Abstract
COUP-TF-interacting protein 2 (CTIP2; also known as Bcl11b) is a transcription factor that plays key roles in the development of the central nervous and immune systems. CTIP2 is also highly expressed in the developing epidermis, and at lower levels in the dermis and in adult skin. Analyses of mice harboring a germline deletion of CTIP2 revealed that the protein plays critical roles in skin during development, particularly in keratinocyte proliferation and late differentiation events, as well as in the development of the epidermal permeability barrier. At the core of all of these actions is a relatively large network of genes, described herein, that is regulated directly or indirectly by CTIP2. The analysis of conditionally null mice, in which expression of CTIP2 was ablated specifically in epidermal keratinocytes, suggests that CTIP2 functions in both cell and non-cell autonomous contexts to exert regulatory influence over multiple phases of skin development, including barrier establishment. Considered together, our results suggest that CTIP2 functions as a top-level regulator of skin morphogenesis.
INTRODUCTION
The development of the skin epidermis begins with the commitment of the primitive ectoderm to the keratinocyte cell fate. The subsequent processes of cellular proliferation, stratification, and differentiation result in formation of the multilayered structure of epidermis. During embryonic development, keratinocytes of the innermost layer of the epidermis, the proliferative, basal cell layer, undergo a program of the terminal differentiation, then exit the basal cell layer and migrate upward to the surface of the skin (Byrne et al., 2003; Mack et al., 2005). These cells initially differentiate into spinous and then granular cells, and finally into the tough, enucleated cells of the cornified layer, the corneocytes (Byrne et al., 2003).
The events of epidermal development are orchestrated by the concerted action of a number of transcription factors that regulate both the proliferative capacity and differentiative potential of epidermal keratinocytes (Segre, 2003, 2006). These include c-Myc, p63, Klf4, GATA3, the AP-1 transcription factors c-Fos and c-Jun, the Id family of proteins, and others (Segre et al., 1999; Langlands et al., 2000; Angel et al., 2001; Arnold and Watt, 2001; Shaulian and Karin, 2002; Koster et al., 2004, 2007; de Guzman Strong et al., 2006). The end product of epidermal development is the formation of the epidermal permeability barrier (EPB), which provides a crucial physical and permeability barrier.
Formation of the EPB is a stepwise and orderly process (Segre, 2003, 2006). In the granular layer of epidermis, lipids are packaged inside lamellar bodies, and structural proteins, such as keratins, loricrin and filaggrin, are assembled into macrofibrils. The cornified envelope (CE) is formed by deposition of precursor proteins on the inner surface of the plasma membrane (Elias, 2005). Transglutaminase enzymes then cross-link the CE proteins, creating a tough sac that holds the keratin fibers (Candi et al., 2005). Finally, cellular organelles, including the nucleus, are degraded and lipids from lamellar bodies are extruded into the intercellular space and onto the CE scaffold, forming a series of extracellular lipid membranes (lamellar bilayers; Candi et al., 2005).
COUP-TF-interacting protein 2 (CTIP2) is a C2H2 zinc finger transcription factor that represses transcription of reporter genes in transiently transfected cells, either by tethering to other promoter-bound transcription factors (Avram et al., 2000) or by direct, sequence-specific DNA-binding activity (Avram et al., 2002; Topark-Ngarm et al., 2006). The hypothesis that CTIP2 functions as a transcriptional regulator has been supported by transcriptome analyses in human neuroblastoma cells (Topark-Ngarm et al., 2006), striatal medium spiny neurons (Arlotta et al., 2008), and mouse thymocytes (Leid M. et al., unpublished data).
CTIP2 is expressed early during mouse development as well as in the adult animal, and in both cases, expression is most predominant in the central nervous system, thymus, and skin and other tissues of ectodermal origin (Leid et al., 2004; Golonzhka et al., 2007). Mice null for expression of CTIP2 exhibit perinatal lethality and severe phenotypes in tissues that express the gene. In the central nervous system, CTIP2 is required for proper axonal projection by corticospinal neurons and normal development of striatal medium spiny neurons (Arlotta et al., 2005, 2008). Germline disruption of CTIP2 results in an arrest of T-cell development at an immature, CD4−CD8− stage(s), with the complete absence of αβ T cells (Wakabayashi et al., 2003).
Here we report a previously unknown function of CTIP2 in skin development. We show that CTIP2 controls epidermal proliferation/differentiation programs and EPB formation. CTIP2−/− mice exhibit a hypoplastic epidermis, defects in EPB development, and increased transepidermal water loss (TEWL). These defects likely arise from large-scale disruption of gene expression in the mutant skin, including genes encoding structural protein and lipid modifying enzymes, as well as those involved in epidermal proliferation and differentiation, and EPB establishment. In the context of EPB development, CTIP2 was found to function cell autonomously. However, the actions of CTIP2 in epidermal proliferation and early differentiation were found to be non-cell autonomous, most likely arising from CTIP2-dependent regulation of paracrine growth factor(s) expression in the dermis, including keratinocyte growth factor (KGF) and/or GM-CSF (Szabowski et al., 2000; Werner and Smola, 2001).
RESULTS
Generation of total (CTIP2−/−) and epidermis-specific (CTIP2ep−/−) CTIP2 knockout mice
CTIP2−/− mice were generated by floxing exon 4 of the CTIP2 locus, which encodes ~75% of the CTIP2 open reading frame and six zinc finger motifs (Figure 1a). The targeting vector used to modify the CTIP2 locus by homologous recombination (HR) is schematically depicted in Figure 1b and c, and HR was confirmed by 5′ and 3′ Southern analyses (Figure 1d and e). Heterozygous mice harboring one wild-type (+) and one mutant CTIP2 allele (−), did not show an overt phenotype, and were viable and fertile. CTIP2+/− mice were bred to generate the CTIP2−/− mouse, and the lack of CTIP2 protein in these mice was confirmed by immunoblotting in skin (Figure 1f). CTIP2−/− mice were born at the expected Mendelian ratio and without obvious skin abnormalities, with the exception of an open eye phenotype (Figure 2a, arrow; compare; Figure S1a and b), as previously described by Kominami and co-workers (Wakabayashi et al., 2003). However, CTIP2−/− mice did not feed and died within 6–8 hours after birth. These phenotypic characteristics, together with severe thymic hypocellularity and complete loss of αβ-T lymphocytes (data not shown), recapitulated previously reported observations of CTIP2-null mice (Wakabayashi et al., 2003).
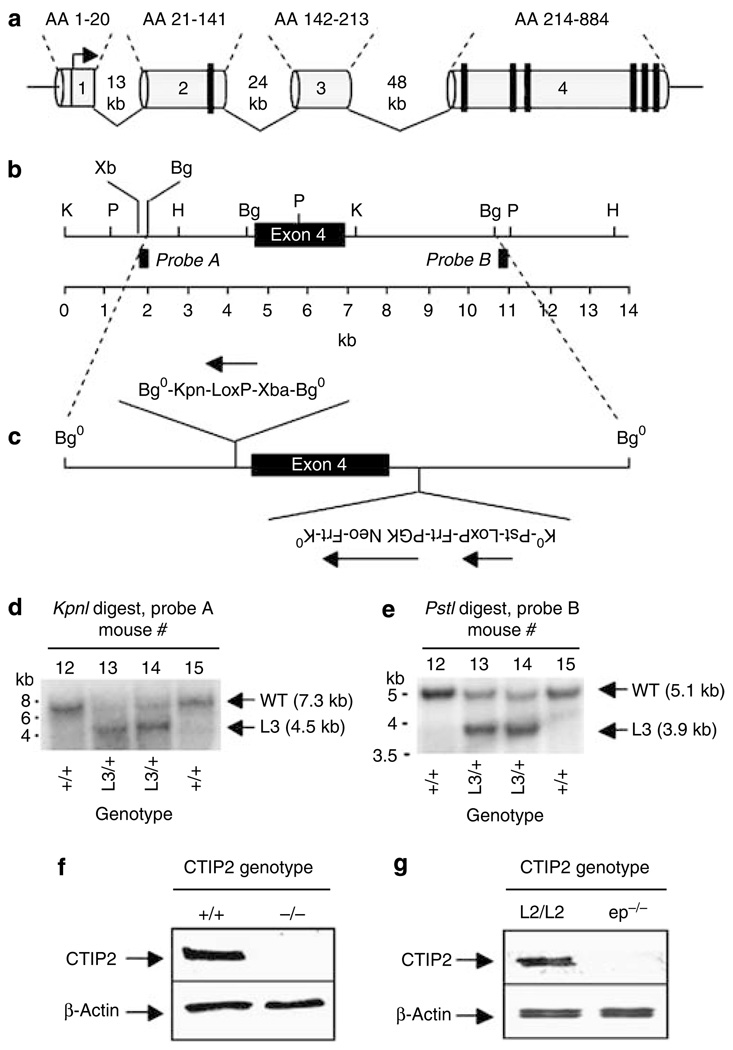
Figure 1. Floxing the CTIP2 locus.
(a) Schematic representation of the mouse CTIP2 locus with exons 1–4 and corresponding amino acids (top). The vertical black bars represent C2HC (exon 2) or C2H2 (exon 4) zinc finger motifs. (b) Magnification of exon 4 and flanking sequences with selected restriction sites and location of Southern probes. (c) Schematic representation of the CTIP2-targeting vector indicating the direction of upstream and downstream LoxP sites and PGK-neo. PGK-neo was flanked by Frt sites for excision by Flp recombinase. Restriction sites were introduced into both the upstream and downstream LoxP sites to facilitate analyses of HR at the CTIP2 locus. (d, e) Southern analyses of HR at the CTIP2 locus using 5′ (Probe A in panel b) and 3′ (Probe B in panel b) probes, respectively. The L3 genotype refers to presence of the entire targeting vector, including the PGK-neo, in the CTIP2 locus by HR. Probe A hybridizes to 7.3 and 4.5 kb KpnI fragments, and Probe B hybridizes to 5.1 and 3.1 kb PstI fragments from wt and L3 alleles, respectively. (f) Immunoblot of whole cell extracts prepared from E18.5 skin from CTIP2+/+ and CTIP2−/− mice using an anti-CTIP2 monoclonal antibody. (g) Immunoblot of the protein extracts from epidermis of E18.5 CTIP2L2/L2 and CTIP2ep−/− mice using an anti-CTIP2 monoclonal antibody.
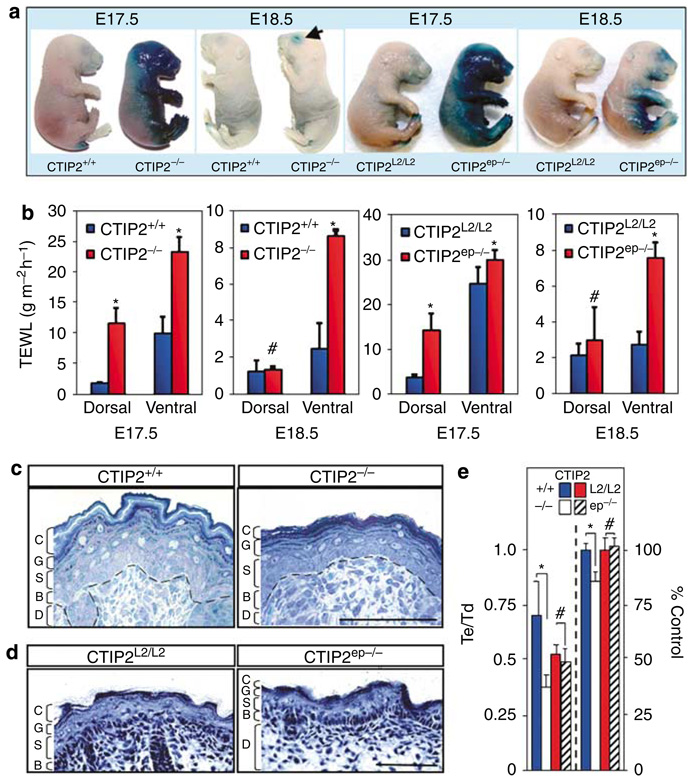
Figure 2. CTIP2 mutant mice exhibit permeability barrier defects and epidermal hypoplasia.
(a) X-gal diffusion assay performed on CTIP2+/+, CTIP2−/−, CTIP2L2/L2, and CTIP2ep−/− fetuses at E17.5 and E18.5 as indicated. (b) Transepidermal water loss measurements from dorsal and ventral skin of CTIP2+/+, CTIP2−/−, CTIP2L2/L2, and CTIP2ep−/− mice at E17.5 and E18.5. (Plotted are mean measurements of three independent mice per genotype ± SEM). *P<0.05, #not statistically significant. (c) Histology of toluidine blue stained dorsal skin biopsies (4-µm thick section) of CTIP2+/+ and CTIP2−/− mice at E18.5. Marked epidermal hypoplasia was observed in CTIP2−/− mice. D, dermis; B, basal layer; S, spinous layer; G, granular layer; C, cornified layer. Scale bar: 50 µm. (d) H&E staining of dorsal skin biopsies (10-µm thick section) of CTIP2L2/L2 and CTIP2ep−/− mice at E18.5. Abbreviations are as in (c). Scale bar: 50 µm. (e) Quantitative histomorphometry of CTIP2−/− and CTIP2ep−/− phenotypes. Epidermal (Te) and dermal (Td) thicknesses were measured on eight independent wt (CTIP2+/+) and null (CTIP2−/−) mice, and on six floxed (CTIP2L2/L2) and conditionally null (CTIP2ep−/−) mice. The ratio of Te to Td for the four genotypes is plotted on the ordinate (left side). Bars on the right represent the number of proliferating Ki67+ cells (as a ratio of DAPI+ cells and expressed as percentage of each, corresponding control). The asterisks and hash denotations are as described in (b), and refer to the comparison of CTIP2+/+ and CTIP2−/−, and CTIP2L2/L2 and CTIP2ep−/− mice, respectively. The results depicted in this (a) to (d) are representative of 6–12 additional studies that have been conducted over the course of 2 years using numerous litters of mice of identical genotypes.
CTIP2 floxed mice (CTIP2L2/L2; see Figure 1) were crossed with a K14-Cre transgenic mouse (Indra et al., 2000) to obtain a mouse line in which CTIP2 was specifically ablated in keratinocytes (epidermis-specific knockout; CTIP2ep−/−). To confirm excision of the floxed CTIP2 allele in CTIP2ep−/− mice, we separated epidermis and dermis and genotyped both tissues for excised (−) and floxed (L2) alleles. The excised, but not L2, allele was detected in the epidermis of E17.5 CTIP2ep−/− fetuses, whereas only the L2 allele was present in the dermis of these mice (Figure S1c), thus demonstrating that CTIP2 was efficiently and selectively ablated in the keratinocytes of the developing epidermis. In agreement with this result CTIP2 protein was not detected in epidermal extracts from CTIP2ep−/− fetuses by immunoblotting, thus validating the CTIP2ep−/− mouse model (Figure 1g). CTIP2ep−/− mice were born without any apparent abnormalities, fed normally, survived into adulthood, and were fertile.
Loss of CTIP2 results in a compromised barrier function
CTIP2 is highly expressed in the mouse ectoderm during development, beginning around embryonic day E10.5 (Golonzhka et al., 2007). To determine if the perinatal lethality of CTIP2−/− mice was due to defects in the EPB, we investigated barrier function in these mice by performing X-gal permeability assays at E17.5 and E18.5. Control embryos showed very little X-gal staining at either E17.5 or E18.5 (Figure 2a, first and second panel). In contrast, CTIP2−/− fetuses stained strongly with X-gal at E17.5, particularly on the ventral surface and head (Figure 2a). The dorsal surface of CTIP2−/− mice did not take up appreciable amounts of X-gal, signifying that dorsal to ventral barrier establishment had commenced in the mutants, but perhaps in a delayed fashion. Similar studies conducted at E18.5 revealed that EPB function in CTIP2−/− and wild-type (wt) mice was indistinguishable, with the exception of blue staining around the eyelids of the CTIP2−/− mice (Figure 2a, arrow in second panel).
To determine if CTIP2 regulates EPB establishment in a cell autonomous manner, we performed X-gal permeability assays on CTIP2L2/L2 (as a control) and CTIP2ep−/− mice at E17.5 and E18.5 (Figure 2a, third and fourth panels). Similar to CTIP2−/− mice, CTIP2ep−/− mice exhibited a delay in barrier establishment. CTIP2ep−/− mice showed extensive blue staining on the ventral aspect of the body, limbs and head at E17.5, and residual X-gal staining at E18.5, whereas control embryos showed very little coloration at either E17.5 or E18.5 (Figure 2a). These results demonstrate that CTIP2, in a cell autonomous manner, regulates EPB formation during fetal development.
We also performed TEWL studies in CTIP2−/− mice to assess the rate of water evaporation from the skin at E17.5 and E18.5. At E17.5, TEWL from CTIP2−/− dorsal skin averaged ~12 gm−2 h−1, which was about six-fold higher than that of wt controls (Figure 2b, first panel). The rate of water loss on the ventral surface of E17.5 CTIP2−/− mice was approximately two-fold greater than that of wt mice (Figure 2b). Although there was no difference in dorsal water loss between wt and CTIP2−/− mice at E18.5, the ventral surface of the mutants continued to lose water at a greater rate than wt controls (Figure 2d, second panel).
The rate of water loss from the dorsal skin of E17.5 CTIP2ep−/− mice was about three-fold higher than that of the control mice (Figure 2b, third panel) but at E18.5 the water loss on the dorsal surface of CTIP2ep−/− mice was not significantly different from controls (Figure 2b, fourth panel). However, TEWL from the ventral side of CTIP2ep−/− mice slightly exceeded that of the control mice at E17.5 and was 2- to 3-fold higher than that of the control mice at E18.5. These findings further demonstrate that CTIP2ep−/− mice exhibit considerably delayed formation of the EPB, recapitulating the cell autonomous barrier phenotype of CTIP2−/− mice. Together, these results indicate that skin barrier establishment is disrupted in the absence of CTIP2, and suggest a previously unreported role of CTIP2 in skin barrier formation.
CTIP2-null mice exhibit altered expression of markers of epidermal proliferation and early differentiation
Histological analyses of dorsal skin at E17.5 and E18.5 revealed that CTIP2−/− epidermis was extremely hypoplastic with decreased numbers of differentiating and cornified cell layers, and profound loss of the normal, cornified layer with the typical “basket weave” appearance (Figure 2c; Figure S1d). The number of proliferating, Ki67+ cells in the epidermis was modestly, but significantly reduced CTIP2−/− mice (~13% reduction relative to wt controls; right panels of Figure 2e), and the thickness of the CTIP2−/− epidermis was approximately 50% that of wt mice, as determined by quantitative histomorphometry (left panels of Figure 2e). All of the epidermal layers were present in the CTIP2ep−/− mice, and both the number of proliferating cells and the epidermal thickness of these conditionally null mice were indistinguishable from that of the corresponding control mice (CTIP2L2/L2; Figure 2e). However, we observed a modest reduction in the number of cornified cell layers in CTIP2ep−/− mice at E17.5 and E18.5 (see Figure 2d; Figure S1e).
Reduced epidermal thickness of the CTIP2−/− mutants could be due to alterations in epidermal proliferation and/or differentiation. To test this, we performed immunohistochemical analyses to assess the expression of: (1) a proliferation marker (Ki67; Schluter et al., 1993), (2) a marker of keratinocyte basal cells (keratin 14, K14; Byrne et al., 1994), and (3) an early keratinocyte differentiation marker (keratin 10, K10; Fuchs and Green, 1980; Roop et al., 1983). Reduced numbers of Ki67+ cells (compare Figure 3a and d) and reduced K14 staining (Figure 3b and e) were both observed in the skin of CTIP2−/− mice. Similarly, the expression of the differentiation marker K10 was strongly reduced in CTIP2−/− mice (Figure 3c and f). Immunoblot analyses of skin protein extracts further confirmed the decrease in the expression of K10 and K14, in the mutant fetuses (Figure 3w). These results suggest that reduced epidermal thickness of the CTIP2−/− fetuses may be due to reduced proliferation and/or altered differentiation in the skin of the mutant mice.
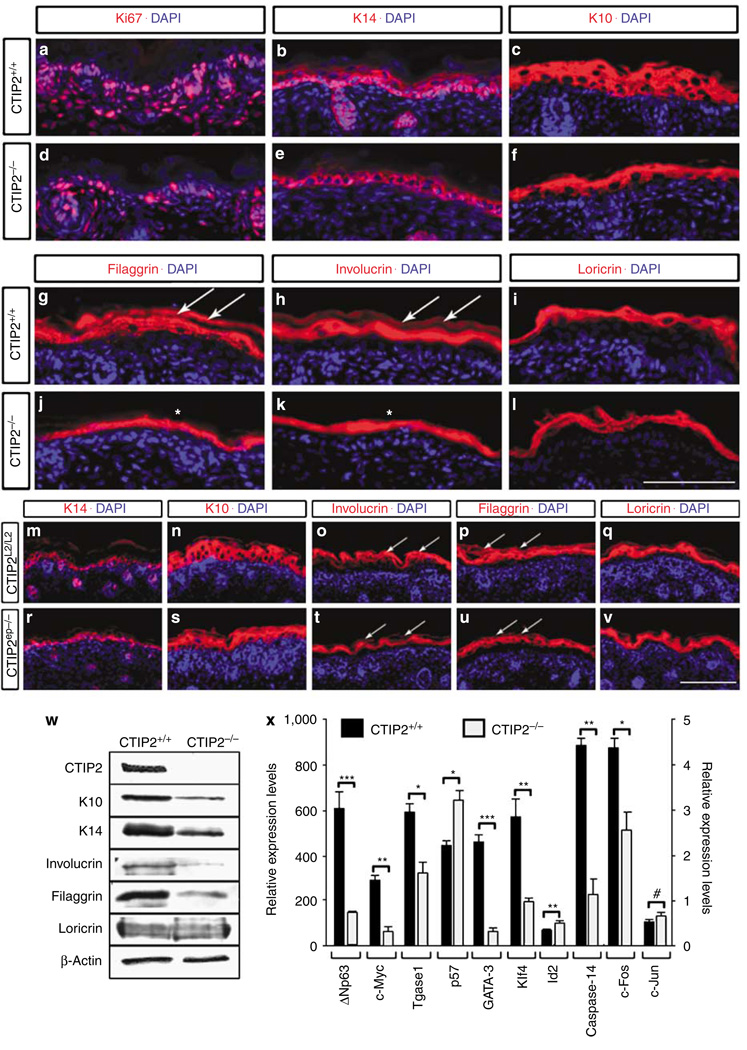
Figure 3. Decreased expression of epidermal markers and genes involved in epidermal barrier establishment in CTIP2 mutant mice.
(a–f) Immunohistochemistry of dorsal skin of CTIP2+/+ and CTIP2−/− mice at E18.5. Genotypes and antibodies (red staining) used are indicated, and all sections were counterstained with DAPI (blue). (g–l) Immunohistochemistry of dorsal skin with late differentiation markers loricrin, involucrin, and filaggrin (all in red) in CTIP2+/+ and CTIP2−/− mice at E18.5. All sections were counterstained with DAPI (blue). White arrows in g and h indicate expression of filaggrin (g) and involucrin (h) in the cornified cell layers. White asterisks in j and k indicate the reduction in filaggrin (j) and involucrin (k) expression. (m–v) Immunohistochemistry of dorsal skin of CTIP2L2/L2 and CTIP2ep−/− mice at E18.5. Genotypes and antibodies (red staining) used are indicated, and all sections were counterstained with DAPI (blue). Scale bar in l: 100 µm for a–l; scale bar in v: 100µm for m–v. (w) Immunoblot of dorsal skin extracts from CTIP2+/+ and CTIP2−/− mice at E18.5 using indicated antibodies. (x) RT-qPCR analysis of dorsal skin from CTIP2+/+ and CTIP2−/− mice at E18.5. Relative expression levels for all genes are shown on y axis to the left, except for c-Fos and c-Jun, which are plotted on the right y axis. Statistical significance denotations: *P<0.05; **P< 0.01; ***P<0.001; #no statistically significant difference between wt and mutant mice. The results depicted in this figure are representative of at least eight additional studies that have been conducted over the course of two years using numerous litters of mice of identical genotypes.
CTIP2 mutant mice exhibit altered epidermal terminal differentiation
We performed immunohistochemical analyses of the late epidermal differentiation markers loricrin, involucrin, and filaggrin. The expression levels of involucrin and filaggrin, but not that of loricrin, were reduced in the E18.5 CTIP2−/− skin (compare Figure 3g and j, h and k, i and l, respectively), and this was further confirmed by immunoblotting (Figure 3w).
Immunohistochemical analyses of the CTIP2ep−/− skin revealed no changes in K14 expression (compare Figure 3m and r) and a slight reduction in the expression of K10 relative to control skin (compare Figure 3n and s). Involucrin and filaggrin expression levels were slightly reduced in the upper, terminally differentiating cell layers in the mutants (white arrows in Figure 3o and t and p and u). However, loricrin levels were comparable in wt and mutant skin (Figure 3q and v).
Our data indicates that CTIP2−/− mice exhibit defects in keratinocytic proliferation, early differentiation, terminal differentiation, and EPB formation. CTIP2ep−/− mutants on the other hand did not demonstrate severe epidermal proliferation or early differentiation defects. However, CTIP2ep−/− mice did exhibit impaired terminal differentiation and EPB establishment, as was observed in the CTIP2−/− mice. This suggests a cell autonomous function of CTIP2 in controlling the terminal differentiation events, and non-cell autonomous influence of CTIP2 on proliferation and early differentiation of keratinocytes.
CTIP2-null mice exhibit altered expression of a subset of genes involved in the epidermal terminal differentiation and barrier formation
We performed RT-qPCR analyses to assess expression levels of several genes implicated in the control of early- and late-differentiation events in the epidermis, as well as genes involved in barrier establishment. Interestingly, although expression of Id2 and p57 was upregulated (or derepressed) in the mutant skin, consistent with the previously demonstrated repressor activity of CTIP2 (Senawong et al., 2003; Topark-Ngarm et al., 2006) expressions of both c-Myc and p63 were downregulated in the mutants at E18.5 (Figure 3x), suggesting that CTIP2 may directly or indirectly activate expression of the latter two genes. RT-qPCR analyses for genes encoding structural proteins and transcription factors involved in late terminal differentiation and barrier formation, such as transglutaminase-1 (Tgase1), GATA3, Klf4, and caspase-14 revealed that all were downregulated at both E17.5 and E18.5 in the mutants (Figure 3x and data not shown). We also observed a significant downregulation of c-Fos, but not c-Jun, suggesting that altered gene expression in CTIP2 mutant skin may be mediated, at least in part, by specific members of AP-1 family of transcription factors (Shaulian and Karin, 2002; Zenz and Wagner, 2006). Altogether, our data suggest that CTIP2 regulates expression of a subset of the keratinocytic genes encoding transcription factors and other proteins implicated in the epidermal homeostasis and EPB formation.
CTIP2 mutant mice exhibit defects in surface lipid distribution and decreased expression of lipid-metabolizing enzymes
To determine if the observed barrier defect in the CTIP2−/− mutants was due to the altered distribution of polar and neutral lipids, we analyzed the surface lipid distribution of mutant and wt epidermis by Nile Red staining. These studies revealed that neutral lipids formed a yellow-colored and dense, continuous ribbon along the top of the cornified layer in control fetuses but these lipids were unevenly distributed along the cornified layer of the CTIP2-null epidermis (compare Figure 4a and b). Nile Red staining performed on E17.5 CTIP2ep−/− skin confirmed that the surface distribution of neutral lipids was impaired (compare Figure S1f and g). These results suggest a possible alteration in the composition of the neutral lipids of the skin in the absence of CTIP2, and this effect seems to be due to a cell autonomous effect of CTIP2 in keratinocytes.
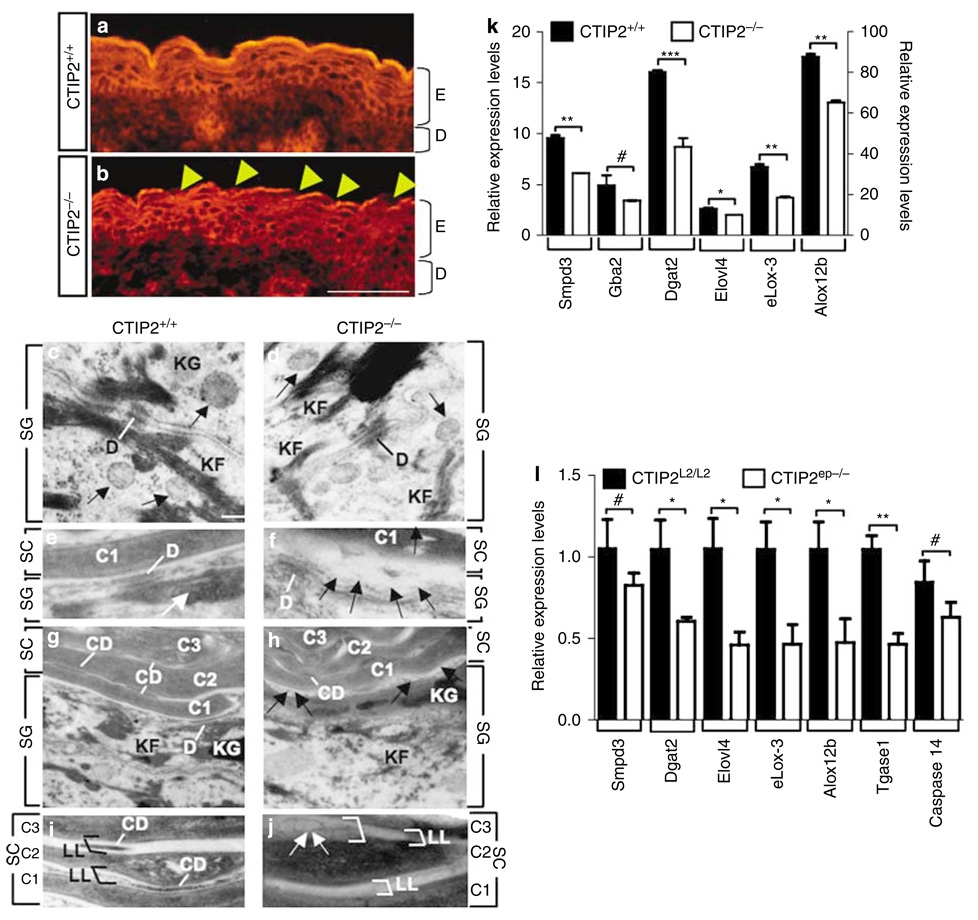
Figure 4. Defects in lipid distribution and expression of lipid-processing enzymes in CTIP2 mutant mice.
(a, b) Nile Red staining of skin from CTIP2+/+ and CTIP2−/− mice at E18.5. Arrowheads indicate an absence of neutral lipids on the skin surface of the mutant mice. Scale bar in b: 50µm for a–b. E, epidermis; D, dermis. (c–j) Transmission electron microscopy of dorsal skin from CTIP2+/+ and CTIP2−/− mice at E17.5 as indicated. SG, Stratum granulosum; SC, stratum corneum; D,desmosomes; CD, corneodesmosomes; KF, keratin filaments; KG, keratohyalin granules; C1, C2, and C3, cornified cell layers; LL, lipid lamellae. Large white arrows in e point toward the lamellar granules (LG) at the SG–SC interface. White and black lines in e–h indicate desmosomes (D) and corneodesmosomes, respectively; black arrows in f and h indicate vesicles at the interface of SG and SC. White arrows in j point toward vesicles. Black and white brackets in i and j indicate LL. Scale bar in c: 0.2µm for c–f; 0.4 µm for g and h; and 0.25 µm for i and j. (k) RT-qPCR analyses for expression of selected genes involved in lipid homeostasis in dorsal skin from CTIP2+/+ and CTIP2−/− mice at E18.5. Relative expression levels for all genes are shown on y axis to the right, except for eLox3 and Alox12b, which are plotted on the right y axis. Statistical significance denotations are as described in the legend of Figure 3x. (l) RT-qPCR validation of CTIP2 target genes involved in lipid metabolism and barrier establishment. Statistical denotations are as described in the legend of Figure 3x. The results depicted in this figure are representative of three (a, b) or two (c–j) additional studies. RT-qPCR results were conducted on at least three independent mice of each genotype.
Ultrastructural analysis of the epidermis from E17.5 fetuses did not reveal differences in basal or spinous cell populations between control and CTIP2 mutants (data not shown). Likewise, similar numbers of desmosomes (D), keratohyaline granules (KG), keratin filaments (KF), and lamellar granules (LG; also called keratinosomes) were present in the granular cells of both control and CTIP2−/− mice (Figure 4c and d; and data not shown). Lipid discs that were extruded from the LGs were uniformly aligned and formed lipid lamellar membranes at the interface of granular and cornified cells in control fetuses (Figure 4e). In contrast, lipid discs were replaced by large vesicles (marked by arrows in Figure 4f), and the intercellular (lipid) lamellar membranes (LL) in cornified layers were disorganized and highly variable in thickness in the mutant epidermis (compare Figure 4g–h, and i–j). Similar numbers of corneodesmosomes (CD) were present between the cornified cells in both control and mutant skin, however, the mutant CDs were smaller in size than the controls, and the mutant corneocytes were loosely packed (Figure 4h and j). These results suggest that the impaired barrier formation in the CTIP2 mutant fetuses could be, at least in part, due to altered lipid metabolism in the developing skin.
The results of Nile Red staining and transmission electron microscopy studies prompted us to perform RT-qPCR analyses for genes encoding proteins that are implicated in lipid homeostasis in the developing skin (Figure 4k). RT-qPCR revealed dysregulated expression of Smpd3 (Mao-Qiang et al., 1996; Gurrieri et al., 2003), Dgat2 (Stone et al., 2004), Elovl4 (Cameron et al., 2007; Li et al., 2007; Vasireddy et al., 2007), eLox3 (Furstenberger et al., 2007), and Alox12b (Moran et al., 2007), but not of Gba2 (Holleran et al., 1994) or Pla2gV in the skin of CTIP2-null mice compared to wt controls (Figure 4k). These results suggest that CTIP2 might regulate the expression of genes implicated in lipid metabolism and in the formation of extracellular lipid matrix.
To gain insights into the molecular mechanisms of permeability barrier defects in the conditional CTIP2ep−/− mice, we performed expression profiling of dorsal skin RNA from E17.5 CTIP2ep−/− and CTIP2L2/L2 mice. We found that out of the total 45,000 probe sets, only 1,688 probe sets, corresponding to 750 unique genes were significantly altered in the conditional mutants (up- or downregulated; Table S1), indicating that epidermal-specific deletion of CTIP2 leads to a highly selective (<1%) alteration in gene expression.
The transcriptome analyses also revealed that a subset of genes implicated in lipid homeostasis was downregulated in CTIP2ep−/− epidermis (Table S1). RT-qPCR analyses of several of the differentially expressed genes confirmed the down-regulation of genes involved in the lipid metabolism (Dgat2, Elovl4, eLox3, Alox12b) as well as Tgase1, which is implicated in the terminal differentiation of skin (Figure 4l).
Approximately half of the genes within the epidermal differentiation complex, which is located in two adjacent bands on chromosome 3 (3qF1 and 3qF2.1; see Figure S2) and encodes structural proteins of the CE, were found to be dysregulated in CTIP2ep−/− mice, and the majority of those were downregulated in mutant skin (Figure S2). Altogether, these findings suggest that CTIP2, in a cell-autonomous manner, regulates expression of a subset of genes implicated in the lipid metabolism and skin barrier formation.
CTIP2 is involved in controlling the mesenchymal–epithelial cross-talk through regulation of secretion of paracrine factors in the dermis
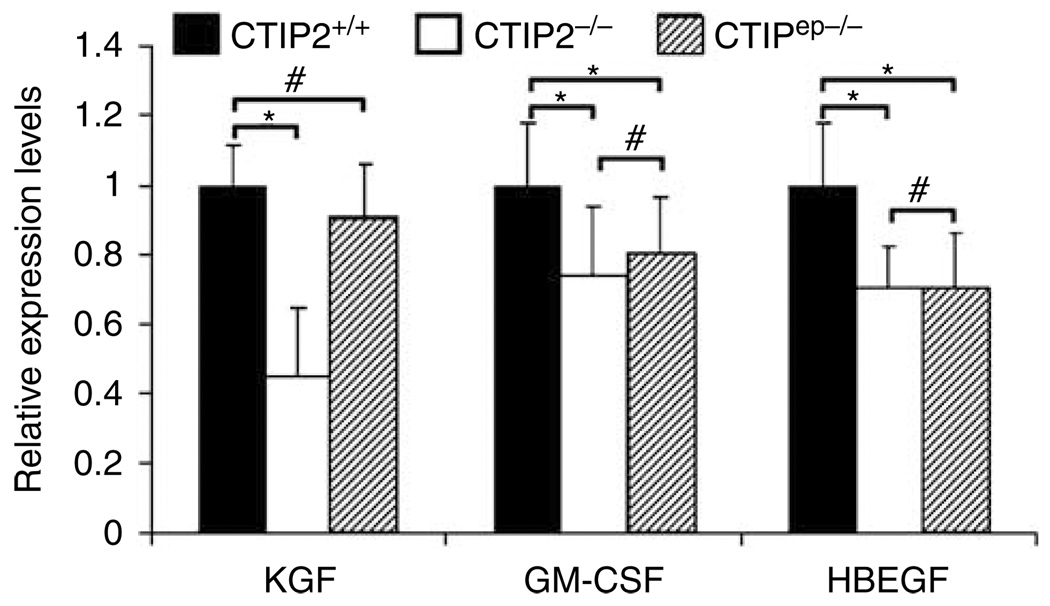
Our data suggest that CTIP2, in a cell autonomous manner, plays a contributory role in the late keratinocyte differentiation events and EPB formation. However, the epidermal proliferation defect seen in CTIP2−/− mice was not recapitulated in CTIP2ep−/− conditional mutants, suggesting that the effect of CTIP2 on keratinocyte proliferation may be the result of non-cell autonomous action(s) of CTIP2. To test this hypothesis we analyzed the expression levels of several diffusible factors known to be involved in the control of keratinocyte proliferation, including KGF, GM-CSF, and heparin-binding EGF-like growth factor (HBEGF), in the skin of wt, CTIP2−/−, and CTIP2ep−/− fetuses. Notably, the expression of KGF was found to be downregulated by almost 60% in the CTIP2−/− mice; however, the expression levels of KGF in wt and CTIP2ep−/− skin were indistinguishable (Figure 5). Reduced expression of KGF in the dermis of the total knockout mice could be partially responsible for the observed hypoplasticity of the CTIP2−/− epidermis (Figure 2c). CTIP2 expression is preserved in the dermis of CTIP2ep−/− mice, and the levels of KGF are normal in these mice, which could explain development of a morphologically normal epidermis (Figure 2d), and underlie the non-cell autonomous action of CTIP2 in epidermal development. We also observed a slight downregulation of GM-CSF and HBEGF in the skin of CTIP2−/− mice, although the levels of these transcripts were similarly downregulated in CTIP2ep−/− mice (Figure 5). Thus, it seems unlikely that GM-CSF and HBEGF contribute to the non-cell autonomous actions of CTIP2 in the epidermis.
Figure 5. CTIP2 regulates expression of growth factors involved in mesenchymal–epithelial cross-talk.
RT-qPCR analysis of genes encoding growth factors KGF, GM-CSF, and HBEGF in the skin (epidermis and dermis) of E18.5 CTIP2+/+, CTIP2−/−, and CTIP2ep−/− fetuses. Statistical denotations are as described in the legend of Figure 3x. These studies were conducted using at least three independent mice of each genotype.
DISCUSSION
We identify herein a key regulator of gene expression and the developmental program responsible for establishment of the EPB during skin organogenesis. In the absence of CTIP2, the epidermal developmental program was perturbed leading to the thinning of the epidermis with a corresponding reduction in a number of differentiated cell layers. Terminal keratinocyte differentiation was also impaired, most likely owing to disrupted gene expression in CTIP2 mutant mice (both null and conditional), contributing to a delay in EPB establishment and increased TEWL. Thus, CTIP2 appears to be a critical regulator of skin developmental processes, in which it functions in both cell and non-cell autonomous contexts.
CTIP2 is required for epidermal barrier establishment
The results of X-gal diffusion assays indicated that barrier formation was severely disrupted in CTIP2−/− mice at E17.5 but this defect was largely overcome by E18.5, suggesting that barrier formation was most likely delayed in CTIP2−/− mice. However, a substantial degree of epidermal hypocellularity was observed in CTIP2−/− mice at E18.5, which led us to conclude that EPB formation and/or function remained disrupted in CTIP2−/− mice at this developmental stage. Indeed, TEWL measurements taken at E18.5 demonstrated an increased rate of water loss in CTIP2−/− mice as compared to wt controls, demonstrating that barrier defects persisted at E18.5 in the mutants. The apparent disparity between results of X-gal and TEWL assays in the mutant mice at E18.5 may be due to the insensitivity of the former assay. Nonetheless, it is unquestionably true that the barrier defect is less severe at E18.5 than at E17.5 in CTIP2−/− mice, and this may be explained by compensatory mechanisms. The highly related CTIP1 is expressed in skin (Figure S1) and shares many functional properties with CTIP2. For example, both CTIP1 and CTIP2 interact with COUP-TF family members (Avram et al., 2000), exhibit identical sequence-specific DNA-binding activity (Avram et al., 2002), and repress transcription via similar mechanisms (Senawong et al., 2003, 2005; Cismasiu et al., 2005; Topark-Ngarm et al., 2006). Although the distribution and function of CTIP1 in skin are unknown, CTIP1 could, in principle, compensate for the lack of CTIP2, which may explain the “recovery” of the barrier defect that we observe at E18.5 in CTIP2−/− and CTIP2ep−/− mice. Consistent with this hypothesis we detected a modest upregulation of CTIP1 expression in the epidermal extracts of CTIP2ep−/− mice as compared to those prepared from wt animals (Figure S1).
Barrier defects can be caused by inappropriate expression of proteins involved in the assembly and cross-linking of the CE, and/or inadequate lipid processing in the granular and cornified layers, which can result in defective lipid lamellae formation. Gene expression profiling performed on CTIP2ep−/− mice revealed that CTIP2, directly or indirectly, regulated the expression of a subset of genes of the epidermal differentiation complex, as well as a number of lipid metabolizing enzymes in skin. Thus, it seems likely that dysregulation expression of these putative CTIP2 target genes contributed to the delay in EPB formation in both CTIP2−/− and CTIP2ep−/− mice.
The irregular distribution of neutral surface lipids (and increased water loss through the breached areas), and abnormal lipid discs and lipid lamellar structures in the mutant mice may be a consequence of the relative deficiency of lipid-metabolizing enzymes in CTIP2 mutant mice. Mutations in and/or inhibition of lipid-metabolizing enzymes, such as Elovl4, Dgat2, Gba2, Smpd3, Alox12b, eLox3, Pla2gV, and Pla2gVI have been linked to alteration in barrier integrity (Mao-Qiang et al., 1996; Gurrieri et al., 2003; Stone et al., 2004; Cameron et al., 2007; Epp et al., 2007; Furstenberger et al., 2007; Vasireddy et al., 2007). Terminal differentiation of keratinocytes and EPB formation were similarly affected in skin of CTIP2−/− and CTIP2ep−/− mice, strongly suggesting a cell-autonomous effect of CTIP2 in the keratinocyte lineage.
CTIP2 is involved in epidermal homeostasis and control of keratinocyte proliferation and differentiation
The hypoplasticity of CTIP2−/− epidermis is most likely the result of insufficient proliferation and/or altered differentiation. CTIP2 is expressed in all basal cells, some suprabasal cells, and a K15+CD34− population of stem cells (Golonzhka et al., 2007). We observed a reduction in epidermal proliferation and early differentiation in the skin of CTIP2−/− mice compared to control mice, which was accompanied by a concomitant downregulation of the expression of several factors that are implicated in the control of epidermal proliferation and differentiation, including p63, c-Myc, GATA3, c-fos, Klf4, and KGF (Byrne et al., 1994; Mills et al., 1999; Pelengaris et al., 1999; Waikel et al., 1999; Yang et al., 1999; Yokota et al., 1999; Grachtchouk et al., 2000; Langlands et al., 2000; Arnold and Watt, 2001; Mill et al., 2003; Koster et al., 2004, 2007; Simbulan-Rosenthal et al., 2005; Yu et al., 2006; de Guzman Strong et al., 2006). These results strongly suggest that CTIP2 directly or indirectly regulates the promoters of all of these genes. Interestingly, expression of Id2 and p57 was upregulated in CTIP2−/− epidermis. p57 (Topark-Ngarm et al., 2006) and Id2 (Leid M. et al., manuscript in preparation) are known, direct targets of CTIP2 in neuroblastoma cells and thymocytes, respectively, and it is very likely that CTIP2 directly regulates expression of both genes in keratinocytes as well.
As was observed in CTIP2−/− mice, conditional ablation of CTIP2 in keratinocytes resulted in delayed EPB formation in the CTIP2ep−/− mice. However, conditional ablation of CTIP2 in keratinocytes did not significantly alter the proliferation or early differentiation programs of these cells, and the epidermis of CTIP2ep−/− mice was similar to that of wt mice. This demonstrates that CTIP2 is dispensable for keratinocyte proliferation and early differentiation events. In that case it would appear that CTIP2 is acting non-cell autonomously, possibly by controlling the expression of secreted dermal factors that regulate epidermal morphogenic events in a paracrine fashion. Indeed, we observed downregulation of KGF, a dermal fibroblast-derived growth factor, in CTIP2−/−, but not in CTIP2ep−/− mice, suggesting that KGF may play a role in the non-cell autonomous actions of CTIP2 in keratinocytes. Interactions between mesenchymal and epithelial cells play an important role in regulating tissue development and homeostasis (Ronnov-Jessen et al., 1996; Angel et al., 2001; Angel and Szabowski, 2002). Similarly, non-cell autonomous effects in skin have been reported for c-Jun and IKK1 (Szabowski et al., 2000; Gareus et al., 2007). KGF and GM-CSF, which are both downstream of c-Jun, were identified as key regulators of a paracrine loop responsible for the finely tuned balance of keratinocyte proliferation and differentiation (Szabowski et al., 2000). c-Jun also regulates expression of HBEGF and EGFR in keratinocytes in a cell autonomous manner, and mice lacking c-Jun specifically in epidermis do not display an obvious skin phenotype (Li et al., 2003; Gareus et al., 2007). Interestingly, we did not observe a significant alteration of c-Jun expression in skin of CTIP2−/− or CTIP2ep−/− mice, suggesting that CTIP2 is either downstream of c-Jun or represents a parallel pathway by which expression of KGF, GM-CSF, and HBEGF is regulated in the skin. Mice harboring a germline deletion of IKK1 exhibit severe defects in epidermal differentiation, defective EPB function, and increased TEWL, much like CTIP2−/− mice (Li et al., 1999). The epidermis of keratinocyte-specific IKK1 knockout mice develops normally in a histological/morphological sense, but this mouse still displays an impaired barrier and increased TEWL (Gareus et al., 2007). Further analysis of IKK1 total knockouts revealed that IKK1 may influence epidermal development in a non-cell autonomous fashion thorough the regulation of expression/release of yet unidentified diffusible factor(s) from the dermis (Gareus et al., 2007). Nonetheless, the parallels between the cell and non-cell autonomous actions of CTIP2 and IKK1 in the epidermis are striking, and suggest a possible convergence in the two pathways within dermal fibroblasts.
In this study, epidermal keratinocyte-specific ablation of CTIP2 gene revealed that CTIP2 cell autonomously controls barrier establishment in a multifaceted fashion, affecting all stages of this process. Every cell derived from basal keratinocytes in the CTIP2ep−/− mice is presumably null for CTIP2 expression, and this was confirmed by immunoblot analysis. However, it is possible that CTIP2 plays a specific role(s) in suprabasal cell population(s) of the epidermis and this can be further addressed by specifically deleting CTIP2 locus in suprabasal cells, as previously described (Calleja et al., 2006).
Finally, expression-profiling studies reported herein identified genes that were both upregulated and downregulated in the absence of CTIP2 in the epidermis. The mechanistic basis of CTIP2-mediated transcriptional repression has been described (Senawong et al., 2003; Cismasiu et al., 2005; Topark-Ngarm et al., 2006), and presumably the upregulated genes were simply derepressed in the absence of CTIP2. However, the majority of the genes identified herein by expression profiling analyses of CTIP2ep−/− mice were found to be downregulated in absence of CTIP2. This could either represent an indirect effect of CTIP2 ablation or a direct effect of the absence of CTIP2 on the promoters of the corresponding genes, the latter of which may involve recruitment of the coactivators p300 and/or CBP to the target promoter templates (Cismasiu et al., 2006). Further analyses will be required to unravel the mechanism(s) of bidirectional transcriptional regulation mediated by CTIP2 in skin in both health and disease.
MATERIALS AND METHODS
Generation of CTIP2−/− mice
A 14 kb fragment of the mouse CTIP2 genomic locus (129SvPAS strain) was used to construct a CTIP2-targeting vector (Figure 1b and c). After electroporation, five ES cell lines (out of 568) were verified to have undergone HR as judged by Southern analysis using 5′ and 3′ probes outside the targeting vector. Two of these clones were injected into blastocysts, and both gave rise to chimeric animals that were bred with C57BL/6 mice to establish germ line transmission of the targeted L3 allele. These mice were bred with transgenic mice expressing the Flp recombinase under the control of the cytomegalovirus promoter (Rodriguez et al., 2000) to excise the neo marker and generate the L2 (floxed) CTIP2 allele. Alternatively, mice harboring the L3 allele (LoxP–Exon 4–Frt-PGKneo–Frt–LoxP) were crossed with transgenic mice expressing Cre recombinase under the control of the cytomegalovirus promoter (Dupe et al., 1997), to excise both the floxed CTIP2 exon 4 (and generate L-allele) as well as the neo marker. These mice were bred to homozygosity (CTIP2L−/L−, referred to as CTIP2−/− hereafter). Approval for the animal experiments was obtained from the OSU Institutional Animal Care and Use Committee.
X-Gal permeability assay and transepidermal water loss
X-Gal diffusion assays were performed as described previously (Hardman et al., 1998) with minor modifications. TEWL was assessed using a vapometer (SWL4142; Delfin Technologies). Data was expressed in gm−2 h−1, and represents the mean ± SEM from three independent animals (three independent measurements per animal) of each genotype. Data were compared using an unpaired Student’s t-test, with corrections for unequal variance.
Antibodies
The following antibodies and their dilutions were used for immunohistochemistry: anti-CTIP2 (Abcam, Cambridge, UK 1:300), anti-K1 (Covance, Denver, PA, 1:1,000), anti-K14 (Covance, 1:1,000), anti-K10 (Covance, 1:1,000), anti-Ki67 (Abcam, 1:500), anti-Involucrin (Covance, 1:1,000), anti-Loricrin (Covance, 1:1,000), Filaggrin (Covance, 1;1,000). For Immunoblotting we used the dilutions mentioned above, except for anti-CTIP2 (1:1,000 dilution), anti-CTIP1 (Abcam) (1:500) and anti-β-Actin (1:4,000).
Immunohistochemistry
Fetuses at E17.5 and E17.5 were collected, fixed with 4% paraformaldehyde, cryopreseved in 30% sucrose and frozen down in optical coherence tomography. Cross-sections (10 µm) were rinsed with phosphate-buffered saline (PBS) three times, permeabilized with ice-cold methanol for 3 minutes, and blocked with blocking buffer (0.3% Boehringer Mannheim blocking reagent, 5% horse serum, 5% fetal calf serum, and 0.1% triton X-100 in PBS). Sections were then incubated with primary antibodies overnight, followed by three washes with PBS+0.01% Tween and incubation with fluorescently labeled secondary antibody (Cy2, 1:250 or Cy3, 1:500; Jackson ImmunoResearch, West Grove, PA) for 2 hours at room temperature. Nuclei were visualized using DAPI. After final washes with PBS+0.01% Tween, sections were dehydrated through a series of ethanol washes, cleared in xylene, and mounted with DPX mounting media. All images were captured at 40 × magnification using Leica DMRA fluorescent microscope and Hamamatsu C4742-95 digital camera and processed using OpenLab software and Adobe Photoshop 7.0.
Ki67+ cells were quantified and used as a measure of epidermal proliferation. For this, the number of Ki67+ cells within a field of fixed size was determined and expressed as a percentage of DAPI+ cells (total cells) in that same field. The percentage value for wt or L2/L2 controls (approximately 60% across several fields and multiple mice) was then set to 100% for the purposes of comparing the number of epidermal proliferating cells in −/− mice or the conditional mutants.
Histological analysis
Toluidine blue and Hematoxylin and Eosin staining of 1.5 and 10 µm-thick skin sections, respectively, were performed as described (Indra et al., 2005a, b). For transmission electron microscopy analysis, 70 nm, ultra-thin sections were processed as described (Indra et al., 2005b).
Nile Red staining
Frozen sections (10 µm) were stained with Nile Red and after 2 minutes examined with a Leitz fluorescence microscope (excitation 489 nm, emission 515 nm; Indra et al., 2005a, b).
RT-qPCR
RNA extraction and cDNA preparation were performed as described (Indra et al., 2005b). Real-time PCR was performed on an ABI 7500 Real-Time PCR system using SYBR green methodology as described (Indra et al., 2005a, b). RT-PCR conditions and primers sequences for most of the genes have been previous described (Indra et al., 2005a, b; Rodius et al., 2007). Other primer sequences are described in the Supplementary Methods section. Statistical analyses for qPCR experiments were conducted as described for TEWL measurements.
Microarray analysis
Total RNA was prepared from whole skin biopsies using Qiagen, Valencia, PA, RNeasy Mini kit. Probe synthesis, hybridization and scanning were conducted by the Center for Genome Research and Biocomputing Core Laboratories at Oregon State University. The integrity and concentration of RNA was assessed using Bioanalyzer 2100 (Agilent Technologies, Santa Clara, CA), and used to prepare the cDNA templates from which the cRNA probes were synthesized. Probes were hybridized to the Affymetrix Mouse Genome 430 2.0 arrays, and the results were analyzed using GeneSpring 7.2 (Agilent Technologies, Santa Clara, CA) software. Genes that differed from the control by at least 1.5-fold (P<0.05 as determined by one-way analysis of variance) were further filtered by confidence level using Benjamini and Hochberg False Discovery Rate (<20%).
Skin protein extraction
Whole skin biopsies were homogenized in RIPA buffer (50mm Tris, pH 7.5, 1% NP-40, 0.5% sodium deoxycholate, 0.1% SDS, 150mm NaCl, 5mm EDTA, proteinase inhibitors) with a tissue homogenizer, and cleared by centrifugation. Supernatants were collected and equal amounts of protein were analyzed by immunoblotting with indicated antibodies. Where indicated, epidermis was separated from the underlying dermis by incubating the whole skin biopsies with the dispase solution (4mgml−1) for 4 hours at room temperature. Epidermal protein extracts were prepared as described above.
Supplementary Material
Supplementary Methods.
Table S1. List of genes dysregulated in CTIP2ep−/− mice.
Figure S1. Characterization of the CTIP2 null and conditional mutant mice.
Figure S2. Epidermal differentiation complex (EDC) genes mapped onto mouse chromosome 3.
ACKNOWLEDGMENTS
We thank Dr S. Dymecki for ACTB:FLPe mice and the IGBMC/ICS ES and animal facility for excellent technical assistance. We also thank Jean-Marie Garnier (IGBMC) for the 129SvPAS genomic DNA library from which the CTIP2 locus was partially cloned, Tulley Long (OSU) for assistance in targeting vector construction and early genotyping, Ann-Marie Girard (OSU Center for Genome Research and Biocomputing) for expression profiling analyses, and Valerie Peterson for assistance with managing mouse colony and latter genotyping. These studies were supported by grants GM60852 (ML) and AR056008 (AI) from the National Institutes of Health, an OHSU Medical Research Foundation grant to GI, and by a NIEHS Center grant (ES00210) to the Oregon State University Environmental Health Sciences Center. We also thank Dr Wayne Kradjan and Dr Gary DeLander of the OSU College of Pharmacy for continuous support and encouragement, and Dr John Ishmael for advice on histopathological analyses. We are grateful to Stephen Hyter and Sophie Lam-Truc for help in the DNA/RNA extraction and cDNA preparation.
Abbreviations
- CE
cornified envelope
- CTIP2
COUP-TF-interacting protein 2
- EPB
epidermal permeability barrier
- HBEGF
heparin-binding EGF-like growth factor
- HR
homologous recombination
- KGF
keratinocyte growth factor
- PBS
phosphate-buffered saline
- TEWL
transepidermal water loss
- wt
wild-type
Footnotes
CONFLICT OF INTEREST
All the authors state no conflict of interests.
REFERENCES
- Angel P, Szabowski A. Function of AP-1 target genes in mesenchymal-epithelial cross-talk in skin. Biochem Pharmacol. 2002;64:949–956. doi: 10.1016/s0006-2952(02)01158-9. [DOI] [PubMed] [Google Scholar]
- Angel P, Szabowski A, Schorpp-Kistner M. Function and regulation of AP-1 subunits in skin physiology and pathology. Oncogene. 2001;20:2413–2423. doi: 10.1038/sj.onc.1204380. [DOI] [PubMed] [Google Scholar]
- Arlotta P, Molyneaux BJ, Chen J, Inoue J, Kominami R, Macklis JD. Neuronal subtype-specific genes that control corticospinal motor neuron development in vivo. Neuron. 2005;45:207–221. doi: 10.1016/j.neuron.2004.12.036. [DOI] [PubMed] [Google Scholar]
- Arlotta P, Molyneaux BJ, Jabaudon D, Yoshida Y, Macklis JD. Ctip2 controls the differentiation of medium spiny neurons and the establishment of the cellular architecture of the striatum. J Neurosci. 2008;28:622–632. doi: 10.1523/JNEUROSCI.2986-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold I, Watt FM. c-Myc activation in transgenic mouse epidermis results in mobilization of stem cells and differentiation of their progeny. Curr Biol. 2001;11:558–568. doi: 10.1016/s0960-9822(01)00154-3. [DOI] [PubMed] [Google Scholar]
- Avram D, Fields A, Pretty On Top K, Nevrivy DJ, Ishmael JE, Leid M. Isolation of a novel family of C(2)H(2) zinc finger proteins implicated in transcriptional repression mediated by chicken ovalbumin upstream promoter transcription factor (COUP-TF) orphan nuclear receptors. J Biol Chem. 2000;275:10315–10322. doi: 10.1074/jbc.275.14.10315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avram D, Fields A, Senawong T, Topark-Ngarm A, Leid M. COUP-TF (chicken ovalbumin upstream promoter transcription factor)-interacting protein 1 (CTIP1) is a sequence-specific DNA binding protein. Biochem J. 2002;368:555–563. doi: 10.1042/BJ20020496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrne C, Hardman M, Nield K. Covering the limb—formation of the integument. J Anat. 2003;202:113–123. doi: 10.1046/j.1469-7580.2003.00142.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrne C, Tainsky M, Fuchs E. Programming gene expression in developing epidermis. Development. 1994;120:2369–2383. doi: 10.1242/dev.120.9.2369. [DOI] [PubMed] [Google Scholar]
- Calleja C, Messaddeq N, Chapellier B, Yang H, Krezel W, Li M, et al. Genetic and pharmacological evidence that a retinoic acid cannot be the RXR-activating ligand in mouse epidermis keratinocytes. Genes Dev. 2006;20:1525–1538. doi: 10.1101/gad.368706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cameron DJ, Tong Z, Yang Z, Kaminoh J, Kamiyah S, Chen H, et al. Essential role of Elovl4 in very long chain fatty acid synthesis, skin permeability barrier function, and neonatal survival. Int J Biol Sci. 2007;3:111–119. doi: 10.7150/ijbs.3.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Candi E, Schmidt R, Melino G. The cornified envelope: a model of cell death in the skin. Nat Rev Mol Cell Biol. 2005;6:328–340. doi: 10.1038/nrm1619. [DOI] [PubMed] [Google Scholar]
- Cismasiu VB, Adamo K, Gecewicz J, Duque J, Lin Q, Avram D. BCL11B functionally associates with the NuRD complex in T lymphocytes to repress targeted promoter. Oncogene. 2005;24:6753–6764. doi: 10.1038/sj.onc.1208904. [DOI] [PubMed] [Google Scholar]
- Cismasiu VB, Ghanta S, Duque J, Albu DI, Chen HM, Kasturi R, et al. BCL11B participates in the activation of IL2 gene expression in CD4+ T lymphocytes. Blood. 2006;108:2695–2702. doi: 10.1182/blood-2006-05-021790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Guzman Strong C, Wertz PW, Wang C, Yang F, Meltzer PS, Andl T, et al. Lipid defect underlies selective skin barrier impairment of an epidermal-specific deletion of Gata-3. J Cell Biol. 2006;175:661–670. doi: 10.1083/jcb.200605057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupe V, Davenne M, Brocard J, Dolle P, Mark M, Dierich A, et al. In vivo functional analysis of the Hoxa-1 3′ retinoic acid response element (3′RARE) Development. 1997;124:399–410. doi: 10.1242/dev.124.2.399. [DOI] [PubMed] [Google Scholar]
- Elias PM. Stratum corneum defensive functions: an integrated view. J Invest Dermatol. 2005;125:183–200. doi: 10.1111/j.0022-202X.2005.23668.x. [DOI] [PubMed] [Google Scholar]
- Epp N, Furstenberger G, Muller K, de Juanes S, Leitges M, Hausser I, et al. 12R-lipoxygenase deficiency disrupts epidermal barrier function. J Cell Biol. 2007;177:173–182. doi: 10.1083/jcb.200612116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs E, Green H. Changes in keratin gene expression during terminal differentiation of the keratinocyte. Cell. 1980;19:1033–1042. doi: 10.1016/0092-8674(80)90094-x. [DOI] [PubMed] [Google Scholar]
- Furstenberger G, Epp N, Eckl KM, Hennies HC, Jorgensen C, Hallenborg P, et al. Role of epidermis-type lipoxygenases for skin barrier function and adipocyte differentiation. Prostaglandins Other Lipid Mediat. 2007;82:128–134. doi: 10.1016/j.prostaglandins.2006.05.006. [DOI] [PubMed] [Google Scholar]
- Gareus R, Huth M, Breiden B, Nenci A, Rosch N, Haase I, et al. Normal epidermal differentiation but impaired skin-barrier formation upon keratinocyte-restricted IKK1 ablation. Nat Cell Biol. 2007;9:461–469. doi: 10.1038/ncb1560. [DOI] [PubMed] [Google Scholar]
- Golonzhka O, Leid M, Indra G, Indra AK. Expression of COUP-TF-interacting protein 2 (CTIP2) in mouse skin during development and in adulthood. Gene Expr Patterns. 2007;7:754–760. doi: 10.1016/j.modgep.2007.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grachtchouk M, Mo R, Yu S, Zhang X, Sasaki H, Hui CC, et al. Basal cell carcinomas in mice overexpressing Gli2 in skin. Nat Genet. 2000;24:216–217. doi: 10.1038/73417. [DOI] [PubMed] [Google Scholar]
- Gurrieri S, Furstenberger G, Schadow A, Haas U, Singer AG, Ghomashchi F, et al. Differentiation-dependent regulation of secreted phospholipases A2 in murine epidermis. J Invest Dermatol. 2003;121:156–164. doi: 10.1046/j.1523-1747.2003.12315.x. [DOI] [PubMed] [Google Scholar]
- Hardman MJ, Sisi P, Banbury DN, Byrne C. Patterned acquisition of skin barrier function during development. Development. 1998;125:1541–1552. doi: 10.1242/dev.125.8.1541. [DOI] [PubMed] [Google Scholar]
- Holleran WM, Ginns EI, Menon GK, Grundmann JU, Fartasch M, McKinney CE, et al. Consequences of beta-glucocerebrosidase deficiency in epidermis. Ultrastructure and permeability barrier alterations in Gaucher disease. J Clin Invest. 1994;93:1756–1764. doi: 10.1172/JCI117160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Indra AK, Dupe V, Bornert JM, Messaddeq N, Yaniv M, Mark M, et al. Temporally controlled targeted somatic mutagenesis in embryonic surface ectoderm and fetal epidermal keratinocytes unveils two distinct developmental functions of BRG1 in limb morphogenesis and skin barrier formation. Development. 2005a;132:4533–4544. doi: 10.1242/dev.02019. [DOI] [PubMed] [Google Scholar]
- Indra AK, Li M, Brocard J, Warot X, Bornert JM, Gerard C, et al. Targeted somatic mutagenesis in mouse epidermis. Horm Res. 2000;54:296–300. doi: 10.1159/000053275. [DOI] [PubMed] [Google Scholar]
- Indra AK, Mohan WS, II, Frontini M, Scheer E, Messaddeq N, Metzger D, et al. TAF10 is required for the establishment of skin barrier function in foetal, but not in adult mouse epidermis. Dev Biol. 2005b;285:28–37. doi: 10.1016/j.ydbio.2005.05.043. [DOI] [PubMed] [Google Scholar]
- Koster MI, Dai D, Marinari B, Sano Y, Costanzo A, Karin M, et al. p63 induces key target genes required for epidermal morphogenesis. Proc Natl Acad Sci USA. 2007;104:3255–3260. doi: 10.1073/pnas.0611376104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koster MI, Kim S, Mills AA, DeMayo FJ, Roop DR. p63 is the molecular switch for initiation of an epithelial stratification program. Genes Dev. 2004;18:126–131. doi: 10.1101/gad.1165104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langlands K, Down GA, Kealey T. Id proteins are dynamically expressed in normal epidermis and dysregulated in squamous cell carcinoma. Cancer Res. 2000;60:5929–5933. [PubMed] [Google Scholar]
- Leid M, Ishmael JE, Avram D, Shepherd D, Fraulob V, Dolle P. CTIP1 and CTIP2 are differentially expressed during mouse embryogenesis. Gene Expr Patterns. 2004;4:733–739. doi: 10.1016/j.modgep.2004.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G, Gustafson-Brown C, Hanks SK, Nason K, Arbeit JM, Pogliano K, et al. c-Jun is essential for organization of the epidermal leading edge. Dev Cell. 2003;4:865–877. doi: 10.1016/s1534-5807(03)00159-x. [DOI] [PubMed] [Google Scholar]
- Li Q, Lu Q, Hwang JY, Buscher D, Lee KF, Izpisua-Belmonte JC, et al. IKK1-deficient mice exhibit abnormal development of skin and skeleton. Genes Dev. 1999;13:1322–1328. doi: 10.1101/gad.13.10.1322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Sandhoff R, Kono M, Zerfas P, Hoffmann V, Ding BC, et al. Depletion of ceramides with very long chain fatty acids causes defective skin permeability barrier function, and neonatal lethality in ELOVL4 deficient mice. Int J Biol Sci. 2007;3:120–128. doi: 10.7150/ijbs.3.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mack JA, Anand S, Maytin EV. Proliferation and cornification during development of the mammalian epidermis. Birth Defects Res C Embryo Today. 2005;75:314–329. doi: 10.1002/bdrc.20055. [DOI] [PubMed] [Google Scholar]
- Mao-Qiang M, Jain M, Feingold KR, Elias PM. Secretory phospholipase A2 activity is required for permeability barrier homeostasis. J Invest Dermatol. 1996;106:57–63. doi: 10.1111/1523-1747.ep12327246. [DOI] [PubMed] [Google Scholar]
- Mill P, Mo R, Fu H, Grachtchouk M, Kim PC, Dlugosz AA, et al. Sonic hedgehog-dependent activation of Gli2 is essential for embryonic hair follicle development. Genes Dev. 2003;17:282–294. doi: 10.1101/gad.1038103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills AA, Zheng B, Wang XJ, Vogel H, Roop DR, Bradley A. p63 is a p53 homologue required for limb and epidermal morphogenesis. Nature. 1999;398:708–713. doi: 10.1038/19531. [DOI] [PubMed] [Google Scholar]
- Moran JL, Qiu H, Turbe-Doan A, Yun Y, Boeglin WE, Brash AR, et al. A mouse mutation in the 12R-lipoxygenase, Alox12b, disrupts formation of the epidermal permeability barrier. J Invest Dermatol. 2007;127:1893–1897. doi: 10.1038/sj.jid.5700825. [DOI] [PubMed] [Google Scholar]
- Pelengaris S, Littlewood T, Khan M, Elia G, Evan G. Reversible activation of c-Myc in skin: induction of a complex neoplastic phenotype by a single oncogenic lesion. Mol Cell. 1999;3:565–577. doi: 10.1016/s1097-2765(00)80350-0. [DOI] [PubMed] [Google Scholar]
- Rodius S, Indra G, Thibault C, Pfister V, Georges-Labouesse E. Loss of alpha6 integrins in keratinocytes leads to an increase in TGFbeta and AP1 signaling and in expression of differentiation genes. J Cell Physiol. 2007;212:439–449. doi: 10.1002/jcp.21040. [DOI] [PubMed] [Google Scholar]
- Rodriguez CI, Buchholz F, Galloway J, Sequerra R, Kasper J, Ayala R, et al. High-efficiency deleter mice show that FLPe is an alternative to Cre-loxP. Nat Genet. 2000;25:139–140. doi: 10.1038/75973. [DOI] [PubMed] [Google Scholar]
- Ronnov-Jessen L, Petersen OW, Bissell MJ. Cellular changes involved in conversion of normal to malignant breast: importance of the stromal reaction. Physiol Rev. 1996;76:69–125. doi: 10.1152/physrev.1996.76.1.69. [DOI] [PubMed] [Google Scholar]
- Roop DR, Hawley-Nelson P, Cheng CK, Yuspa SH. Keratin gene expression in mouse epidermis and cultured epidermal cells. Proc Natl Acad Sci USA. 1983;80:716–720. doi: 10.1073/pnas.80.3.716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schluter C, Duchrow M, Wohlenberg C, Becker MH, Key G, Flad HD, et al. The cell proliferation-associated antigen of antibody Ki-67: a very large, ubiquitous nuclear protein with numerous repeated elements, representing a new kind of cell cycle-maintaining proteins. J Cell Biol. 1993;123:513–522. doi: 10.1083/jcb.123.3.513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segre J. Complex redundancy to build a simple epidermal permeability barrier. Curr Opin Cell Biol. 2003;15:776–782. doi: 10.1016/j.ceb.2003.10.001. [DOI] [PubMed] [Google Scholar]
- Segre JA. Epidermal barrier formation and recovery in skin disorders. J Clin Invest. 2006;116:1150–1158. doi: 10.1172/JCI28521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segre JA, Bauer C, Fuchs E. Klf4 is a transcription factor required for establishing the barrier function of the skin. Nat Genet. 1999;22:356–360. doi: 10.1038/11926. [DOI] [PubMed] [Google Scholar]
- Senawong T, Peterson VJ, Avram D, Shepherd DM, Frye RA, Minucci S, et al. Involvement of the histone deacetylase SIRT1 in chicken ovalbumin upstream promoter transcription factor (COUP-TF)-interacting protein 2-mediated transcriptional repression. J Biol Chem. 2003;278:43041–43050. doi: 10.1074/jbc.M307477200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senawong T, Peterson VJ, Leid M. BCL11A-dependent recruitment of SIRT1 to a promoter template in mammalian cells results in histone deacetylation and transcriptional repression. Arch Biochem Biophys. 2005;434:316–325. doi: 10.1016/j.abb.2004.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaulian E, Karin M. AP-1 as a regulator of cell life and death. Nat Cell Biol. 2002;4:E131–E136. doi: 10.1038/ncb0502-e131. [DOI] [PubMed] [Google Scholar]
- Simbulan-Rosenthal CM, Trabosh V, Velarde A, Chou FP, Daher A, Tenzin F, et al. Id2 protein is selectively upregulated by UVB in primary, but not in immortalized human keratinocytes and inhibits differentiation. Oncogene. 2005;24:5443–5458. doi: 10.1038/sj.onc.1208709. [DOI] [PubMed] [Google Scholar]
- Stone SJ, Myers HM, Watkins SM, Brown BE, Feingold KR, Elias PM, et al. Lipopenia and skin barrier abnormalities in DGAT2-deficient mice. J Biol Chem. 2004;279:11767–11776. doi: 10.1074/jbc.M311000200. [DOI] [PubMed] [Google Scholar]
- Szabowski A, Maas-Szabowski N, Andrecht S, Kolbus A, Schorpp-Kistner M, Fusenig NE, et al. c-Jun and JunB antagonistically control cytokine-regulated mesenchymal-epidermal interaction in skin. Cell. 2000;103:745–755. doi: 10.1016/s0092-8674(00)00178-1. [DOI] [PubMed] [Google Scholar]
- Topark-Ngarm A, Golonzhka O, Peterson VJ, Barrett B, Jr, Martinez B, Crofoot K, et al. CTIP2 associates with the NuRD complex on the promoter of p57KIP2, a newly identified CTIP2 target gene. J Biol Chem. 2006;281:32272–32283. doi: 10.1074/jbc.M602776200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasireddy V, Uchida Y, Salem N, Jr, Kim SY, Mandal MN, Reddy GB, et al. Loss of functional ELOVL4 depletes very long-chain fatty acids (>or =C28) and the unique omega-O-acylceramides in skin leading to neonatal death. Hum Mol Genet. 2007;16:471–482. doi: 10.1093/hmg/ddl480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waikel RL, Wang XJ, Roop DR. Targeted expression of c-Myc in the epidermis alters normal proliferation, differentiation and UV-B induced apoptosis. Oncogene. 1999;18:4870–4878. doi: 10.1038/sj.onc.1203040. [DOI] [PubMed] [Google Scholar]
- Wakabayashi Y, Watanabe H, Inoue J, Takeda N, Sakata J, Mishima Y, et al. Bcl11b is required for differentiation and survival of alphabeta T lymphocytes. Nat Immunol. 2003;4:533–539. doi: 10.1038/ni927. [DOI] [PubMed] [Google Scholar]
- Werner S, Smola H. Paracrine regulation of keratinocyte proliferation and differentiation. Trends Cell Biol. 2001;11:143–146. doi: 10.1016/s0962-8924(01)01955-9. [DOI] [PubMed] [Google Scholar]
- Yang A, Schweitzer R, Sun D, Kaghad M, Walker N, Bronson RT, et al. p63 is essential for regenerative proliferation in limb, craniofacial and epithelial development. Nature. 1999;398:714–718. doi: 10.1038/19539. [DOI] [PubMed] [Google Scholar]
- Yokota Y, Mansouri A, Mori S, Sugawara S, Adachi S, Nishikawa S, et al. Development of peripheral lymphoid organs and natural killer cells depends on the helix-loop-helix inhibitor Id2. Nature. 1999;397:702–706. doi: 10.1038/17812. [DOI] [PubMed] [Google Scholar]
- Yu Z, Lin KK, Bhandari A, Spencer JA, Xu X, Wang N, et al. The Grainyhead-like epithelial transactivator Get-1/Grhl3 regulates epidermal terminal differentiation and interacts functionally with LMO4. Dev Biol. 2006;299:122–136. doi: 10.1016/j.ydbio.2006.07.015. [DOI] [PubMed] [Google Scholar]
- Zenz R, Wagner EF. Jun signalling in the epidermis: From developmental defects to psoriasis and skin tumors. Int J Biochem Cell Biol. 2006;38:1043–1049. doi: 10.1016/j.biocel.2005.11.011. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Methods.
Table S1. List of genes dysregulated in CTIP2ep−/− mice.
Figure S1. Characterization of the CTIP2 null and conditional mutant mice.
Figure S2. Epidermal differentiation complex (EDC) genes mapped onto mouse chromosome 3.