Abstract
Somitogenesis is the process of division of the anterior–posterior vertebrate embryonic axis into similar morphological units known as somites. These segments generate the prepattern which guides formation of the vertebrae, ribs and other associated features of the body trunk. In this work, we review and discuss a series of mathematical models which account for different stages of somite formation. We begin by presenting current experimental information and mechanisms explaining somite formation, highlighting features which will be included in the models. For each model we outline the mathematical basis, show results of numerical simulations, discuss their successes and shortcomings and avenues for future exploration. We conclude with a brief discussion of the state of modeling in the field and current challenges which need to be overcome in order to further our understanding in this area.
I. Introduction
The dramatic advances made in genetic and molecular biology in recent years have allowed us to produce detailed descriptions of a number of complex processes that arise on many different spatial and temporal scales during development. This unparalleled flood of experimental data may well enable us to understand how genes and proteins work collectively in a cell, and from this how multicellular organisms develop. But, therein exists one of the great challenges of modern science; all too often our knowledge remains in isolated pockets, lacking a conceptual framework tying the fragmented data together and allowing ideas and hypotheses to be explored. This is where mathematical modeling can play a fundamental role, comparable with any laboratory research tool: it allows us to combine the effects of multiple nonlinear processes into a coherent structure which can be used for hypothesis testing and making experimentally testable predictions.
Somitogenesis, the segmentation of the vertebrate anteroposterior (AP) axis, is a perfect paradigm for studying such issues (Gossler and Hrabě de Angelis, 1998); it has a long history of experimental investigation at the cellular level, and we have observed recently a huge explosion in the rate of identification of the underlying molecular components. Somitogenesis serves as a model process for studying pattern formation during embryogenesis, encompassing a wide range of mechanisms and processes, such as the role of biological clocks (Dale et al., 2003, 2006; Dale and Pourquié, 2000; Maroto and Pourquié, 2001; Pourquié 1998, 2001b), positional information gradients (Diez del Corral et al., 2002, 2003; Diez del Corral and Storey, 2004; Dubrulle et al., 2001; Dubrulle and Pourquié, 2002, 2004b), cell migration and adhesion (Drake and Little, 1991; Duband et al., 1987; Duguay et al., 2003; Kulesa and Fraser, 2002), all of which are tightly coupled, both spatially and temporally.
Somites are tightly bound blocks of cells that lie along the AP axis of vertebrate embryos (Gossler and Hrabě de Angelis, 1998; McGrew and Pourquié, 1998; Pourquié, 2001b). They are transient structures, and further differentiation of the somites gives rise to the vertebrae, ribs and other associated features of the trunk. Somites segment from the presomitic mesoderm (PSM); thick bands of tissue that lie on either side of the notochord, along the AP axis. At regular time intervals, groups of cells at the anterior ends of the PSM undergo changes in their adhesive and migratory behavior and condense together to form an epithelial block of cells known as a somite (Stockdale et al., 2000). As a result, somites form in a very strict AP sequence. As the body axis extends, the budding of cells anteriorly is compensated by the addition of cells at the posterior ends of the PSM (Pourquié, 2003). Each band of the PSM stays approximately constant in length throughout the process of somite formation and a wave of patterning appears to sweep along the AP axis leaving the somites in its wake (Collier et al., 2000).
From the molecular point of view, formation of the somites begins with the establishment of a prepattern of gene expression. In mouse and zebrafish, members of the Mesp family are periodically activated in regions corresponding to the future somites (Haraguichi et al., 2001; Morimoto et al., 2006; Saga et al., 2001; Takahashi et al., 2000; Takashi et al., 2005). Overt signs of segmentation occur during subsequent cell cycles when the prepattern is converted to morphological somites. Cells undergo changes in their adhesive and migratory properties and a series of extensive rearrangements and shape-changes, mediated by cell adhesion molecules (CAMs), take place (Drake and Little, 1991; Duband et al., 1987; Duguay et al., 2003; Foty and Steinberg, 2004; Kalcheim and Ben-Yair, 2005).
Although the physical somite boundaries form at the anterior-most end of the PSM, early scanning microscope images showed that the PSM is not a homogeneous tissue (Meier, 1979; Tam and Meier, 1982). Prior to somite formation early signs of segmentation were seen: the posterior PSM was shown to display metameric arrangements of cells known as somitomeres, which seem to be precursors of the somites (Gossler and Hrabě de Angelis, 1998; Jacobson and Meier, 1986). The existence of this prepattern was confirmed by microsurgical experiments in which isolated parts of the PSM formed somites in strict isolation (Chernoff and Hilfer, 1982; Packard, 1976). However, this somitomeric prepattern was shown to be undetermined and susceptible to modification by external agents (Packard et al., 1993). More recently, existence of the prepattern has been further contested by microsurgical experiments conducted by Dubrulle and co-workers: AP inversions of somite-length regions of the posterior PSM resulted in normal segmentation whilst inversions of the anterior PSM resulted in somites with reversed polarity (Dubrulle et al., 2001). These results suggest that the anterior-most portion of the PSM is determined with regard to its segmentation program, whilst the posterior-most portion is labile in this respect. The different regions of the PSM were found to correspond to regions of varying FGF signaling; we discuss this more in following paragraphs.
There are several genes expressed dynamically in the PSM. Some are known as cyclic genes, because they oscillate with the same frequency it takes to form one somite (Dale et al., 2006; Ishikawa et al., 2005; McGrew et al., 1998; Palmeirim et al., 1997; Pourquié, 2001a, 2003). In the chick, gene expression bands of c-hairy1 and l-fng sweep along the AP axis: beginning as wide bands in the tail end of the embryo, they narrow as they travel in an anterior direction, until they come to rest in the newly forming somite. Expression is considered to arise as a result of an underlying segmentation clock in the PSM cells: several cell-autonomous components of the clock have been identified (Aulehla and Johnson, 1999), together with other molecular pathways which couple the oscillations in neighboring PSM cells (del Barco Barrantes et al., 1999; Jiang et al., 1998, 2000; Jiang and Lewis, 2001; Pourquié, 1999; Rida et al., 2004).
An additional gene with dynamic expression in the PSM is fgf8. A gradient of FGF8 exists along the AP axis, peaking at the posterior end of the embryo (Dubrulle et al., 2001 Dubrulle and Pourquié, 2002, 2004a, 2004b). As the body axis elongates, the gradient of FGF8 moves in a posterior direction so that it stays at a constant axial level, relative to the PSM. Cells are initially part of a region where FGF8 signaling prevails, but they gradually exit this region as they move up through the PSM. Dubrulle and co-workers have shown that downregulation of FGF signaling is necessary for cells to become part of a somite, and they term the threshold level of FGF signaling at which this ability is gained the determination front (Dubrulle et al., 2001).
Following the experimental discoveries described above, Pourquié and co-workers have postulated that it is the interaction between the segmentation clock and the FGF8 gradient that gates the cells into somites: the segmentation clock defines when somite boundaries form, whilst the FGF8 gradient defines where they form (Dubrulle et al., 2001; Tabin and Johnson, 2001). This is now the generally accepted “model” for somite formation.
II. Models for Somite Formation
In this section we will present a number of mathematical models which describe different stages of the formation of somites. We begin by presenting a mathematical formulation of the Clock and Wavefront model, which integrates information from the segmentation clock and the FGF8 wavefront to produce a somitic prepattern. Next we consider individual models for the segmentation clock and FGF8 gradient, which may be integrated into the Clock and Wavefront model. Finally we present a model which includes the role of cell adhesion molecules and the cell rearrangements which take place during the generation of epithelial somites. In each case we introduce the model and its mathematical formulation. We show the results of numerical simulations, discuss the successes and shortcomings of the model and avenues for extension and refinement.
A. The Clock and Wavefront Model
With the recent experimental findings concerning the existence of a segmentation clock and a wavefront of FGF8 along the AP of vertebrate embryos, the Clock and Wavefront model has received a large amount of support. First proposed by Cooke and Zeeman (Cooke, 1975, 1998; Cooke and Zeeman, 1976; Zeeman, 1974), the model assumed the existence of a longitudinal positional information gradient along the AP axis which interacts with a smooth cellular oscillator (the clock) to set the times at which cells undergo a catastrophe. In this context, Cooke and Zeeman were referring to the changes in adhesive and migratory behavior of cells as they form somites.
Pourquié and co-workers recently proposed a revised version of the model which involves the interaction of the segmentation clock and FGF8 wavefront in gating the cells into somites (Dubrulle et al., 2001; Tabin and Johnson, 2001). Specifically, they assume that the clock sets the times at which new somite boundaries form whilst the position of the determination front sets where they form (Dubrulle et al., 2001). We developed a mathematical formulation of the Clock and Wavefront model based on these assumptions (Baker et al., 2006a, 2006b). We also made the following further assumptions: (i) upon reaching the determination front, a cell acquires the ability to segment by becoming competent to produce a somitic factor—which could be, for example, Mesp2 (Morimoto et al., 2006; Saga et al., 2001; Takashi et al., 2000, 2005)—in response to the presence of a signaling molecule; (ii) one clock oscillation after reaching the determination front, cells become able to produce the signaling molecule; (iii) after responding to the signaling molecule, a cell is specified as somitic and becomes refractory to FGF8 signaling; (iv) a cell forms part of a somite with nearby cells which produce somitic factor at a similar time.
Our mathematical model is based on the signaling model for somite formation, originally proposed by Primmett and co-workers (Primmett et al., 1988, 1989), and then developed by Maini and co-workers (Baker et al., 2003; Collier et al., 2000; McInerney et al., 2004; Schnell et al., 2002). In this model (see Fig. 1), a small group of cells at the anterior end of the PSM will reach the determination front and undergo a whole cycle of the segmentation clock. These pioneer cells produce and emit a diffusible signaling molecule which spreads along the PSM. Any other cell which has reached the determination front, but not yet been specified as somitic, will respond to the signal by producing somitic factor. At this point the cell becomes allocated to a somite along with other cells which are behaving in a similar manner. The process begins once again when the new cohort of cells at the anterior end of the PSM becomes able to produce signaling molecule.
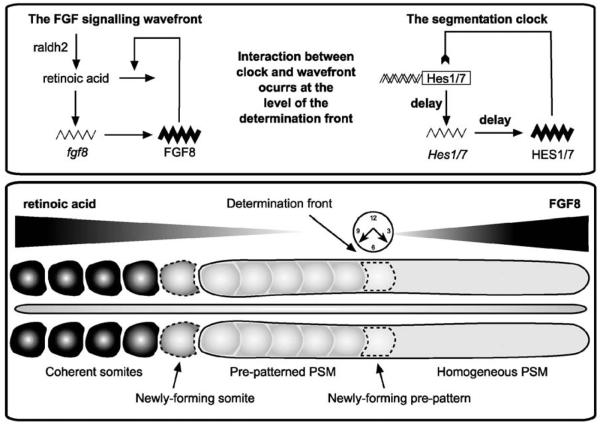
Figure 1.
The mechanisms underlying somite formation. The lower box illustrates the AP axis and the various stages of somite formation. The posterior PSM (gray) is homogeneous and cells are undetermined with respect to their developmental pathway. At the level of the determination front, the interaction between the clock and wavefront specifies the chemical prepattern. Cells of the PSM which lie anterior to the determination front will follow a specific developmental pathway which cannot be altered by subsequent perturbation of the clock or wavefront. At the anterior end of the PSM cells undergo changes in their morphological properties and condense to form coherent somites (black segments). The upper box shows schematics of the networks underlying the clock and the gradient. On the left, FGF acts in a negative feedback loop with retinoic acid to control gradient progression whilst on the right a negatively-regulating transcriptional delay network consisting of Hes1/7 controls clock oscillations.
The mathematical model consists of a system of three nonlinear partial differential equations (PDEs). The variables are somitic factor concentration (only cells with a high level of somitic factor can go on to be specified as somitic), signaling molecule levels and FGF8 concentration. We choose to model the gradient by assuming that FGF8 is produced only in the tail of the embryo and that it undergoes linear decay (see Baker et al., 2006a, 2006b, for more details). As the embryo extends the gradient regresses posteriorly, conferring the ability on cells to be able to produce somitic factor. A time ts later cells gain the ability to produce signaling molecule; ts is the period of the segmentation clock. Somitic factor production is activated in response to a pulse in signaling molecule, which is emitted from cells at the anterior end of the PSM. However, rapid inhibition of signal production means that the peaks in signal concentration are transient and produced at regular intervals: this ensures that a series of coherent somites is generated (McInerney et al., 2004).
The system of nonlinear PDEs can be written as follows:
| (1) |
where u represents the concentration of somitic factor, v the concentration of signaling molecule and w the concentration of FGF8. μ, γ , κ, ε, η, Dv, and Dw are positive constants and production of u, v, and w are controlled by the respective Heaviside functions1
| (2) |
where w* is the level of FGF8 at the determination front, t*(w*, t) is the time at which a cell at x reaches the determination front (i.e., w(x, t*) = w*), ts is the period of the segmentation clock, xn is the initial position of the tail and cn represents the rate at which the AP axis is extending. In short, the system of PDEs describes the following behavior: (i) somitic factor production is activated by the signaling molecule and is self-regulating; (ii) signaling molecule is produced rapidly in areas where somitic factor concentration is low and it is able to diffuse along the AP axis; (iii) FGF8 is produced in the tail, which is constantly regressing along the x axis.
We solve the mathematical model on the interval (−∞,∞), where x → ∞ corresponds to the tail of the embryo and x → −∞ the head of the embryo. We assume that: (i) the levels of somitic factor, signaling molecule and FGF8 are bounded as x → ±∞; (ii) initially the region x < 0 has already been specified as somitic; and (iii) the system is between pulses in the signaling molecule.
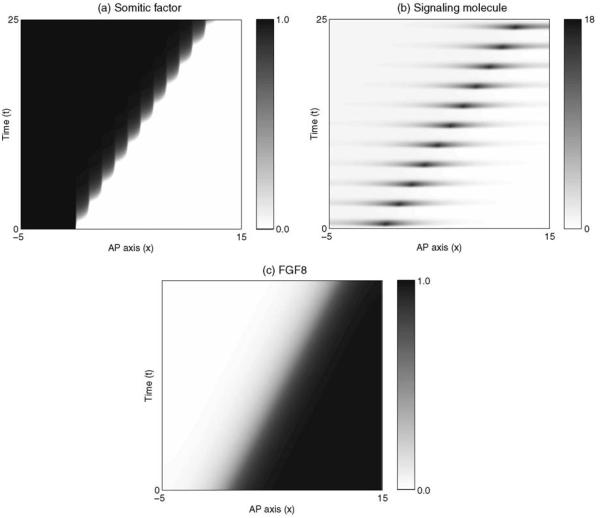
The mathematical model can be solved numerically using the NAG library routine D03PCF and the results plotted using the Matlab function [monospace]imagesc[/monospace]. The results are shown in Fig. 2. As time progresses, the region of high FGF8 signaling regresses along the AP axis with constant speed. In its wake a series of pulses in signaling molecule result in a series of coherent rises in somitic factor, enabling the formation of discrete somites.
Figure 2.
Numerical solution of the clock and wavefront model in one spatial dimension given by Eqs. (1) and (2). Continuous regression of the wavefront (c), is accompanied by a series of pulses in the signaling molecule (b), and coherent rises in the level of somitic factor (a). Parameters are as follows: μ = 10−4, γ = 10−3, κ = 10, ε = 10−3, η = 1.0, ϕ = 0.0, Dv = 50, Dw = 20, xn = 0.0, cn = 0.5, xb = 7.5 and ξ =0.0.
The model has been tested against experimental data. Dubrulle and co-workers used experimental techniques to make local perturbations to the FGF8 wavefront (Dubrulle et al., 2001). By implanting beads soaked in FGF8 alongside or into the PSM they generated a series of somite anomalies. Typical anomaly patterns consisted of series of small somites ahead of the bead, followed by a large somite behind the bead and normal segmentation thereafter. The mathematical model can be adapted to take this into account by adding a term into the equation for FGF8, which represents a stationary source of FGF8 (Baker et al., 2006a, 2006b):
| (3) |
where the term
| (4) |
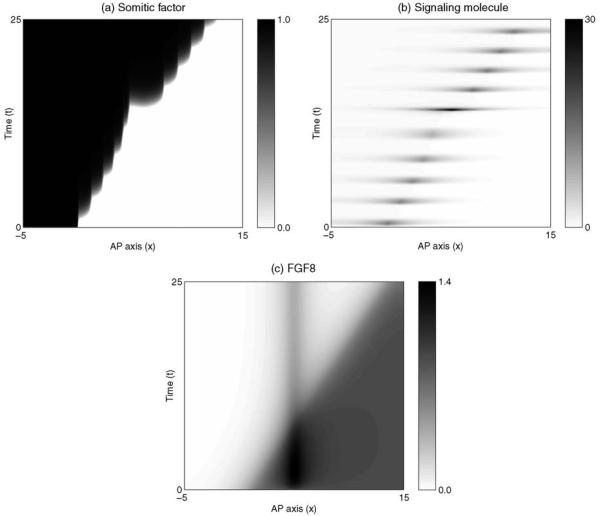
represents a source of FGF8 from a bead implant. ϕ is a measure of the strength of the bead source, xb is the position of the bead and ξ is a measure of the width of the bead. The model can be solved as before, and the results are shown in Fig. 3. As time progresses, the region of high FGF8 signaling regresses along the AP axis as in the control case, but the speed of wavefront progression is disrupted around the bead implant. This results in a series of small somites ahead of the bead and a large somite behind the bead, as observed experimentally.
Figure 3.
Numerical solution of the clock and wavefront model in one spatial dimension given by Eqs. (1)-(4). Continuous regression of the wavefront (c), is accompanied by a series of pulses in the signaling molecule (b), and coherent rises in the level of somitic factor (a). With a source of FGF8 implanted in the PSM the somite anomalies are obvious. Parameters are as follows: μ = 10−4, γ = 10−3, κ = 10, ε = 10−3, η = 1.0, ϕ = 1.5, Dv = 50, Dw = 20, xn = 0.0, cn = 0.5, xb = 7.5 and ξ = 0.5.
The model can also be adapted to include other possible experimental perturbations, such as downregulation of FGF8 signaling and perturbation of the segmentation clock (see Baker et al., 2006a, 2006b, for more details). Testing of these kinds of hypotheses is vital for furthering our understanding of the mechanisms underlying somite formation.
At present the drawback to this kind of modeling approach is that the clock and gradient are modeled at a very high (phenomenological) level. We incorporate the segmentation clock by using a single parameter to represent the period of the clock. Likewise, it is simply assumed that a generic FGF8 molecule makes up the gradient controlling the position of the determination front. Whilst this approach has many benefits—we can make simplifications and predictions regarding the effects of many experimental perturbations (Baker et al., 2003, 2006a, 2006b)—we are unable to infer exactly how each biological perturbation affects somite formation. For example, vitamin A deficient embryos have somites which are reduced in size (Diez del Corral et al., 2003; Molotkova et al., 2005; Moreno and Kinter, 2004). In our model we could explain this observation by assuming that the determination front progresses more slowly along the AP axis. This requires us to decrease the parameter cn, which controls the rate at which the Heaviside function controlling FGF8 production moves [see Eq. (2)]. However from our model, we cannot deduce why this change in cn occurs. Explicit modeling of the segmentation clock and FGF8 wavefront will allow us to address these kinds of questions.
B. Transcriptional Regulation Models for the Segmentation Clock
The segmentation clock has been modeled using networks of transcriptional regulatory modules (Giudicelli and Lewis, 2004; Hirata et al., 2004; Horikawa et al., 2006; Lewis, 2003; Monk, 2003; Pourquié and Goldbeter, 2003). In each model, the simplest possible feedback loop is considered: binding of a transcription factor to the regulatory region of its own gene inhibits expression (see Fig. 1). Such a model can be written in the form
| (5) |
where p represents protein (transcription factor) concentration and m represents mRNA concentration. The models incorporate delays of length τp and τm to reflect the time taken for synthesis of mature protein and mRNA, respectively. a is the rate of protein production, b and c are the decay rates of protein and mRNA, respectively, and the function f(·) is the rate of production of new mRNA molecules. To satisfy the requirement that protein binding inhibits transcription, f(·) should be a decreasing function of p, which we assume to be
| (6) |
where k, n, and p0 are positive constants. In general we assume n = 2; this represents the inhibitory action of a protein which acts as a dimer (Lewis, 2003).
To consider a more realistic model for the behavior of a single cell, Lewis also includes a characteristic property of gene regulation: noise (Lewis, 2003). Binding and dissociation of the protein to/from the regulatory DNA site is considered to behave in a stochastic manner (Turner et al., 2004). The regulatory site makes transitions between bound and unbound states with a certain probability; these probabilities can be calculated by considering the rates of binding and unbinding and the numbers of protein molecules in the cell.
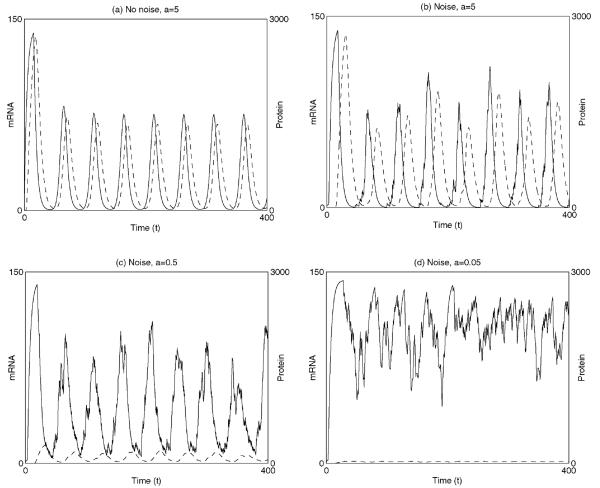
Fig. 4 shows the results of simulation of the system. In Fig. 4a, we plot the results of numerical simulation of the deterministic system using the Matlab solver [monospace]dde23[/monospace]. After initial large-amplitude oscillations in protein and mRNA levels, the system settles down to oscillate in a regular fashion with a period of approximately 50 minutes. The remaining plots, Figs. 4b-4d, show the results of numerical simulation of the noisy system. Simulations were carried out in the following fashion: at each time step, a random number drawn from a uniform distribution was used to determine whether binding or unbinding events occurred and then numbers of protein and mRNA molecules were updated accordingly. Fig. 4b is the stochastic equivalent of Fig. 4a—the system oscillates in much the same manner as the deterministic case, with a similar period but a small amount of variability in peak levels of mRNA and protein. As the protein synthesis rate is decreased the stochastic nature of gene regulation becomes more prominent. The period of oscillation remains robust even at a tenth of its original value [see Fig. 4c] but the peaks become increasingly noisy. At 100th of its original value, the oscillations are hard to discern and protein numbers fluctuate randomly and persistently (see Fig. 4d) (Lewis, 2003).
Figure 4.
Numerical solution of the segmentation clock model given by Eqs. (5) and (6). (a) The case with no noise. (b–d) The case in which binding of the protein to and from its site on the DNA are stochastic processes. As the rate of protein synthesis is gradually decreased the oscillations become more irregular, becoming almost undetectable as synthesis rates reach 1/100th of their original value. In each case protein concentration/number is indicated by the solid line and mRNA concentration/number by the dashed line. Parameters are as follows: dt = 0.1 min, τm = 12.0 min, τp 2.8 min, p0 = 40, k = 33 min−1, koff = 1 min−1, b = 0.23 and c = 0.23.
These models neatly capture the dynamics of the segmentation clock and reproduce oscillations with a period relatively close to that observed experimentally. The period of oscillation is robust in the sense that it remains practically unchanged even when the rate of protein synthesis is drastically reduced (Pourquié and Goldbeter, 2003). However, this generally requires the Hill coefficient, n, to be higher than that reported experimentally (Monk, 2003). In other words, one must assume that rather than a dimer, regulation of mRNA transcription requires binding of a multimer, such as a tetramer or larger, to the regulatory DNA site.
The models also assume that (i) translation is nonsaturating, (ii) the movement of protein molecules between the cytoplasm and cell nucleus is instantaneous and (iii) the delays in transcription and translation take discrete values. Monk (2003) suggests that a more appropriate assumption would be that the delay is uniformly distributed on some finite interval; this results in a revised equation for mRNA dynamics of the form
| (7) |
where
| (8) |
One prediction of the model is that oscillations are crucially dependent on instability of the protein. Hirata and co-workers (Hirata et al., 2004) generated mice expressing mutant Hes7 with a half-life of approximately 30 minutes (compared to approximately 22 minutes in the wild-type mouse). Somite segmentation and oscillatory gene expression became severely disorganized after a few cycles. This observation is consistent with the results observed in their mathematical models and provides support for this mechanism in the segmentation clock. In a similar manner, Lewis uses his stochastic models to postulate that the effect of reduced protein synthesis may be to cause the progressively irregular oscillations seen in mutant embryos (Jiang et al., 2000; Lewis, 2003). It would be interesting to see if this could be tested experimentally.
The models discussed in this section consider oscillations in protein and mRNA numbers in a single cell, where the environment has been assumed to be spatially homogeneous. The reaction kinetics of gene transcription can vary substantially in spatially heterogeneous environments (Schnell and Turner, 2004; Turner et al., 2004). Although Lewis and Horikawa and co-workers briefly discuss models which incorporate the role of the Notch–Delta pathway in coupling adjacent PSM cells (Horikawa et al., 2006; Lewis, 2003), future work in this area will need to consider the possibilities of coupling large numbers of cells, as is the case with the segmentation clock.
C. Models for the FGF8 Gradient
Less well studied have been the mechanisms underlying maintenance of the FGF8 gradient, which has been shown to control the position of the determination front (Dubrulle et al., 2001). Our mathematical formulation of the Clock and Wavefront model (Baker et al., 2006a, 2006b) assumed simply that FGF8 is produced in the tail of the embryo, which is extending at a constant rate. In our model, diffusion and linear decay are combined to set up a monotonic gradient, which regresses along the AP axis as time proceeds and hence is a constantly moving determination front. In this case, the rate of progression of the determination front is solely controlled by a parameter chosen in the simulations.
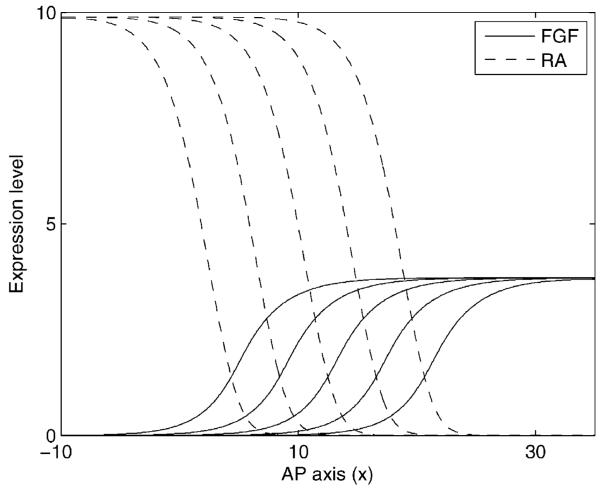
We consider a revised mechanism for FGF8 gradient formation (Baker and Maini, 2007) based on the finding that FGF8 acts in a negative feedback loop with RA (Diez del Corral et al., 2002, 2003; Diez del Corral and Storey, 2004). We show a schematic representation of this feedback in Fig. 1. In the negative feedback loop, it is possible that FGF8 might downregulate RA production or upregulate RA decay, or both. The same occurs for RA. In order to account for both possibilities, we investigated the phenomenon using the following system of nonlinear PDEs:
| (9) |
where F denotes the concentration of FGF8, A the concentration of RA and rf , ra, λf , λa, ηf , ηa, sf , sa, and D are positive constants. In briefly, the first equation assumes: (i) the rate of production of FGF8 is decreased by RA; (ii) FGF8 production saturates for high FGF8 concentration; (iii) FGF8 undergoes linear decay; (iv) RA augments FGF8 decay; and (v) FGF8 diffuses at rate D compared to RA. Similar statements can be made about the dynamics of RA.
The system of equations describing RA and FGF8 dynamics is mathematically intractable and so we carry out numerical simulation of the system using the Matlab function [monospace]pdepe[/monospace], in order to analyze the behavior. Fig. 5 shows the results of numerical simulation for a specific set of parameter values. The initial conditions are chosen so that FGF8 obtains its maximal value on the positive x axis and is zero elsewhere and vice versa for RA. In this case, traveling waves (constant shape) form, with the region of high FGF8 signaling moving in a positive x direction at a constant rate. Variation of the model parameters produces changes in both wave speed and shape (Baker and Maini, 2007).
Figure 5.
Numerical solution of the FGF8/RA model given by Eq. (9) over the series of successive time steps t = 10, 20, 30, 40, 50. The fronts move from left to right as time proceeds. Parameters are as follows: rf = 2.0, ra = 5.0, λf = 1.0, λa = 1.0, ηf = 0.5, ηa = 0.5, sf = 0.5, sa = 0.5, βa = 1.0, βf = 1.0, and D = 5.0.
Despite the fact that analytical solution of the system is impossible, it can be shown that the presence of RA, acting in negative feedback loop with FGF8, makes the model more robust. Robustness is meant in the sense that it increases the parameter range for which the wave moves in a positive x direction (since we wish the determination front to regress along the AP axis over time).
Goldbeter and co-workers have also recently proposed a model for the FGF8 gradient which considers the negative interactions between RA and FGF8 (Goldbeter et al., 2007). In this model, negative feedback results in windows of bistability: regions along the PSM in which both high and low levels of FGF8 signaling are possible. Goldbeter and co-workers postulate that the action of the clock within this window could result in a shift from high to low levels of FGF signaling and the specification of a presumptive somite.
At present the model of Goldbeter and co-workers does not consider extension of the AP axis or movement of the bistability window along the AP axis. Also it does not explicitly consider interaction of the clock with this window in gating cells into somites. On the other hand, our model does not explicitly couple gradient formation to the rate of AP axis extension. Moreover it needs to be coupled to our previous formulation of the Clock and Wavefront model in order to predict somite patterns.
D. Models Including Cell Adhesion
All the models discussed thus far are concerned purely with the formation of a prepattern in gene expression, which is later converted to physical somites by a series of changes in cell morphology. However, it is possible to write down a model capturing the essential interactions between cells and cell adhesion molecules (CAMs), which leads to formation of a series of peaks in cell density—the somites. To date, the only mathematical model attempting to describe the bulk movement of PSM cells to form a somite is by Schnell and co-workers (Schnell et al., 2002). This is an extension of the signaling model for somite formation by Maini and co-workers, which we have discussed previously. Here we present a simpler model, which can explain the bulk movements of cells to form a somite.
Letting n denote cell density and a denote the concentration of a morphogen that stimulates expression of CAMs, the following system of nonlinear PDEs can be used to describe somite formation:
| (10) |
The first term on the right-hand side of the equation for cell density represents cell diffusion and the second term the tendency of cells to move up gradients in CAM activity (resulting in cell aggregations). f(n) is a decreasing function of n which for illustrative purposes we take to be χn(N – n). The constant N is chosen to be large enough to ensure that f(n) does not become negative. The equation for morphogen concentration also contains a diffusion term, with diffusion of CAM-triggering morphogen occurring at rate D relative to PSM cells. Production of CAM-triggering morphogen occurs at a rate proportional to the number of cells present, whilst we assume that no cell birth or death occurs.
In this model we have assumed the presence of a diffusive morphogen that triggers CAM response. In this simplified model we have combined the effects of both morphogen and CAMs into one term, a, in order to facilitate mathematical analysis.
Once again, it is not possible to solve the model directly: however, it is possible to undertake a linear stability analysis of the system to find parameter regions in which patterns of cell aggregations may arise. The condition for spatial patterns in cell density is
| (11) |
where n0 is the initial (uniform) cell density.
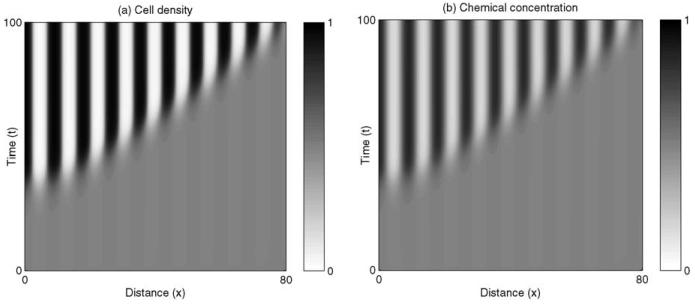
The system can be solved numerically using the Matlab solver pdepe. We assume that initially both PSM cells and CAM-triggering morphogen are distributed uniformly along the AP axis and that a small perturbation is made to the cell density at the anterior end of the PSM (in our simulations this corresponds to x = 0). Fig. 6 shows the results of numerical simulation for a set of parameter values which satisfy Eq. (10). From an initially homogeneous field we see the sequential development of a series of peaks and troughs in cell density with equivalent patterns in CAM-triggering morphogen concentration. The patterns arise on a timescale and with a wavelength that is dependent on the choice of parameters. In other words, each new somite forms at a specific time interval after the last somite and with a defined length, which depends upon the model parameters.
Figure 6.
Numerical solution of the cell-chemotaxis model for somite formation in one spatial dimension given by Eq. (10). Initially the field is supposed to be homogeneous throughout, with a small perturbation made to the cell density at x = 0. The pattern of somites propagates across the field over time, from left to right. Parameters are as follows: D = 1.0, χ = 10.0, N = 1.0 and n0 = 0.5.
An extension of this model needs to address a number of issues. Firstly, how can the model be made consistent with the observations of genetic prepatterns along the AP axis and how can we incorporate the role of the segmentation clock? Secondly, mathematical analysis shows that the initial (unpatterned) field of cells is unstable to perturbation: should a disturbance arise in a region ahead of the somite pattern, a second patterning envelope would initiate from the point of disturbance. Due to the inherently stochastic nature of gene transcription and translation it is likely this will occur—extension of the model to take account of this is the subject of current work. It is also important to point out that the major drawback of this model is that it does not take into account the intercellular mechanical forces involved in the process of somite formation. As a consequence, it cannot account for the morphological changes observed during somitogenesis.
III. Discussion
We began this article by outlining the processes involved in somite formation, from creation of a genetic prepattern to the mechanisms involved in generating morphological somites. We described the patterns of oscillating gene expression which are readouts of the segmentation clock and the traveling patterns of gene expression which control segmental determination. In the middle section we described a series of mathematical models which aim to capture various aspects of somite formation. The mathematical formulation of the Cooke–Zeeman–Pourquié Clock and Wavefront model integrates simple readouts from the clock and gradient to describe the formation of a genetic prepattern which seems to guide development of the physical somites. Models for the segmentation clock assume an underlying regulatory transcription network, characterized by delays in transcription and translation, drives oscillations in mRNA and protein levels. Models for the FGF8 gradient assume similar types of negative feedback (but without delay) to describe the formation of traveling gradients of RA and FGF8. Finally we postulated a model which, rather than characterizing the formation of a genetic prepattern, describes the formation of cellular aggregations—the physical somites.
The current challenge is to build on these models in order to drive forward knowledge in this area. Development of more sophisticated models for both clock and gradient will allow us to elucidate the crucial mechanisms underlying each system. These models can be integrated back into a generalized “Clock and Wavefront” framework and used to make predictions and test hypotheses. Below we outline the key challenges in each area.
Current models for the segmentation clock generally consist of simple transcriptional oscillators with delay terms allowing the intermediate synthesis steps, such as transport, elongation and splicing, to be encompassed by a single parameter. Such models are easily solved numerically and allow predictions on the period of the clock to be made based on model parameters. However, there is now a wealth of information available regarding the different molecular players involved in regulation of the segmentation clock (Dequéant et al., 2006): how they interact and what happens when embryos mutant for crucial components are generated. Larger scale models are required in order to integrate this new data into a concrete framework. This will entail developing a “Systems Biology” approach with the deployment of, for example, Boolean and network models, and multiscale simulation techniques (Schnell et al., 2007). Another important aspect of the segmentation clock that has not yet been fully explored is the coupling between adjacent PSM cells that arises from signaling via the Notch–Delta pathway. A mean-field approach generally assumes equal coupling between all cells of a network, which is clearly not the case in the somitogenesis oscillator. Development of a theoretical approach for dealing with large systems of coupled oscillators is clearly important here, and also in a wider biological context.
Modeling of the gradient which underlies the determination front has only recently begun to acquire attention and models for this are still at an early stage. It seems highly probable that the formation and maintenance of such gradients are tightly coupled to the events surrounding axial elongation. It has recently been shown, in mouse, that the FGF8 signaling gradient is mirrored by a gradient in Wnt3a, and that in fact, Wnt3a acts upstream of FGF8 (Aulehla and Herrmann, 2004; Aulehla et al., 2003). Wnt3a has also been shown to play a major role in controlling cell proliferation, axis elongation and oscillations of Axin2 in the PSM (Aulehla et al., 2003; Galli et al., 2004). This yields the exciting possibility that Wnt3a may provide an underlying link between the segmentation clock, specification of positional information and body axis elongation (Aulehla and Herrmann, 2004; Dubrulle and Pourquié, 2004a).
The final piece of the story is the transition from genetic prepattern to epithelial somites. Most current models for somitogenesis are concerned with the formation of a genetic prepattern along the AP axis and few have attempted to model the series of cell rearrangements and shape changes that accompany the transition from prepattern to coherent somite. Probably this is linked to the enormity of the task: our understanding of the biological processes involved is far from complete—numerous complex, interacting processes are taking place—and theoretical tools have not yet been developed to answer some of the questions which need to be addressed. The cell–CAM model presented here is a simple attempt to address this process. It neglects many of the salient features of the epithelialization process and cannot yet replicate many experimental observations. However, it is a first step on the road to constructing a framework for this complex process and development of the model is the subject of current work. Glazier et al. (in this volume) developed a computational model of the spatiotemporal pattern of adhesive and repulsive forces at work during somite formation. Sherratt and co-workers are also interested in applying their theoretical studies of differential cell adhesion (Armstrong et al., 2006) to somite formation, using the genetic prepattern to drive the process (Armstrong et al., 2007), whilst Schnell and Grima have proposed a model in which chemotaxis, differential cell adhesion and minimization of tissue surface tension drive the cell sculpting process (Grima and Schnell, 2007).
IV. Perspective
The goal of multiscale modeling is to investigate events happening on the microscopic scale and integrate results into models on a macroscopic level (Schnell et al., 2007). Although most models for somite formation are still firmly based on events taking place at the macroscopic level, the wealth of new experimental data becoming available and the generation of new experimental techniques is beginning to allow the generation of microscale models, thereby paving the way for truly multiscale models in the future.
The future of interdisciplinary research in developmental biology lies in the attitudes of both theoretical and experimental communities. Commitment to communication across the boundaries of increasingly specialist research areas, alongside the development of tools and methods to facilitate the achievement of common goals, is crucial. Mathematical modeling should be used as a tool to drive and expand knowledge. It has the power to integrate multiple hypotheses and ideas into a rigorous framework in which they can be submitted to analysis and in silico experiments, used to elucidate pertinent questions and devise future experiments. In turn, results from these experiments can be applied to adjust and refine models. This feedback loop of modeling, testing and fine-tuning is essential for the development of biologically-accurate models and the acceleration of our understanding in the biomedical sciences.
Acknowledgments
R.E.B. thanks Research Councils UK for an RCUK Fellowship in Mathematical Biology, Lloyds Tercentenary Foundation for a Lloyds Tercentenary Foundation Fellowship, Microsoft for a European Postdoctoral Research Fellowship, St. Hugh's College, Oxford for a Junior Research Fellowship and the Australian Government, Department for Science, Education and Training for an Endeavor Postdoctoral Research Fellowship to visit the Department of Mathematics and Statistics, University of Melbourne. S.S. acknowledges support from NIH Grant R01GM76692. P.K.M. was partially supported by a Royal Society Wolfson Merit Award.
Footnotes
The Heaviside function H(x) is equal to unity if x > 0 and zero otherwise: it acts as a switch.
References
- Armstrong NJ, et al. A continuum approach to modeling cell–cell adhesion. J. Theor. Biol. 2006;243:98–113. doi: 10.1016/j.jtbi.2006.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armstrong NJ, et al. Modeling the role of cell adhesion in somite formation (work in progress) 2007 [Google Scholar]
- Aulehla A, Herrmann BG. Segmentation in vertebrates: Clock and gradient finally joined. Genes Dev. 2004;18:2060–2067. doi: 10.1101/gad.1217404. [DOI] [PubMed] [Google Scholar]
- Aulehla A, Johnson RL. Dynamic expression of lunatic fringe suggests a link between notch signaling and an autonomous cellular oscillator driving somite segmentation. Dev. Biol. 1999;207:49–61. doi: 10.1006/dbio.1998.9164. [DOI] [PubMed] [Google Scholar]
- Aulehla A, et al. Wnt3a plays a major role in the segmentation clock controlling somitogenesis. Dev. Cell. 2003;4:395–406. doi: 10.1016/s1534-5807(03)00055-8. [DOI] [PubMed] [Google Scholar]
- Baker RE, Maini PK. Traveling gradients in interacting morphogen systems. Math. Biosci. 2007 doi: 10.1016/j.mbs.2007.01.006. doi:10.1016/j.mbs.1007.01.006 (to appear) [DOI] [PubMed] [Google Scholar]
- Baker RE, et al. Formation of vertebral precursors: Past models and future predictions. J. Theor. Med. 2003;5:23–35. [Google Scholar]
- Baker RE, et al. A clock and wavefront mechanism for somite formation. Dev. Biol. 2006a;293:116–126. doi: 10.1016/j.ydbio.2006.01.018. [DOI] [PubMed] [Google Scholar]
- Baker RE, et al. A mathematical investigation of a clock and wavefront model for somitogenesis. J. Math. Biol. 2006b;52:458–482. doi: 10.1007/s00285-005-0362-2. [DOI] [PubMed] [Google Scholar]
- Chernoff EAG, Hilfer SR. Calcium dependence and contraction in somite formation. Tiss. Cell. 1982;14:435–449. doi: 10.1016/0040-8166(82)90038-6. [DOI] [PubMed] [Google Scholar]
- Collier JR, et al. A cell cycle model for somitogenesis: Mathematical formulation and numerical solution. J. Theor. Biol. 2000;207:305–316. doi: 10.1006/jtbi.2000.2172. [DOI] [PubMed] [Google Scholar]
- Cooke J. Control of somite number during morphogenesis of a vertebrate, Xenopus laevis. Nature. 1975;254:196–199. doi: 10.1038/254196a0. [DOI] [PubMed] [Google Scholar]
- Cooke J. A gene that resuscitates a theory—somitogenesis and a molecular oscillator. Trends Genet. 1998;14:85–88. doi: 10.1016/s0168-9525(98)01396-1. [DOI] [PubMed] [Google Scholar]
- Cooke J, Zeeman EC. A clock and wavefront model for control of the number of repeated structures during animal morphogenesis. J. Theor. Biol. 1976;58:455–476. doi: 10.1016/s0022-5193(76)80131-2. [DOI] [PubMed] [Google Scholar]
- Dale KJ, Pourquié O. A clock-work somite. BioEssays. 2000;22:72–83. doi: 10.1002/(SICI)1521-1878(200001)22:1<72::AID-BIES12>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- Dale JK, et al. Periodic notch inhibition by lunatic fringe underlies the chick segmentation clock. Nature. 2003;421:275–278. doi: 10.1038/nature01244. [DOI] [PubMed] [Google Scholar]
- Dale JK, et al. Oscillations of the Snail genes in the presomitic mesoderm coordinate segmental patterning and morphogenesis in vertebrate somitogenesis. Dev. Cell. 2006;10:355–366. doi: 10.1016/j.devcel.2006.02.011. [DOI] [PubMed] [Google Scholar]
- del Barco Barrantes I, et al. Interaction between Notch signaling and Lunatic Fringe during somite boundary formation in the mouse. Curr. Biol. 1999;9:470–480. doi: 10.1016/s0960-9822(99)80212-7. [DOI] [PubMed] [Google Scholar]
- Dequéant M-L, et al. A complex oscillating network of signaling genes underlies the mouse segmentation clock. Science. 2006;314:1595–1598. doi: 10.1126/science.1133141. [DOI] [PubMed] [Google Scholar]
- Diez del Corral R, et al. Onset of neural differentiation is regulated by paraxial mesoderm and requires attenuation of FGF8 signaling. Development. 2002;129:1681–1691. doi: 10.1242/dev.129.7.1681. [DOI] [PubMed] [Google Scholar]
- Diez del Corral R, et al. Opposing FGF and retinoid pathways control ventral neural pattern, neuronal differentiation and segmentation during body axis extension. Neuron. 2003;40:65–79. doi: 10.1016/s0896-6273(03)00565-8. [DOI] [PubMed] [Google Scholar]
- Diez del Corral R, Storey K. Opposing FGG and retinoid pathways: A signaling switch that controls differentiation and patterning onset in the extending vertebrate body axis. BioEssays. 2004;26:957–969. doi: 10.1002/bies.20080. [DOI] [PubMed] [Google Scholar]
- Drake CJ, Little CD. Integrins play an essential role in somite adhesion to the embryonic axis. Dev. Biol. 1991;143:418–421. doi: 10.1016/0012-1606(91)90092-h. [DOI] [PubMed] [Google Scholar]
- Duband JL, et al. Adhesion molecules during somitogenesis in the avian embryo. J. Cell Biol. 1987;104:1361–1374. doi: 10.1083/jcb.104.5.1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubrulle J, Pourquié O. From head to tail: Links between the segmentation clock and anteroposterior patterning of the embryo. Curr. Opin. Genet. Dev. 2002;12:519–523. doi: 10.1016/s0959-437x(02)00335-0. [DOI] [PubMed] [Google Scholar]
- Dubrulle J, Pourquié O. Coupling segmentation to axis formation. Development. 2004a;131:5783–5793. doi: 10.1242/dev.01519. [DOI] [PubMed] [Google Scholar]
- Dubrulle J, Pourquié O. fgf8 mRNA decay establishes a gradient that couples axial elongation to patterning in the vertebrate embryo. Nature. 2004b;427:419–422. doi: 10.1038/nature02216. [DOI] [PubMed] [Google Scholar]
- Dubrulle J, et al. FGF signaling controls somite boundary position and regulates segmentation clock control of spatiotemporal Hox gene activation. Cell. 2001;106:219–232. doi: 10.1016/s0092-8674(01)00437-8. [DOI] [PubMed] [Google Scholar]
- Duguay D, et al. Cadherin-mediated cell adhesion and tissue segregation: Qualitative and quantitative determinants. Dev. Biol. 2003;253:309–323. doi: 10.1016/s0012-1606(02)00016-7. [DOI] [PubMed] [Google Scholar]
- Foty RA, Steinberg MS. Cadherin-mediated cell–cell adhesion and tissue segregation in relation to malignancy. Int. J. Dev. Biol. 2004;48:397–409. doi: 10.1387/ijdb.041810rf. [DOI] [PubMed] [Google Scholar]
- Galli LM, et al. A proliferative role for Wnt-3a in chick somites. Dev. Biol. 2004;269:489–504. doi: 10.1016/j.ydbio.2004.01.041. [DOI] [PubMed] [Google Scholar]
- Giudicelli F, Lewis J. The vertebrate segmentation clock. Curr. Opin. Genet. Dev. 2004;14:407–414. doi: 10.1016/j.gde.2004.06.014. [DOI] [PubMed] [Google Scholar]
- Goldbeter A, Gonze D, Pourquie O. Sharp developmental thresholds defined through bistability by antagonistic gradients of retinoic acid and FGF signaling. Dev. Dyn. 2007;236:1495–1508. doi: 10.1002/dvdy.21193. [DOI] [PubMed] [Google Scholar]
- Gossler A, Hrabě de Angelis M. Somitogenesis. Curr. Top. Dev. Biol. 1998;38:225–287. [PubMed] [Google Scholar]
- Grima R, Schnell S. Can tissue surface tension drive somite formation? Dev. Biol. 2007;307:248–257. doi: 10.1016/j.ydbio.2007.04.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haraguichi S, et al. Transcriptional regulation of Mesp1 and Mesp1 genes: Differential usage of enhancers during development. Mech. Dev. 2001;108:59–69. doi: 10.1016/s0925-4773(01)00478-6. [DOI] [PubMed] [Google Scholar]
- Hirata H, et al. Instability of Hes7 protein is crucial for the somite segmentation clock. Nat. Genet. 2004;36:750–754. doi: 10.1038/ng1372. [DOI] [PubMed] [Google Scholar]
- Horikawa K, et al. Noise-resistant and synchronized oscillation of the segmentation clock. Nature. 2006;441:719–723. doi: 10.1038/nature04861. [DOI] [PubMed] [Google Scholar]
- Ishikawa A, et al. Mouse Nkd1, a Wnt antagonist, exhibits oscillatory gene expression in the PSM under the control of Notch signaling. Mech. Dev. 2005;121:1443–1453. doi: 10.1016/j.mod.2004.08.003. [DOI] [PubMed] [Google Scholar]
- Jacobson A, Meier S. Somitomeres: The primordial body segments. In: Bellairs R, et al., editors. Somites in Developing Embryos. Plenum; New York: 1986. pp. 1–16. [Google Scholar]
- Jiang Y-J, et al. Notch signaling and the synchronization of the segmentation clock. Nature. 2000;408:475–479. doi: 10.1038/35044091. [DOI] [PubMed] [Google Scholar]
- Jiang Y-J, Lewis L. Notch as a synchronizer in somite segmentation. In: Ordahl CP, editor. Proceedings of the NATO Advanced Research Workshop on the Origin and Fate of Somites. Vol. 329. IOS Press; Amsterdam: 2001. pp. 71–79. [Google Scholar]
- Jiang YJ, et al. Vertebrate segmentation: The clock is linked to Notch signaling. Curr. Biol. 1998;8:R868–R871. doi: 10.1016/s0960-9822(07)00547-7. [DOI] [PubMed] [Google Scholar]
- Kalcheim C, Ben-Yair R. Cell rearrangements during development of the somite and its derivatives. Curr. Opin. Genet. Dev. 2005;15:371–380. doi: 10.1016/j.gde.2005.05.004. [DOI] [PubMed] [Google Scholar]
- Kulesa PM, Fraser SE. Cell dynamics during somite boundary formation revealed by time-lapse analysis. Science. 2002;298:991–995. doi: 10.1126/science.1075544. [DOI] [PubMed] [Google Scholar]
- Lewis J. Autoinhibition with transcriptional delay: A simple mechanism for the zebrafish somitogenesis oscillator. Curr. Biol. 2003;13:1398–1408. doi: 10.1016/s0960-9822(03)00534-7. [DOI] [PubMed] [Google Scholar]
- Maroto M, Pourquié O. A molecular clock involved in somite segmentation. Curr. Top. Dev. Biol. 2001;51:221–248. doi: 10.1016/s0070-2153(01)51007-8. [DOI] [PubMed] [Google Scholar]
- McGrew MJ, Pourquié O. Somitogenesis: Segmenting a vertebrate. Curr. Op. Genet. Dev. 1998;8:487–493. doi: 10.1016/s0959-437x(98)80122-6. [DOI] [PubMed] [Google Scholar]
- McGrew MJ, et al. The Lunatic Fringe gene is a target of the molecular clock linked to somite segmentation in avian embryos. Curr. Biol. 1998;8:979–982. doi: 10.1016/s0960-9822(98)70401-4. [DOI] [PubMed] [Google Scholar]
- McInerney D, et al. A mathematical formulation for the cell cycle model in somitogenesis: Parameter constraints and numerical solutions. IMA J. Math. Appl. Med. & Biol. 2004;21:85–113. doi: 10.1093/imammb/21.2.85. [DOI] [PubMed] [Google Scholar]
- Meier S. Development of the chick embryo mesoblast: Formation of the embryonic axis and establishment of the metameric pattern. Dev. Biol. 1979;73:24–45. doi: 10.1016/0012-1606(79)90135-0. [DOI] [PubMed] [Google Scholar]
- Molotkova N, et al. Requirement of mesodermal retinoic acid generated by Raldh2 for posterior neural transformation. Mech. Dev. 2005;122:145–155. doi: 10.1016/j.mod.2004.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monk NAM. Oscillatory expression of Hes1, p53, and NF-κB driven by transcriptional time delays. Curr. Biol. 2003;13:1409–1413. doi: 10.1016/s0960-9822(03)00494-9. [DOI] [PubMed] [Google Scholar]
- Moreno TA, Kinter C. Regulation of segmental patterning by retinoic acid signaling during Xenopus somitogenesis. Dev. Cell. 2004;6:205–218. doi: 10.1016/s1534-5807(04)00026-7. [DOI] [PubMed] [Google Scholar]
- Morimoto M, et al. Cooperative Mesp activity is required for normal somitogenesis along the anterior–posterior axis. Dev. Biol. 2006;300:687–698. doi: 10.1016/j.ydbio.2006.08.043. [DOI] [PubMed] [Google Scholar]
- Packard DS. The influence of axial structures on chick formation. Dev. Biol. 1976;53:36–48. doi: 10.1016/0012-1606(76)90207-4. [DOI] [PubMed] [Google Scholar]
- Packard DS, et al. Somite pattern regulation in the avian segmental plate mesoderm. Development. 1993;117:779–791. doi: 10.1242/dev.117.2.779. [DOI] [PubMed] [Google Scholar]
- Palmeirim I, et al. Avian hairy gene expression identifies a molecular clock linked to vertebrate segmentation and somitogenesis. Cell. 1997;91:639–648. doi: 10.1016/s0092-8674(00)80451-1. [DOI] [PubMed] [Google Scholar]
- Pourquié O. Clocks regulating developmental processes. Curr. Opin. Neurobiol. 1998;8:665–670. doi: 10.1016/s0959-4388(98)80097-4. [DOI] [PubMed] [Google Scholar]
- Pourquié O. Notch around the clock. Curr. Opin. Gen. Dev. 1999;9:559–565. doi: 10.1016/s0959-437x(99)00011-8. [DOI] [PubMed] [Google Scholar]
- Pourquié O. The vertebrate segmentation clock. J. Anat. 2001a;199:169–175. doi: 10.1046/j.1469-7580.2001.19910169.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pourquié O. Vertebrate somitogenesis. Ann. Rev. Cell Dev. Biol. 2001b;17:311–350. doi: 10.1146/annurev.cellbio.17.1.311. [DOI] [PubMed] [Google Scholar]
- Pourquié O. The segmentation clock: Converting embryonic time into spatial pattern. Science. 2003;301:328–330. doi: 10.1126/science.1085887. [DOI] [PubMed] [Google Scholar]
- Pourquié O, Goldbeter A. Segmentation clock: Insights from computational models. Curr. Biol. 2003;13:R632–R634. doi: 10.1016/s0960-9822(03)00567-0. [DOI] [PubMed] [Google Scholar]
- Primmett DRN, et al. Heat-shock causes repeated segmental anomalies in the chick-embryo. Development. 1988;104:331–339. doi: 10.1242/dev.104.2.331. [DOI] [PubMed] [Google Scholar]
- Primmett DRN, et al. Periodic segmental anomalies induced by heat-shock in the chick-embryo are associated with the cell-cycle. Development. 1989;105:119–130. doi: 10.1242/dev.105.1.119. [DOI] [PubMed] [Google Scholar]
- Rida PC, et al. A Notch feeling of somite segmentation and beyond. Dev. Biol. 2004;265:2–22. doi: 10.1016/j.ydbio.2003.07.003. [DOI] [PubMed] [Google Scholar]
- Saga Y, et al. Mesp2: A novel mouse gene expressed in the presegmented mesoderm and essential for segmentation initiation. Genes Dev. 2001;2:835–845. doi: 10.1101/gad.11.14.1827. [DOI] [PubMed] [Google Scholar]
- Schnell S, et al. Multiscale modeling in biology. Am. Sci. 2007;95:134–142. [Google Scholar]
- Schnell S, Turner TE. Reaction kinetics in intracellular environments with macromolecular crowding: Simulations and rate laws. Prog. Biophys. Mol. Biol. 2004;85:235–260. doi: 10.1016/j.pbiomolbio.2004.01.012. [DOI] [PubMed] [Google Scholar]
- Schnell S, et al. Models for pattern formation in somitogenesis: A marriage of cellular and molecular biology. C. R. Biol. 2002;325:179–189. doi: 10.1016/s1631-0691(01)01418-4. [DOI] [PubMed] [Google Scholar]
- Stockdale FE, et al. Molecular and cellular biology of avian somite development. Dev. Dyn. 2000;219:304–321. doi: 10.1002/1097-0177(2000)9999:9999<::AID-DVDY1057>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- Tabin CJ, Johnson RL. Developmental biology: Clocks and Hox. Nature. 2001;412:780–781. doi: 10.1038/35090677. [DOI] [PubMed] [Google Scholar]
- Takahashi Y, et al. Mesp2 initiates somite segmentation through the Notch signaling pathway. Nat. Gen. 2000;25:390–396. doi: 10.1038/78062. [DOI] [PubMed] [Google Scholar]
- Takashi Y, et al. Differential contributions of Mesp1 and Mesp2 to the epithelializations and rostrocaudal patterning of somites. Development. 2005;132:787–796. doi: 10.1242/dev.01597. [DOI] [PubMed] [Google Scholar]
- Tam PPL, Meier S. The establishment of a somitomeric pattern in the mesoderm of the gastrulating mouse embryo. Am. J. Anat. 1982;164:209–225. doi: 10.1002/aja.1001640303. [DOI] [PubMed] [Google Scholar]
- Turner TE, et al. Stochastic approaches for modeling in vivo reactions. Comput. Biol. Chem. 2004;28:165–178. doi: 10.1016/j.compbiolchem.2004.05.001. [DOI] [PubMed] [Google Scholar]
- Zeeman EC. Primary and secondary waves in developmental biology: Some mathematical questions in biology, VI; Proc. Eighth Symp., Mathematical Biology; San Francisco, CA. 1974; 1974. pp. 69–161. Lectures on Mathematics in the Life Sciences. Am. Math. Soc., Providence, RI. [Google Scholar]