Abstract
The syntheses and applications of two metallointercalators, Rh(bpy)2(chrysi)3+ and Rh(bpy)2(phzi)3+, that target single base mismatches in DNA are described. The complexes selectively bind thermodynamically destabilized mismatched DNA sites and, upon photoactivation, promote strand scission neighboring the mismatch. Owing to their high specificity, independent of sequence context, targeting mismatches with these complexes offer an effective and convenient alternative to current mismatch- and SNP-detection methodologies. Syntheses of these complexes are described as well as protocols for the application of these complexes in marking mismatched sites. In the first, the irradiation of [32P]-labeled duplex DNA with either intercalator followed by denaturing PAGE allows for the detection of mismatches in oligonucleotide sequences. The second protocol describes a method for the efficient detection of single nucleotide polymorphisms (SNPs) in larger genes or plasmids. Pooled genes are denatured and re- annealed to form heteroduplexes, incubated with Rh(bpy)2(chrysi)3+ or Rh(bpy)2(phzi)3+, irradiated, and analyzed by capillary electrophoresis for fragments indicative of mismatch and thus SNP sites. The synthesis of the metallointercalators requires about five to seven days. Both the mismatch and SNP detection experiments require approximately three days.
INTRODUCTION
The synthesis and study of octahedral metal complexes that bind DNA have long been pursuits of our laboratory1. The unique modularity of metal complexes has allowed us to investigate systematically the factors that contribute to DNA site recognition. For example, the site selectivity of rhodium complexes bearing the 5,6-phenanthrenequinone diimine (phi) intercalating ligand varies dramatically with the identity of the ancillary ligands: Rh(bpy)2(phi)3+ shows little site selectivity while Rh(R,R-dimethyltrien)(phi)3+ binds specifically to 5′-TGCA-3′ sites1,2.
In recent years, we have applied our understanding of molecular recognition elements to the development of complexes that selectively bind mispaired sites in DNA3,4,5. DNA mismatches occur in the cell as a result of polymerase errors or DNA damage6,7. In order to preserve the fidelity of its genome, the cell has developed a complex mismatch repair (MMR) machinery to find and correct these mismatches, thus preventing consequent mutations8,9. Abnormalities in this machinery, however, can lead to the accumulation of mismatches, and thus mutations, in the genome with a likelihood of cancerous transformation. Indeed, mutations in MMR genes have been identified in 80% of hereditary non-polyposis colon cancers, and 15–20% of biopsied solid tumors have shown evidence of somatic mutations associated with MMR10,11,12. Mismatch detection may provide an early indicator of these cancerous transformations13.
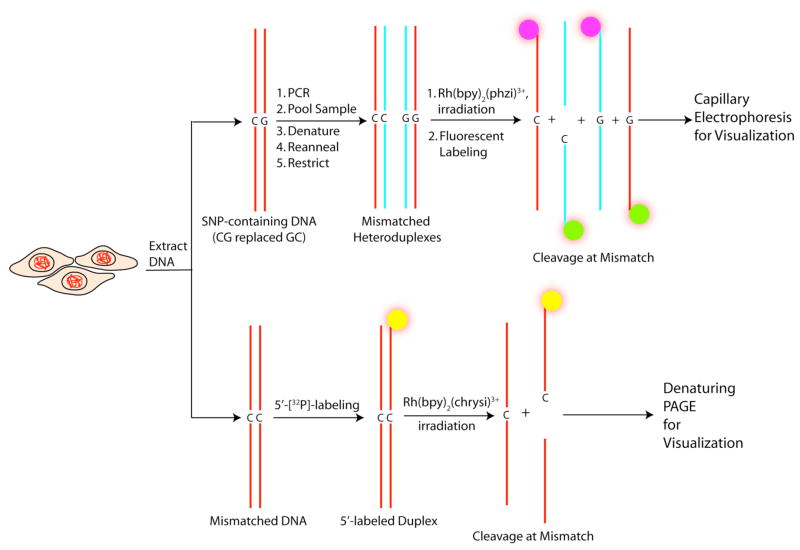
Given the wealth of sequence information available through the human genome project, much attention has turned to the discovery of single nucleotide polymorphisms (SNPs), the single base differences that lead to variations in one’s disposition to disease or response to pharmaceuticals14. Importantly, DNA mismatches can also serve as important intermediates in the search for SNPs. When a test gene fragment is mixed with the wild type fragment, and the DNA heated and reannealed, 50% of the reannealed duplexes will contain single base mismatches if the test DNA contains a SNP. These mismatches thus provide the target for SNP detection15. Figure 1 schematically illustrates how mismatch targeting can be usefully applied in both situations: the discovery of SNPs and the detection of deficiencies in MMR.
Figure 1.
Schematic of the use of mismatch-selective metallointercalators in detection of both single nucleotide polymorphisms (SNPs) (top) and single base mismatches (bottom).
In our laboratory, two families of mismatch-specific metallointercalators have been developed based on a pair of bulky intercalating ligands, 5,6-chrysenequinone diimine (chrysi) and 3,2-benzo[a]phenazine quinone diimne (phzi) (Fig. 2)3,13. The preparation of the simplest complexes in each family, Rh(bpy)2(chrysi)3+ and Rh(bpy)2(phzi)3+, are described here. In both cases, the sterically expansive ligand is too large to intercalate easily into the base stack of regular B-form DNA. However, each binds with high affinity to the thermodynamically destabilized mismatched sites. Binding affinities are on the order of 106 M−1 for Rh(bpy)2(chrysi)3+ and 108 M−1 for Rh(bpy)2(phzi)3+ 10,13. Importantly, affinities correlate with the destabilization associated with a mismatch. Our structural model is shown in Figure 3. The correlation between affinity and mismatch stabilization can be understood based upon the ease of extruding the mismatched bases upon insertion of the metal complex into the base pair stack (Pierre, Kaiser, Barton submitted). Those sites that are most destabilized are most easily bound by the metal complexes. In all, the compounds bind > 80% of mismatch sites in all possible sequence contexts5.
Figure 2.
Δ-[Rh(bpy)2(chrysi)]3+ (left) and Δ-[Rh(bpy)2(phzi)]3+ (right). In some cases (ref. 15, for example), these complexes are referred to as Rh(chrysi) and Rh(phzi).
Figure 3.

Structural model of Rh(bpy)2(chrysi)3+ bound to a CA mismatch (adapted from Pierre, Kaiser, Barton, submitted). The bulky metal complex (red) inserts into the DNA base stack (gray) and the mismatched bases (blue) are extruded.
Despite their differences in binding strength, both complexes exhibit ≥ 1000-fold selectivity for mismatched DNA sites over Watson-Crick base paired DNA sites. Site-selectivity in solution can be readily discerned through DNA photocleavage experiments. In addition to tightly and selectively binding mismatches, the complexes promote direct strand scission adjacent to the mismatch site with photoactivation3,13. The selectivity of the complexes is a tremendous asset; Rh(bpy)2(chrysi)3+, for example, is capable of binding and cleaving a single CC mismatch in a 2725 base pair plasmid4. Because the complexes target only those sites that are thermodynamically destabilized in the base stack, however, more stable mismatches, for example, those containing guanines, are not readily detected based upon photocleavage. Interestingly, the Δ-enantiomers of the complexes bind far better than the Λ-enantiomers. Thus, while racemic complexes may be used for all the experiments described here, only the Δ-enantiomer is required.
This protocol first describes the synthesis of Rh(bpy)2(chrysi)3+ and Rh(bpy)2(phzi)3+ and the enantiomeric separation of Δ- and Λ-Rh(bpy)2(chrysi)3+ 16. First explained are the syntheses of the two intercalating ligands (Fig. 4 and 5). The two compounds, then, are synthesized identically from RhCl3 until the last step, in which the ligand diones are condensed onto Rh(bpy)2(NH3)23+ under basic conditions (Fig. 6). It should be pointed out that other methods for the formation of the diimine complexes are known. All others, however, require the cumbersome anaerobic coordination of the ligand diamine followed by the subsequent oxidation of the diamine to the diimine;17 this alternative methodology is further limited by low product yields and the necessity of using the ligand diamine precursor. In contrast, the condensation method we employ is far more synthetically facile and produces the desired products in high yield. The UV-visible spectra for the complexes are given in Figure 7. The enantiomeric separation of Δ- and Λ-Rh(bpy)2(chrysi)3+ is achieved through the somewhat unorthodox use of 0.15 M (+)-potassium antimonyl tartrate as a chiral eluant for a cation exchange column (Fig. 8). Other methods, including HPLC using a chiral column, have been attempted but have yielded poorer enantiomeric separation. The circular dichrosim spectra are provided in Figure 9.
Figure 4.
Synthesis of 2,3-benzo[a]phenazine quinone.
Figure 5.
Synthesis of 5,6-chrysene quinone.
Figure 6.
Synthetic route to rac-Rh(bpy)2(chrysi)3+ and rac-Rh(bpy)2(phzi)3+.
Figure 7.
UV-Visible spectra of Rh(bpy)2(chrysi)3+ (red) and Rh(bpy)2(phzi)3+ (blue) in water. Extinction coefficients for Rh(bpy)2(chrysi)3+: 303 nm (ε=57,000 M−1), 315 nm (ε=52,200 M−1), 391 nm (ε=10,600 M−1). Extinction coefficients for Rh(bpy)2(phzi)3+: 304 nm (ε=65,800 M−1), 314 nm (ε=67,300 M−1), 343 nm (ε=39,300 M−1).
Figure 8.
Column set-up for enantiomeric separation of Rh(bpy)2(chrysi)3+.
Figure 9.
Circular dichroism spectra of Δ- and Λ-[Rh(bpy)2(chrysi)](Cl)3 in water (blue and red, respectively). Δε values for Δ-[Rh(bpy)2(chrysi)](Cl)3: 233 (34), 264 (26), 286 (−12), 308 (−42), 318 (−100), 341 (6). Δε values for Λ- [Rh(bpy)2(chrysi)](Cl)3: 233 (−34), 264 (−26), 286 (12), 308 (42), 318 (100), 341 (−6).
In addition to the syntheses and enantiomeric separation, two experimental applications of the mismatch-selective complexes are included. In the first, a PAGE sequencing gel can be used to visualize the presence and location of mismatched photocleavage sites in oligonucleotide DNA sequences. In this method, samples of duplex DNA, in which a very small amount of one of the strands has been 5′-[32P]-labeled, are irradiated in the presence of Rh(bpy)2(chrysi)3+ or Rh(bpy)2(phzi)3+. The radioactive samples are then eluted through a denaturing polyacrylamide gel and visualized via phosphorimagery. If the original duplex strand contains mismatches, bands corresponding to shorter fragments created by photocleavage at the mismatched site are evident in addition to the full-length parent band (see Fig. 10 for a sample PAGE experiment). While this methodology may be somewhat limited by the DNA length, it provides information on both the presence (or absence) of a mismatched site and, with the help of parallel sequencing ladders, the exact location of the mismatched site in the sequence5. Of course, other procedures have been developed to search DNA for mismatches using enzymatic or chemical methods, such as RNAase cleavage and chemical cleavage with osmium tetroxide or hydroxylamine18. However, none of these combines the accuracy, selectivity, robustness, and ease desired in a clinical testing procedure.
Figure 10.
Phosphorimagery of a 20% polyacrylamide denaturing gel showing mismatch-specific photocleavage by Δ-[Rh(bpy)2(chrysi)]3+ of a 5′32P-labeled 36-mer containing a CC-mismatch. Conditions employed: 1 μM duplex DNA, 50 mM NaCl/10mM NaPi pH 7.1 buffer, 1 μM Rh complex when applicable, irradiations performed on an Oriel Instruments solar simulator (320 nm–450 nm) for 15 min. Lanes 1 and 2: matched DNA, Maxam Gilbert sequencing reactions. Lane 3: matched DNA, sample irradiated without rhodium. Lane 4: matched DNA, sample with rhodium but no irradiation. Lane 5: matched DNA irradiated with Δ-Rh(bpy)2(chrysi)3+. Lane 6: mismatched DNA, sample irradiated without rhodium. Lane 7: mismatched DNA, sample with rhodium but no irradiation. Lane 8: mismatched DNA irradiated with Δ-Rh(bpy)2(chrysi)3+. Lanes 9 and 10: mismatched DNA, Maxam Gilbert Sequencing reactions.
The mismatch-specificity of our complexes can also be used to detect single nucleotide polymorphisms (SNPs) 15. In this second application, a regions of the genome, perhaps containing an SNP, is amplified, denatured, and re-annealed in the presence of a pooled sample to create heteroduplexes that may contain a mismatch at the site of the original SNP. After annealing, the samples are incubated with Rh(bpy)2(chrysi)3+ or Rh(bpy)2(phzi)3+, irradiated to promote photocleavage, end-labeled with a fluorophore, and analyzed by capillary gel electrophoresis. If an SNP is present in one of the original segments amplified, the capillary electrophoresis trace will display both a parent peak for the full, uncleaved strand and a peak that corresponds to a fragment resulting from photocleavage at the mismatched site (see Fig. 11 schematic of the procedure). Importantly, this technique allows for the detection of SNPs with allele frequencies as low as 5% from a single pooled sample. This method greatly reduces the cost of discovering new SNPs. Perhaps the greatest advantage of our method is the ease of use; specifically, fewer polymerase chain reactions (PCRs), and no cycle sequencing are required.
Figure 11.
Schematic of SNP detection with Rh(bpy)2(phzi)3+. Adapted from ref. 15.
An alternative method for SNP detection, resequencing, is known19–22. However, resequencing is expensive with respect to materials, labor, and data processing. Further, while the current technique must scan a particular region of the genome many times to detect a single SNP, even then the false positive rate is high23. Resequencing does, however, hold certain advantages over our method, namely that it can provide a clear picture of both the sequence identity of the SNP and its frequency. We recommend using first our SNP protocol for discovery followed by resequencing of the specific region marked by the metal complex for characterization. Perhaps, then, in an ideal situation metallointercalators and resequencing could be used in concert to discover new SNPs and create extensive genomic SNP maps.
The possible uses of our mismatch-specific metallointercalators extend well beyond the two applications outlined here. For example, trisheteroleptic complexes bearing both a mismatch-binding intercalating ligand and a linker-modified bipyridine ligand have been synthesized for the development of mismatch-directed bifunctional conjugates. To date, this strategy has been employed to make bifuvntional conjugates for platination24 and alkylation25 near mismatched sites as well as conjugates for fluorescence-based mismatch detection26, and to improve nuclear uptake of the mismatch-specific complexes27. Moreover, recent experiments have revealed that Rh(bpy)2(chrysi)3+ and Rh(bpy)2(phzi)3+ may have chemotherapeutic value. Both complexes have been shown to inhibit selectively the cell proliferation of MMR-deficient cells over MMR-proficient cells28.
MATERIALS
REAGENTS
Chrysene (Aldrich Zone Purified)
Glacial acetic acid
Sodium dichromate (Aldrich)
2,3-Dichloro-1,4-napthoquinone (Aldrich)
o-Phenylene diamine (Fluka)
Pyridine
Concentrated nitric acid (not fuming yellow or red)
Water (deionized and filtered, i.e. Millipore purified to > 18Ω resistivity)
Ethanol
Diethyl ether
RhCl3 hydrate (Pressure Chemicals)
Hydrazine monohydrate (Aldrich)
2,2′-Bipyridyl (Aldrich)
Triflic acid (5g ampule, Fluka)
Concentrated ammonium hydroxide
Sodium hydroxide
Acetonitrile
Methanol
Trifluoroacetic acid (TFA)
Potassium nitrate (Aldrich)
Sephadex® SP-C25 ion exchange resin (Aldrich)
Magnesium chloride
(+)-KSb-tartrate (Aldrich)
Sodium chloride
Sodium phosphate (NaH2PO4, referred to as NaPi)
Polynucleotide kinase (Roche, 10 activity units/μL)
Polynucleotide kinase buffer (Roche, supplied with PNK)
[32P]-ATP (End-labeling grade, MP Biomedicals)
10X Tris-borate-EDTA (TBE) buffer (National Diagnostics)
Ultrapure SequaGel® Sequencing System Diluent (National Diagnostics)
Ultrapure SequaGel® Sequencing System Concentrate (National Diagnostics)
Ultrapure SequaGel® Sequencing System Buffer (National Diagnostics)
Ultrapure TEMED (Invitrogen)
Ammonium persulfate (MP Biomedicals)
Denaturing formamide loading dye (see REAGENT SET-UP)
Saran Wrap® (or equivalent product)
ExoI (New England Biolabs)
Calf Intestinal Alkaline Phosphtase (Roche)
QIAquick® PCR purification kit (Qiagen)
Tris Hydrochloride
Dithioerythritol
XhoI (Roche)
ClaI (Roche)
SNaPshot® Fluorescent Labeling Kit (Applied Biosystems)
Chloroform
Ethyl acetate
Magnesium chloride
EQUIPMENT
Assortment of round-bottom flasks (50 mL, 100 mL, 250 mL, 500 mL)
Reflux condensers (14/20 and 24/40 joints)
Magnetic stir bar and stir plate
Heating mantle
500 mL Beaker
60 mL Medium glass frit
500 mL Vacuum flask
50 mL Schlenk flask
Pasteur pipettes
Silica TLC plates (Baker)
Rotary evaporator
Flash column (about 20 cm × 1.5 cm)
Very long glass column (about 1.7 m × 1.5 cm)
2 Medium-sized glass columns (about 0.5 m × 1.5 cm)
Tubing (Tygon)
Lyophilizer
Razor blades
Tweezers
X-ray film (Kodak BioMax XAR film, 35 cm × 43 cm)
X-ray developer (i.e. Konica Minolta SRX-101A)
1.7 mL Centrifuge tubes
Microcentrifuge (i.e. Eppendorf 5415C)
Lucite sample boxes for 1.7 mL centrifuge tubes
Reverse phase C18 cartridges (i.e. Waters Sep-Pak®)
Micro Bio-Spin® 6 Chromatography Column (Bio-rad)
Evacuated centrifuge (SpeedVac)
Glass gel plates, notched and un-notched (35 cm × 45 cm, FisherBiotech)
Gel running apparatus (23 cm × 40 cm × 50 cm, FisherBiotech)
Power supply for gel running apparatus (Fisher Biotech, FB-EC-4000P)
Adjustable heat block
Scintillation counter
Assortment of micropipettes (10 μL, 20 μL, 200 μL, 1000 μL)
Light source (see EQUIPMENT SET-UP)
Phosphorimager screen (35 × 43 cm, Molecular Dynamics)
Phosphorimager (Storm 820, GE Healthcare)
Photoshop Software (Adobe)
ImageQuant Software (Molecular Dynamics)
Thermocycler
Peristaltic Pump
REAGENT SET-UP
Denaturing Formamide Loading Dye: 80% formamide, 10 mM NaOH, 0.025% xylene cyanol, 0.025% bromophenol blue, in 1X TBE buffer.
EQIPMENT SET-UP
Both Rh(bpy)2(chrysi)3+ and Rh(bpy)2(phzi)3+ cleave DNA at a variety of wavelengths; while this allows for a number of light source options, the cleavage efficiency is highly wavelength-dependent. Typically, irradiations are performed using either a solar simulator (Oriel Instruments, wavelength output 320–450 nm) equipped with a UV filter or at 340 or 442 nm on a 1,000 W Hg/Xe arc lamp equipped with a monochromator, a 295-nm UV-cutoff filter, and an IR filter (Oriel Instruments). Typical irradiation times are 15 min on the solar simulator and 30–60 min on the lamp. Other options exist, however. Cleavage can also be induced using a 302-nm transilluminator or a 365-nm “black-light” situated 3–4 cm above an open sample tube. However, in these two cases, about 10X more irradiation time (when compared to the more powerful lamp and solar simulator) is required to prompt substantial photocleavage. Neither white fluorescent light nor incandescent light provide sufficient UV power for strand cleavage with Rh(bpy)2(chrysi)3+ or Rh(bpy)2(phzi)3+. Regardless of the source, all irradiations are performed in 1.7 mL centrifuge tubes. In a case where the light shines vertically from above, transparent Lucite® samples boxes are simple and effective sample holders. In a case where the light shines horizontally, sample holders more typical of laser set-ups can be employed.
While other options exist for capillary electrophoresis, this procedure has been optimized for the use of an ABI Prism 310 instrument (Applied Biosystems). A sample instrument configuration is shown below:
capillary: 47 cm × 50 μm
SNaPshot dye: Hex®
polymer: POP-4
dye set: E5
molecular weight marker: Genescan 500 TAMRA
module: GS STR POP4 (1 mL)
injection seconds: 5 seconds
injection kV: 15.0 V
run kV: 15.0 V
run temperature: 60°C
run time: 35 min
analysis matrix: matrix E5
analysis size standard: gs-500liz
analysis parameters: 0–500 base pairs
PROCEDURE
General note of caution for syntheses
5,6-chrysene quinone, 3,2-benzo[a]phenazine quinone, and their metal complexes are DNA intercalators. While their exact toxicity profile is unknown, they are almost certainly highly toxic and most likely carcinogenic. These compounds should be handled with extreme care.
Synthesis of 5,6-chrysene quinone
-
1 Combine 10.0 grams (44 mmol) of chrysene and 110 mL of glacial acetic acid in a 250 mL round bottom flask and stir vigorously.
CAUTION: Chrysene is a known carcinogen.
2 Slowly add 46 g sodium dichromate to the stirring slurry.
-
3 Affix a reflux condenser and heat the slurry to reflux for 24 h. Over the course of the heating the color of the solution will slowly change from bright orange with white solid to dark green/brown with orange precipitate.
CRITICAL STEP: The reaction can be monitored by color change. The reaction is complete when no more white solid can be seen in the refluxing mixture.
4 After 24 h, stop heating. Remove the reflux condenser once boiling has stopped, and before the mixture reaches room temperature, pour it into 100 mL of rapidly boiling water in a beaker.
-
5 While still hot, filter the solution through a medium glass frit.
CAUTION: Hot-filtering, while necessary in this case, is always dangerous. Be careful.
-
6 Wash the collected red solid 3 times with 100 mL boiling water.
CRITICAL STEP: The filtration MUST be performed when the solution is boiling or near-boiling. Any cooling will result in the precipitation of insoluble chromium byproducts which are very difficult to remove.
-
7 No purification is required. Very small amounts of chrysene may be observed via 1H NMR, though these will not react in subsequent steps and thus will be eliminated.
PAUSE POINT: The product can be stored as a solid indefinitely.
Synthesis of 3,2-benzo[a]phenazine quinone
8 Dissolve 4.5 g (20 mmol) 2,3-dichloro-1,4-napthoquinone and 2.0 g (20 mmol) o-phenylene diamine in 150 mL pyridine. This must be done by placing both materials into the flask and then adding pyridine.
9 Attach a reflux condenser and bring the solution to reflux.
10 After 1 hour, allow the solution to cool to room temperature.
-
11 Filter the cooled solution. This should yield a dark brown-red solid. The solid can be recrystallized from hot pyridine or carried on as is.
PAUSE POINT: This intermediate can be stored for up to a week before further use. Longer periods of storage may be possible.
-
12 After weighing the brown-red solid, place it in a second round bottom flask and add to it 10 mL glacial acetic acid and 1 mL concentrated nitric acid. Subsequently add 0.66 mL water per 1 g solid.
CAUTION: Concentrated nitric acid is caustic and an oxidizer. Handle with care.
-
13 Heat the resultant solution in a boiling water bath for 1 hour.
CRITICAL STEP: The reaction is complete when only yellow-orange precipitate remains.
14 Filter this solution. The solid should be yellow.
-
15 Rinse the resulting solid with 15 mL ethanol and 15 mL diethyl ether.
CAUTION: Do not let the washes contact the nitric acid solution. Use a new flask.
-
16 The final product can be purified by recrystallization from 7:3 chloroform:ethyl acetate, but further purification is usually not necessary.
PAUSE POINT: The product can be stored as a solid indefinitely.
Synthesis of [Rh(bpy)2Cl2]Cl
17 Dissolve 0.64 g rhodium chloride hydrate (2.8 mmol) and 50 mg hydrazine monohydrochloride in 12.5 mL deionized water in a 50 mL round bottom flask.
18 Add a solution of 0.85 g (5.6 mmol) 2,2′-bipyridyl in 20 mL ethanol and de-oxygenate the resultant solution by the repeated application of vacuum followed by backfilling with Ar (g).
19 Bring the reaction mixture to reflux and heat until all materials have dissolved and formed a yellow-orange solution.
-
20 While still hot, filter the reaction mixture through a medium glass frit.
CAUTION: Hot-filtering, while necessary in this case, is always dangerous. Be careful.
21 Chill the filtrate overnight to promote crystallization.
-
22 Isolate the yellow product crystals via filtration with a medium glass frit.
PAUSE POINT: The product can be stored as a solid indefinitely.
Synthesis of [Rh(bpy)2(OTf)2]OTf
23 Add 500 mg Rh(bpy)2(Cl)2+ (1.0 mmol) to a 50 mL Schlenk flask with a 14/20 joint on top and a side-arm and de-oxygenate the flask by evacuating it and refilling with Ar(g) three times.
-
24 Carefully crack open the ampule of triflic acid and use a glass Pasteur pipette to add 5 g (excess) trflic acid to the reaction vessel under positive argon pressure.
CAUTION: Triflic acid is very reactive and pyrophoric. Handle quickly and with care.
25 After adding the triflic acid, place a close the flask with a rubber septum, pierce the septum with a needle, and purge the flask with argon for 30–60 seconds.
26 Allow the reaction mixture to stir for 16 h. Purge occasionally (every 4 to 6 h) to remove HCl generated by the reaction.
27 After 16 h, cool 300 mL diethyl ether to −78° C in a dry ice/acetone bath. Place the bath on a stir plate and add a stir bar to the flask.
-
28 Using a Pasteur pipette, add the reaction mixture drop-wise to the rapidly stirring, cold diethyl ether. A yellowish-white powder will precipitate. No purification is necessary.
CRITICAL STEP: It is imperative that the diethyl ether solution is stirring, so that each new drop of reaction mixture falls in to cold diethyl ether and thus prompts product precipitation.
PAUSE POINT: While it is best to procedure directly to the next step, this product can be stored for short periods of time in a dessicator.
Synthesis of [Rh(bpy)2(NH3)2](X)3 (where X = PF6− or OTf−)
29 Combine 500 mg Rh(bpy)2(OTf)2+ (0.6 mmol) and 20–50 mL concentrated NH4OH in a 250 mL round bottom flask fitted with a reflux condenser. The starting material should be relatively insoluble in NH4OH.
30 Bring the mixture to reflux and boil it until all the material has gone into solution (5–10 min).
-
31 Depending on the desired counter-ion, the product can be isolated using option A or option B:
-
Precipitate from NH4OH solution with NH4PF6.
Add excess NH4PF6 to the ammonia solution.
Chill overnight at 4°C to facilitate precipitation.
Filter with a medium frit to isolate the product.
-
Remove solvent under vacuum.
Remove all NH4OH with rotary evaporation. This will yield the triflate salt of the product.
PAUSE POINT: The product can be stored as a solid indefinitely.
-
Synthesis of [Rh(bpy)2(chrysi)](Cl)3
32 In a 100 mL round bottom flask, dissolve 195 mg [Rh(bpy)2(NH3)2](X)3 (approximately 0.2 mmol) and 57 mg 5,6-chrysene quinone (0.22 mmol) in 50 mL acetonitrile with rapid stirring under ambient conditions.
33 Add 2 mL aqueous sodium hydroxide (0.4 M) and close the vessel to prevent evaporation.
34 After 3 h, halt the reaction by bringing the pH of the solution to 7 by adding a stoichiometric amount of HCl. By this point the reaction should have changed color dramatically, from orange/yellow to dark red. Alternatively, one can monitor the reaction by TLC using Silica F plates in a solvent system of 3:1:1 MeCN:H2O:MeOH with 0.1 M KNO3.
35 Remove acetonitrile in vacuo by rotary evaporation.
-
36 Redissolve the reaction mixture in a minimum volume of water.
PAUSE POINT: The reaction mixture can stand at room temperature while the ion exchange column is being prepared.
37 Equilibrate 20 g of Sephadex SP-C25 ion exchange resin with MgCl2 by making a slurry of the resin in approximately 500 mL 0.05 M MgCl2.
38 With this equilibrated mixture, fill a 1–1.5 inch diameter column with about 5 inches of resin.
39 Flush the excess MgCl2 solution out of the column with 250 mL H2O.
40 Flush the column with 500 mL deionized water.
41 Load the Rh(bpy)2(chrysi)3+ by pushing the diluted reaction mixture through the column. The rhodium complex should stick to the uppermost layer of resin, creating a red band at the top of the column.
42 Once all the reaction mixture has been loaded onto the column, wash the column with 250 mL deionized H2O.
-
43 Elute the metal complex by slowly increasing the MgCl2 concentration of the eluant in 500 mL batches. Start with 0.01 M and increase by 0.01 M increments until 0.10 M MgCl2; at this point, switch to 0.05 M increases until the metal complex (red band) has been eluted (most likely around 0.3 M MgCl2).
CRITICAL STEP: It is very important that the MgCl2 concentration gradient is shallow. Too steep a gradient will cause the ion exchange column to “crack”, leading to band smearing and poor separation.
44 Concentrate the product-containing fractions on a reverse-phase cartridge (i.e. Waters 2 g C18 Sep-Pak Cartridge) primed with methanol.
45 Wash the cartridge thoroughly with water.
46 Elute the product with 1:1:0.001 H2O:MeCN:TFA.
-
47 Remove the solvent by rotary evaporation or lyophilization.
PAUSE POINT: The finished product can be stored as a solid indefinitely. NOTE: A representative UV-Visible spectrum of the product is shown in Fig. 7.
Synthesis of [Rh(bpy)2(phzi)](Cl)3
48 In a 100 mL round bottom flask, dissolve 100 mg [Rh(bpy)2(NH3)2](X)3 (approximately 0.1 mmol) and 35 mg 3,2-benzo[a]phenazine quinone (0.125 mmol) in 50 mL acetonitrile with rapid stirring under ambient conditions.
49 Add 2 mL aqueous sodium hydroxide (0.4 M) and close the vessel to prevent evaporation.
50 After three h, halt the reaction by bringing the pH of the solution to 7 by adding water and a stoichiometric amount of HCl.
51 Remove the acetonitrile in vacuo by rotary evaporation.
-
52 Redissolve the product in a minimum volume of water.
PAUSE POINT: The reaction mixture can stand at room temperature while the ion exchange column is being prepared.
53 Equilibrate 20 g of Sephadex SP-C25 ion exchange resin with MgCl2 by making a slurry of the resin in approximately 500 mL 0.05 M MgCl2.
54 With this equilibrated mixture, fill a 1–1.5 inch diameter column with about 5 inches of resin.
55 Flush the excess MgCl2 solution out of the column with 250 mL water.
56 Flush the column with 500 mL deionized water.
57 Load the Rh(bpy)2(phzi)3+ by pushing the diluted reaction mixture through the column. The rhodium complex should stick, yielding a dark yellow/brown band at the top of the ion exchange column.
58 Once all the reaction mixture has been loaded onto the column, wash the column with 250 mL deionized H2O.
-
59 Elute the metal complex by slowly increasing the MgCl2 concentration of the eluant in 500 mL batches. Start with 0.01M and increase by 0.01M increments until 0.10M MgCl2; at this point, switch to .05M increases until the metal complex (dark band) has been eluted (most likely around 0.3 M MgCl2).
Critical Step: It is very important that the MgCl2 concentration gradient is shallow. Too steep a gradient will cause the ion exchange column to “crack”, leading to band smearing and poor separation.
60 Concentrate the product-containing fractions on a reverse phase cartridge (i.e. Waters 2 g C18 Sep-Pak Cartridge) primed with methanol.
61 Wash the cartridge thoroughly with water.
62 Elute the product with 1:1:0.001 water:MeCN:TFA.
-
63 Remove the solvent by rotary evaporation or lyophilization.
PAUSE POINT: The finished product can be stored as a solid indefinitely.
NOTE: A representative UV-Visible spectrum of the product is shown in Fig. 7.
Enantiomeric Separation of Rh(bpy)2(chrysi)3+
64 Fill one very long column (1.70 m × 1.5 cm) and two smaller columns (0.5 m × 1.5 cm, hereafter referred to as guard columns) with Sephadex® SP-C25 ion exchange resin equilibrated with water.
-
65 Prepare 4 L of a solution of 0.15 M (+)-KSb-tartrate in water. Filter this solution.
CAUTION: (+)-KSb-tartrate is toxic. Handle with care and dispose of properly.
-
66 Elute all three columns thoroughly with the 0.15 M (+)-KSb-tartrate solution. Do not recycle this eluant.
CRITICAL STEP: Upon elution of the KSb-tartrate solution, the resin will compact a little bit (20%); this may necessitate the addition of more resin to fill the columns all the way.
68 Set aside on of the guard columns and set up the other two columns and eluant-recycling pump as shown in Figure 8 . Make sure all three columns are full of both the chiral eluant and ion exchange resin.
67 Dissolve 0.4 g rac-[Rh(bpy)2(chrysi)]Cl3 in 5 mL water.
-
68 Carefully load the rhodium solution onto the top of the large column.
CRITICAL STEP: To obtain good enantiomeric separation, it is very important that the initial rhodium band on the column is very small. To ensure this, use a minimum of water when first dissolving the rhodium complex and be careful when first loading the rhodium solution onto the top of the large column.
-
69 Turn on the pump and allow the eluant to cycle. The separation will take about 2–3 d. During cycling, monitor the column regularly (every 4 h during the day).
CRITICAL STEP: Make sure all connections are secured with clamps or wire to prevent leaks.
-
70 After about one day, separation should begin to become apparent. The first (lower, faster) band is Λ-Rh(bpy)2(chrysi)3+; the second (upper, slower) band is Δ-Rh(bpy)2(chrysi)3+. Detach the first guard column after it “catches” the lower band and replace it with the second guard column. Cap the first guard column to prevent it from drying.
CRITICAL STEP: Do not let the guard columns or the main column dry out during the separation. This will cause the compound to stick permanently to the resin.
71 Continue eluting the column (using the pump) until the second guard column fully catches the second band.
-
72 After the second guard column has fully caught the second band, detach and cap it to prevent any evaporation.
CAUTION: All of the eluant and resin used will contain antimony. Elute the large column with water and dispose of the antimony-containing eluant appropriately. Likewise, the resin will be contaminated with antimony and should be disposed of properly.
-
73 Wash the two guard columns with 0.05 M MgCl2 solution to remove the remaining antimony tartrate.
CAUTION: Be sure to dispose of the antimony-containing eluant properly.
CRITICAL STEP: Be careful to keep all columns and product-containing eluants well-labeled and separate during product isolation to prevent inadvertent re-racemization.
74 Remove the compound from the guard columns by washing the columns with 0.5 M MgCl2 until all the compound has been eluated.
75 Concentrate the two product fractions on 5 g Waters Sep-Pak C18 cartridges previously primed with 2 × 10 mL methanol and 1 × 10 mL water.
76 Wash the cartridges with 200 mL water.
77 Elute the products with a mixture of 1:1:0.001 MeCN:H2O:TFA.
78 Freeze the eluants with liquid N2 and lyophilize to dryness. Due to some permanent product adhesion to the ion exchange resin, only about 80% mass yield can be expected from the separation. The two enantiomers should be obtained in about 1:1 ratio. NOTE: Representative circular dichroism spectra of both the Λ- and Δ-enantiomers are shown in Figure 9.
Mismatch detection via photocleavage with 5′-[32P]-labeled DNA and denaturing PAGE
79 Make a 100 μM stock solution of the single stranded DNA sequences (forward and complement) to be investigated.
-
80 Label the 5′ end of each of the strands with [32P]-ATP: combine 15 μL water, 2 μL polynucleotide kinase buffer (provided), 1 μL ssDNA stock solution, 1 μL polynucleotide kinase solution, and 1 μL 32P-ATP solution in a 1.7 μL microcentrifuge tube. Vortex and spin the reaction mixture in a microcentrifuge.
CAUTION: Be sure to follow all standard radioactivity safety procedures to limit your exposure.
CRITICAL STEP: The mismatch-specific metallointercalators –Rh(bpy)2(chrysi)3+, in this case – will only cleave one strand of the mismatched DNA. Therefore, cleavage will only be evident using one of the two labeled strands. While the location of cleavage has been determined for most sequence contexts,5 it is best to label both and determine cleavage on both strands.
81 Incubate the labeling reaction for 2 h at 37°C.
-
82 After 2 h, add 80 μL water, vortex, and purify using a Micro Bio-Spin® 6 Chromatography Column.
CAUTION: Do not centrifuge the spin columns at greater than 3000 rpm; they will break.
83 Dry the samples on a lyophilizer or vacuum centrifuge.
84 Purify the 5′-labeled oligonucleotides via 20% PAGE:
A Pour a 20% polyacrylamide gel.
B Take up the dried oligonucleotide samples in 10 μL denaturing formamide loading dye.
C Load the sample onto the gel, and run the gel for 60–90 min at 90 Watts using 1X TBE as the running buffer.
D Remove one gel plate, leaving the gel affixed to the other gel plate.
E Visualize the gel via X-ray
F Cut out the parts of the gel that correspond to full-length, labeled DNA with a clean razor blade.
G With tweezers, place these gel bits into a clean centrifuge tube.
H Add 1 mL 100 mM triethylammonium acetate pH 7.0 to each centrifuge tube containing gel.
I Incubate at 37°C overnight.
J Remove triethylammonium solution and place in a clean 1.7 mL centrifuge tube.
K Speedvac to remove solvent.
L Take up dried sample in 100 μL water and purify with a Micro Bio-Spin® 6 Chromatography Column.
M Dry the samples on a vacuum centrifuge.
-
N Take up labeled oligonucleotide in 50 μL of 10 mM NaPi buffer, pH 7.1.
PAUSE POINT: Labeled DNA can be stored for weeks at 4°C. However, it is best to perform the experiments before the label decays too much.
85 Prepare an aqueous buffer solution of 100 mM NaCl and 20 mM NaPi, pH 7.1
86 Make a 2 μM duplex stock solution for the irradiation experiments: combine 2 μL of each unlabeled DNA single strand (forward and complement), 4 μL of one of the two labeled oligonucleotide solutions, and 92 μL 100 mM NaCl/20 mM NaPi buffer.
-
87 Anneal the 2 μM duplex stock solution via heating to 90°C followed by gradual cooling.
CRITICAL STEP: It is important that the solution cool to room temperature slowly to ensure that proper hybridization has taken place. Our suggested method is placing the centrifuge tube containing the solution on a 90°C heat block for 5 min, removing the entire heat block, and allowing the block and sample tube to cool to room temperature together over the course of about 90 min.
88 Prepare a stock of 2 μM Rh(bpy)2(chrysi)3+ in water.
-
89 Prepare irradiation samples: in 1.7 mL centrifuge tubes, combine 10 μL DNA stock solution (with either forward or reverse strand 5′-labeled) and 10 μL rhodium stock solution to create a final 20 μL sample containing 1 μM DNA (with radiolabel) and 1 μM Rh(bpy)2(chrysi)3+ in a buffer of 50 mM NaCl and 10 mM NaPi, pH 7.1.
NOTE: In addition to samples containing both DNA and Rh(bpy)2(chrysi)3+, it is also important to make control samples under various conditions: for example, DNA and intercalator in the absence of irradiation, DNA alone in the presence of light, and DNA alone in the absence of light.
90 Irradiate the samples (for more information see EQUIPMENT SET-UP).
91 Dry all samples under vacuum.
92 Determine the counts per minute (cpm) for each sample using a scintillation counter.
-
93 Dissolve each sample in enough denaturing formamide dye such that there are 10,000 cpm/μL (e.g. dissolve a sample containing 100,000 counts in 10 μL loading dye).
PAUSE POINT: Samples dissolved in denaturing formamide dye can be stored at room temperature for days.
94 Load 5 μL of each sample onto a 20% denaturing polyacrylamide gel.
95 Run the gel at 90 Watts for approximately 1 hour (or the amount of time it takes the parent band, estimated using the running dyes, to travel about 2/5 of the way down the plate).
96 Separate the gel plates, being careful that gel stays on just one of the plates.
97 Remove the gel from the plate using a large piece of film.
98 Cover the gel with Saran Wrap®.
99 Place the gel in a blanked phosphoroimager screen and expose it for 4 h.
100 After 4 h, remove the gel from phosphorimager screen and develop it on a phosphorimager.
101 Analyze the gel as a *.gel file in the ImageQuant program (Molecular Dynamics) or as a *.tiff file in Photoshop (Adobe). NOTE: See Figure 10 for the result of a typical PAGE experiment.
SNP detection with Rh(bpy)2(phzi)3+
NOTE: For the sake of simplicity, the following protocol is specific to the synthetic plasmid described earlier15. The procedure, however, can be easily adapted to any biologically derived gene. For example, the paper mentioned above also discusses the application of the procedure to SNP detection in the tumor necrosis factor (TNF) promoter region. For a schematic of the technique described below, see Figure 10.
102 Amplify both genes in question, one known to have the correct sequence and one suspected to contain a SNP, by PCR using primers containing restriction enzyme sites (in this case, sites for ClaI and XhoI).
103 Add 6 units of ExoI and 20 units of calf alkaline phosphatase to each PCR reaction and incubate for 1 h at 37°C to degrade excess primers and dNTPs.
104 To determine the success and purity of the PCR reaction, run out a small amount of both PCR products on a 2% agarose gel pre-stained with ethidium bromide. Only a single band should be present. The purity of this initial PCR is very important to the success of the overall assay, so optimization may be required if the reaction appears messy or frequent stops are visible.
-
105 Purify both PCR products by using a QIAquick PCR purification column eluting with 10 mM Tris•HCl (pH 8.5).
PAUSE POINT: At this point, the PCR products can be stored for up to a month at 4°C.
106 Thermally denature the DNA under these low ionic strength conditions by heating to 99 °C for 30 min followed by immediate and rapid cooling to 4°C.
107 Combine equimolar concentrations of each PCR product in a buffer of 60 mM Tris•HCl (pH 7.5), 10 mM MgCl2, 100 mM NaCl, and 1 mM dithioerythritol.
108 Re-anneal the pooled sample by heating to 95°C for 10 min and linearly decreasing the temperature to 4°C over the course of 150 min. This procedure generates the heterozygous duplexes that contain the photocleavable mismatch.
109 Add 5 units ClaI and 5 units XhoI to the pooled sample and incubate the duplex with the restriction enzymes for 1 hour at 37°C followed by denaturing the enzymes at 80 °C for 20 min.
110 Prepare a sample of 400 nM Rh(bpy)2(phzi)3+ in water.
111 In a 1.7 mL centrifuge tube, combine 10 μL pooled DNA stock and 10 μL Rh(bpy)2(phzi)3+ stock to yield a final reaction mixture containing 200 nM Rh(bpy)2(phzi)3+, 30 mM Tris•HCl (pH 7.5), 5 mM MgCl2, 50 mM NaCl, and 0.5 mM dithioerythritol.
112 Irradiate the samples (for more information on irradiation see EQUIPMENT SETUP).
-
113 Dry the samples in vacuo.
PAUSE POINT: Dried samples can be left overnight at room temperature or for a few days at 4°C.
114 Add fluorescent tags to the strands by single base extension using the Applied Biosystems SNaPshot® kit. The two strands, forward and reverse, will receive different fluorescent tags, resulting in two different “color” signals on the capillary electrophoresis trace, one “color” for the forward strand and a different “color” for the reverse strand.
115 Add 2 units calf intestinal alkaline phosphatase to each sample and incubate 1 hour at 37°C followed by denaturing the enzyme at 80°C for 20 min.
116 Ethanol precipitate each sample and remove left-over solvent in vacuo.
117 Re-dissolve each sample in 1 μL water and 1 μL molecular weight standard (see EQUIPMENT SETUP for more information on the molecular weight standard).
118 Add 24 μL deionized formamide to each sample and heat to 95°C for 5 min.
119 Cool the samples to 4°C for 5 min.
-
120 Load samples on an ABI Prism 310 capillary electrophoresis instrument and analyze results. For more information on the configuration of the ABI instrument, see EQUIPMENT SETUP.
NOTE: Sample capillary electrophoresis traces for assayed wildtype and SNP-containing genes are shown in Figure 12.
Figure 12.
Sample SNP Detection Gel Electrophoresis Traces. The trace above (a) illustrates the result obtained using two homozygous plasmids. Without an SNP, no mismatch is formed upon denaturing and re-annealing the two plasmids. Therefore, only full-length products (in this case 436 bases), the parent band, can be observed. In contrast, the trace below (b) shows the result when two different templates, one containing an SNP, are mixed to create a mismatched site. In this case, the denaturing and re-annealing process creates a photocleavable CC mismatch. Therefore, after irradiation at 442 nm for 30 min with 200 nM Rh(bpy)2(phzi)3+, a fragment corresponding to the photocleaved strand, 170 bases in length, marking the SNP site, can be seen in addition to the full-length parent band. Adapted from ref. 15.
TIMING
Synthesis of 5,6-chrysene quinone: Steps 1–3, 24 h; Steps 4–7, 45 min. Total: 24.75 h.
Synthesis of 3,2-benzo[a]phenazine quinone: Steps 8–10, 90 min.; 11–16, 2 h. Total: 3.5 h.
Synthesis of [Rh(bpy)2Cl2]Cl: Steps 17–18, 30 min.; Steps 19–20, 45 min.; Steps 21–22, 14 h. Total: 15.25 h.
Synthesis of [Rh(bpy)2(OTf)2]OTf: Step 23, 10 min.; Step 24–26, 16 h; Step 27–28, 1 h. Total: 17 h.
Synthesis of [Rh(bpy)2(NH3)2](X)3: Step 29–30, 15 min.; Steps 31Ai–31Aiii, 16 h; Step 31B, 45 min. Total: 1 h or 16.25 h depending on Step 31.
Synthesis of [Rh(bpy)2(chrysi)](Cl)3: Steps 32–35, 3 h; Steps 36–43, 5 h; Steps 44–46, 10 min.; Step 47, 12 h. Total: 20 h.
Synthesis of [Rh(bpy)2(phzi)](Cl)3: Steps 48–51, 3h; Steps 52–59, 5h; Steps 60–62, 10 min.; Step 63, 12 h. Total: 20 h.
Enantiomeric Separation of Rh(bpy)2(chrysi)3+: Steps 64–68, 3 h; Steps 69–71, 3 d; Steps 73–78, 3 h. Total: 3.5 d.
Mismatch detection with Rh(bpy)2(chrysi)3+ analyzed by PAGE: Step 79, 10 min.; Steps 80–83, 2.5 h; Steps 84, 1 d; Steps 85–86, 30 min.; Step 87, 1.5 h; Steps 88–90, 1.5 h; Step 91, 1 h; Steps 92–93, 30 min.; Steps 94–95, 1.5 h; Steps 96–101, 5 h. Total: 3 d.
SNP detection with Rh(bpy)2(phzi)3+ analyzed by capillary electrophoresis: Step 102, 4 h; Step 103, 1 h; Step 104, 3 h; Step 105, 30 min.; Step 106, 30 min.; Step 107–108, 3 h; step 109, 1 h; Step 110–113, 2 h; Step 114, 30 min.; Step 115, 1.5 h; Step 116–119, 30 min.; Step 120, 2 h. Total: 3 d.
TROUBLE-SHOOTING
For troubleshooting advice, see Table 1.
Table 1.
Troubleshooting Table
| Problem | Possible Explanation | Solution |
|---|---|---|
| Chrysene quinone synthesis yields green solid | Chromium byproducts are contaminating the solid | Re-suspend solid in boiling water and re-filter while water is still boiling |
| Low yields for phzi quinone synthesis | Nitric acid too strong, resulting in over- oxidation of product | Do not use fuming yellow or red nitric acid. Use normal concentrated (68–70%) nitric acid instead. |
| Over 100% yield of [Rh(bpy)2(NH3)2](PF6)3 | Precipitation of excess PF6− salts along with desired product | Desalt on a reverse phase cartridge (i.e. Waters C18 Sep-Pack) or isolate product by rotary evaporation instead |
| Rh(bpy)2(chrysi)3+ or Rh(bpy)2(phzi)3+ does not stick to ion- exchange column | Column still contains MgCl2 pre-equilibration solution | Flush column thoroughly with deionized water; reload product and try again. |
| Ion exchange column cracked, yielding uneven bands | MgCl2 concentration gradient too steep | Elute everything with a high salt eluant, desalt via reverse phase cartridge, and try again with a more gradual salt gradient |
| No cleavage observed in denaturing gel | DNA not properly annealed | Try annealing the DNA again; be sure to cool the sample slowly |
| Not enough irradiation time or power | Turn up power on light source or irradiate for a longer amount of time. | |
| Mismatch not cleaved by Rh(bpy)2(chrysi)3+ | ||
| Switch to Rh(bpy)2(phzi)3+, it recognizes a few more mismatches than its Rh(chrysi) counterpart. | ||
| Mismatch or mismatch sequence not cleaved by either intercalator | Check reference 5 to see if mismatch is one of those that aren’t cleaved | |
| No cleavage observed in denaturing gel | Ran gel too long, cleavage band ran out the bottom of the gel | Check to see if mismatched site is near the end of the sequence; run gel for a shorter amount of time. |
| Gel looks smeared | Ran gel too soon after polymerization | Pour a new gel. Try again. Wait longer between pouring and running. |
| Photocleavage apparent, but parent band disappears | Too much photocleavage | Reduce the amount of rhodium intercalator or the irradiation time. |
| Lots of peaks in the capillary electrophoresis trace | Polymerase stops from the original PCR | Optimize initial PCR by changing primers, changing polymerase from Taq to FastStart HiFi (Roche), etc.; be sure to check for PCR success and stops using agarose gels. |
| One of two tag colors missing in capillary electrophoresis trace | Restriction not working properly | Use a fresh batch of restriction enzyme |
| Low signal intensity on capillary electrophoresis trace | Too much salt in samples | Desalt samples, analyze again |
| Deionized formamide old/now longer fully deionized. | Make or purchase fresh deionized formamide | |
| Strange (trailing) peak shape | Sample not denatured | Denature sample again |
| Capillary or gel too old | Replace gel; if problem persists, replace capillary | |
| Standards do not line up as anticipated with sample peaks on capillary electrophoresis trace | Sample not denatured properly | Denature sample again |
| Capillary or gel too old | Replace gel; if problem persists, replace capillary | |
| Large peaks on c.e. trace that correspond to primer lengths | ExoI or alkaline phosphatase not functioning properly | Replace ExoI and calf intestinal alkaline phosphtase |
ANTICIPATED RESULTS
Typical Synthetic Yields
5,6-chrysene quinone (85%); 3,2-benzo[a]phenazine quinone (60%); [Rh(bpy)2(Cl)2]Cl (60%); [Rh(bpy)2(OTf)2]OTf (30–40%); [Rh(bpy)2(NH3)2](X)3, (30% via precipitation, 95% via rotary evaporation); [Rh(bpy)2(chrysi)](Cl)3 (80%); [Rh(bpy)2(phzi)](Cl)3 (50%).
Product Appearance
chrysene quinone, orange powder; 3,2-benzo[a]phenazine quinone intermediate, red powder; 3,2-benzo[a]phenazine quinone, yellow solid; [Rh(bpy)2Cl2]Cl, yellow crystalline solid; [Rh(bpy)2(OTf)2]OTf, yellow-white powder; [Rh(bpy)2(NH3)2](X)3, white powder; [Rh(bpy)2(chrysi)](Cl)3, brownish-red powder; [Rh(bpy)2(phzi)](Cl)3, brownish-yellow powder; Λ-[Rh(bpy)2(chrysi)](Cl)3, browinish-red powder; Δ-[Rh(bpy)2(chrysi)](Cl)3, browinish-red powder.
Analytical Data
5,6-Chrysene quinone: 1H NMR (CD2Cl2) δ 9.4 (d, 1H), 8.85 (d, 1H), 8.8 (d, 1H), 8.5–7.5 (m, 7H); MS (m/z): 259 (M+H+).
3,2-Benzo[a]phenazine quinone: 1H NMR (CD2Cl2) δ 8.8 (d, 1H), 8.3–8.1 (m, 3H), 8.1–7.8 (m, 3H), 7.7 (t, 1H); MS (m/z): 261 (M+H+).
[Rh(bpy)2(Cl)2]Cl: 1H NMR (d6-DMSO): δ 9.71 (d, 2H), 9.01 (d, 2H), 8.90 (d, 2H), 8.63 (t, 2H), 8.33 (t, 2H), 8.17 (t, 2H), 7.82 (d. 2H), 7.59 (t, 2H); FAB-MS (m/z): 485.96.
[Rh(bpy)2(OTf)2]OTf: 1H NMR (d6-DMSO): δ 9.17 (d, 2H), 9.08 (d, 2H), 8.90 (d, 2H), 8.80 (t, 2H), 8.4 (m, 4H), 7.8 (d, 2H), 7.7 (t. 2H); FAB-MS (m/z): 712.9.
[Rh(bpy)2(NH3)2](X)3: 1H NMR (d6-acetone): δ 9.44 (d, 2H), 9.05 (d, 2H), 8.89 (d, 2H), 8.75 (t, 2H), 8.45 (t, 2H), 8.30 (t, 2H), 8.03 (d. 2H), 7.75 (t, 2H), 5.06 (broad singlet, 6H); FAB-MS (m/z): 449.
[Rh(bpy)2(chrysi)](Cl)3: 1H NMR (d4-methanol): δ 8.94 (t, 2H), 8.86 (t, 2H), 8.80 (d, 1H), 8.77 (d, 1H), 8.56 (split t, 2H), 8.44 (m, 5H), 8.4 (d, 1H), 8.15 (m, 1H), 8.03 (m, 1H), 7.95 (m, 3H), 7.86 (d, 1H), 7.81 (d, 1H), 7.64 (m, 5H); FAB-MS (m/z): 671; UV-Vis (Figure 7): λmax: 303 nm (ε=57,000 M−1), 315 nm (ε=52,200 M−1), 391 nm (ε=10,600 M−1).
Δ -[Rh(bpy)2(chrysi)](Cl)3: Circular Dichroism (H2O, see Figure 9): 233 (34), 264 (26), 286 (−12), 308 (−42), 318 (−100), 341 (6).
Λ -[Rh(bpy)2(chrysi)](Cl)3: Circular Dichroism (H2O, see Figure 9): 233 (−34), 264 (−26), 286 (12), 308 (42), 318 (100), 341 (−6).
[Rh(bpy)2(phzi)](Cl)3: 1H NMR (d6-DMSO): δ 14.88 (s, 1H), 14.70 (s, 1H), 9.03 (m, 4H), 8.90 (d, 2H), 8.72 (d, 1H), 8.6 (t, 2H), 8.54 (d, 1H), 8.47 (t, 2H), 8.32 (d, 1H), 8.2 (d, 1H), 8.11 (t, 1H), 8.03 (m, 3H), 7.94 (t, 1H), 7.84 (t, 1H), 7.75 (t, 3H), 7.69 (d, 1H); ESI-MS (m/z): 671 (m-2H+); UV-Vis (Figure 7): λmax: 304 nm (ε=65,800 M−1), 314 nm ( ε=67,300 M−1), 343 nm (ε=39,300 M−1).
Acknowledgments
We are grateful to the National Institutes of Health (GM33309) for their financial support. We also thank ABI for their support. Additionally we are grateful to our coworkers, J. Hart, I. Lau, V. Pierre, and R. Ernst, for their help in working out experimental details.
Footnotes
COMPETING FINANCIAL INTERESTS: The authors have no competing financial interests regarding this chemistry.
References
- 1.Erkkila KE, Odom DT, Barton JK. Recognition and reaction of metallointercalators with DNA. Chem Rev. 1999;99:2777–2795. doi: 10.1021/cr9804341. [DOI] [PubMed] [Google Scholar]
- 2.Kielkopf CL, Erkkila KE, Hudson BP, Barton JK, Rees DC. Structure of a photoactive rhodium complex intercalated into DNA. Nat Struct Bio. 2000;7:117–121. doi: 10.1038/72385. [DOI] [PubMed] [Google Scholar]
- 3.Jackson BA, Barton JK. Recognition of DNA base mismatches by a rhodium intercalator. J Am Chem Soc. 1997;119:12986–12987. [Google Scholar]
- 4.Jackson BA, Alekseyev VY, Barton JK. A versatile mismatch recognition agent: specific cleavage of a plasmid DNA at a single base mispair. Biochem. 1999;38:4655–4662. doi: 10.1021/bi990255t. [DOI] [PubMed] [Google Scholar]
- 5.Jackson BA, Barton JK. Recognition of base mismatches in DNA by 5,6-chrysenequinone diimine complexes of rhodium(III): a proposed mechanism for preferential binding in destabilized regions of the double helix. Biochem. 2000;39:6176–6182. doi: 10.1021/bi9927033. [DOI] [PubMed] [Google Scholar]
- 6.Modrich P. Mechanisms and biological effects of mismatch repair. Ann Rev Genet. 1991;25:229–253. doi: 10.1146/annurev.ge.25.120191.001305. [DOI] [PubMed] [Google Scholar]
- 7.Kolodner R. Biochemistry and genetics of eukaryotic mismatch repair. Genes Dev. 1996;10:1433–1442. doi: 10.1101/gad.10.12.1433. [DOI] [PubMed] [Google Scholar]
- 8.Harfe BD, Jinks-Robertson S. DNA mismatch repair and genetic instability. Ann Rev Genet. 2000;34:359–399. doi: 10.1146/annurev.genet.34.1.359. [DOI] [PubMed] [Google Scholar]
- 9.Modrich P. J. Mechanisms in eukaryotic mismatch repair. Biol Chem. 2006;41:30305–30309. doi: 10.1074/jbc.R600022200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kolodner RD. Mismatch repair: mechanisms and relationship to cancer susceptibility. Trends Biochem Sci. 1995;20:397–401. doi: 10.1016/s0968-0004(00)89087-8. [DOI] [PubMed] [Google Scholar]
- 11.Arzimanoglou II, Gilbert F, Barber HRK. Microsatellite instability in human solid tumors. Cancer. 1998;82:1808–1820. doi: 10.1002/(sici)1097-0142(19980515)82:10<1808::aid-cncr2>3.0.co;2-j. [DOI] [PubMed] [Google Scholar]
- 12.Loeb LA, Loeb KR, Anderson JP. Multiple mutations and cancer. Proc Natl Acad Sci. 2003;100:776–781. doi: 10.1073/pnas.0334858100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Junicke H, Hart JR, Kisko J, Glebov O, Kirsch IR, Barton JK. A rhodium(III) complex for high-affinity DNA base-pair mismatch recognition. Proc Nat Acad Sci. 2003;100:3737–3742. doi: 10.1073/pnas.0537194100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Syvänen A. Acessing genetic variation: genotyping single nucleotide polymorphisms. Nat Rev. 2001;2:930–942. doi: 10.1038/35103535. [DOI] [PubMed] [Google Scholar]
- 15.Hart JR, Johnson MD, Barton JK. Single-nucleotide polymorphism discovery by targeted DNA photocleavage. Proc Nat Acad Sci. 2004;101:14040–14044. doi: 10.1073/pnas.0406169101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mürner H, Jackson BA, Barton JK. A versatile synthetic approach to rhodium(III) diimine intercalators: condensation of ortho-quinones with coordinated cis-ammines. Inorg Chem. 1998;37:3007–3012. [Google Scholar]
- 17.Krotz AH, Kuo LY, Barton JK. Metallointercalators: syntheses, structures, and photochemical characterizations of phenanthrenequinone diimine complexes of rhodium(III) Inorg Chem. 1993;32:5963–5974. [Google Scholar]
- 18.Ferrari M, Carrera P, Cremonesi L. Different approaches to molecular scanning of point mutations in genetic diseases. Pure Appl Chem. 1996;68:1913–1918. [Google Scholar]
- 19.Kwok PY, Deng Q, Zakeri H, Taylor AL, Nickerson DA. Increasing the information content of STS-based genome maps: identifying polymorphisms in mapped STSs. Genomics. 1996;31:123–126. doi: 10.1006/geno.1996.0019. [DOI] [PubMed] [Google Scholar]
- 20.Taillon-Miller P, Piernot EE, Kwok PY. Efficient approach to unique single nucleotide polymorphism discovery. Genome Res. 1999;9:499–505. [PMC free article] [PubMed] [Google Scholar]
- 21.Zhou W. Mapping genetic alterations in tumors with single nucleotide polymorphisms. Curr Opin Onc. 2003;15:50–54. doi: 10.1097/00001622-200301000-00007. [DOI] [PubMed] [Google Scholar]
- 22.Schork NJ, Fallin D, Lanchbury JS. Single nucleotide polymorphisms and the future of genetic epidemiology. Clinical Gentics. 2000;58:250–264. doi: 10.1034/j.1399-0004.2000.580402.x. [DOI] [PubMed] [Google Scholar]
- 23.Rider MJ, Tobe VT, Taylor SL, Nickerson DA. Automating the identification of DNA variations using quality-based fluorescence re-sequencing: analysis of the human mitochondrial genome. Nucleic Acids Res. 1998;26:967–973. doi: 10.1093/nar/26.4.967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Petitjean A, Barton JK. Tuning the DNA-reactivity of cisplatin: conjugatin to a mismatch-specific metallointercalator. J Am Chem Soc. 2004;126:14728–14729. doi: 10.1021/ja047235l. [DOI] [PubMed] [Google Scholar]
- 25.Schatzschneider U, Barton JK. Bifunctional rhodium intercalator conjugates as mismatch-directing DNA alkylating agents. J Am Chem Soc. 2004;126:8630–8631. doi: 10.1021/ja048543m. [DOI] [PubMed] [Google Scholar]
- 26.Zeglis BM, Barton JK. A mismatch-selective bifunctional rhodium-oregon green conjugate: a fluorescent probe for mismatched DNA. J Am Chem Soc. 2006;128:5654–5655. doi: 10.1021/ja061409c. [DOI] [PubMed] [Google Scholar]
- 27.Brunner J, Barton JK. Targeting DNA mismatches with rhodium intercalators functionalized with a cell-penetrating peptide. Biochem. 2006;45:12295–12302. doi: 10.1021/bi061198o. [DOI] [PubMed] [Google Scholar]
- 28.Hart JR, Glebov O, Ernst RJ, Kirsch IR, Barton JK. DNA mismatch-specific targeting and hypersensitivity of mismatch-repair-deficient cells to bulky rhodium(III) intercalators. Proc Nat Acad Sci. 2006;103:15359–15363. doi: 10.1073/pnas.0607576103. [DOI] [PMC free article] [PubMed] [Google Scholar]