polα (0.5 U) was incubated with 32P-labeled primer:template substrate (25 nM) and increasing concentrations (0–500 µM) of a single deoxynucleotide (dCTP, dGTP, dATP or dTTP) in 50 mM Tris–HCl buffer (pH 8.0) containing 10 mM MgCl2, 20 nM (NH4)2SO4, 2 mM dithiothreitol (DTT) and 0.5 µg/µl BSA; Polβ (0.2 U) was incubated in the same buffer without ammonium sulfate; Polη (25 nM) was incubated in 40 mM Tris–HCl buffer (pH 8.0) containing 10 mM MgCl2, 60 mM KCl, 0.25 µg/µl BSA, 10 mM DTT and 2.5% glycerol. All reaction mixtures were incubated at 30°C for 1 min in a total volume of 5 µl, heated to 90°C to inactivate the enzyme and then subjected to 20% polyacrylamide gel electrophoresis. Bands in gels were visualized and their intensities were measured using a BAS 2000 scanner (Fuji, Japan). The percentage of primer extended was plotted as a function of the dNTP concentration and the kinetic parameters were calculated from the non-linear regression fit to a rectangular hyperbola [v = (Vmax × [dNTP] / (Km + [dNTP])], as described (30). Average values obtained from two to five independent experiments are presented.
Extension assay
Polα (0.5 U), Polβ (0.2 U) or Polη (25 nM) was incubated with primer:template substrates and increasing concentrations (0–100 µM) of a dNTPs mixture for 5 h, as described above. The reaction time was adjusted in the series of preliminary experiments using Polα and Polβ and the control unmodified template T0. Five hours was found to be the shortest incubation time necessary to obtain clear partial extension products at 1 µM dNTPs and the band of full-length product at 100 µM dNTPs (lowest and highest concentration used). Reaction products were subjected to gel electrophoresis and visualized as described above.
RESULTS
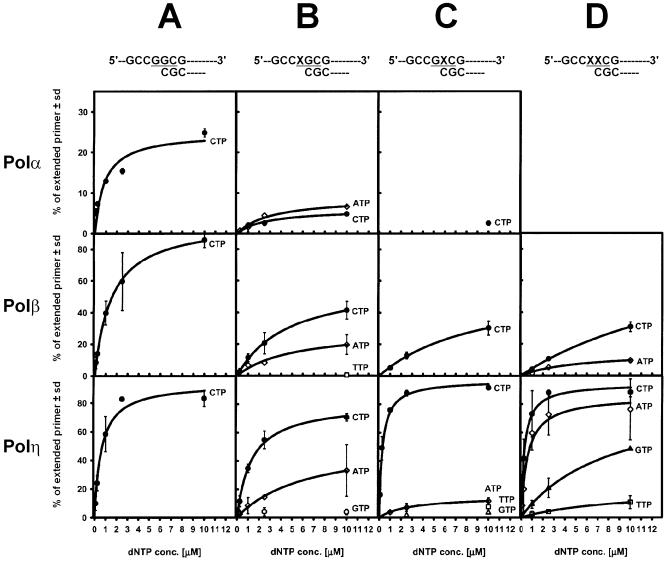
In preliminary insertion experiments using the unmodified control template T0 (oligonucleotide sequences used in the study are listed in Fig. 1), we found that dNTP concentrations of over 10 µM (100 and 500 µM were tested) caused incorporation of small amounts of dTMP by Polα and Polβ, and dAMP, dGMP and dTMP by Polη opposite guanine (data not shown). With between 0 and 10 µM dNTPs all polymerases incorporated dCMP exclusively and specifically opposite G (Fig. 2A). Therefore, incorporation at higher concentrations was regarded as non-specific.
Figure 2.
Single nucleotide insertion curves for Polα, Polβ and Polη. Individual reaction mixtures containing template:primer substrate and increasing concentrations of one of each dNTP were incubated with Polα, Polβ and Polη, reaction products were separated on polyacrylamide gels, and the percent of extended primer was calculated from band intensities; a non-linear fit was applied to obtain rectangular hyperbolic insertion curves, as described in Materials and Methods. Mean values from two to five independent experiments are shown, and the position of codon 12 is highlighted. (A) Replication of unmodified control template T0. (B) Replication in the direct mutagenesis system: template T1 containing 8-OH-G in the first position of codon 12. (C) ‘Action-at- a-distance’ mutagenesis system: replication of template T2 containing G in the first position of codon 12 and 8-OH-G in the second position. (D) Replication of template T3 containing two adjacent 8-OH-G residues in codon 12.
Inhibition of replication
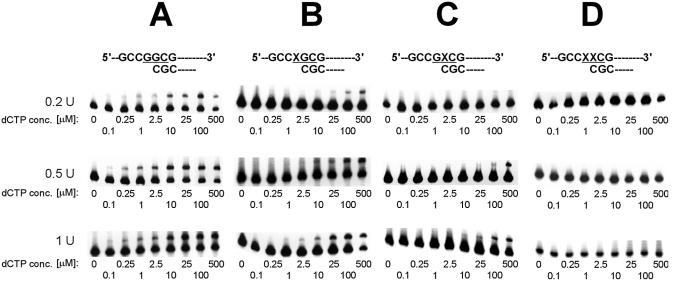
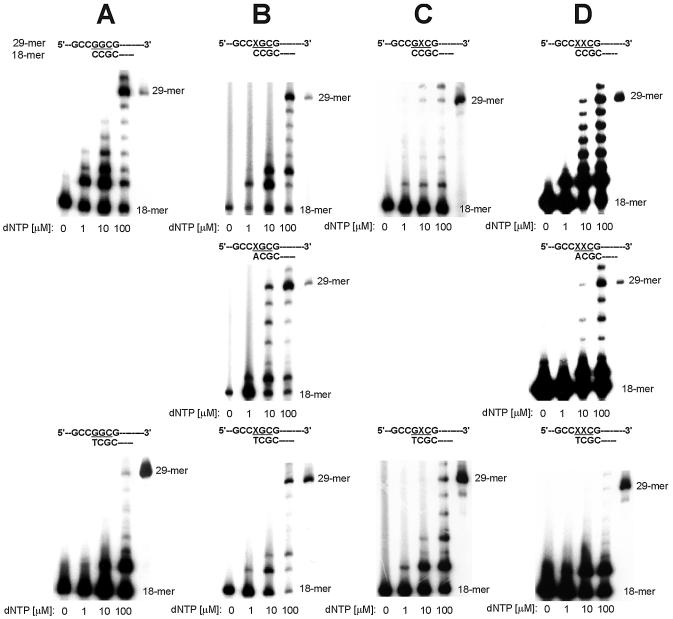
In preliminary experiments using 0.2 U of Polα, the presence of 8-OH-G inhibited replication to such an extent that the products of insertion of dCMP were observed only at the highest concentrations of dCTP used (100 and 500 µM), making calculated values of kinetic parameters uncertain. Therefore, this effect was further studied using various activities of Polα (0.2–1 U), and the results are shown in Figure 3. When the first replicated nucleotide was 8-OH-G in the first position of codon 12, 3′-flanked by unmodified G (template T1), relatively inefficient insertion of dCMP was observed (Fig. 3B), compared to that with unmodified template T0 (Fig. 3A). At similar kcat values (Table 1), the Km of incorporating dCMP opposite 8-OH-G was much higher than opposite G (7.29 ± 0.11 and 0.28 ± 0.02 µM, respectively), which shows a strong inhibition of Polα-catalyzed replication caused by oxidized guanine. Comparison of kcat/Km ratios revealed that the efficiency of correct replication by Polα decreased 27 times, compared with the unmodified template. Interestingly, when the first replicated base was unmodified guanine, 3′-flanked by 8-OH-G (template T2), no replication products were obtained when 0.2 U of Polα was used, and only a very inefficient insertion of dCMP was observed with higher activities of the enzyme (Fig. 3C). The efficiency of the correct replication in this case was decreased 82.8 times compared with that of the unmodified template, and seven times lower than when 8-OH-G was the first replicated base. When 8-OH-G was the first replicated base, and also the adjacent 3′ guanine in the second position of codon 12 was oxidized (template T3), incorporation of dCMP by Polα was completely inhibited within the activity range used (Fig. 3D). Moreover, in an extension assay, when a mixture of dNTPs was used to extend from C paired with 8-OH-G 3′-flanked by another 8-OH-G:C pair, no traces of replication products were observed (Fig. 4D).
Figure 3.
Inhibitory effect of 8-OH-G on replication catalyzed by Polα. Reaction mixtures containing template:primer substrate and increasing concentrations of dCTP were incubated with the indicated activity units of Polα; products were separated on polyacrylamide gels, as described in Materials and Methods. The position of codon 12 is highlighted. (A) Replication of unmodified control template T0. (B) Replication of template T1 containing 8-OH-G in the first position of codon 12. (C) Replication of template T2 containing G in the first position of codon 12 and 8-OH-G in the second position. (D) Replication of template T3 containing two adjacent 8-OH-G residues in codon 12.
Table 1. Kinetic parameters of insertion reactions.
| Polymerase | DNA substrate | dNMP incorporated | Km ± SDa (µM) | kcat ± SDa (s–1) | kcat/Km |
|---|---|---|---|---|---|
| Polα | 5′-GCCGGCGCGC- | dCMP | 0.28 ± 0.02 | 3.71 ± 0.02 | 13.25 |
| 5′-GCCXGCGCGC- | dCMP | 7.29 ± 0.11 | 3.64 ± 0.09 | 0.49 | |
| dAMP | 1.09 ± 0.12 | 3.74 ± 0.01 | 3.43 | ||
| 5′-GCCGXCGCGC- | dCMP | 8.60 ± 0.10 | 1.35 ± 0.15 | 0.16 | |
| Polβ | 5′-GCCGGCGCGC- | dCMP | 0.16 ± 0.08 | 9.75 ± 0.02 | 60.94 |
| 5′-GCCXGCGCGC- | dCMP | 1.71 ± 0.61 | 5.34 ± 6.1 | 3.12 | |
| dAMP | 6.78 ± 4.10 | 1.04 ± 0.04 | 0.15 | ||
| dTMP | 9.59 ± 0.64 | 1.16 ± 0.43 | 0.12 | ||
| 5′-GCCGXCGCGC- | dCMP | 5.05 ± 4.60 | 7.41 ± 1.53 | 1.47 | |
| 5′-GCCXXCGCGC- | dCMP | 2.92 ± 0.04 | 1.04 ± 0.01 | 0.36 | |
| dAMP | 5.46 ± 1.19 | 1.48 ± 0.10 | 0.27 | ||
| Polη | 5′-GCCGGCGCGC- | dCMP | 0.05 ± 0.01 | 8.71 ± 0.59 | 174.20 |
| 5′-GCCXGCGCGC- | dCMP | 0.15 ± 0.05 | 8.36 ± 0.41 | 55.73 | |
| dAMP | 1.34 ± 0.50 | 8.79 ± 1.59 | 6.56 | ||
| dGMP | 5.49 ± 2.05 | 4.45 ± 0.30 | 0.81 | ||
| 5′-GCCGXCGCGC- | dCMP | 0.03 ± 0.01 | 9.33 ± 0.02 | 311.00 | |
| dAMP | 1.46 ± 0.62 | 6.79 ± 1.58 | 4.65 | ||
| dGMP | 5.12 ± 0.50 | 2.02 ± 0.26 | 0.39 | ||
| dTMP | 6.21 ± 2.53 | 5.07 ± 4.75 | 0.92 | ||
| 5′-GCCXXCGCGC- | dCMP | 0.03 ± 0.01 | 9.15 ± 0.05 | 305.00 | |
| dAMP | 0.07 ± 0.04 | 8.79 ± 0.68 | 125.57 | ||
| dGMP | 0.32 ± 0.31 | 6.47 ± 0.70 | 20.22 | ||
| dTMP | 8.74 ± 1.95 | 1.04 ± 0.01 | 0.12 |
aMean values calculated from two to five independent insertion experiments are presented, as described in Materials and Methods.
Figure 4.
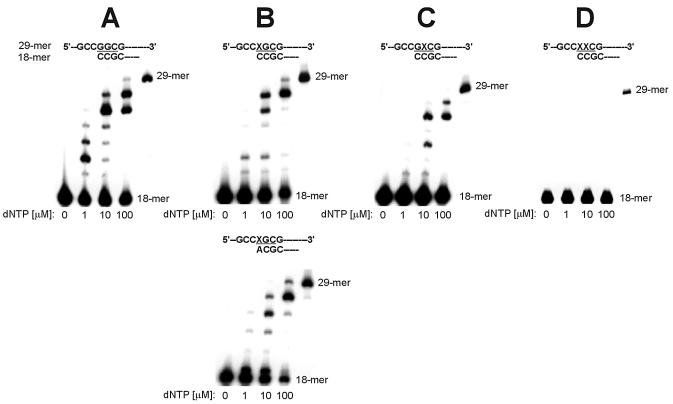
Extension by Polα from nucleotides incorporated in the insertion assay. Primers identical with all reaction products obtained in the insertion assay were used. Reaction mixtures containing template:primer substrate and increasing concentrations of a dNTPs mixture were incubated with Polα; products were separated on polyacrylamide gels, as described in Materials and Methods. The position of codon 12 is highlighted; positions of primer (18mer) and full-length extension product (29mer) are indicated. (A) Replication of unmodified control template T0. (B) Replication of template T1 containing 8-OH-G in the first position of codon 12. (C) Replication of template T2 containing G in the first position of codon 12 and 8-OH-G in the second position. (D) Replication of template T3 containing two adjacent 8-OH-G residues in codon 12.
Incorporation of the correct nucleotide was also strongly inhibited when Polβ was used to incorporate dCMP opposite 8-OH-G in template T1 (Fig. 2B). The kcat/Km ratio for dCMP insertion opposite 8-OH-G on unmodified template T0 was 60.94, compared with 3.12 for dCMP insertion opposite 8-OH-G on template T1. These values show that correct replication in the presence of 8-OH-G by Polβ was 19.5 times less efficient than with the unmodified template. The presence of adjacent 8-OH-G 3′-flanking the first replicated guanine in template T2 inhibited incorporation of dCMP more strongly (Fig. 2C). The comparison of kcat/Km ratios indicated that the efficiency of the correct replication in this case was decreased 41.5 times compared to that with the unmodified template, and 2.1 times lower than when 8-OH-G was the first replicated base. In the case when 8-OH-G was the first replicated base, and the 3′-adjacent base in the second position of the codon was also 8-OH-G (template T3), very strong inhibition of the insertion of dCMP was observed (Fig. 2D). The efficiency of the correct replication in this case was decreased 169.3 times compared with unmodified template T0, and 4.1 times when compared with template T2.
In the case of replication catalyzed by Polη, the kcat values for incorporation of dCMP opposite G in unmodified template T0 and opposite 8-OH-G in template T1 were comparable, but the difference between the Km constants (0.05 ± 0.01 and 0.15 ± 0.05 µM, respectively) indicated decreased efficiency of correct insertion caused by the presence of 8-OH-G. However, when kcat/Km ratios were compared, it was found that 8-OH-G inhibited the correct replication by Polη only 3.1-fold. Interestingly, in the case of templates T2 and T3, nearly 2-fold more efficient insertion of dCMP was observed, as compared to that with unmodified template T0. In the presence of 8-OH-G in a position penultimate to the first replicated unmodified guanine (template T2, Fig. 2C), the correct replication was 1.8 times more efficient, as concluded from comparison of kcat/Km ratios; a similar increase in the efficiency (1.75 times) was observed when the first replicated 8-OH-G was 3′-flanked by 8-OH-G (template T3, Fig. 2D).
Direct mutagenesis
Using template T1 containing 8-OH-G in the first position of codon 12, we found that Polα incorporated dCMP and misincorporated dAMP (Fig. 2B). The kcat values for the insertion of both nucleotides were comparable (3.64 ± 0.09 s–1 for dCMP and 3.74 ± 0.01 s–1 for dAMP), however, clearly different Km constants contributed to a much lower kcat/Km ratio for incorporation of the correct nucleotide than that obtained for insertion of dAMP (0.49 and 3.43, respectively), suggesting the higher efficiency of misinsertion. In an extension assay, Polα was able to catalyze efficient extension from misincorporated A opposite 8-OH-G (Fig. 4B), in a way similar to extension from C opposite 8-OH-G, i.e. giving partial products at 1 µM of a dNTPs mixture and a clear band of the full-length extension product at 100 µM.
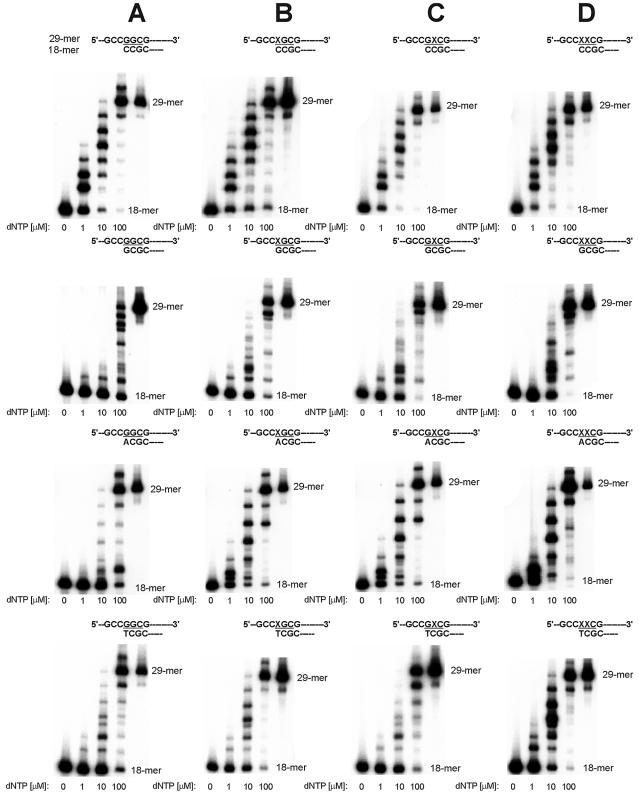
Polβ incorporated dCMP, dAMP and very small amounts of dTMP opposite 8-OH-G (Fig. 2B). The insertion of dCMP was favored over that of dAMP, and dAMP over that of dTMP, with kcat/Km ratios of 3.12, 0.15 and 0.12, respectively. Extension from the misincorporated A was relatively more efficient than from C opposite 8-OH-G (Fig. 5B), giving the full-length product not only at 100 µM of a dNTPs mixture, as in the case of the 8-OH-G:C pair, but also at 10 µM. Products of extension from the pair 8-OH-G:T were also formed; the full-length products being observed only at 100 µM (Fig. 5B).
Figure 5.
Extension by Polβ from nucleotides incorporated in the insertion assay. Primers identical with all reaction products obtained in the insertion assay were used. Reaction mixtures containing template:primer substrate and increasing concentrations of a dNTPs mixture were incubated with Polβ; products were separated on polyacrylamide gels, as described in Materials and Methods. The position of codon 12 is highlighted; positions of primer (18mer) and full-length extension product (29mer) are indicated. (A) Replication of unmodified control template T0. (B) Replication of template T1 containing 8-OH-G in the first position of codon 12. (C) Replication of template T2 containing G in the first position of codon 12 and 8-OH-G in the second position. (D) Replication of template T3 containing two adjacent 8-OH-G residues in codon 12.
Polη incorporated dCMP, dAMP and, unlike Polβ, small amounts of dGMP opposite 8-OH-G in the template T1 (Fig. 2B). At very high concentrations of dTTP (100 and 500 µM), very small amounts of dTMP were also incorporated (data not shown). The insertion of dCMP was favored over that of dAMP, and dAMP over that of dGTP, with kcat/Km ratios of 55.73, 6.56 and 0.81, respectively. In the extension assay, Polη showed an ability to extend from all incorporated nucleotides efficiently (Fig. 6B).
Figure 6.
Extension by Polη from nucleotides incorporated in the insertion assay. Primers identical with all reaction products obtained in the insertion assay were used. Reaction mixtures containing template:primer substrate and increasing concentrations of a dNTPs mixture were incubated with Polη; products were separated on polyacrylamide gels, as described in Materials and Methods. The position of codon 12 is highlighted; positions of primer (18mer) and full-length extension product (29mer) are indicated. (A) Replication of unmodified control template T0. (B) Replication of template T1 containing 8-OH-G in the first position of codon 12. (C) Replication of template T2 containing G in the first position of codon 12 and 8-OH-G in the second position. (D) Replication of template T3 containing two adjacent 8-OH-G residues in codon 12.
‘Action-at-a-distance’ mutagenesis
To study the potential influence of 3′-flanking 8-OH-G on the replication from unmodified guanine, the system using template T2 was applied. When the first replicated base was G in the first position of codon 12, and 8-OH-G was present in the second position, Polα and Polβ incorporated only the correct dCMP (Fig. 2C). However, at the highest concentration of dTTP used, Polβ also inserted very small amounts of dTMP (data not shown) and extension from G:T 3′-flanked by the 8-OH-G:C pair was relatively efficient (Fig. 5C). Different effects were found when Polη was used (Fig. 2C). Besides efficient insertion of the correct dCMP, Polη was able to misincorporate dAMP, dTMP and dGMP (however, with relatively low efficiencies). Adenine was favored over guanine and guanine over thymine (with kcat/Km ratios of 4.65, 0.39 and 0.92, respectively). Efficient extension from all incorporated nucleotides was observed (Fig. 6C).
Effect of two adjacent 8-OH-G residues
To analyze the effect of two adjacent 8-OH-G residues on the replication, we used a system where the first replicated base was 8-OH-G in the first position of codon 12, 3′-flanked by 8-OH-G in the second position (template T3). In this arrangement, Polα was completely inhibited (Fig. 2D, empty panel). Polβ incorporated dCMP and dAMP (Fig. 2D), in addition to a very small amount of dTMP inserted when very high concentrations of dTTP were used (data not shown). Incorporation of dCMP was favored over that of dAMP, with kcat/Km ratios of 0.36 and 0.27, respectively. The extension from misinserted A and T was distinctly less efficient than that from C (Fig. 5D). However, the full-length product was obtained at 10 and 100 µM dNTPs for extension from the 8-OH-G:A pair. In the case of extension from the 8-OH-G:T pair, an accumulation of a single nucleotide incorporation product was observed, with only very faint bands of longer products formed at the highest concentration of dNTPs used.
Polη was able to incorporate all four nucleotides, with high efficiencies of misinsertion of dAMP (kcat/Km = 125.57) and dGMP (kcat/Km = 20.22); dTMP was inserted with a very low kcat/Km = 0.12. In the extension assay, efficient elongation was observed for all incorporated nucleotides, without any particular stall sites (Fig. 6D).
DISCUSSION
Using the c-Ha-ras gene system containing 8-OH-G in the well-known mutational hot spot replicated by three eukaryotic DNA polymerases, we observed a relatively complex array of biochemical events. In agreement with expectations based on data in the literature, the predominating mutation caused directly by 8-OH-G in the first position of codon 12 was G→T transversion. This substitution was a result of the misincorporation of dAMP opposite the lesion by all three polymerases tested. Additionally, Polβ was able to misincorporate dTMP at low rates, causing G→A transitions; and Polη misinserted dGMP, causing G→C transversions. All these substitutions were previously observed in DNA sequences containing 8-OH-G replicated in in vitro systems and also in mammalian cells [recently reviewed by Kamiya (9)]. In our study, 100 and 500 µM dNTPs in reaction mixtures caused unspecific incorporation of small amounts of dTMP by Polα and Polβ, and dAMP, dGMP and dTMP by Polη opposite guanine in the unmodified control template. Also, only at such high concentrations, was Polη in the direct mutagenesis system able to insert very small amounts of dTMP, and Polβ inserted dTMP when two adjacent 8-OH-G residues in codon 12 were present. The physiological concentrations of dNTPs in normal cells are: 1.5 ± 1.0 µM dGTP, 2.1 ± 2.7 µM dCTP, 3.2 ± 3.4 µM dATP and 5.4 ± 6.4 µM dTTP; in tumor cells these values are several times higher, ranging from 7.2 ± 4.4 µM for dGTP to 32 ± 22 µM for dATP [thoroughly reviewed by Traut (31)]. The above values are total cellular concentrations; certainly localized concentrations may be substantially higher, and may affect the outcome of DNA synthesis. However, dNTP concentrations of 100–500 µM can be regarded as unusually high, and, from the mutagenic effects observed in the unmodified template, we conclude that this misreplication was non-specific and forced by the very high amount of a single dNTP present in the reaction mixture. A wide spectrum of mutagenic events associated with various DNA modifications may be explained by different mutagenic properties of the variety of mammalian DNA polymerases taking part in DNA replication and repair.
A system requiring the sequential action of two DNA polymerases for translesion DNA repair was recently postulated (32). This model assumes that one polymerase serves as an inserter to incorporate a nucleotide opposite the lesion, and the other one acts as an extender, elongating the DNA chain from the inserted base. This view is based on the observation that some polymerases are able to insert a nucleotide opposite some kinds of DNA lesions, but the newly formed DNA base pair is an inefficient starting site for further reaction, or it completely blocks the replication. On the other hand, some polymerases are highly tolerant to mispaired DNA bases and can efficiently extend from the lesion site. In general, replicative DNA polymerases can be considered highly accurate but relatively intolerant to geometrical distortions caused by some DNA lesions (32–34). One of the well-known lesions inhibiting replication is thymine glycol, which blocks DNA synthesis by bacterial and mammalian polymerases (35–39). In the case of Polα it was clearly shown that the replication block takes place at the insertion step (40). Inhibited insertion of the correct nucleotide opposite the lesion was also reported in the case of the Klenow fragment of E.coli pol I and Polα used to replicate past 8-OH-G (7). In contrast, Polη was able to insert a nucleotide opposite such lesions as thymine glycols, platinum adducts, ethenoadenine or 8-OH-G, and also had efficient extending activity (25,26,40). This suggested the important role Polη may play in the efficient bypass of blocking DNA lesions under in vivo conditions, a critical event for cell survival. Both error-proof and error-prone insertion by Polη was reported, depending on the kind of DNA lesion or the experimental system used. In the cases of an abasic site analog, N-2-acetylaminofluorene-guanine and cis-platin G–G crosslinks, the error-free synthesis was assured by a mechanism blocking extension by Polη only from incorrect base pairs (28). When the correct base was incorporated, Polη was able to extend from the resulting pair. Altogether, the above data suggest that when the major polymerase involved in the DNA replication or repair encounters a blocking DNA lesion, Polη or another DNA polymerase with similar properties may be responsible for the lesion bypass. However, a very low-fidelity Polη (24), and error-prone translesion DNA synthesis shown for several types of lesions, may lead to increased mutagenicity of DNA replication catalyzed by this enzyme. Error-prone replication was observed when Polη was used to replicate past the abasic site, benzo[a]pyrene adduct and 8-OH-G (27), in contrast to results obtained by Haracska et al. (21), who reported accurate DNA synthesis on templates containing 8-OH-G. Other enzymes possibly involved in the sequential, two-polymerase translesion DNA synthesis are Polζ and Polκ. Polζ was reported to be an efficient extender of mismatched termini; similar data exist for Polκ, as discussed by Prakash and Prakash (32). However, both enzymes have been studied with UV-damaged DNA or templates containing abasic sites, and kinetic parameters of DNA synthesis past 8-OH-G catalyzed by Polζ and Polκ remain unknown.
Our results show that repair Polβ, and especially replicative Polα, were strongly inhibited by the presence of 8-OH-G. The efficiency of incorporation of dCMP opposite 8-OH-G in the first position of codon 12 of the c-Ha-ras gene fragment by Polα was decreased 27 times, and by Polβ nearly 20 times, as compared with that of the unmodified template. Interestingly, the effect of 8-OH-G in the second position of codon 12 on replication from 5′-flanking unmodified guanine was even stronger. In this case, the insertion of dCMP by Polα was inhibited nearly 83 times, and that by Polβ 41.5 times. Moreover, when the first replicated 8-OH-G in the first position was 3′-flanked by another 8-OH-G in the second position of codon 12, the inhibitory effect on replication was very great. No insertion products were detected when Polα was used, even when the concentration of the enzyme was increased up to 1 U per reaction mixture, and the reaction catalyzed by Polβ was inhibited 169.3 times, compared with the insertion of dCMP on the unmodified template. However, Polη seemed to be resistant to this inhibitory effect. The incorporation of dCMP opposite 8-OH-G 3′-flanked by G was only 3.1 times less efficient than the reaction on the control template. The presence of 8-OH-G in the second position of codon 12, and two adjacent 8-OH-G residues, slightly increased the efficiency of insertion of dCMP by Polη (1.8 and 1.75 times, respectively). These effects were relatively weak and it can be concluded that 8-OH-G only slightly affects correct replication catalyzed by Polη, in particular cases even being able to increase the replication efficiency. The above data support the two-polymerase affair model. When Polα or Polβ encounter 8-OH-G during DNA replication or repair, the efficient bypass of the lesion may require dissociation of the enzyme from the damaged template, and insertion of one or more nucleotides by Polη or other DNA polymerase with similar properties. All studied polymerases were able to extend from the first inserted nucleotide in the system used in our study, suggesting that Polη may be necessary only to insert a single nucleotide opposite the blocking site. This mechanism may play a critical role when the main polymerase involved in DNA synthesis is completely inhibited by the lesion, and cannot bypass the damaged template, as in the case of two adjacent 8-OH-G residues and Polα in our study. The sensitivities of Polα and Polβ to the inhibitory effect of 8-OH-G, and the resistance displayed by Polη, can be explained by structure. The active site of high-fidelity polymerases fits tightly to the templating base, a few base pairs in the DNA adjacent to the incorporation site and to the incoming dNTP. Moreover, only a single template base is held in the active site, a number of minor-groove interactions between DNA and the enzyme stabilize the position of the polymerase, and the next 5′-template base is precisely positioned (41–45). The geometrical distortions caused by 8-OH-G account for inhibition or blocking of replication past the lesion by this type of DNA polymerase. In contrast, Polη is relatively insensitive to geometrical distortions, because its active site can hold two template nucleotides, and there is little or only weak evidence for any specific contact of the enzyme with an incoming nucleotide or template-forming base and for any specific minor-groove interactions of any base pairs in the template with the enzyme molecule (32,46).
When the first replicated base was unmodified guanine in the first position of codon 12, and the 3′-adjacent template base in the second position was 8-OH-G, some mutations were also detected. Polη showed ‘action-at-a-distance’ mutagenic potency, being able to incorporate dCMP, dAMP, dGMP and dTMP (in decreasing order of efficiency) opposite G. Moreover, Polη efficiently extended from all inserted nucleotides. Polα and Polβ incorporated exclusively dCMP opposite guanine in this experimental system. Very small amounts of dTMP were also inserted by Polβ, but only at the highest concentration of dTTP used. However, when such an event happens, Polβ can extend from the misinserted thymine. The ability of 8-OH-G to stimulate mispairing at an adjacent template site was first reported by Kuchino et al. (17), who showed this effect for the Klenow fragment of E.coli pol I. Efrati et al. (18) observed a similar phenomenon for Polβ, but not for Polα replicating past 8-OH-G. Studies using vectors containing c-Ha-ras gene fragments replicated in NIH3T3 cells were also used to show mutagenesis opposite undamaged sites stimulated by neighboring 8-OH-G (13,16). These authors observed incorporation of all four nucleotides opposite guanine in the first position of codon 12, 3′-flanked by 8-OH-G. In this case, the polymerase involved in the mutagenesis was not known. However, the similar broad spectrum of mutations caused by Polη in our system suggests that this low-fidelity enzyme is a serious candidate.
Interesting mutagenic patterns were obtained when the first replicated base was 8-OH-G in the first position of codon 12, and guanine in the second position was also modified. Polα was not able to replicate past such a damaged sequence, and therefore could not account for mutagenesis. The remaining polymerases were able to insert incorrect nucleotides. Polβ showed an ability to incorporate dAMP with an efficiency comparable with that observed in the template containing a single 8-OH-G in the first position. However, the specificity of Polη was found to be highly relaxed in this system. The efficiency of misinsertion of dAMP was nearly 20 times higher, and misincorporation of dGMP 25 times higher than in the case of a single 8-OH-G, when kcat/Km ratios were compared. Moreover, a relatively high misinsertion of dTMP was also observed, whereas very small amounts of this nucleotide were incorporated in the single 8-OH-G system, and only at the highest dTTP concentrations. No data suggesting this striking relaxation of the specificity of Polη by two residues of 8-OH-G in tandem arrangement exist in the literature, and this phenomenon may have serious biological consequences. The presence of 8-OH-G in the second position of the mutational hotspot of the c-Ha-ras gene remarkably increases the efficiency of mutagenesis at the first position. Mutations in the first position of codon 12, in turn, have a potency to activate the gene. Under in vivo conditions the probability of oxidation of two adjacent 8-OH-G residues is hard to estimate, but this event may be expected to be rare. However, when such an arrangement occurs, it may play a crucial role in proto-oncogene activation by its ability to inhibit at least some high-fidelity replicative and repair polymerases, and to stimulate remarkably efficient mutagenicity when replicated by Polη. There is no direct evidence that Polη is responsible for such an effect in vivo, but this polymerase can be considered a good candidate. It may be assumed that the particular hypermutagenic properties of codon 12 of the c-Ha-ras gene may be partly due to the presence of two adjacent guanines, which, when oxidized, may promote a dramatic increase in the frequency of mutations. This finding is especially important when the critical role of this mutational hotspot for gene activation is taken into account. Our data may help in understanding the processes modulating the hypermutagenicity of some particular gene sequences, where a single mutation may trigger mechanisms leading to cancer development.
Acknowledgments
ACKNOWLEDGEMENTS
We express sincere thanks to Dr Tsuyoshi Arai and Dr Vincent Kelly for valuable discussions.
REFERENCES
- 1.Barbacid M. (1985) Oncogenes in human cancers and in chemically induced animal tumors. Prog. Med. Virol., 32, 86–100. [PubMed] [Google Scholar]
- 2.Barbacid M. (1987) Ras genes. Rev. Biochem., 51, 655–693. [DOI] [PubMed] [Google Scholar]
- 3.White S.I. and Balmain,A.G. (1988) G to T mutation in codon 12 of the human Harvey ras oncogene derived from a basal cell carcinoma. J. Invest. Dermatol., 91, 407. [Google Scholar]
- 4.van der Schoef G., Even,L.M., Boot,A.J.M. and Bos,J.L. (1990) Ras oncogene mutations in basal cell carcinomas and squamous cell carcinomas of human skin. J. Invest. Dermatol., 94, 423–425. [DOI] [PubMed] [Google Scholar]
- 5.Pierceall W., Goldberg,L.H., Tainsky,M.A., Mukhopadhyay,T. and Ananthaswamy,H.N. (1991) Ras gene mutations and amplifications in human nonmelanoma skin cancers. Mol. Carcinog., 4, 203–209. [DOI] [PubMed] [Google Scholar]
- 6.Du M.-Q., Carmichael,P.L. and Phillips,D.H. (1994) Induction of activating mutations in the human c-Ha-ras-1 proto-oncogene by oxygen free radicals. Mol. Carcinog., 11, 170–175. [DOI] [PubMed] [Google Scholar]
- 7.Shibutani S., Takeshita,M. and Grollman,A.P. (1991) Insertion of specific bases during DNA synthesis past the oxidation-damaged base 8-oxodG. Nature, 349, 431–434. [DOI] [PubMed] [Google Scholar]
- 8.Cheng K.C., Cahill,D.S., Kasai,H., Nishimura,S. and Loeb,L.A. (1992) 8-Hydroxyguanine, an abundant form of oxidative DNA damage, causes G→T and A→C substitutions. J. Biol. Chem., 267, 166–172. [PubMed] [Google Scholar]
- 9.Kamiya H. (2003) Mutagenic potentials of damaged nucleic acids produced by reactive oxygen/nitrogen species: approaches using synthetic oligonucleotides and nucleotides. Nucleic Acids Res., 31, 517–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sekiguchi M. and Tsuzuki,T. (2002) Oxidative nucleotide damage: consequences and prevention. Oncogene, 21, 8895–8904. [DOI] [PubMed] [Google Scholar]
- 11.Kamiya H., Sakaguchi,T., Murata,N., Fujimuro,M., Ishikawa,H., Shimizu,M., Inoue,H., Nishimura,S., Matsukage,A., Masutani,C., Hanaoka,F. and Ohtsuka,E. (1992) In vitro replication study of modified bases in ras sequences. Chem. Pharm. Bull., 40, 2792–2795. [DOI] [PubMed] [Google Scholar]
- 12.Kamiya H., Murata-Kamiya,N., Fujimuro,M., Kido,K., Inoue,H., Nishimura,S., Masutani,C., Hanaoka,F. and Ohtsuka,E. (1995) Comparison of incorporation and extension of nucleotides in vitro opposite 8-hydroxyguanine (7,8-dihydro-8-oxoguanine) in hot spots of the c-Ha-ras gene. Jpn J. Cancer Res., 86, 270–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kamiya H., Murata-Kamiya,N., Koizume,S., Inoue,H., Nishimura,S. and Ohtsuka,E. (1995) 8-Hydroxyguanine (7,8-dihydro-8-oxoguanine) in hot spots of the c-Ha-ras gene: effects of sequence contexts on mutation spectra. Carcinogenesis, 16, 883–889. [DOI] [PubMed] [Google Scholar]
- 14.Tan X., Grollman,A. and Shibutani,S. (1999) Comparison of the mutagenic properties of 8-oxo-7,8-dihydro-2′-deoxyadenosine and 8-oxo-7,8-dihydro-2′-deoxyguanosine DNA lesions in mammalian cells. Carcinogenesis, 20, 2287–2292. [DOI] [PubMed] [Google Scholar]
- 15.Le Page F., Margot,A., Grollman,A., Sarasin,A. and Gentil,A. (1995) Mutagenicity of a unique 8-oxoguanine in human Ha-ras sequence in mammalian cells. Carcinogenesis, 16, 2779–2784. [DOI] [PubMed] [Google Scholar]
- 16.Kamiya H., Miura,K., Ishikawa,H., Inoue,H., Nishimura,S. and Ohtsuka,E. (1992) c-Ha-ras containing 8-hydroxyguanine at codon 12 induces point mutations at the modified and adjacent positions. Cancer Res., 52, 3483–3485. [PubMed] [Google Scholar]
- 17.Kuchino Y., Mori,F., Kasai,H., Inoue,H., Iwai,S., Miura,K., Ohtsuka,E. and Nishimura,S. (1987) Misreading of DNA templates containing 8-hydroxydeoxyguanosine at the modified base and at adjacent residues. Nature, 327, 77–79. [DOI] [PubMed] [Google Scholar]
- 18.Efrati E., Tocco,G., Eritja,R., Wilson,S.H. and Goodman,M.F. (1999) “Action-at-a-distance” mutagenesis. J. Biol. Chem., 274, 15920–15926. [DOI] [PubMed] [Google Scholar]
- 19.Johnson R.E., Kondratick,C.M., Prakash,S. and Prakash,L. (1999) hRAD30 mutations in the variant form of xeroderma pigmentosum. Science, 285, 263–265. [DOI] [PubMed] [Google Scholar]
- 20.Masutani C., Kusumoto,R., Yamada,A., Dohmae,N., Yokoi,M., Yuasa,M., Araki,M., Iwai,S., Takio,K. and Hanaoka,F. (1999) The XPV (xeroderma pigmentosum variant) gene encodes human DNA polymerase η. Nature, 399, 700–704. [DOI] [PubMed] [Google Scholar]
- 21.Haracska L., Yu,S.-L., Johnson,R.E., Prakash,L. and Prakash,S. (2000) Efficient and accurate replication in the presence of 7,8-dihydro-8-oxoguanine by DNA polymerase η. Nature Genet., 25, 458–462. [DOI] [PubMed] [Google Scholar]
- 22.Johnson R.E., Washington,M.T., Prakash,S. and Prakash,L. (2000) Fidelity of human DNA polymerase η. J. Biol. Chem., 275, 7447–7450. [DOI] [PubMed] [Google Scholar]
- 23.Yu S.-L., Johnson,R.E., Prakash,S. and Prakash,L. (2001) Requirement of DNA polymerase η for error-free bypass of UV-induced CC and TC photoproducts. Mol. Cell. Biol., 21, 185–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Matsuda T., Bebenek,K., Masutani,C., Hanaoka,F. and Kunkel,T.A. (2000) Low fidelity DNA synthesis by human DNA polymerase-η. Nature, 404, 1011–1013. [DOI] [PubMed] [Google Scholar]
- 25.Veisman A., Masutani,C., Hanaoka,F. and Chaney,S.G. (2000) Efficient translesion replication past oxaliplatin and cisplatin GpG adducts by human DNA polymerase η. Biochemistry, 39, 4575–4580. [DOI] [PubMed] [Google Scholar]
- 26.Levine R.L., Miller,H., Grollman,A., Ohashi,E., Ohmori,H., Masutani,C., Hanaoka,F. and Moriya,M. (2001) Translesion DNA synthesis catalyzed by human Pol η and Pol κ across 1, N6-ethenodeoxyadenosine. J. Biol. Chem., 276, 18717–18721. [DOI] [PubMed] [Google Scholar]
- 27.Zhang Y., Yuan,F., Wu,X., Rechkoblit,O., Taylor,J.-S., Geacintov,N.E. and Wang,Z. (2000) Error-prone lesion bypass by human DNA polymerase η. Nucleic Acids Res., 28, 4717–4724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Masutani C., Kusumoto,R., Iwai,S. and Hanaoka,F. (2000) Mechanisms of accurate translesion synthesis by human DNA polymerase η. EMBO J., 19, 3100–3109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bebenek K., Matsuda,T., Masutani,C., Hanaoka,F. and Kunkel,T.A. (2001) Proofreading of DNA polymerase η-dependent replication errors. J. Biol. Chem., 276, 2317–2320. [DOI] [PubMed] [Google Scholar]
- 30.Creighton S., Bloom,L.B. and Goodman,M.F. (1995) Gel fidelity assay measuring nucleotide misinsertion, exonucleolytic proofreading and lesion bypass efficiencies. Methods Enzymol., 262, 232–256. [DOI] [PubMed] [Google Scholar]
- 31.Traut T.W. (1994) Physiological concentrations of purines and pyrimidines. Mol. Cell. Biochem., 140, 1–22. [DOI] [PubMed] [Google Scholar]
- 32.Prakash S. and Prakash,L. (2002) Translesion DNA synthesis in eukaryotes: a one- or two-polymerase affair. Genes Dev., 16, 1872–1883. [DOI] [PubMed] [Google Scholar]
- 33.Echols H. and Goodman,M.F. (1991) Fidelity mechanisms in DNA replication. Annu. Rev. Biochem., 60, 477–511. [DOI] [PubMed] [Google Scholar]
- 34.Goodman M.F., Creighton,S., Bloom,L.B. and Petruska,J. (1993) Biochemical basis of DNA replication fidelity. Crit. Rev. Biochem. Mol. Biol., 28, 83–126. [DOI] [PubMed] [Google Scholar]
- 35.Ide H., Kow,Y.W. and Wallace,S.S. (1986) Thymine glycols and urea residues in M13 DNA constitute replicative blocks in vitro. Nucleic Acids Res., 13, 8035–8052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rouet P. and Essigmann,J.M. (1985) Possible role for thymine glycol in the selective inhibition of DNA synthesis on oxidized DNA templates. Cancer Res., 45, 6113–6118. [PubMed] [Google Scholar]
- 37.Clark J.M. and Beardsley,G.P. (1986) Thymine glycol lesions terminate chain elongation by DNA polymerase I in vitro. Nucleic Acids Res., 14, 737–749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Clark J.M. and Beardsley,G.P. (1987) Functional effects of cis-thymine glycol lesions on DNA synthesis in vitro. Biochemistry, 26, 5398–5403. [DOI] [PubMed] [Google Scholar]
- 39.McNulty J.M., Jerkovic,B., Bolton,P.H. and Basu,A.K. (1998) Replication inhibition and miscoding properties of DNA templates containing a site-specific cis-thymine glycol or urea residue. Chem. Res. Toxicol., 6, 666–673. [DOI] [PubMed] [Google Scholar]
- 40.Kusumoto R., Masutani,C., Iwai,S. and Hanaoka,F. (2002) Translesion synthesis by human DNA polymerase η across thymine glycol lesions. Biochemistry, 41, 6090–6099. [DOI] [PubMed] [Google Scholar]
- 41.Doublie S., Tabor,S., Long,A.M., Richardson,C.C. and Ellenberger,T. (1998) Crystal structure of a bacteriophage T7 DNA replication complex at 2.2 Å resolution. Nature, 391, 251–258. [DOI] [PubMed] [Google Scholar]
- 42.Kiefer J.R., Mao,C., Braman,J.C. and Beese,L.S. (1998) Visualizing DNA replication in a catalyticallly active Bacillus DNA polymerase crystal. Nature, 391, 304–307. [DOI] [PubMed] [Google Scholar]
- 43.Li Y., Korolev,S. and Waksman,G. (1998) Crystal structures of open and closed forms of binary and ternary complexes of the large fragment of Thermus aquaticus DNA polymerase I: structural basis for nucleotide incorporation. EMBO J., 17, 7514–7525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pelletier H., Sawaya,M.R., Kumar,A., Wilson,S.H. and Kraut,J. (1994) Structure of ternary complexes of rat DNA polymerase β, a DNA template-primer and ddCTP. Science, 264, 1891–1903. [PubMed] [Google Scholar]
- 45.Eom S.H., Wang,J. and Steitz,T.A. (1996) Structure of Taq polymerase with DNA at the polymerase active site. Nature, 382, 278–281. [DOI] [PubMed] [Google Scholar]
- 46.Trincao J., Johnson,R.E., Escalante,C.R., Prakash,S., Prakash,L. and Aggarwal,A.K. (2001) Structure of the catalytic core of S. cerevisiae DNA polymerase η: implications for translesion DNA synthesis. Mol. Cell, 8, 417–426. [DOI] [PubMed] [Google Scholar]