Abstract
We recently developed an oblique-incidence reflectivity difference (OI-RD) microscope, a form of polarization-modulated imaging ellipsometer, for label-free/high-throughput detection of biomolecular reactions on DNA and protein microarrays. We present examples of application of this technique to end-point and real-time investigations of DNA-DNA hybridization, antibody-antigen capture, and protein-small-molecule binding reactions. Compared to a conventional imaging ellipsometer based on the PCSA scheme and under the off-null condition, a polarization-modulated OI-RD microscope is inherently more sensitive by at least one order of magnitude to thickness changes on a solid surface. Compared to imaging surface plasmon resonance microscopes based on reflectance change on falling or rising slopes of the surface plasmon resonance, the OI-RD microscope (1) has a comparable sensitivity; (2) is applicable to conventional microscope glass slides; (3) easily covers a field of view as large as the entire surface of a 1” × 3” microscope slide.
1. Introduction
Optical techniques have played an instrumental role at almost every stage of advancement in life sciences. With the advent of lab-on-chips, namely, microarrays of biological macromolecules and individual cells immobilized on solid supports, we are in an exciting era of life sciences when molecular-level and cellular level chemistry are being explored and characterized in a highly parallel fashion [1,2]. This approach complements conventional molecular and cellular biochemistry approaches. Given the multitude and cooperative aspects of interactions among biological molecular complexes at cellular and sub-cellular levels, parallel detection of tens or thousands of biochemical reactions in microarray format will accelerate the process of discovery. Fluorescence-labeling is commonly used in detection of biochemical reactions in microarray format [1,2]. Typically one of the reaction partners is tagged with a fluorescent molecule or a quantum dot (through either genetic engineering such as incorporation of green fluorescence protein or a direct reaction with the host molecule). Fluorescence-labeling enables the detection of as few as a single macromolecule and drives the field of single-molecule detection in molecular/cellular biology.
However, an extrinsic tag such as a fluorescent molecule or a quantum dot always changes the properties of a host macromolecule. The significance of the change is often not known a priori. This is particularly relevant when studying properties of proteins [3]. Subtle changes in binding affinities and associated kinetics of protein molecules, by added physical properties of an extrinsic tag or through tag-induced conformational changes in protein molecules, can have a profound influence on some functions of protein molecules. Recognition of stereo-chemically modified double stranded DNA by specialized proteins in a living system is an example [4]. It is thus sensible to develop label-free detection techniques with adequate sensitivities to complement the fluorescence-based detection methods.
Imaging Surface Plasmon Resonance Spectroscopy (SPR) [5–7], Imaging Optical ellipsometry (OE) [8–11], and Reflectometric Interference Spectroscopy (RIFS) [12] are some of the optical techniques that have been explored to meet such a need. In their respective ways, these three label-free optical techniques in essence measure the same optical dielectric response of a thin film, consisting of molecules of biological significance on a solid substrate. As s result they detect same physical or chemical properties of the thin film such as thickness and mass density (including surface coverage) or changes of them during biochemical reactions. So far only Imaging SPR and Imaging Ellipsometry have been explored for high-throughput detection of microarrays of biological molecules [5–11].
In an SPR microscope equipped with a CCD camera, one typically uses the reflectance of a collimated, monochromatic light at a fixed incidence angle as the contrast and obtains an SPR image of an illuminated area on a solid surface covered with microarrays of biomolecules of interest. To maximize the sensitivity, the incidence angle is chosen to be on the falling or rising slope of the surface plasmon resonance peak. Over 2 orders of magnitude, the reflectance changes linearly with the thickness and the mass density (through the optical dielectric constant or refractive index of the film). The proportionality constant can vary from one gold-coated substrate to another and thus needs to be calibrated if quantitative information is required from such an image. Shumaker-Parry and Campbell were able to quantify the performance of such an imaging SPR microscope and achieved a detection limit of 2 × 10−5 RIU (refractive index unit) or 0.01 nm in detected protein thickness simultaneously over an area of 5mm × 5mm. It remains to be explored how such a microscope may be extended to examine microarrays over a much larger area.
Optical ellipsometry, in one form or another, measures changes in magnitude and phase of complex optical reflectivity (i.e., Fresnel reflection coefficient) in response to changes on solid or liquid surfaces [8]. If one is not concerned with magnetic and chiral properties of the surface layer, relevant reflectivity changes are those for p-polarized (transverse magnetic) and s-polarized (transverse electric) components of an optical beam. At oblique incidence, in response to a surface-bound change such as a biochemical reaction, the complex reflectivity changes disproportionately for p- and s-polarized light at a fixed optical frequency. As a result the magnitude and the phase of the ratio of the Fresnel coefficient for p-polarized light to that for s-polarized light, rp/rs ≡ ρ ≡ tanψ exp(iδ), change. Optical ellipsometry measures such changes.
In a typical imaging ellipsometer, one uses the polarizer-compensator-sample-analyzer (PCSA) scheme in which the phase compensator (C) is fixed and the polarizer (P) and the analyzer (A) are variable. The light beam reflected from an illuminated area on the sample surface (S) passes through the analyzer (A) and is subsequently imaged onto a CCD camera. P and A are rotated until the photocurrents are more or less minimized across the image region on the CCD to yield ψ and δ maps before for example a biochemical reaction takes place on the sample surface. During a subsequent biochemical reaction on the sample surface that results in a thickness change Δd in a surface-bound microarray feature, the corresponding change in the photocurrent under this off-null condition is proportional to (Δψ)2 and (Δδ)2, thus proportional to (Δd/λ)2. The off-null photocurrents are monitored in real-time to detect the biochemical reactions. However the quadratic dependence of the off-null photocurrent on already small quantities Δψ and Δδ sets the detection limit of this type of imaging ellipsometer to roughly Δψ ∼ 0.01° and Δδ ∼ 0.01° (i.e., ∼ 0.0002 radians).
A more sensitive form of ellipsometry is the oblique-incidence reflectivity difference (OI-RD) technique [6,7]. It is a polarization-modulated nulling ellipsometry in which the harmonics of modulated photocurrents are measured under suitable nulling conditions and are directly proportional Δψ and Δδ. This makes an imaging ellipsometer based upon the measurement of OI-RD signals at least an order of magnitude more sensitive than the aforementioned imaging ellipsometer, namely, with the detection limit to Δψ ∼ 0.001° and Δδ ∼ 0.001°. Such sensitivity is required for high-throughput affinity detection of low molecular weight analytes. The detection limit of Δδ ∼ 0.001° corresponds to 0.01 nm in detected protein thickness, similar to that of an imaging SPR microscope [6].
2. Oblique-incidence reflectivity difference (OI-RD): - a polarization-modulated nulling ellipsometry
Let rp0 = |rp0| exp(iΦp0) and rs0 = |rs0| exp(iΦs0) be the reflectivity for p- and s-polarized light from a bare substrate surface, respectively. Let rp = |rp| exp(iΦp) and rs = |rs| exp(iΦs) be the reflectivity when an ultrathin film is deposited on the substrate or when the surface layer of the substrate is modified. The fractional reflectivity change is defined as Δp = (rp – rp0)/rp0 and Δs = (rs – rs0)/rs0. The difference in fractional reflectivity change is then Δp - Δs. When it is small, Re{Δp - Δs} = (|rp| – |rp0|)/|rp0| - (|rs| – |rs0|)/|rs0| is simply the differential magnitude change, Im{Δp - Δs} = (Φp - Φp0) – (Φs - Φs0) is the differential phase change. In terms of ρ = rp/rs = tanψ exp(iδ), Δp - Δs ≈ (ρ – ρ0)/ρ with Re{Δp - Δs} ≈ (ψ – ψ0)/sinψ0cosψ0 = Δψ/sinψ0cosψ0 and Im{Δp - Δs} = δ – δ0 = Δδ [8]. The OI-RD technique has been successfully applied to detection of a wide variety of ultrathin films and surface modifications ranging from vapor-phase deposited rare gas films and perovskite oxide films in vacuum [13,14], electrochemically deposited metallic films at liquid-solid interfaces [15], to microarrays of biological molecules on functionalized glass (i.e., gene chips and protein chips) [11].
To relate the structural information on an ultrathin film or the modified surface layer on a substrate to the experimentally measured Δp - Δs, we use a classical three-layer model to describe the optical response from the surface of a homogeneous substrate covered with an ultrathin film (or a modified surface layer) [16],
| (1) |
θinc is the incidence angle. ε0, εd, and εs are the optical dielectric constants of the ambient, the film (or the modified surface layer), and the substrate, respectively. d is the thickness of the film. Θ is the coverage of the film on the substrate, i.e., the ratio of the covered area to the total area. Changes other than thickness and coverage such as mass density, chemical make-up, and morphology are represented by corresponding changes in εd. In addition to the dependence on structural properties of the ultrathin film (d, Θ, and εd), Δp - Δs also depends on θinc. It is maximized when θinc is close to the Brewster angle θB on a transparent substrate or its equivalent on an opaque substrate [17]. From Eq. (1), it is clear that at the interface of two transparent media (ε0 and εs being real), the presence of non-absorbing biomolecules with εd also real, only Im{Δp - Δs} is non-zero. This is what we observe for un-labeled DNA and protein microarrays. It is noteworthy that the surface-plasmon resonance (SPR) technique (another label-free optical detection) measures same properties (thickness and density) of a surface layer from the shift in SPR angle, δθSPR ≈ (3πd/λ)(εd - ε0)/εd [18,19]. In addition to being as sensitive as the SPR technique when operated near θB [19], the OI-RD technique is applicable to all flat solid substrates and in this respect more versatile since high-quality metal coatings are not required and the total internal reflection condition is also not necessary. As a result it is suited for high-throughput detection of microarrays fabricated on conventional solid substrates such as microscope slides. We have developed optical scanning microscopes using Δp - Δs as contrasts and performed a number of proof-of-principle experiments on DNA microarrays and protein microarrays.
3. Applications of OI-RD microscopes to detection of biomolecular reactions on microarrays
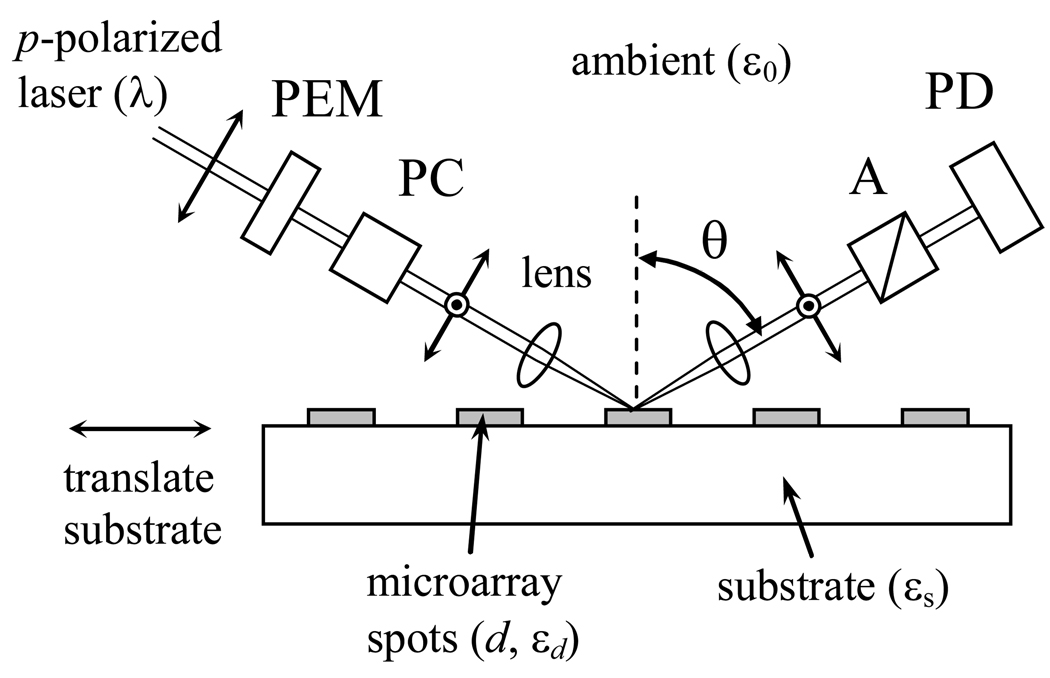
The arrangement of an OI-RD microscope is sketched in Fig. 1. The procedures for obtaining Δp - Δs have been described in details by Thomas and coworkers [13]. In this case, the ambient is air with ε0 = 1, the substrate is glass with εs = 2.31. In a microscope configuration, a microarray-covered glass slide is mounted on a dual-axis translation stage underneath fixed illumination and detection optics. The stage is driven by computer-controlled stepping motors and is movable along two perpendicular directions of the glass slide surface.
Fig. 1.
An OI-RD microscope for imaging biomolecular microarrays on functionalized glass substrate. The substrate is on a translation stage that is movable along the x and y directions. PEM: photoelastic modulator for polarization modulation; PC: Pockel cell for initial phase shift adjustment; A: polarizing analyzer; PD: single-element or multi-element photo-detector.
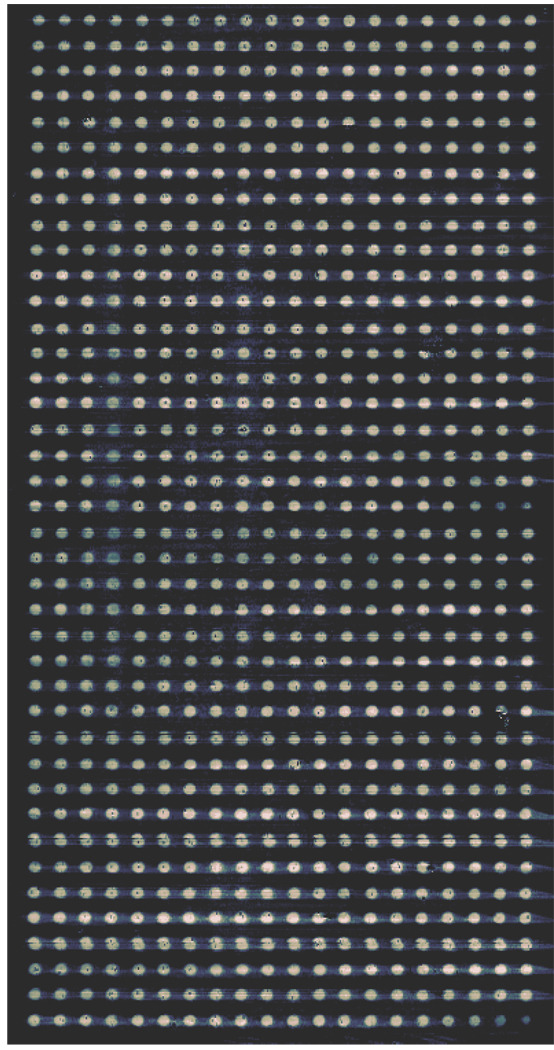
For high-resolution imaging, we focus the illumination beam to a spot of 2.4 microns (the full width at 1/e2 of the maximum of the intensity profile) on the microarray-covered surface and image the reflected beam from the spot onto a single photodiode detector. Using Rayleigh’s criterion, the image resolution is nominally 1.7 microns. To obtain a 2-D image of a microarray using Δp - Δs as contrasts, we mechanically move the stage in both x- and y-directions and record the values of Δp - Δs at each spot. The scan time is long in this configuration. For high-speed imaging with a spatial resolution of 15-micron to capture both the end-point and kinetics of biomolecular reactions on 1000-feature or 10000-feature microarrays (with the feature size in the range of 100 microns and feature separation in the range of 300 microns), we use cylindrical optics to focus the illumination beam into a line on the microarray-covered surface and image the reflected beam from the line onto a multi-element photodiode array (instead of a single detector). The scan along the line direction is then achieved by electronically interrogating the elements of the photodiode array at a rate at least 1000 times faster than the mechanical scan. This has enabled us to obtain an end-point image of 800-feature microarrays in less than 14 minutes as shown in Fig. 2. We should note that in application to microarrays, the spatial resolution in the range of 1.7 microns to 15 microns is more than enough since typical printed microarray features are between 80 to 150 microns, and typical separation between neighboring features are between 200 to 500 microns [1]. There is no observable edge effect arising from the finite size of the microarray features. In our present OI-RD microscopes, we have achieved the sensitivity of 0.1 Å. In term of resonance unit (RU) routinely used in SPR biosensors, we have achieved the sensitivity of 1 RU or 10−4 deg. Such sensitivity is adequate for our current microarray applications in high-throughput small molecule library screening for protein ligands and in high-throughput small molecule drug screening. And it is comparable to surface plasmon resonance microscopy as reported by Shumaker-Parry and Campbell [6].
Fig. 2.
An OI-RD image of a 20 × 40 bovine serum albumin (BSA) microarray of one monolayer in thickness, obtained with a high-speed OI-RD microscope using a combination of line illumination and 152-element photodiode array detector. The contrast mechanism is Im{Δp - Δs}. The contrast shown in the figure is on average what we have expected of a full BSA monolayer covering each of the 800 features. The pixel dimension of the image is 15 microns by 15 microns. There are 410×760 pixels in the image. The feature is 130 microns in diameter, and the center-to-center separation between neighboring features is 300 microns. The total scan time for this 800-featuer image is 14 minutes. The numerical aperture (N.A.) of this microscope is 0.15.
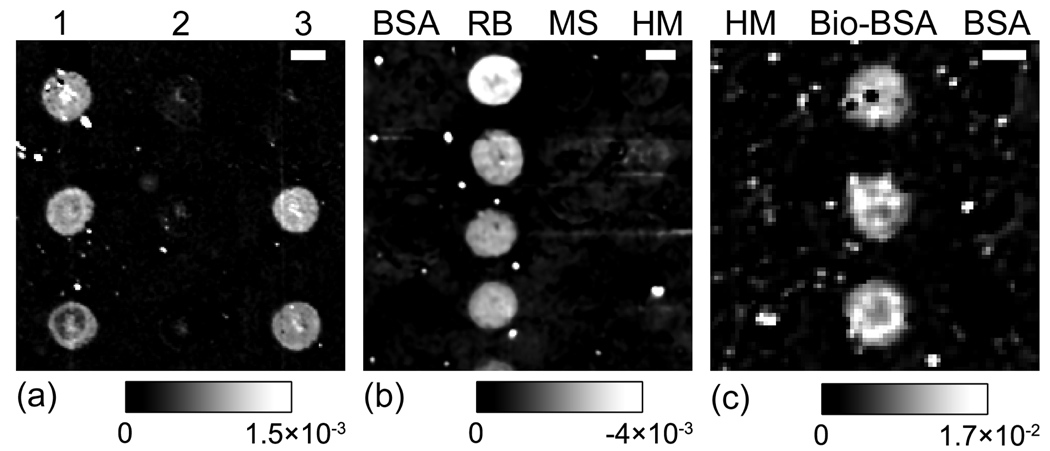
In Fig. 3a, we show an Im{Δp - Δs} image of a 3×3 60-nucleotide (nt) oligomer microarray after it has reacted with a mixture of unlabeled 60-nt oligomers complementary to Column 1 and Cy5-labeled 60-nt oligomers complementary to Column 3. Each column is a triplicate of 120-micron spots of oligomers with a well defined sequence: 5'-TCACAAACCC GTCCTACTCT ACTAGCTGCA GTAGCCCCAC TGGTTCCCGT TTCCGATGTT-3' for Column 1; 5'-CCTTGTACCG CTGAGTTCAC ACCGACACAC CTCACCACAC TTACACCGTC CACAAAGAGA-3' for Column 2; and 5'-TTTCCATGCG GACCTACCAC CGTAGTACCT CGCAATGCCA GTGCAACAAG TACACCTGGA-3' for Column 3. The oligomer microarray is printed on a commercial poly-L-lysine functionalized glass slide. The printed oligomers lie flat on the glass surface, due to the electrostatic interaction between the negatively charged DNA backbone and the positively charged amine group at neutral pH. The excess oligomers on top of the saturated oligomer monolayer are removed by a washing step. The rest of the poly-L-lysine functionalized surface is blocked with succinic anhydride in borate-buffered 1-methyl-2-pyrolidinone before the microarray is subjected to the hybridization reaction. The image shown in Fig. 3a is the difference between the image taken after the reaction and the image taken before the reaction. The scale bar is 100 microns in length. The average optical signal change is 1.5×10−3 in Column 1 and indicates that 60% (Θ = 0.6) of the surface-immobilized oligomers have reacted with complementary partners. The hybridization is well resolved without extrinsic labeling. The average optical signal change in Column 3 (the positive control column) is also 1.5×10−3 [6]. This is expected since the wavelength of the He-Ne laser used for illumination of the OI-Rd microscope is far from the absorption peaks for Cy5 dye.
Fig. 3.
(a) Image in Im{Δp - Δs} of a 3×3 60-nt DNA microarray after reaction with a mixture of unlabeled DNA complementary to the first column and Cy5-labeled DNA complementary to the third column. The incidence angle θinc is 45°. (b) Image in Im{Δp - Δs} of a 4×4 antigen microarray after reaction with unlabeled goat anti-rabbit IgG. θinc is again 45°. (c) Image in Im{Δp - Δs} of a 3×3 protein microarray after reaction with unlabeled streptavidin. θ inc in this case is 59°. The spatial resolution of the microscope that we used to obtain these images is 3 microns.
In Fig. 3b, we display an Im{Δp - Δs} image of a 4×4 protein microarray after it has been exposed to un-labeled goat antibody against rabbit IgG. Each column of the microarray is a titration series of one type of proteins: bovine serum albumin (BSA) for Column 1; rabbit IgG (RB) for Column 2; mouse IgG (MS) for Column 3; and human IgG (HM) for Column 4. They are printed as 150-micron spots on an epoxy-functionalized glass slide. The excess of the printed proteins is removed by washing steps and the rest of the epoxy-functionalized surface is blocked with BSA. Again, the image shown in Fig. 3b is the difference between the one taken after the reaction and the one taken before the reaction. The scale bar is 100 microns. Without fluorescent labeling, the differential image reveals clearly the specific antibody-antigen capture with a good signal-to-noise ratio. The change in Im{Δp - Δs} (4×10−3) indicates that roughly 20% (Θ = 0.2) of a saturated monolayer of rabbit IgG have reacted with the goat anti-rabbit IgG.
In Fig. 3c, we show an Im{Δp - Δs} image of another 3×3 protein microarray after it has been exposed to un-labeled streptavidin. Each column is a triplicate of 150-micron spots of same protein: human IgG for the first column; BSA-biotin complex for the second column; and BSA alone for the third column. The BSA-biotin complex is synthesized for the purpose of “immobilizing” small molecules such as biotin to epoxy-functionalized glass slide with BSA as the anchor. A linker molecule is inserted between BSA and biotin to minimize the effect of BSA on the affinity of biotin. The image shown in Fig. 3c is the difference between the one taken after the reaction and the one taken before the reaction. The specific reaction of streptavidin with BSA-biotin complex is clearly shown in the differential OI-RD image. The bright spots in the images shown in Fig. 3 are present in the microarrays instead of noise. They are dust particles from the ambient and residuals from the processing of the microarray. Streptavidin is roughly a spherical molecule with a 5-nm diameter [20, 21] and a mass volume density of ρd = 1.35 g/cm3. When packed in square lattice to a full monolayer, the surface number density of streptavidin is 4.0 × 1012 molecules/cm2, and the surface mass density is 4 ng/mm2. Assume that the optical dielectric constant for streptavidin at λ = 532 nm is εd(streptavidin) = 2.51 (nd(streptavidin) = 1.584), we can deduce the surface coverage Θ of the reacted streptavidin using Eq. (1) from the change in the OI-RD signal. After taking into account of the effect of the incidence angle at 59°, the change in Im{Δp - Δs} (1.7×10−2) shows that the coverage of the streptavidin is 1.4 × 1012 molecules/cm2, namely, Θ = 0.35. We should note here that the surface coverage obtained this way is an approximate to the true value as the former depends on the assumption of εd = 2.51, the packing geometry, and the validity of Eq. (1). The true relation between the OI-RD signal and the surface coverage can be established in our future investigation by a calibration against a fluorescence method or a nuclear method.
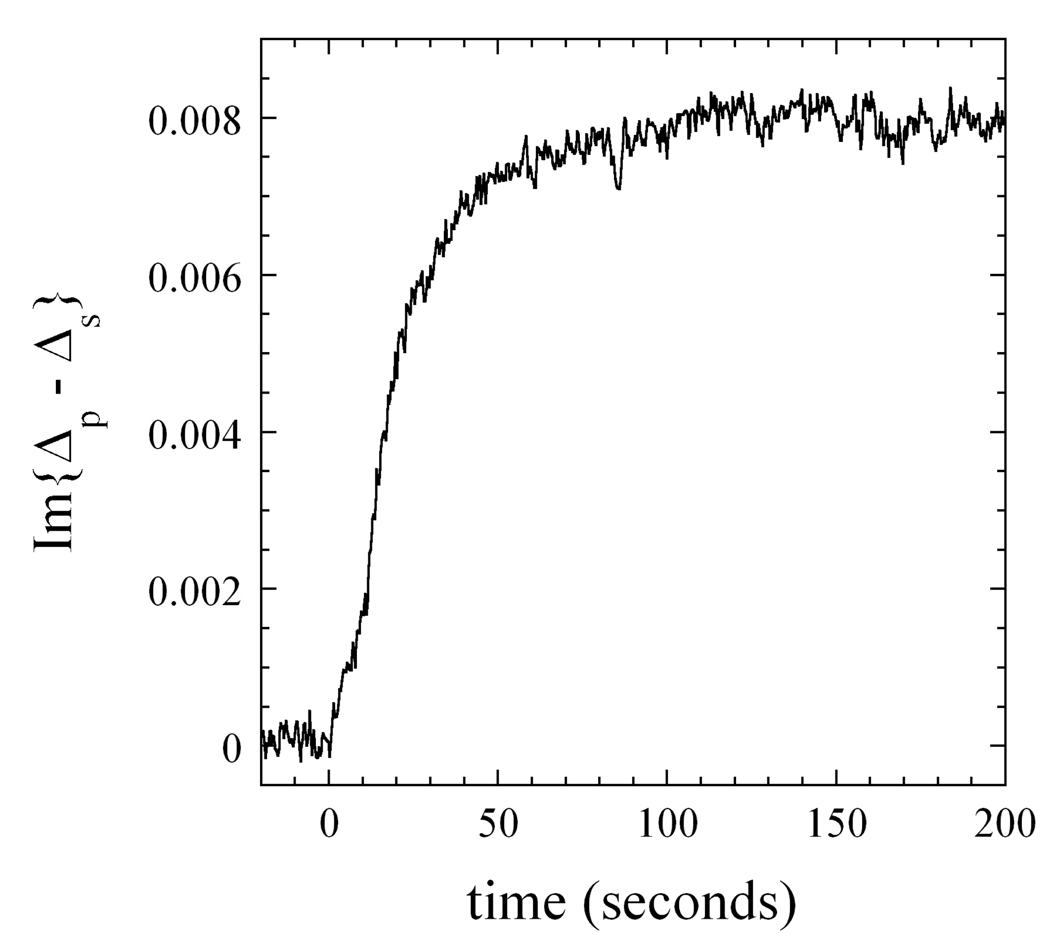
To illustrate that the OI-RD technique is capable of measuring protein-small molecule binding reactions on functionalized glass surfaces in real time, we have performed a series of measurements on bovine serum albumin (BSA) on epoxy-coated glass slide. Because of the small dielectric contrast or refractive index difference between glass and water, the reflection from the interface is very weak, making ellipsometry measurements of biomolecular processes at the interface a difficult task. In our experiments, the functionalized side of the glass slide is in contact with the buffer or the BSA solution as a part of a fluid cell. The other side of the slide is in air. The illumination laser beam is incident on the functionalized surface through the air side and the reflected beam is detected as illustrated in Fig. 1 with a single detector. The experiment begins with 1× phosphate buffered saline (PBS) solution in the fluid cell, and then BSA is added and quickly mixed with the 1×PBS (using a magnetic stir in the cell) to make it a 7.2 µM BSA solution (1.0 mg/ml) in less than 6 seconds. In Fig. 4, we show Im{Δp - Δs} from the glass-solution interface before and after BSA is added (at t = 0) to the 1×PBS. Except for the first 6 seconds when the mixture is being homogenized, the OI-RD signal shows the uptake of one monolayer of BSA that fully covers the epoxy-functionalized glass. The saturation level at 0.008 is not changed when the BSA solution is replaced with 1×PBS, indicating that the uptake or adsorption of BSA is irreversible. This uptake curve compares well with the observation of BSA adsorption from aqueous buffer on a gold-coated substrate reported by Jung et al. using an SPR microscope [7]. By subsequently exposing the BSA-covered glass slide with Cy5-labeled IgG molecules, we were able to confirm that the saturation level at 0.008 in Im{Δp - Δs} corresponds to one full monolayer of BSA that covers 98% of the epoxy-coated surface. From Eq. (1), we find that the signal level of 0.008 corresponds to a uniform layer of BSA with thickness of 1.4 nm and an effective dielectric constant of εd(BSA) = 2.5 or a refractive index of nd(BSA) = 1.58. It is difficult to determine independently the refractive index and the thickness of the BSA layer. By keeping the magnetic stir on during the entire experiment, we maintain a constant BSA flux toward the glass surface (namely, 6 seconds after the BSA is added.) Fig. 4 shows that the uptake follows the Langmuir kinetics, namely, (a) the uptake rate is proportional to the probability of an impinging BSA to strike an open epoxy-coated surface; (b) the probability of a striking BSA molecule to bind to the open epoxy-coated surface is a constant.
Fig. 4.
Im{Δp - Δs} from the interface between an epoxy-functionalized glass slide and the aqueous solution of bovine serum albumin (BSA) in 1× phosphate buffered saline (1×PBS). At t = 0, the BSA is added to an initial 1×PBS to make it a 7.2 µM BSA solution 1×PBS in less than 6 seconds. The saturated signal level at 0.008 corresponds to a full monolayer of BSA that covers 98% of the epoxy-functionalized surface. A magnetic stir is inside the fluid cell to continuously mix the solution during the experiment, ensuring a constant flux of BSA toward the glass surface.
The result shown in Fig. 4 demonstrates that even with a small dielectric contrast between glass slide and an aqueous solution, the biochemical reaction at the interface can be captured in real-time with a very good sensitivity using the OI-RD technique. By employing a multi-element array detector such as the one used to obtain the image in Fig. 2, we can further remove the effect of systemic changes in an OI-RD set-up and in a fluid cell on the measured signal and achieve an even better sensitivity than that displayed in Fig. 4. More importantly, we can simultaneously measure multiple biochemical reactions on a microarray with a high-speed OI-RD microscope.
4. Conclusion
We demonstrated that the oblique-incidence reflectivity difference (OI-RD) as a special form of polarization modulated ellipsometry is a most sensitive, versatile optical platform for label-free detection of biomolecular reactions, particularly in microarray format. In microscope configurations, this ellipsometric technique is suited for very high throughput screening of small molecule libraries for protein ligand candidates and for high throughput search for biomarkers. Because an OI-RD microscope is capable of both end-point and real-time measurements in a highly parallel fashion, we expect it to be instrumental in discovery-oriented and function-oriented proteomic research from molecular to cellular levels.
Acknowledgments
This work was supported by the Graduate Research and Education in Adaptive bio-Technology (GREAT) program under Grant#2004–09, administered by the University of California System-wide Biotechnology Research and Education Program, by NSF Center for Biophotonics Science and Technology, and by National Institute of Health under NIH-R01-HG003827-01.
Contributor Information
X.D. Zhu, Email: xdzhu@physics.ucdavis.edu, Department of Physics, University of California, Davis, California 95616.
J. P. Landry, Department of Physics, University of California, Davis, California 95616
Y.S. Sun, Department of Physics, University of California, Davis, California 95616
J.P. Gregg, School of Medicine, University of California at Davis, Sacramento, California 95817
K.S. Lam, School of Medicine, University of California at Davis, Sacramento, California 95817
X.W. Guo, Clinical Diagnostics Group, Bio-Rad Laboratories, 4000 Alfred Nobel Drive, Hercules, California 94547
References
- 1.Schena M. Microarray Analysis. Hoboken: John Wiley and Sons; 2003. [Google Scholar]
- 2.Zhu H, Bilgin M, Bangham R, Hall D, Casamayor A, Bertone P, Ning Lan R, Jansen S, Bidlingmaier T, Moufek T, Mitchell P, Miller RA, Dean M, Gerstein, Scheider M. Global analysis of protein activities using proteome chips. Science. 2001;293:2101–2105. doi: 10.1126/science.1062191. [DOI] [PubMed] [Google Scholar]
- 3.MacBeath G. Protein microarrays and proteomics. Nature Genet. 2002;32:526–532. doi: 10.1038/ng1037. [DOI] [PubMed] [Google Scholar]
- 4.Lindahl T, Wood RD. Quality Control by DNA Repair. Science. 1999;286 doi: 10.1126/science.286.5446.1897. 18971905. [DOI] [PubMed] [Google Scholar]
- 5.Nelson BP, Frutos AG, Brockman JM, Corn RM. Near-Infrared Surface Plasmon Resonance Measurements of Ultrathin Films. 1 Angle Shift and SPR Imaging Experiments. Anal. Chem. 1999;71 3928-2934. [Google Scholar]
- 6.Shumaker-Parry JS, Campbell CT. Quantitative Methods for Spatially Resolved Adsorption/Desorption Measurements in Real Time by Surface Plasmon Resonance Microscopy. Anal. Chem. 2004;76:907–917. doi: 10.1021/ac034962a. [DOI] [PubMed] [Google Scholar]
- 7.Jung LS, Campbell CT, Chinowsky TM, Mar MN, Yee SS. Quantitative Interpretation of the Response of Surface Plasmon Resonance Sensors to Adsorbed Films. Langmuir. 1998;14:5636–5648. [Google Scholar]
- 8.Azzam RMA, Bashara NM. Ellipsometry and Polarized Light. New York: Elsevier Science; 1987. [Google Scholar]
- 9.Jin G, Jansson R, Arwin H. Imaging ellipsometry revisited: Development for visualization of thin transparent layers on silicon substrates. Rev. Sci. Instrum. 1996;67:2930–2936. [Google Scholar]
- 10.Wang ZH, Jin G. A Label-Free Multi-sensing Immuno-sensor Based on Imaging Ellipsometry. Anal. Chem. 2003;75:6119–6123. doi: 10.1021/ac0347258. [DOI] [PubMed] [Google Scholar]
- 11.Landry JP, Gregg JP, Zhu XD. Label-free detection of microarrays of biomolecules by oblique-incidence reflectivity difference microscopy. Opt. Lett. 2004;29:581–583. doi: 10.1364/ol.29.000581. [DOI] [PubMed] [Google Scholar]
- 12.Piehler J, Brecht A, Gauglitz G. Affinity Detection of Low Molecular Weight Analytes. Anal. Chem. 1996;68:139–143. doi: 10.1021/ac9504878. [DOI] [PubMed] [Google Scholar]
- 13.Thomas P, Nabighian E, Bartelt MC, Fong CY, Zhu XD. An oblique-incidence optical reflectivity difference and LEED study of rare-gas growth on a lattice-mismatched metal substrate. Appl. Phys. A. 2004;79:131–137. [Google Scholar]
- 14.Fei YY, Zhu XD, Liu LF, Liu HB, Chen ZH, Yang GZ. Oscillations in oblique-incidence optical reflection from a growth surface during layer-by-layer epitaxy. Phys. Rev. B. 2004;69 233405-4. [Google Scholar]
- 15.Schwarzacher W, Gray J, Zhu XD. Oblique Incidence Reflectivity Difference as an In-Situ Probe of Electrodeposition: Co on Au, Electrochem. Solid-State Lett. 2003;6:C73–C76. [Google Scholar]
- 16.Zhu XD. Oblique-incidence optical reflectivity difference from a rough film of crystalline material. Phys. Rev. B. 2004;69 115407-5. [Google Scholar]
- 17.Landry JP, Gray J, O’Toole MK, Zhu XD. Incidence-angle dependence of optical reflectivity difference from an ultrathin film on solid surface. Opt. Lett. 2006;31:531–533. doi: 10.1364/ol.31.000531. [DOI] [PubMed] [Google Scholar]
- 18.Liedberg B, Nylander C, Lundstrom I. Surface Plasmon Resonance for Gas Detection and Biosensing. Sensors and Actuators. 1983;4:299–304. [Google Scholar]
- 19.Zhu XD. Comparison of two optical techniques for label-free detection of biomolecular microarrays on solids. Opt. Commun. 2006;259:751–753. [Google Scholar]
- 20.Hendrickson WA, Paehler A, Smith JL, Satow Y, Merritt EA, Phizackerley RP. Crystal structure of core streptavidin determined from multiwavelength anomalous diffraction of synchrotron radiation. Proc. Natl. Acad. Sci. 1989;86:2190–2194. doi: 10.1073/pnas.86.7.2190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Weber P, Ohlendorf D, Wendoloski J, Salemme F. Structural origins of high-affinity biotin binding to streptavidin. Science. 1989;243:85–88. doi: 10.1126/science.2911722. [DOI] [PubMed] [Google Scholar]