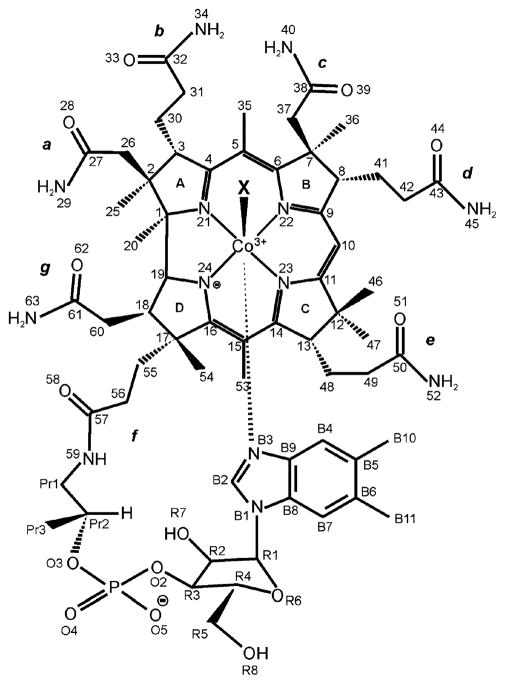
Abstract
The structure of nitrosylcobalamin (NOCbl) in solution has been studied by NMR spectroscopy and the 1H and 13C NMR spectra have been assigned. 13C and 31P NMR chemical shifts, the UV-vis spectrum of NOCbl and the observed pK base-off value of ~5.1 for NOCbl provide evidence that a significant fraction of NOCbl is present in the base-off, 5,6-dimethylbenzimidazole (DMB) deprotonated, form in solution. NOE-restrained molecular mechanics modelling of base-on NOCbl gave annealed structures with minor conformational differences in the flexible side chains and the nucleotide loop position compared with the X-ray structure. A molecular dynamics simulation at 300 K showed that DMB remains in close proximity to the α face of the corrin in the base-off form of NOCbl. Simulated annealing calculations produced two major conformations of base-off NOCbl. In the first, the DMB is perpendicular to the corrin and its B3 nitrogen is about 3.1 Å away from and pointing directly at the metal ion; in the second the DMB is parallel to and tucked beneath the D ring of the corrin.
Introduction
Two vitamin B12-dependent enzyme reactions occur in mammals – methylcobalamin-dependent methionine synthase and adenosylcobalamin-dependent L-methylmalonyl-CoA mutase.1 The early stages of B12 deficiency in humans lead to hyperhomocysteinaemia and/or methylmalonic acidaemia followed by megaloblastic anaemia and/or neurological disorders.2 The nitric oxide (NO) derivative of vitamin B12, nitrosylcobalamin (NOCbl, also referred to as nitroxylcobalamin or nitrosocobalamin in the literature), is also of potential interest in mammalian biochemistry. It has recently been proposed that cobalamins (Cbls, vitamin B12 derivatives) scavenge nitric oxide to form NOCbl in vivo,3,4 and that Cbl is beneficial in treating pathological disorders associated with high NO, including sepsis.4,5 NO regulates vasodilation, neurotransmission, and the immune response and is an inhibitor of platelet aggregation and cell proliferation.6,7 Aquacobalamin suppresses NO-induced relaxation of smooth muscle,8–10 NO-induced vasodilation11 and NO-mediated inhibition of cell proliferation.3 Vitamin B12 (cyanocobalamin) also reverses NO-induced neural tube defects.12 Furthermore, both MeCbl-dependent methionine synthase and adenosylcobalamin-dependent methylmalonyl-CoA mutase are inhibited by NO or NO donors.13–17
The extremely air sensitive NOCbl can be prepared by bubbling NO(g) through anaerobic solutions of cob(II)alamin or glutathionylcobalamin, by photoreduction of an aquacobalamin (H2OCbl+) solution in the presence of NO(g) using a laser, or reacting aquacobalamin with NO donors.18–23 There has been some controversy in the literature concerning whether H2OCbl+ reacts directly with NO to form NOCbl, although it is now generally accepted that this is not the case.22,24 There has also been controversy as to whether H2OCbl+ reacts directly with NO to form NOCbl in acidic solution (pH < 4); however commercial NO(g) is contaminated with nitrogen dioxide which reacts with NO to ultimately produce NO+ and nitrite in aqueous solution,24 and a recent study provides convincing evidence that the initial reaction step instead involves the formation of base-off NO2Cbl.25
NOCbl has been characterized in solution by UV-vis, 1H NMR (aromatic region only) and 15N NMR spectroscopy.18,19,23 Although the ν(N–O) stretch of NOCbl was not observed in solution by resonance Raman spectroscopy, a Co–N(O) band was observed at 514 cm−1.19 Recently some of us determined the structure of NOCbl·15H2O by X-ray diffraction.23 On the basis of the Co–N–O angle (117.4–121.4°) it was concluded that the oxidation state of the Co centre in NOCbl is +3 in the solid state, since the Co–N–O group of low spin NO−–CoIII complexes is bent (~120°) as a consequence of the lone pair on the N atom of NO−,26 whereas Co–N–O is essentially linear for CoII–NO complexes.26 A similar conclusion was reached for NOCbl in solution based on resonance Raman spectroscopy measurements, the observation of well defined 1H and 15N NMR resonances indicative of a diamagnetic complex and the 15N NMR chemical shift of 15NOCbl.18,19
Some of us have previously successfully applied a molecular mechanics (MM) force field to address a variety of questions in the structural chemistry of corrins.27–31 We have also used NMR-derived distance restraints in molecular dynamics (MD) and simulated annealing (SA) calculations to explore the solution structure of a number of corrins, including those that fail to crystallize.32–37 In the present work we have investigated the structure of NOCbl in solution using 2-D NMR spectroscopy techniques combined with NOE-restrained molecular dynamics calculations. The 1H and 13C NMR chemical shifts of NOCbl were completely assigned by ROESY, TOCSY, HSQC, and HMBC experiments. Molecular mechanics modelling of NOCbl was performed using the parameters derived specifically to model the cobalt corrins38,39 as an extension of Allinger’s MM2 force field.40 Our results suggest that the electronic effects exerted by the NO ligand of NOCbl are closer to alkyl ligands rather than other inorganic ligands such as CN−, NO2− and H2O and that, consequently, a significant fraction of NOCbl in neutral solution exists in the DMB-deprotonated, base-off form.
Experimental
Synthesis of NOCbl
Both the synthesis and handling of NOCbl were carried out inside a glove box under an argon atmosphere. NOCbl was synthesized according to our published procedure,23 with minor modifications. A freshly prepared anaerobic solution of HOCbl·HCl (35 mg HOCbl·HCl in 1 mL H2O) was added to solid DEA-NONOate (8.73 mg, 2.5 equiv.). The product solution was shaken gently to ensure complete mixing and the reaction left to proceed at room temperature for 12 h. The final pH of the mixture was 10.8. Formation of the desired product was checked by UV-vis and 1H NMR spectroscopy.23 The product was precipitated by drop-wise addition to cold acetone (20 mL, −20 °C), filtered and dried under vacuum (2 × 10−2 mbar) for 3 h. We observed that both the unreacted DEA-NONOate and the reaction product DEA-NO co-precipitated with NOCbl, with their corresponding 1H NMR signals in the aliphatic region of the spectrum interfering with our measurements. Therefore, the acetone precipitation step (and dissolution back into H2O) was repeated two more times, resulting in removal of most of the interfering compounds. NOCbl remained stable during the procedure, yielding a final purity of ~97% by 1H NMR spectroscopy.
Preparation of the NOCbl NMR spectroscopy sample
NOCbl (35 mg) was dissolved in 0.80 mL of anaerobic phosphate buffer (0.15 M, pH 7.40) made up with 90% H2O/10% D2O, and containing 1 mg of TSP. The final pH of the sample was 7.70. The NMR tube was flame-sealed under vacuum and the purity of the sample checked by 1H NMR spectroscopy. The sample was stable for at least 3 months at room temperature, in the dark, as assessed by 1H NMR and UV-vis spectroscopy (the latter experiment requiring the NMR tube to be broken open and the sample diluted with anaerobic buffer prior to transferring to a Schlenk cuvette for a UV-vis spectroscopy measurement).
31P NMR spectroscopy experiments
SO3Cbl and NO2Cbl were prepared using published procedures.41 31P NMR spectroscopy measurements were made at 25 °C on a Bruker Avance 400 MHz spectrometer operating at 161.975 MHz. Samples (~6–7 mM) were dissolved in D2O (1 ml), and chemical shifts externally referenced to an NMR tube containing 85% H3PO4 equipped with a concentric insert containing D2O.
NOCbl 2-D NMR spectroscopy experiments
Two-dimensional homonuclear (TOCSY and ROESY) experiments with Watergate solvent suppression and heteronuclear (HSQC and HMBC) experiments for resonance assignment were carried out as previously described30 on a Brucker DMX 600 NMR spectrometer. Assignments were made as described elsewhere.42
For molecular modelling distance restraints, ROESY spectra were obtained on a Brucker DRX 800 NMR spectrometer at mixing times of 40, 80, 120, and 160 ms.
Attempted determination of pKbase-off for NO2Cbl
1H NMR spectra of samples of NO2Cbl (5 × 10−3 M) in acidic solution (H2SO4 + D2O) were recorded at 25 °C on a Bruker Avance 400 MHz spectrometer with TSP used as an internal reference.
Molecular modelling calculations
All modelling calculations (energy minimizations, MD runs and SA calculations) were performed with HYPERCHEM v. 7.0343 using the potential energy functions of the MM+ force field (as MM2 is called in this suite of programs) and the parameters derived for the cobalt corrins.38,39 Parameters for NO coordinated to Co(III) in NOCbl were not available. We therefore developed preliminary parameters based upon the crystal structures of NOCbl23 and four Co(III)NO porphyrins.44–47 NO is disordered over three positions in NOCbl; the mean N–O bond length is 1.18(3) Å. It is not significantly different in the CoNO porphyrins (1.14(6) Å). We used a value of ks = 10.0 mdyn Å−1 for the bond force constant and lo = 1.175 Å for the strain-free bond length for modelling the bond between N (atom type n1) and O (atoms type o1). In NOCbl, the Co–N bond length is 1.927 Å and shorter in CoNO porphyrins (1.839(8) Å). We used ks = 2.0 mdyn Å−1 and lo = 1.865 Å in the force field. NO coordinates to Co(III) in a bent manner (Co–N–O = 120(2)° in NOCbl and is similar (122.9(9)°) in CoNO porphyrins. The angle was modelled using an angle bending force constant kb = 1.0 mdyn Å rad−2 and a strain free bond angle of θo = 116°.
A total of 64 NOE cross peaks (Table S1 †), not including those resulting from geminal hydrogens, could be resolved and assigned in the ROESY spectra in H2O at the longest mixing time (160 ms) for NOCbl. These cross peaks were classified48 as strong, medium, weak, or very weak, depending on whether they first appeared in the ROESY spectra at mixing times of 40, 80, 120, or 160 ms, respectively.
The crystal structure of NOCbl,23 from which we excluded all solvent molecules, was used as starting point for the modelling. Energy minimization was performed using a Polak-Ribiere conjugate gradient algorithm with a convergence criterion of 0.01 kcal Å−1 mol−1 r.m.s. gradient.
We used a set of parabolic potential energy functions of the form UNOE(rij) = kNOE(rij −rijo)2 to restrain the distance rij between protons i and j determined from the 2D ROESY spectra. Following the definition of Clore and co-workers,49 and as we have explained elsewhere,33 we used values of the restraining force constant, kNOE = 1.2, 0.52, 0.3, and 0.075 kcal mol−1 Å−2 at 300 K for strong, medium, weak and very weak NOE’s, respectively. Violations of these distance restraints were taken when rij ≥ 3.0, 3.8, 4.8 and 5.3 Å, respectively.
The assignments of all pro-chiral protons were made on a trial and error basis until the minimum number of violations of distance criteria were obtained during a series of 100 ps MD simulations at 300 K. Where a number of NOE’s to the same achiral centre occur, care was taken to choose for each restraint the proton pair with the smallest inter-proton distance. The leapfrog algorithm50,51 was used to solve the Newtonian equations of motion that describe the dynamics trajectory for MD simulation calculations. A small time step of 0.5 fs was used. Temperature scaling with coupling to an external heat sink52 (using a temperature relaxation time of 0.1 ps), was used to control the temperature. Initial velocities for MD simulations were assigned to fully energy minimized structures using a random number generator. In a typical simulation, the molecule was heated from 0 K to the run temperature (300 or 800 K) during a 20 ps heating phase. The run phase at 800 K was varied between 1 and 750 ps for simulations designed to find stable conformations while a run phase of 500 ps was used to determine inter-proton distances at 300 K. The run phase was followed by an annealing phase of 20 ps to 0 K for simulations designed to find stable conformations, followed by full energy minimization. A consensus structure was obtained by averaging the coordinates of each atom of each of 25 annealed structures and then energy minimizing again.
Results and discussion
NMR spectroscopy measurements for NOCbl
2-D NMR experiments (TOCSY, ROESY, HSQC, and HMBC) allowed the complete assignment of the 1H and 13C NMR resonances for NOCbl in solution (pH 7.70, 25 °C). Table 1 summarizes the final 1H and 13C chemical shift assignments for NOCbl and Table S1† shows the 2-D NMR correlations which support these assignments. A labelling scheme for NOCbl is given in Fig. 1. Significant (≥1.0 ppm) 13C chemical shift differences between NOCbl and two “normal” Co(III)Cbls with nitrogen-linked upper axial ligands, nitrocobalamin (NO2Cbl) and amminocobalamin (NH3Cbl), are summarized in Table 2. As 19 of 62 (31%) chemical shifts are significantly different, it is reasonable to conclude that NOCbl is not a typical Co(III)Cbl with a nitrogenous upper axial ligand. In particular, the chemical shift differences at the corrin ring carbons C15, C14, C9, C16, and C4 suggest significantly different corrin ring electronic effects in NOCbl.
Table 1.
1H and 13C NMR assignments for NOCbla
| Group | δ 13C (ppm) | δ 1H (ppm) | Group | δ 13C (ppm) | δ 1H (ppm) |
|---|---|---|---|---|---|
| C53 | 17.87 | 2.42 | R1 | 88.51 | 6.26e |
| C35 | 18.03 | 2.51 | C10 | 97.40 | 6.34 |
| C25 | 19.39 | 1.39 | C15 | 107.60 | |
| C54 | 20.34 | 1.30 | C5 | 109.91 | |
| Pr3 | 21.42b | 1.24c | B7 | 113.43 | 7.19 |
| C36 | 22.09 | 1.82 | B4 | 121.29 | 6.77 |
| C47 | 22.09 | 1.50 | B5 | 133.99 | |
| B10 | 22.24 | 2.11 | B8 | 134.21 | |
| B11 | 22.24 | 2.11 | B6 | 135.75 | |
| C20 | 24.48 | 0.54 | B9 | 142.20 | |
| C30 | 28.50 | 1.89, 2.01 | B2 | 144.53 | 7.44 |
| C41 | 28.91 | 1.40, 2.04 | C14 | 166.30 | |
| C48 | 29.25 | 1.88, 2.03 | C6 | 166.40 | |
| C46 | 34.33 | 0.98 | C9 | 174.53 | |
| C55 | 34.78 | 1.81, 2.37 | C38 | 177.36 | |
| C42 | 34.78 | 1.72, 2.06 | C57 | 177.91 | |
| C31 | 34.95 | 2.47, 2.73 | C16 | 178.23 | |
| C56 | 34.95 | 1.87, 2.47 | C27 | 178.71 | |
| C49 | 36.04 | 2.20, 2.35 | C11 | 178.71 | |
| C60 | 37.86 | 2.52, 2.56 | C4 | 178.71 | |
| C18 | 41.88 | 2.75 | C32 | 178.71 | |
| C26 | 45.83 | 2.28, 2.37 | C43 | 180.37 | |
| C37 | 46.51 | 1.98, 2.44 | C50 | 180.73 | |
| Pr1 | 47.46 | 3.26, 3.49 | C61 | 180.73 | |
| C2 | 49.12 | dHS | 6.52 | ||
| C12 | 49.86 | cHS | 6.75 | ||
| C7 | 53.31 | dHA | 6.81 | ||
| C13 | 55.31 | 3.18 | eHS | 6.85 | |
| C8 | 57.93 | 3.60 | bHS | 6.94 | |
| C3 | 58.18 | 4.11 | aHS | 7.09 | |
| C17 | 61.22 | gHS | 7.13 | ||
| R5 | 63.55 | 3.79, 3.89 | eHA | 7.52 | |
| R2 | 72.91 | 4.50 | cHA | 7.57 | |
| Pr2 | 75.33d | 4.35 | bHA | 7.66 | |
| R3 | 76.46 | 4.77 | aHA | 7.84 | |
| C19 | 77.27 | 4.12 | gHA | 7.99 | |
| R4 | 85.40 | 4.32 | f H | 8.05 | |
| C1 | 88.24 |
In 0.15 M phosphate buffer, pH 7.70, 90% H2O/10% D2O, 25 °C. Chemical shifts are relative to internal TSP. The syn and anti amide H’s are designated aHS, aHA, bHS, bHA, etc., respectively, where a, b, etc., refer to the standard side chain lettering.
3JPC = 4.2 Hz.
JPr2-Pr3 = 6.1 Hz.
2JPC = 5.4 Hz.
JR1,R2 = 3.2 Hz.
Fig. 1.
Labeling scheme for cobalamins. X = Me, Ado, H2O, CN, NO, NH3, NO2, SO3 etc.
Table 2.
| Group | δ NOCbl | δ NO2Cbl | (Δδ) | δNH3Cbl | (Δδ) |
|---|---|---|---|---|---|
| C54 | 20.34 | 18.31 | (2.03) | 19.16 | (1.18) |
| C20 | 24.48 | 22.45 | (2.00) | 22.73 | (1.75) |
| C49 | 36.04 | 37.33 | (−1.29) | 37.36 | (−1.32) |
| C60 | 37.86 | 34.05 | (3.81) | 34.01 | (3.85) |
| C37 | 46.51 | 44.80 | (1.71) | 48.09 | (−1.58) |
| R2 | 72.91 | 71.29 | (1.62) | 71.28 | (1.63) |
| R1 | 88.51 | 89.64 | (−1.13) | 89.77 | (−1.26) |
| C15 | 107.60 | 106.05 | (1.55) | 106.41 | (1.19) |
| B4 | 121.29 | 119.07 | (2.22) | 118.81 | (2.48) |
| B5 | 133.99 | 135.64 | (−1.65) | 135.88 | (−1.89) |
| B8 | 134.21 | 132.05 | (2.16) | 132.09 | (2.12) |
| B6 | 135.75 | 137.80 | (−2.05) | 138.07 | (−2.32) |
| B9 | 142.20 | 139.17 | (3.03) | 139.11 | (3.09) |
| C14 | 166.30 | 167.96 | (−1.66) | 168.23 | (−1.93) |
| C9 | 174.53 | 175.89 | (−1.36) | 177.01 | (−2.48) |
| C16 | 178.23 | 180.73 | (−2.50) | 182.50 | (−4.27) |
| C4 | 178.71 | 182.47 | (−3.76) | 183.73 | (−5.02) |
| C32 | 178.71 | 180.44 | (−1.73) | 180.42 | (−1.71) |
| C61 | 180.73 | 178.23 | (2.50) | 177.63 | (3.10) |
Only chemical shifts which differ by ≥1.00 ppm from those for NOCbl are shown.
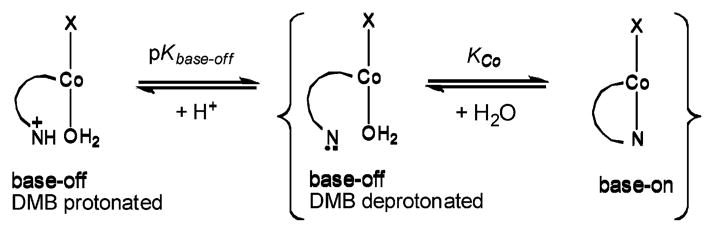
As depicted in Scheme 1, in aqueous solution Cbls exist in three forms: base-on, base-off with the 5,6-dimethylbenzimidazole (DMB) deprotonated, and base-off with the DMB protonated. Under more acidic conditions, Cbl species that are mono and diprotonated at the phosphodiester also exist (pKa ~ −0.1 and −1.653). The ground state structural and thermodynamic trans influence of the β-axial ligand of Cbls is well established, with strong σ donor ligands resulting in long Co-NB3 bond distances, small KCo values (Scheme 1) and higher pKbase-off values.54–56
Scheme 1.
Cobalamin forms present in aqueous solution.
A pK base-off value of 5.1 was recently reported for NOCbl using UV-vis and 1H NMR spectroscopy,18 which is the highest reported value for any Cbl (pKbase-off for other Cbls are in the −2.1 to 4.1 range53). We also obtained a similar value using 1H NMR spectroscopy. Using pKaBz = 5.56,57 a value of KCo = 1.9 is calculated; that is, under neutral conditions approximately one third of NOCbl exists in a DMB deprotonated, base-off form, Scheme 1.
It was of interest to us to determine pKbase-off for NO2Cbl for comparison purposes. To our knowledge, this value has not been reported. Upon conversion of Cbls to their base-off, protonated forms, large changes are observed in the aromatic region of the 1H NMR spectrum.18 However, measuring 1H NMR spectra for acidic solutions of NO2Cbl at varying pD values showed that hydrolysis of NO2Cbl to H2OCbl+ occurs prior to formation of significant amounts of protonated, base-off NO2Cbl. For example, at pD 0.8, only ~18% NO2Cbl remains, with chemical shifts identical within experimental error to those for base-on NO2Cbl at pD 6.7. Assuming that at this pD condition ≥90% NO2Cbl is base-on, a value of pKbase-off ≤ −0.15 is calculated. Hence unlike NOCbl, pKbase-off NO2Cbl is within the range expected for Cbls with inorganic ligands at the β-axial site (pKbase-off (H2OCbl+) =−2.13, pKbase-off (CNCbl) = 0.153,58).
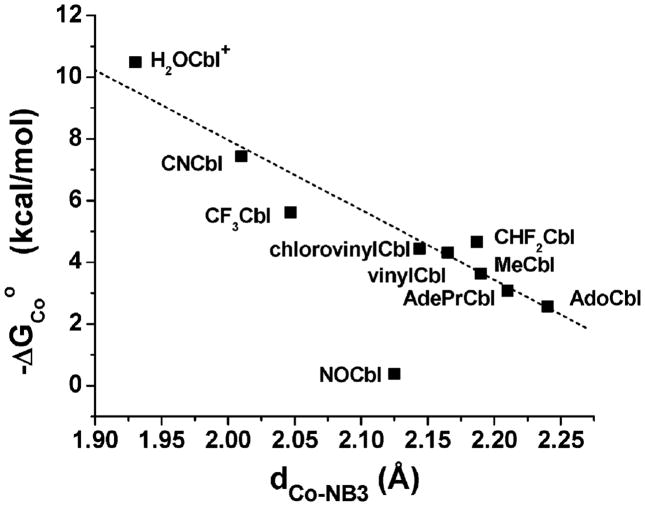
A linear relationship between the apparent Gibbs free energy for coordination of 5,6-dimethylbenzimidazole to cobalt, −ΔGCoo, and the Co-NB3 bond distance, dCo–NB3, has previously been established.59,60 Using KCo = 1.9, a value of 0.38 kcal mol−1 is calculated for NOCbl. Fig. 2 gives a plot of −ΔGCoo versus dCo–NB3 for a series of Cbls, including NOCbl. −ΔGCoo is remarkably small for NOCbl, in accordance with the high pKbase-off value. It is apparent from this plot that NOCbl deviates significantly from the line of best fit for other non-alkyl and alkylcobalamins, and is closer to that expected for an alkylcobalamin.
Fig. 2.
Plot of −ΔGCoo versus the Co–NB3 bond distance for Cbls. The data has been fitted to a straight line (r2 = 0.93; data for NOCbl excluded) with slope = −22.6 ± 2.4 and intercept 53.2 ± 5.1. Co-NB3 bond lengths and −ΔGCoo values are taken from the literature.23,53,59,60,66,67,70,79,80–82
Given that the pKbase-off value of 5.1 for NOCbl is unexpectedly high, it is informative to evaluate the importance of the DMB deprotonated, base-off conformation by other means. The chemical shift of the DMB carbons have been shown to be sensitive to the strength of coordination of the axial DMB to the cobalt atom in base-on Cbls.61,62 Table 3 shows a comparison of the 13C DMB chemical shifts of adenosylcobalamin (AdoCbl), methylcobalamin (CH3Cbl), cyanocobalamin (CNCbl), and aquacobalamin (H2OCbl+) to those of NOCbl and those of two base-off (but DMB deprotonated) Cbls, dicyanocobalamin, (CN)2Cbl− and α-AdoCbl (Ado (= 5′-deoxyadenosyl) ligand in the lower or α-axial position). Clearly, the DMB of NOCbl more closely resembles that of AdoCbl or CH3Cbl, where the equilibrium constant for formation of the base-on species from the base-off, DMB deprotonated species, KCo, is of the order of 101 to 102, than that of CNCbl or H2OCbl+ where KCo is >105. Indeed, the 13C NMR chemical shifts of the DMB of NOCbl are quite close to those of the base-off species (CN)2Cbl− and α-AdoCbl, Table 3, which again suggests that the base-off, DMB deprotonated conformation of NOCbl (Scheme 1) is important in neutral, aqueous solution.
Table 3.
| Carbon | AdoCbl | CH3Cbl | CNCbl | H2OCbl+ | NOCbl | (CN)2Cbl− | α-AdoCbl |
|---|---|---|---|---|---|---|---|
| B7 | 113.14 | 113.12 | 114.04 | 114.50 | 113.43 | 113.63 | 113.89 |
| B4 | 120.98 | 120.83 | 119.00 | 118.00 | 121.29 | 121.59 | 121.38 |
| B8 | 133.03 | 133.06 | 132.49 | 131.82 | 134.21 | 134.15 | 134.29 |
| B5 | 136.27 | 136.28 | 135.60 | 136.41 | 133.99 | 135.07 | 135.91 |
| B6 | 136.40 | 136.37 | 137.67 | 137.52 | 135.75 | 135.92 | 135.09 |
| B9 | 140.62 | 140.75 | 139.25 | 138.70 | 142.20 | 142.96 | 142.40 |
| B2 | 144.20 | 144.40 | 144.39 | 144.70 | 144.53 | 145.22 | 145.20 |
| KCoa | 76.6 | 467 | 2.88 × 105 | 4.90 × 107 | 0b | 0b |
One of us has previously shown that for a series of 10 Cbls, the 13C NMR chemical shift of the DMB carbons of the nucleotide loop also correlates directly with ΔGCoo.61 Table 4 shows attempts to use the correlations of the DMB chemical shifts with the Gibbs free energy of formation of the base-on species, ΔGCoo, from this work to estimate the value of KCo and pKbase-off for NOCbl. The first three columns show the slope, intercept, and correlation coefficient of the correlation of each DMB carbon with ΔGCoo for the 10 Cbls studied. The B7 carbon was omitted, since the chemical shifts of the alkylcobalamins do not fall between those of XCbls (X = CN−, H2O) and the base-off Cbls. The B2 carbon was also omitted, since its chemical shift shows little dependence on ΔGCoo. The correlations for B4, B5, and B6 all give values of KCo for NOCbl on the order of 10, i.e., similar to an alkylcobalamin with a strongly electron donating alkyl ligand such as nPrCbl. The B8 and B9 correlations give KCo values ≪ 1 for NOCbl, characteristic of a base-off Cbl. The value of 1.9 determined from pKbase-off = 5.1 lies in between these two extremes. Importantly, although the lack of consistency of these estimates is somewhat disconcerting, it is consistent with NOCbl being an outlier, as observed in the −ΔGCo versus dCo–NB3 correlation (Fig. 2), and in the 31P NMR chemical shift versus dCo–NB3 correlation (Fig. 3, see below), albeit to a much lesser extent. Furthermore all of the correlations suggest that in solution NOCbl behaves more like an alkylcobalamin, with a covalent upper axial Co–X bond, than an inorganic Cbl with a dative Co–X bond, consistent with the deprotonated base-off conformation of NOCbl being an important species in neutral aqueous solution.
Table 4.
Estimates of KCo and pKbase-off for NOCbl from the DMB 13C NMR chemical shiftsa
| Correlation |
Calculated for NOCbl |
|||||
|---|---|---|---|---|---|---|
| Carbon | Slope (ppm kcal−1) | Intercept (ppm) | r2 | ΔGCoo (kcal mol−1) | KCo | pKbase-off |
| B4 | 0.380 ± 0.025 | 122.10 ± 0.14 | 0.966 | −2.14 | 37.1 | 3.98 |
| B5 | −0.306 ± 0.016 | 133.087 ± 0.084 | 0.980 | −2.17 | 39.0 | 3.96 |
| B6 | −0.282 ± 0.014 | 135.437 ± 0.076 | 0.981 | −1.11 | 6.56 | 4.68 |
| B8 | 0.151 ± 0.010 | 133.420 ± 0.055 | 0.966 | 5.22 | 1.49 × 10−4 | 5.56 |
| B9 | 0.277 ± 0.020 | 141.64 ± 0.11 | 0.960 | 2.67 | 1.10 × 10−2 | 5.56 |
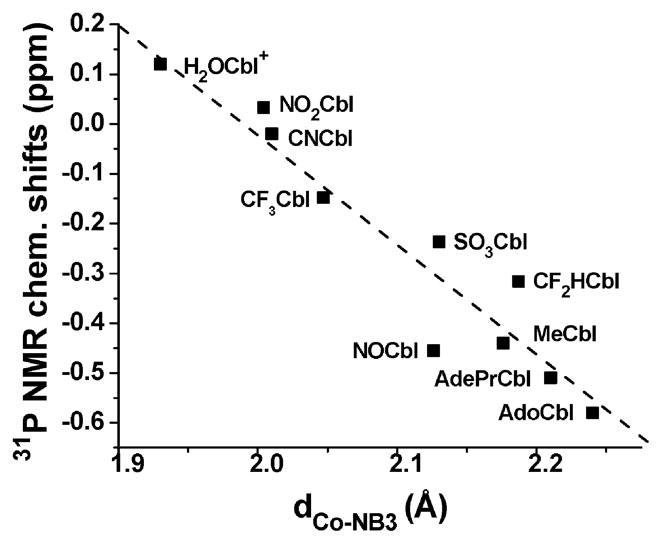
Fig. 3.
Plot of 31P NMR chemical shift versus the Co–NB3 bond distance for Cbls (in D2O or H2O, referenced to 85% H3PO4, 25 °C). The data has been fitted to a straight line (r2 = 0.91) with slope = −2.26 ± 0.26 and intercept 4.51 ± 0.54. 31P NMR chemical shifts for SO3Cbl (−0.24 ppm) NO2Cbl (0.03 ppm) and NOCbl (−0.45 ppm, in excellent agreement with a literature value18) were determined in this study. All other 31P NMR chemical shift data taken from the literature.53,59 Co–NB3 bond lengths taken from reported structures.23,64,66–68,70,73,79–82
It has also previously been shown that the 31P NMR chemical shift of the nucleotide phosphodiester moiety of Cbls correlates directly with the Co–NB3 bond distance.53,59,63,64 The 31P NMR chemical shift reflects changes in the phosphodiester bond angles, which become less strained as the Co–NB3 bond lengthens.53,63 A plot of 31P NMR chemical shift versus Co–NB3 bond distance, dCo–NB3, is given in Fig. 3. In line with the −ΔGCoo versus dCo–NB3 plot (Fig. 2), the 31P NMR chemical shift of NOCbl is unexpectedly low and is closer to 31P NMR shifts of alkylcobalamins rather than other Cbls with inorganic ligands. A single 31P NMR resonance is observed for NOCbl under neutral and acidic conditions; hence on the NMR time scale, the base-on and base-off forms of NOCbl are in fast exchange, as observed for alkylcobalamins (CH3Cbl, nPrCbl, CF3CHCbl, CF2HCbl, CF3Cbl and NCCH2Cbl53). The same is not true for CNCbl and H2OCbl+, for which two distinct peaks corresponding to the base-on and base-off, DMB protonated forms are observed in acidic solution at pH(pD) values close to pK base-off53,58
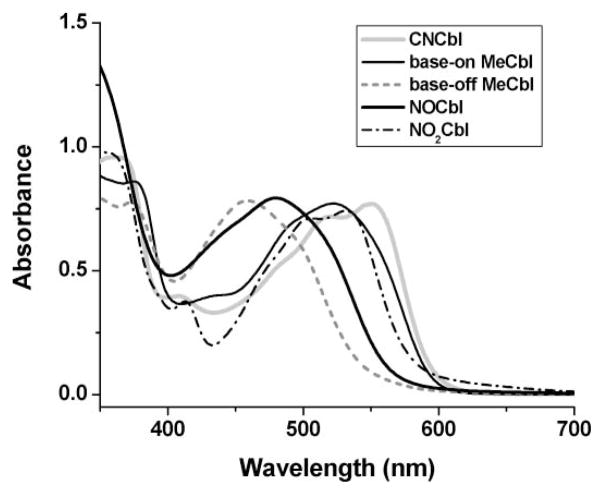
Large spectral shifts of the αβ band region (400–600 nm) of the UV-visible spectrum to higher energy accompanied by a colour change from red to yellow occur upon conversion of base-on XCbls to their deprotonated (or protonated) base-off forms.65 In Fig. 4, the UV-vis spectrum of NOCbl is compared with MeCbl, protonated base-off MeCbl, CNCbl and NO2Cbl. The αβ band region of NOCbl is noticeably shifted towards shorter wavelengths compared with the other XCbls with inorganic ligands (X = CN−or NO2−), providing further compelling evidence that the base-off, DMB-deprotonated form is especially important for NOCbl under neutral pH conditions. The spectral differences observed in Fig. 4 are also reflected in the colours of the solutions. Whilst CNCbl (KCo > 105, Scheme 1) and NO2Cbl are red, MeCbl (KCo = 467) is red-orange, NOCbl (KCo = 1.9) is bright orange and base-off, protonated MeCbl is yellow.
Fig. 4.
Visible region spectra of selected Cbls (6.0 × 10−5 M) in H2O or at pH 1.37 (base-off MeCbl only), 25°C.
Importantly, there was no evidence for a deprotonated, base-off NOCbl species in the X-ray diffraction electron density map for NOCbl;23 hence it appears that significant amounts of this form occurs in solution only. However, the finding that the NO ligand behaves electronically more like an alkyl ligand rather than a typical inorganic ligand is supported by the X-ray structural data. For NOCbl, the Co–NB3 bond length is 2.13 Å,23 which is similar in length to that reported for MeCbl (2.16 Å66). The Co–NB3 bond lengths for Cbls with typical inorganic ligands such as CN−, NO2−, H2O and N-acetyl-L-cysteine are 2.01–2.04, 1.99–2.01, 1.93, and 2.06 Å, respectively.41,67–70 In pentammine complexes of Co, [CoX(NH3)5]n+, NO was found to exert an even stronger trans influence than Me.71 Interestingly, the Co–NB3 bond distance for AdoCbl (2.24 Å72) is considerably longer than that for MeCbl (2.16 Å66). Finally, like NOCbl, sulfitocobalamin (SO3Cbl) also has a longer Co–NB3 bond distance (2.13–2.15 Å73,74) compared with other Cbls with β-axial inorganic ligands. The longer Co–NB3 for SO3Cbl and NOCbl can be attributed to the strong σ donor properties of these ligands, in addition to NO being a moderately good π acceptor ligand.75
Molecular dynamics and simulated annealing
Table S2† lists all the NOE’s, their assigned strength, and the average, standard deviation, maximum, minimum and median H–H distances during a 500 ps molecular dynamics simulation at 300 K of the base-on form of NOCbl. Given the clear importance of a base-off form of NOCbl in solution (vide supra) we then broke the Co–NB3 bond and repeated the simulation for the base-off form; the data are also listed in Table S2.
The modelling accords well with the solution structure of NOCbl. Of the 64 NOE’s only 3 (4.7%) violate the distance criteria outlined above in both the base-on and the base-off forms of NOCbl. The distance between C26′ and C3 (2.55 and 2.56 Å on average for the base-on and base-off forms, respectively) accords with a strong NOE, but that between C3 and C26″ is too long (3.53 Å for both forms of NOCbl); this is in line with the relative inflexibility we see in the a side chain (vide infra). The C53 and C18 protons are well separated (the average modelled distance is 5.08 Å for base-on NOCbl and 5.24 Å for base-off NOCbl); that this gives rise to a clear, albeit weak NOE, is puzzling. The average modelled distance between the B7 proton and the proton on R4 is long even for a very weak NOE (5.67 and 5.69 Å for base-on and base-off NOCbl, respectively) but we note the wide range of inter-proton distances (6.41 to 2.32 Å, and 6.41 to 4.42 Å, respectively) and the relatively large standard deviation of the distance (0.21 and 0.25 Å, respectively) and conclude that the violation is marginal.
Two NOE’s violate the distance criteria in the base-on form of NOCbl. The distance violation between C56′ and C18 is marginal in the base-on form (the average distance is 3.36 Å) but is very well accounted for in the base-off form (average 2.56 Å). The distance between the fH proton and that on R3 (5.09 Å in the base-on form) is too long for even a weak NOE, but is also well accounted for in the modelling of the base-off form (3.04 Å). There is a single NOE that is compatible only with the base-on form. The weak NOE between C56″ and the R4 proton cannot be due to the base-off form (average 5.92 Å) but is within the expected distance in the base-on form (average 4.53 Å).
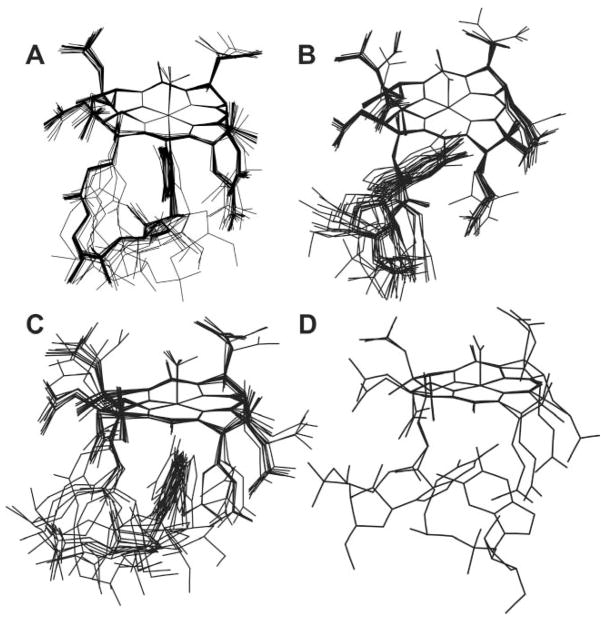
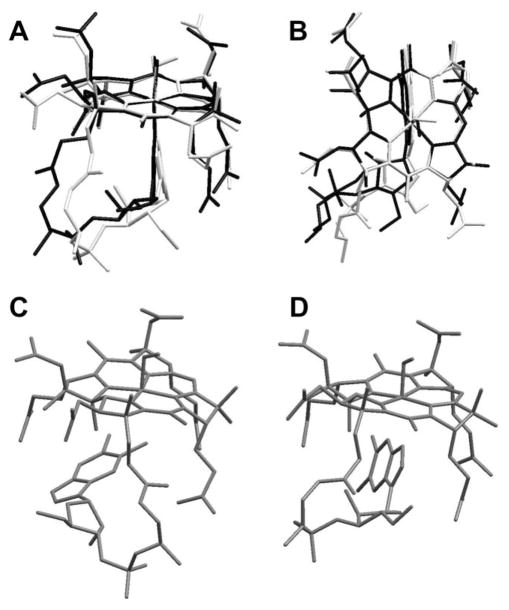
Simulated annealing calculations were performed on both the base-on form of NOCbl (omitting the C56′–C18 and the fH–R3 NOE’s) and the base-off form (omitting the C56″–R4 NOE). Fig. 5A shows 25 annealed structures for base-on NOCbl overlaid at the Co ion and four corrin N donor atoms. Two views of the consensus structure of base-on NOCbl are shown in Fig. 6A and 6B (in black) overlaid with the crystal structure of NOCbl (white). Base-off NOCbl annealed into two principal conformations. In the first (Fig. 5C; the consensus structure is shown in Fig. 6D) the base is perpendicular to the corrin and the B3 nitrogen is about 3.1 Å away from and pointing directly at the metal ion. In the second structure (Fig. 5B, consensus structure in Fig. 6C), the base is virtually parallel to and tucked beneath the D ring of the corrin. Two other annealed structures in which the base is more remote from the metal were also identified and are shown in Fig. 5D.
Fig. 5.
Superposition at the Co centre and the four corrin N atoms of (A) 25 annealed structures of base-on NOCbl; (B) 13 annealed structures of base-off NOCbl with DMB parallel to corrin and anti to ribose; (C) 10 annealed structure of base-off NOCbl with DMB perpendicular to corrin; and (D) 2 annealed structures of base-off NOCbl with DMB parallel to corrin and syn to ribose.
Fig. 6.
(A) and (B) show two views of the consensus structure obtained from MD/SA calculations of base-on NOCbl (dark lines) overlaid with the crystal structure of NOCbl (light lines). (C) and (D) show the consensus structures of the two main classes of structures into which base-off NOCbl annealed.
Structural metrics
In Table S3† we compare some structural metrics of the base-on consensus structure and the crystal structure. The O atom of the NO ligand in the modelled structures, as in the crystal structure, occupies three different positions. When we developed the force field parameters for modelling the corrins38 we surveyed the structures of the then available Co(III) corrins and based our force field parameters on the averages of the structural metrics. We found that the average Co–N bond length to N21 and N24 is 1.885(38) Å, whilst that to N22 and N23 is longer, 1.924(44) Å. This is well-reproduced in the modelling of NOCbl (Table S3). The Co–N bond length to coordinated NO and the N–O bond lengths are well-reproduced (1.175 versus an average of 1.176 Å in the X-ray structure). The Co–NB3 bond length to coordinated DMB in the consensus structure (2.063 Å) is significantly shorter than that observed in the X-ray structure of NOCbl (2.123 Å). It is well established that the Co–NB3 bond length is influenced by the electronic donor power of the trans β ligand.54,55 Since MM methods are insensitive to such electronic effects, when modelling alkylcobalamins27 we use a different parameter for the Co–NB3 bond (ks = 2.8 mdyn Å−1, lo = 2.138 Å) than when modelling cobalt corrins where the β ligand is an “inorganic” moiety such as CN−, H2O or S2O32− (ks =2.8 mdyn Å−1, lo = 2.010 Å) The Co–NB3 bond length of 2.063 Å (or 0.06 Å too short) was produced using the “inorganic” parameter; it increased to 2.173 Å (i.e., 0.05 Å too long) when we used the parameter appropriate for modelling alkylcobalamins. Thus, NO behaves in NOCbl somewhat like an alkyl ligand of moderate trans influence. For convenience, all modelling was performed with the “inorganic” parameters and the axial bond lengths to coordinated DMB are too short by about 0.06 Å.
The C–N bond lengths observed in the X-ray structure of NOCbl are within the range for cobalt corrins (N21–C1, N24–C19, 1.49(4) Å; N21–C4, N24–C16, 1.31(4) Å; N22, N23–C, 1.37(5) Å) and are well reproduced in the modelling. The same is true for the C–C bonds with double bond character (crystallographic mean 1.40(7) Å) and the C–C single bonds (crystallographic mean 1.55(6) Å).
Bond angles are generally well-reproduced (Table S3†); the average difference between the base-on consensus structure and the solid state structure is 1.5° and 85% of the bond angles reported in Table 2 are reproduced to better than 2.5°.
The corrin fold angle is defined as the angle between the mean planes through N21, C4, C5, C6, N22, C9 and C10, and through N24, C16, C15, C14, N23, C11 and C10.76 In the X-ray structure of NOCbl the corrin has a fold angle of 15.9°. The consensus structure is slightly more folded (16.8°) and changing the parameters for modelling the Co–NB3 bond thus elongating that bond (vide supra) changes the fold to 15.3°, lower than the X-ray structure value. The fold angles of the 25 annealed structures from which the consensus structure was derived vary between 9.8° and 18.9° (mean 14.4(3.1)°).
The annealed structures
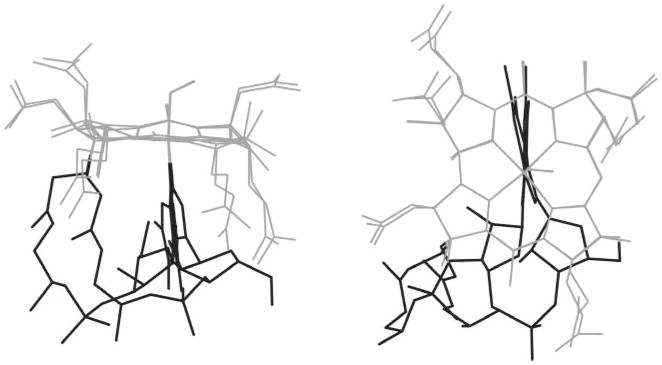
The annealed structures illustrated in Fig. 5 provide insight into the considerable flexibility of NOCbl. In both the base-on and base-off forms, the O atom of the NO ligand occupies one of three positions, very similar to the positions observed in the crystal structure. In the base-on form the a, d and g side chains are all in approximately the same orientation, whilst the b, c and e side chains occupy a number of orientations. In the base-off form the a and d side chains are more flexible. In the base-on form the greatest variability is seen in the f side chain and the nucleotide loop; this leads to a variety of dispositions of the ribose and DMB relative to the corrin. Fig. 7 shows the extremes; at one extreme DMB is parallel to the C5 ···C15 line and the ribose is beneath the C15–C16 bond and at the other extreme DMB has rotated by 17° in a counter clockwise fashion towards C4 when viewed from above the corrin and the ribose is nearly beneath the C ring. We have previously noted this considerable flexibility in the f side chain in the modelling of other cobalamins.37
Fig. 7.
Superposition of base-on NOCbl showing the extremes of the orientation of the f side chain and the nucleotide loop in the annealed structure. This results in a range of orientations of the ribose and the DMB relative to the corrin.
There is little variation in the A ring of the corrin in the base-on form. C1, N21 and C4 are planar with C2 and C3 below the plane. Similarly, in the B ring C6, N22 and C9 are planar, C7 is above the plane and C8 below. On the other hand, both the C and D rings show considerable flexibility. The C ring adopts a range of conformations. At one extreme the ring is virtually planar, and both the C46 and C47 methyl groups are in a pseudo equatorial position. At the other, C11, N23 and C14 are planar, C12 is above the plane so that the C46 methyl group is in an axial position whilst the C47 methyl is equatorial, and C13 is below the plane. The D ring adopts a range of conformations between two extremes. In the first, C16, N24, C17 and C19 are virtually coplanar and C18 is displaced towards the α face of the corrin, whilst in the second C17, C18, C19 and N24 are coplanar and C16 is displaced towards the β face.
As might be expected, untethering of the nucleotide loop imparts greater flexibility to the corrin, and especially the f side chain; however, in all annealed structures of the base-off form of NOCbl that we obtained DMB remains close to the α face of the corrin. Annealing produced structures that fit broadly into one of three classes. In the first (13 of the 25 structures, 52%, Fig. 5B) DMB is anti to the ribose, and nearly parallel to the corrin beneath the A ring (Fig. 6C shows the consensus structure). The distance between NB3 and Co ranges between 7.4 and 8.3 Å (average 7.9(2) Å; 7.78 Å in the consensus structure). In the second (10 structures, 30%, Fig. 5C), DMB is perpendicular to the corrin with the distance between Co and NB3 between 2.9 and 3.8 Å (3.11 Å in the consensus structure, Fig. 6D). In two structures (8%, Fig. 5D) DMB annealed syn to ribose and is nearly parallel to the corrin. As only two structures were found we could not determine a consensus structure for what is likely to be a minor conformation of base-off NOCbl.
In a 250 ps dynamics simulation at 300 K, in which the distance between Co and NB3 and between C20 and NB3 was monitored every 10 fs, the largest distance between Co and NB3 was 8.35 Å and that between C20 and NB3 was 5.94 Å (see Figure S1 of the ESI†). If all the NOE’s are removed, the nucleotide loop swung out by rotating about the Pr1–Pr2 bond (see Fig. 1) and the structure energy-minimised, the distance between C20 and NB3 is over 17 Å whilst the Co ···NB3 distance is over 18 Å. Hence the 300 K dynamics simulation shows that DMB remains tucked in near the α face throughout the dynamics trajectory. Removing the weak NOE between C56″H and R4H, which is unique to the base-off species, and repeating the dynamics trajectory does not significantly alter the results. It is clear that, probably because of the sentinel-like effect of the three propionamide side chains and the C20 methyl that point downward towards the α face, the DMB ligand does not readily move from the vicinity of the corrin’s α face. These results are in broad agreement with those we obtained previously when we modelled a base-off cobalamin, methyl-3,5,6-trimethylbenzimidazolylcobamide (CH3Me3BzmCba+), in which the B3 nitrogen of DMB is blocked by a methyl group.33 In that case we found that in the absence of tethering by the metal ion, the nucleotide loop has considerable motional freedom but that in an extended dynamics simulation at 300 K the base also remains near the α face of the corrin.
Conclusion
1H and 13C NMR chemical shifts of NOCbl have been assigned using 2-D NMR techniques. Interproton distances obtained by NOE experiments have been used in conjunction with molecular mechanics to obtain the structure of NOCbl in solution, with over 95% agreement between the resulting structures and the NOE data. 13C and 31P NMR chemical shifts, pKbase-off/−ΔGCoo and UV-vis spectra of NOCbl compared with other Cbls suggest that the NO ligand has electronic properties significantly differing from inorganic ligands such as CN− or H2O, closer to the covalently bound alkyl ligands of alkylcobalamins. Furthermore, pKbase-off, 13C NMR shifts and the visible spectrum of NOCbl are consistent with the base-off DMB deprotonated form being much more important for NOCbl in solution compared with all other known Cbl species, including alkylcobalamins. Molecular modelling was performed separately on a base-on and on a base-off form of NOCbl. The solution structure of the base-on form is similar to that observed by X-ray diffraction, with minor differences in the conformationally flexible side chains and in the position of the nucleotide loop. Modelling of the base-off form shows that DMB can adopt a range of positions relative to the corrin, but remains in close proximity of the macrocycle’s α face. It was not possible to obtain a reliable estimate of the Co–NB3 bond distance from the molecular mechanics calculations, since these calculations do not take into account electronic effects experienced by the Co–NB3 from the β-axial NO (the trans influence); however MM calculations using parameters typical for either β-axial inorganic or alkyl ligands provide further support that NO has electronic properties between those of inorganic and alkyl ligands.
Supplementary Material
Acknowledgments
We wish to acknowledge funding of this research from Kent State University (NEB), the South African Department of Science and Technology and the National Research Foundation, Pretoria, (HMM), the University of the Witwatersrand (HMM), and the Egyptian Ministry of Higher Education (HAH).
Footnotes
Electronic supplementary information (ESI) available: Table S1–S3 and Fig. S1. See DOI: 10.1039/b810895a
References
- 1.Banerjee R, editor. Chemistry and Biochemistry of B12. Wiley; New York: 1999. [Google Scholar]
- 2.Carmel R. In: Homocysteine in Health and Disease. Jacobsen DW, Carmel R, editors. Cambridge University Press; Cambridge: 2001. pp. 289–305. [Google Scholar]
- 3.Brouwer M, Chamulitrat W, Ferruzzi G, Sauls DL, Weinberg JB. Blood. 1996;88:1857–1864. [PubMed] [Google Scholar]
- 4.Wheatley C. Med Hypotheses. 2006;67:124–142. doi: 10.1016/j.mehy.2006.01.036. [DOI] [PubMed] [Google Scholar]
- 5.Wheatley C. J Nutr Environ Med. 2007;16:181–211. doi: 10.1080/10520290701791839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Valko M, Rhodes CJ, Moncol J, Izakovic M, Mazur M. Chem Biol Interact. 2006;160:1–40. doi: 10.1016/j.cbi.2005.12.009. [DOI] [PubMed] [Google Scholar]
- 7.McInnes IB, Liew FY. In: Nitric Oxide in Bone and Joint Disease. Hukkanen MVJ, Polak JM, Hughes SPF, editors. Cambridge University Press; Cambridge: 1998. pp. 8–20. [Google Scholar]
- 8.Rand MJ, Li CG. Eur J Pharmacol. 1993;241:249–254. doi: 10.1016/0014-2999(93)90210-9. [DOI] [PubMed] [Google Scholar]
- 9.Greenberg SS, Xie J, Zatarain JM, Kapusta DR, Miller MJ. J Pharmacol Exp Ther. 1995;273:257–265. [PubMed] [Google Scholar]
- 10.Schubert R, Krien U, Wulfsen I, Schiemann D, Lehmann G, Ulfig N, Veh RW, Schwarz JR, Gago H. Hypertension. 2004;43:891–896. doi: 10.1161/01.HYP.0000121882.42731.6b. [DOI] [PubMed] [Google Scholar]
- 11.Jiang F, Li CG, Rand MJ. Eur J Pharmacol. 1997;340:181–186. doi: 10.1016/s0014-2999(97)01381-2. [DOI] [PubMed] [Google Scholar]
- 12.Weil M, Abeles R, Nachmany A, Gold V, Michael E. Cell Death Differ. 2004;11:361–363. doi: 10.1038/sj.cdd.4401371. [DOI] [PubMed] [Google Scholar]
- 13.Nicolaou A, Kenyon SH, Gibbons JM, Ast T, Gibbons WA. Eur J Clin Invest. 1996;26:167–170. doi: 10.1046/j.1365-2362.1996.122254.x. [DOI] [PubMed] [Google Scholar]
- 14.Nicolaou A, Ast T, Garcia CV, Anderson MM, Gibbons JM, Gibbons WA. Biochem Soc Trans. 1994;22:296S. doi: 10.1042/bst022296s. [DOI] [PubMed] [Google Scholar]
- 15.Danishpajooh IO, Gudi T, Chen Y, Kharitonov VG, Sharma VS, Boss GR. J Biol Chem. 2001;276:27296–27303. doi: 10.1074/jbc.M104043200. [DOI] [PubMed] [Google Scholar]
- 16.Kambo A, Sharma VS, Casteel DE, Woods VL, Jr, Pilz RB, Boss GR. J Biol Chem. 2005;280:10073–10082. doi: 10.1074/jbc.M411842200. [DOI] [PubMed] [Google Scholar]
- 17.Nicolaou A, Waterfield CJ, Kenyon SH, Gibbons WA. Eur J Biochem. 1997;244:876–882. doi: 10.1111/j.1432-1033.1997.00876.x. [DOI] [PubMed] [Google Scholar]
- 18.Wolak M, Zahl A, Schneppensieper T, Stochel G, van Eldik R. J Am Chem Soc. 2001;123:9780–9791. doi: 10.1021/ja010530a. [DOI] [PubMed] [Google Scholar]
- 19.Zheng D, Birke RL. J Am Chem Soc. 2001;123:4637–4638. doi: 10.1021/ja015682k. [DOI] [PubMed] [Google Scholar]
- 20.Zheng D, Birke RL. J Am Chem Soc. 2002;124:9066–9067. doi: 10.1021/ja017684a. [DOI] [PubMed] [Google Scholar]
- 21.Zheng D, Yan L, Birke RL. Inorg Chem. 2002;41:2548–2555. doi: 10.1021/ic010802a. [DOI] [PubMed] [Google Scholar]
- 22.Wolak M, Stochel G, van Eldik R. Inorg Chem. 2006;45:1367–1379. doi: 10.1021/ic051300q. [DOI] [PubMed] [Google Scholar]
- 23.Hannibal L, Smith CA, Jacobsen DW, Brasch NE. Angew Chem Int Ed Engl. 2007;46:5140–5143. doi: 10.1002/anie.200701131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wolak M, Stochel G, Hamza M, van Eldik R. Inorg Chem. 2000;39:2018–2019. doi: 10.1021/ic991266d. [DOI] [PubMed] [Google Scholar]
- 25.Roncaroli F, Shubina TE, Clark T, van Eldik R. Inorg Chem. 2006;45:7869–7876. doi: 10.1021/ic061151r. [DOI] [PubMed] [Google Scholar]
- 26.Holleman-Wiberg . In: Inorganic Chemistry. 34. Wiberg N, editor. Academic Press, Walter de Gruyter; Berlin: 2001. pp. 1582–1587. [Google Scholar]
- 27.Marques HM, Brown KL. Inorg Chem. 1995;34:3733–3740. [Google Scholar]
- 28.Brown KL, Marques HM. Polyhedron. 1996;15:2187–2197. [Google Scholar]
- 29.Brown KL, Cheng S, Marques HM. Inorg Chem. 1995;34:3038–3049. [Google Scholar]
- 30.Brown KL, Cheng S, Marques HM. Polyhedron. 1998;17:2213–2224. [Google Scholar]
- 31.Brown KL, Cheng S, Zou X, Chen G, Valente EJ, Zubkowski JD, Marques HM. Biochemistry. 1998;37:9704–9715. doi: 10.1021/bi980707m. [DOI] [PubMed] [Google Scholar]
- 32.Marques HM, Hicks RP, Brown KL. J Chem Soc Chem Commun. 1996:1427–1428. [Google Scholar]
- 33.Brown KL, Zou X, Marques HM. J Mol Struct (Theochem) 1998;453:209–224. [Google Scholar]
- 34.Marques HM, Brown KL. J Mol Struct. 2000;520:75–95. [Google Scholar]
- 35.Brown KL, Zou X, Chen GD, Xia ZP, Marques HM. J Inorg Biochem. 2004;98:287–300. doi: 10.1016/j.jinorgbio.2003.10.016. [DOI] [PubMed] [Google Scholar]
- 36.Brown KL, Zou X, Banka RR, Perry CB, Marques HM. Inorg Chem. 2004;43:8130–8142. doi: 10.1021/ic040079z. [DOI] [PubMed] [Google Scholar]
- 37.Perry CB, Brown KL, Zou X, Marques HA. J Mol Struct. 2005;737:245–258. [Google Scholar]
- 38.Marques HM, Brown KL. J Mol Struct (Theochem) 1995;340:97–124. [Google Scholar]
- 39.Marques HM, Warden C, Monye M, Shongwe MS, Brown KL. Inorg Chem. 1998;37:2578–2581. [Google Scholar]
- 40.Allinger NL. J Am Chem Soc. 1977;99:8127–8134. [Google Scholar]
- 41.Suarez-Moreira E, Hannibal L, Smith CA, Chavez RA, Jacobsen DW, Brasch NE. Dalton Trans. 2006:5269–5277. doi: 10.1039/b610158e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Brown KL. Chapter 7:197. Ref. 1. [Google Scholar]
- 43.HYPERCHEM. Hypercube, Inc.; Gainesville, FL: 2002. [Google Scholar]
- 44.Kadish KM, Ou Z, Tan X, Boschi T, Monti D, Fares V, Tagliatesta P. J Chem Soc, Dalton Trans. 1999:1595–1602. [Google Scholar]
- 45.Ellison MK, Scheidt WR. Inorg Chem. 1998;37:382–383. doi: 10.1021/ic971109j. [DOI] [PubMed] [Google Scholar]
- 46.Godbout N, Sanders LK, Salzmann R, Havlin RH, Wojdelski M, Oldfield E. J Am Chem Soc. 1999;121:3829–3844. [Google Scholar]
- 47.Jene PG, Ibers JA. Inorg Chem. 2000;39:5796–5802. doi: 10.1021/ic0006753. [DOI] [PubMed] [Google Scholar]
- 48.Clore GM, Gronenborn AM. Science. 1991;252:1390–1399. doi: 10.1126/science.2047852. [DOI] [PubMed] [Google Scholar]
- 49.Clore GM, Brünger AT, Karplus M, Gronenborn AM. J Mol Biol. 1986;191:553–561. doi: 10.1016/0022-2836(86)90146-4. [DOI] [PubMed] [Google Scholar]
- 50.Allen MP, Tildesley DJ. Computer Simulation of Liquids. Claredon; Oxford: 1987. [Google Scholar]
- 51.Saiz E, Tarazona MP. J Chem Educ. 1997;74:1350–1354. [Google Scholar]
- 52.Berendsen HJC, Postma JPM, van Gunsteren WF, DiNola A, Haak JR. J Chem Phys. 1984;81:3684–3690. [Google Scholar]
- 53.Brown KL, Hakimi JM, Jacobsen DW. J Am Chem Soc. 1984;106:7894–7899. [Google Scholar]
- 54.Kratky C, Kraeutler B. Chapter 2:9. Ref. 1. [Google Scholar]
- 55.Pratt JM. Inorganic Chemistry of Vitamin B12. Academic Press; London, New York: 1972. [Google Scholar]
- 56.Hayward GC, Hill HAO, Pratt JM, Vanston NJ, Williams RJP. J Chem Soc. 1965:6485–6493. [PubMed] [Google Scholar]
- 57.Brown KL, Hakimi JM, Nuss DM, Montejano YD, Jacobsen DW. Inorg Chem. 1984;23:1463–1471. [Google Scholar]
- 58.Brown KL, Hakimi JM. Inorg Chem. 1984;23:1756–1764. [Google Scholar]
- 59.Brown KL, Evans DR, Zubkowski JD, Valente EJ. Inorg Chem. 1996;35:415–423. doi: 10.1021/ic950951r. [DOI] [PubMed] [Google Scholar]
- 60.McCauley KM, Pratt DA, Wilson SR, Shey J, Burkey TJ, van der Donk WA. J Am Chem Soc. 2005;127:1126–1136. doi: 10.1021/ja048573p. [DOI] [PubMed] [Google Scholar]
- 61.Brown KL, Hakimi JM. J Am Chem Soc. 1986;108:496–503. doi: 10.1021/ja00263a023. [DOI] [PubMed] [Google Scholar]
- 62.Note that in this paper the assignments of the B5 and B6 carbons are interchanged as this work was completed prior to the availability of NMR methods for the unambiguous assignment of 13C resonances in Cbls.
- 63.Brown KL. Inorg Chem. 1986;25:3111–3113. [Google Scholar]
- 64.Rossi M, Glusker JP, Randaccio L, Summers MF, Toscano PJ, Marzilli LG. J Am Chem Soc. 1985;107:1729–1738. [Google Scholar]
- 65.Chemaly SM, Pratt JM. J Chem Soc, Dalton Trans. 1980:2267–2273. [Google Scholar]
- 66.Randaccio L, Furlan M, Geremia S, Slouf M, Srnova I, Toffoli D. Inorg Chem. 2000;39:3403–3413. doi: 10.1021/ic0001199. [DOI] [PubMed] [Google Scholar]
- 67.Kraeutler B, Konrat R, Stupperich E, Faerber G, Gruber K, Kratky C. Inorg Chem. 1994;33:4128–4139. [Google Scholar]
- 68.Garau G, Geremia S, Marzilli LG, Nardin G, Randaccio L, Tauzher G. Acta Crystallogr Sect B: Struct Sci. 2003;B59:51–59. doi: 10.1107/s0108768102019353. [DOI] [PubMed] [Google Scholar]
- 69.Perry CB, Fernandes MA, Brown KL, Zou X, Valente EJ, Marques HM. Eur J Inorg Chem. 2003:2095–2107. [Google Scholar]
- 70.Kratky C, Faerber G, Gruber K, Wilson K, Dauter Z, Nolting HF, Konrat R, Kraeutler B. J Am Chem Soc. 1995;117:4654–4670. [Google Scholar]
- 71.Randaccio L, Geremia S, Nardin G, Wuerges J. Coord Chem Rev. 2006;250:1332–1350. [Google Scholar]
- 72.Ouyang L, Rulis P, Ching WY, Nardin G, Randaccio L. Inorg Chem. 2004;43:1235–1241. doi: 10.1021/ic0348446. [DOI] [PubMed] [Google Scholar]
- 73.Randaccio L, Geremia S, Nardin G, Slouf M, Srnova I. Inorg Chem. 1999;38:4087–4092. [Google Scholar]
- 74.Randaccio L, Geremia S, Stener M, Toffoli D, Zangrando E. Eur J Inorg Chem. 2002:93–103. [Google Scholar]
- 75.Bultitude J, Larkworthy LF, Mason J, Povey DC, Sandell B. Inorg Chem. 1984;23:3629–3633. [Google Scholar]
- 76.Glusker JP. In: B12. Dolphin D, editor. Vol. 1. Wiley-Interscience; New York: 1982. pp. 23–107. [Google Scholar]
- 77.Brown KL, Zou X. J Am Chem Soc. 1992;114:9643–9651. [Google Scholar]
- 78.Brown KL, Brooks HB, Gupta BD, Victor M, Marques HM, Scooby DC, Goux WJ, Timkovich R. Inorg Chem. 1991;30:3430–3438. [Google Scholar]
- 79.Zou X, Brown KL. Inorg Chim Acta. 1998;267:305–308. [Google Scholar]
- 80.Bouquiere JP, Finney JL, Savage HFJ. Acta Crystallogr Sect B: Struct Sci Section B: Structural Science. 1994;B50:566–578. [Google Scholar]
- 81.Pagano TG, Marzilli LG, Flocco MM, Tsai C, Carrell HL, Glusker JP. J Am Chem Soc. 1991;113:531–542. [Google Scholar]
- 82.Wagner T, Afshar CE, Carrell HL, Glusker JP, Englert U, Hogenkamp HPC. Inorg Chem. 1999;38:1785–1794. doi: 10.1021/ic980751q. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.