Abstract
Mitogen-activated protein kinase (MAPK) signaling pathways play an essential role in the transduction of environmental stimuli to the nucleus, thereby regulating a variety of cellular processes, including cell proliferation, differentiation and programmed cell death. The components of the MAPK extracellular activated protein kinase (ERK) cascade represent attractive targets for cancer therapy as their aberrant activation is a frequent event among highly prevalent human cancers. To understand how MAPKs recognize and phosphorylate their targets is key to unravel their function. However, these events are still poorly understood due to the lack of complex structures of MAPKs with their bound targets in the active site. Here, we have modeled the interaction of ERK with a target peptide and analyzed the specificity towards Ser/Thr-Pro motifs. By using a Quantum Mechanics/Molecular Mechanics (QM/MM) approach we propose a mechanism for the phosphoryl transfer catalyzed by ERK that offers new insights into MAPK function. Our results suggest that 1) the proline residue has a role both in specificity and phospho transfer efficiency; 2) the reaction occurs in one step with ERK2 Asp147 acting as the catalytic base; 3) a conserved Lys in the kinase superfamily usually mutated to check kinase activity strongly stabilizes the transition state; and 4) the reaction mechanism is similar with either one or two Mg2+ ions in the active site. Taken together, our results provide a detailed description of the molecular events involved in the phosphorylation reaction catalyzed by MAPK and contributes to the general understanding of kinase activity.
Introduction
Mitogen-activated protein kinase (MAPK) signaling cascades1 play an essential role in the transduction of environmental stimuli to the nucleus, regulating the expression of genes involved in a variety of cellular processes, including cell proliferation, differentiation, programmed cell death, and neoplastic transformation2–4. MAPKs have been classified into at least six subfamilies, among which the extracellular activated protein kinases ERK/MAPKs (ERK1 and ERK2) are the most extensively studied. The aberrant activation of the ERK pathway has been shown to be a frequent event in many human tumor types5, 6. As an attractive target for cancer chemotherapy, interest in the components of the ERK signaling pathway has exploded in the past few years (see review7 and references therein).
MAPKs are characterized by their requirement of a dual phosphorylation at conserved threonine (Thr) and tyrosine (Tyr) residues for activation8–10 and their specific activity towards serine (Ser)/Thr residues followed by proline (Pro)11. Structural and biochemical studies have extensively characterized the activation process and the catalytic activity of MAPKs; in this sense there is compelling evidence that MAPKs facilitate substrate recognition through docking interactions outside the active site12. However, the origin of Pro selectivity and the phosphotransfer mechanism are not clearly understood, mainly due to a lack of structures of these kinases with targets bound in the active site. Here, we have used computational tools to model the complex of ERK2 with a target peptide and analyzed in detail the reaction mechanism. Based on the wealth of structural information on MAPKs and other kinases we have assembled the catalytic site of ERK2, which enabled us to study the phospho transfer process by means of hybrid quantum-chemical/molecular mechanics (QM/MM) computational schemes 13–15.
Our results indicate that specificity for Pro in the phospho acceptor site may be due to binding restrictions and the fixed positioning of the target residue, suggesting a role in the MAPK catalytic activity besides being important for substrate recognition. The reaction mechanism of members of the kinase superfamily has been studied extensively16, 17. Nevertheless, the detailed chemical mechanism of catalysis remains a matter of controversy. Analysis of the MAPK reaction mechanism using QM/MM tools shows that (1) the ERK2 catalyzed reaction occurs in one step, (2) a conserved aspartic acid (Asp) (Asp147 in ERK2) acts as the catalytic base, and (3) a lysine (Lys) residue conserved in the kinase superfamily (Lys149 in ERK2), forms strong interactions with the γ-phosphate along the reaction pathway, thereby providing the right positioning of the ATP and stabilizing the transition state. Our computational model also enabled us to investigate the contribution of different residues within the MAPK active site to the catalytic efficiency. Overall, our emerging results are aligned with previous experimental studies of MAPKs17–21. Together, the results presented here provide a detailed description of the phosphorylation reaction catalyzed by ERK2 which enhances our ability to understand MAPK function and inhibition.
Materials and Methods
Complex preparation
There is currently no experimental structure of active WT ERK with ATP and Mg2+ cations in a conformation suitable for catalysis. For our study we started by reconstructing the ERK/ATP/Mg2+ complex followed by docking the target peptide and relaxing the whole complex. We have used the crystal structures of the protein ERK2 [Protein Data Bank (PDB22) code 2ERK10] as the starting point for our calculations. The structures of other MAPK-ATP-Mg2+ complexes (P38-γ 23 and JNK324) and biochemical experiments suggest that ERK2 can bind two Mg2+ ions20. Accordingly, we generated a complex of activated ERK2 with ATP and two Mg2+ cations in the active site. For the positioning of ATP and the two Mg2+ ions we used the ERK2-ATP complex (PDB code 1GOL21) and the P38-γ ATP complex (PDB code 1CM8 23). The 1GOL structure21 has a distorted ATP because of the K52R mutation and the presence of only one Mg2+ ion, whereas 1CM8 has two Mg2+ ions and the ATP in a conformation suitable for catalysis. All classical simulations were performed with the AMBER-99 force field 25 and the AMBER 8 package26. The system was solvated with an isometric truncated octahedron of TIP3P 27 water molecules and neutralized by the addition of K+ cations. The parameters for phosphotyrosine and phosphoserine were taken from Craft et al.28 and the ATP parameters from Meagher et al.29. The time step was 2 fs. The particle-mesh Ewald method30 was used with a non-bonded cutoff of 12 Å and the Berendsen thermostat31 was used to maintain the desired temperature. All hydrogen bonds were kept rigid by using the SHAKE algorithm 32.
The structure of ERK2-ATP was first equilibrated at 300 K for 100 ps while keeping the whole protein fixed to allow the water and the ligand molecules to relax. As a restraint we used a harmonic potential with a force constant of 4 kcal/(mol Å2). In a subsequent 500-ps molecular dynamics (MD) simulation without restraints the entire system was allowed to relax and equilibrate. The final snapshot was used as starting point for modeling the peptide-protein interaction.
The complex of ERK2-ATP with a model tripeptide CH3CO-Ala-Thr-Pro-NHCH3 as substrate was assembled first by replacing a water molecule that during the dynamics remain located near the γ-phosphate of ATP by the OH group of Thr, a similar position to the one found for Ser in other kinase-ligand complexes33. It is important to remark that longer peptides than the tripeptide modeled in this study are used in the experimental measurement of MAPKs’ enzymatic activity. This is because peptides lacking the docking motives have only low affinity towards MAPKs. However these docking motives affect binding affinity but not catalytic activity, as they bind to docking-motive binding sites that are far away from the catalytic site. In this sense, for understanding the MAPK reaction mechanism, there is no gain in modeling longer peptides. Among several different trial conformers for the Pro residue following Thr, only trans-Pro left the OH of the Thr in the right position and did not produce clashes with other residues of ERK2. We conducted a 200 ps MD simulation at 300 K while keeping the whole protein and peptide fixed, followed by 500 ps allowing the whole system to relax, which helps accommodate the ATP, the peptide and the C-terminal and N-terminal domains for efficient catalysis. We energy minimized the resulting structure by simulated annealing and conjugate gradient search. The QM/MM calculations were initiated from the resulting complex structure, including water molecules within a sphere of 45 Å from the center of the Mg cations.
To explore the role of the two Mg2+ cations in the reaction process, we performed additional calculations in which one of the ions was removed from the active site to the water solvent. To characterize the specific effect of the Mg2+ ion and maintain electroneutrality, the ion was exchanged with a classical water molecule located near the active site, while remaining part of the QM subsystem. We relaxed the system by conducting short 200-ps MD simulations with restraints acting on residues more than 8 Å away from the active site.
QM/MM methods
The QM computations were performed with the SIESTA code34 at the density functional theory (DFT) level. For all atoms in the QM subsystem, basis sets of double plus polarization quality were employed, with a pseudo-atomic orbital energy shift of 25 meV and a grid cutoff of 150 Ry. Basis functions consist of localized (numerical) pseudo-atomic orbitals, projected on a real space grid to compute the Hartree potential and exchange correlation potentials matrix elements. Calculations were performed using the generalized gradient approximation functional proposed by Perdew, Burke and Ernzerhof35. The classical subsystem was treated using the parameterization from the AMBER force field36. The interface between the QM and MM portions of the system has been treated by the scaled position link atom method37, 38 adapted to the SIESTA code 39.
We used different quantum subsystems in our study. The smallest quantum subsystem consisted of the triphosphate part of ATP, the two Mg2+ cations, the side chain of Asp147, the CH2NH3 + part of the side chain of Lys149 and the side chain of the Thr of the target peptide for a total of 43 atoms. As a bigger subsystem we considered the small subsystem plus all the target tripeptide atoms giving a total of 84 atoms. To explore possible differences between Ser and Thr phosphorylation we mutated the Thr in the peptide to a Ser and used the smaller quantum subsystem to evaluate the differences. The rest of the ERK2 protein, together with water molecules, was treated classically. To separate the QM and classical subsystems, link atoms have been used in the side chains of Asp147, Lys149, the ATP triphosphate moiety and the side chains of the target Thr or Ser.
The complexes obtained from the MD simulations were further relaxed by hybrid QM/MM geometry optimizations using a conjugate gradient algorithm. Only residues located within 8 Å of the any QM atom were allowed to move freely in the QM/MM runs. We then computed the potential energy profiles using restrained energy minimizations along the selected reaction paths. For this purpose, an additional term is added to the potential energy, V(ε) = k (ε − ε0)2/2, where k is an adjustable force constant, ε is the value of the reaction coordinate in the particular system configuration and ε0 is the target value of the reaction coordinate. By varying ε0, the system is forced to follow a low-energy reaction path along the given coordinate.
Results
Target binding specificity
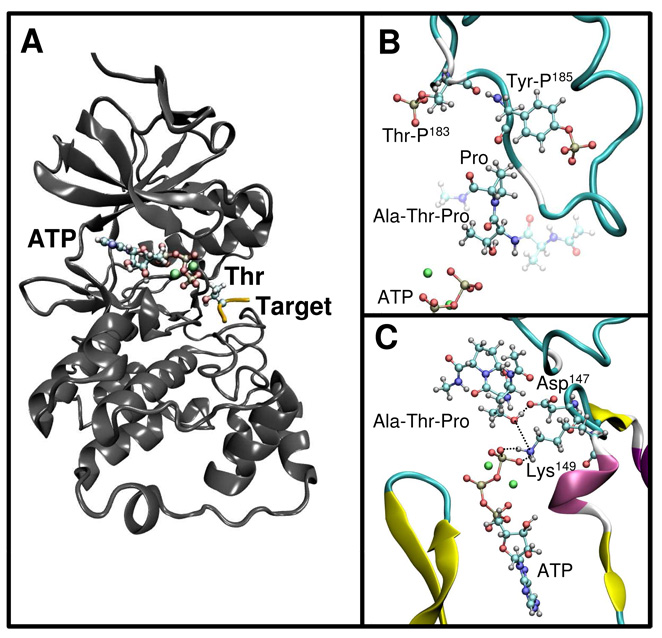
MAPKs are composed of two domains40, an N-terminal domain formed largely by β-sheets and a C-terminal domain that is mostly helical, with four short β-strands [see Figure 1A depicting the complex of active ERK2 (PDB code 2ERK10) with ATP and two Mg2+ cations and a tripeptide CH3CO-Ala-Thr-Pro-NHCH3 as substrate]. The catalytic site, where the ATP and one or two Mg ions bind, is at the junction between these two domains (Figure 1A)40. Upon phosphorylation a conformational change in the activation lip of ERK2 opens up the catalytic site and exposes a hydrophobic pocket to the solvent10. In our model, the small peptide is bound in this hydrophobic pocket, making specific interactions with ERK2.
Figure 1.
(A)The structure of active ERK2 (gray ribbon rendering) in complex with ATP, two Mg2+ cations (ball and stick rendering) and the target peptide (yellow) with the Thr shown in ball and stick representation. (B) Detail of the complex structure showing the Pro conformation and the phosphor-Ser and phsopho-Tyr residues (ball and stick rendering). (C) Another view of the complex showing the interactions of the target Thr with the conserved Asp147 and Lys149.
Our model of the complex provides insights into the molecular basis of the sequence specificity for the peptide target. We found that the consensus Pro is in a trans conformation that helps position the target Thr deep into the catalytic site. Pro interacts with the aromatic side chain of the phosphorylated tyrosine (Tyr185) (Figure 1B) and the side chain of leucine (Leu168). With backbone dihedral angles ω= −170°, φ= −78° and ψ=30°, Pro has an α-helical conformation, as previously observed in an isolated TSPI peptide41. To explore the molecular basis of the specificity of MAPKs toward Pro, we tested the possibility of having different amino acids at the Pro position C-terminal to the phosphorylation site. While keeping the target Thr near the ATP, introduction of bulkier amino acids at the Pro site resulted in clashes with the phosphorylated Tyr, which is tightly bound to Arg189 and Arg192. Even Val, that is structurally similar to Pro, did not fit in. When we mutated Pro to Val in the docked peptide, one of the methyl groups of Val clashed with the side chain of the phosphotyrosine. In contrast, the smaller amino acids glycine (Gly) or alanine (Ala) lack the rigidity to keep the Thr deeply bound into the binding pocket, resulting in significant Thr movement away from the ATP upon unrestrained minimization of the mutated peptides. Indeed, in our QM/MM simulations (see below), we found that the distance between the target oxygen and the ATP is critical for the barrier height. These differences affect the positioning of the peptide in the active site, which helps explain previous experiments in which mutations of Pro to Ala and Gly in the ERK substrate EtsΔ138 produced 5 and 20 fold reductions, respectively, in the rate of catalysis kcat 42.
Comparison of our model with CDK2-peptide complexes shows similar features overall. However, one important difference is the presence of the phospho Tyr185 in ERK2 that makes a smaller binding site for Pro. This difference could reflect the fact that CDK2 also recognizes a positive residue (Lys or Arg) apart from Pro33, a feature that MAPKs lack. Another important characteristic of Pro is the lack of the backbone NH group; interestingly, there is no polar group of ERK2 that enables the formation of a hydrogen bond in that position, which suggests that the binding of amino acids other than Pro would be energetically unfavorable as has been shown in CDK2 and suggested for ERK210, 33. Ala187 in ERK2 has a similar left-handed conformation than Val164 in CDK2 which results in the carbonyl oxygen atom being directed away from the substrate. In summary our results for the complex structure suggest that Pro has two roles to ensure MAPKs specificity, binding deep into the pocket in a rigid structure for efficient catalysis and recognition by specific interactions.
The target peptide and ATP interact with several key residues of ERK2. The OH group of Thr forms a tight hydrogen bond with Asp147, with an O-O distance of 2.6 Å. Lys149 is strongly bound to the γ-phosphate of ATP via two salt-bridges and also interacts with the target Thr as the basic nitrogen is only 3.1 Å apart from the hydroxyl oxygen (Figure 1C). Two possible binding sites for Mg2+ ions in Ser/Thr kinases have been proposed; M1, where the cation is coordinated by two oxygen atoms from the β and γ phosphates of ATP, the acidic oxygen atoms of a conserved Asp, and two water molecules; M2, where the Mg2+ cation is coordinated by two oxygen atoms from the α and γ phosphates of ATP, one oxygen from the conserved Asp, one oxygen from a conserved Asn and a water molecule43. In our model of ERK2 Mg2+ binding is similar to that observed in P3823, Asp165 is coordinating both Mg2+ cations and Asn152 is interacting with the M2 Mg2+ site.
Phosphorylation mechanism
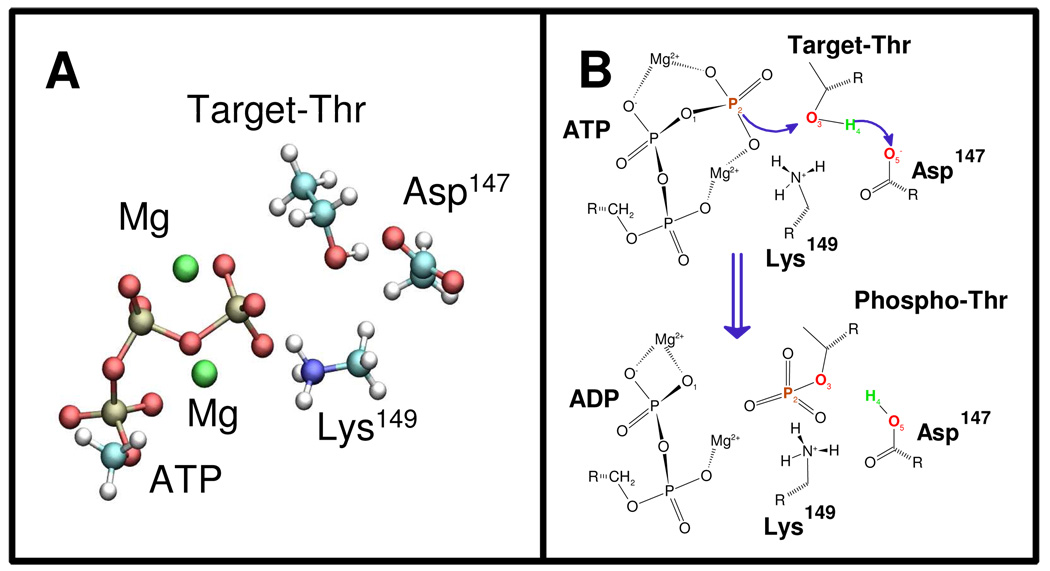
We performed QM/MM calculations to explore the mechanism of the phospho transfer reaction from ATP to the Thr of the bound peptide, catalyzed by the phosphorylated (active) ERK2. We considered the quantum subsystem depicted in Figure 2A; the system includes the triphosphate part of ATP, two Mg2+ atoms and the side chains of Asp147, Lys149 and Thr of the target peptide, resulting in a total of 43 atoms.
Figure 2.
(A) The small quantum subsystem used in the QM/MM calculations. (B) Scheme depicting the movements of the atoms in the phosphoryl-transfer process.
We considered two possible reaction mechanisms, with either the ATP γ-phosphate or Asp147 acting as the proton acceptor from the target Thr, As the reaction involves the rupture of the ATP γ-phosphate bond and the OH bond of the Thr, and the formation of the phosphate Thr bond and the hydrogen (H4) Asp147 or γ-phosphate bond, a natural reaction coordinate should be the simple coordinate, d(O1-P2) - d(P2-O3). However, we found that with this coordinate H4 was not transferred and the energy went continuously up, resulting in an unproductive reaction (data not shown). We then tried the more complex reaction coordinate d(O1-P2) + d(O3-H4) − d(P2-O3) − d(H4-O5) (Figure 2B), where Asp147 acts as the proton acceptor. With this coordinate, we obtained a complete reaction profile with a reasonable activation barrier. However, when we examined the transfer of the Thr OH hydrogen atom to an oxygen atom of the γ-phosphate group of ATP instead of Asp147 we failed to obtain stable products (not shown), which is probably caused by a high energy barrier.
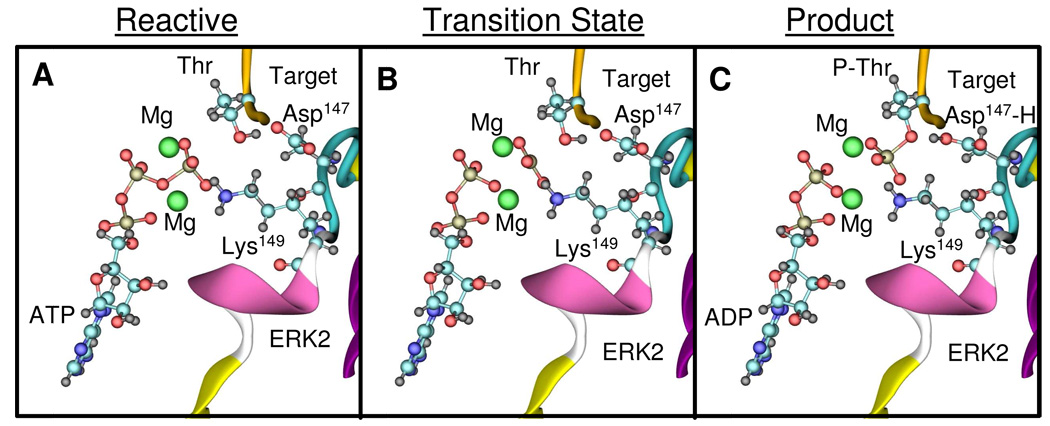
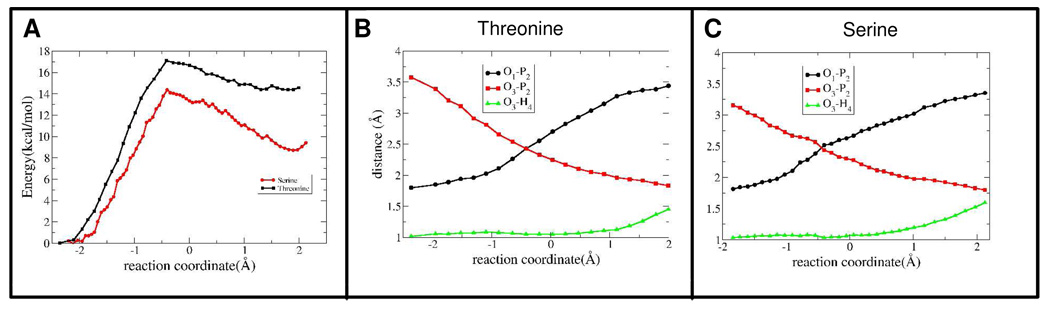
For the reaction with Asp147 as acceptor, the conformations of the reactant, transition, and product states are depicted in Figures 3A–C. The energy as a function of the reaction coordinate is shown in Figure 4A, with an activation barrier of 17.1 kcal/mol. The changes of the most relevant distances as a function of the reaction coordinate are shown in Figure 4B and summarized in Table 1. The reaction pathway involves the breaking of the O1-P2 bond and the concomitant formation of the P2-O3 bond. The O1-P2 bond lengthens from 1.80 Å to 2.43 Å when going from the reactant state to the transition state, while the P2-O3 distance is reduced from 3.60 Å to 2.43 Å (Figure 4B). At the beginning of the reaction path, the Thr moves towards the ATP accompanied by only a modest increase in the O1-P2 distance; only as the Thr OH moiety is brought within ~3 Å of the γ-phosphate does the O1-P2 bond start to break. At the transition state the γ-phosphate PO3 becomes planar and symmetric, with identical P2-O1 and P2- O3 distances of ~2.4 Å (Table 1). By using the analysis suggested by Mildvan44 we found that the reaction has ~90% dissociative character, which is in agreement with similar studies on PKA16, 45.
Figure 3.
Structure of the reactants (A), transition state (B) and products (C) determined for the ERK2 catalyzed reaction. ATP, Mg2+ cations and the conserved Asp147 and Lys149 are depicted in ball and stick rendering.
Figure 4.
(A) Change in potential energy as determined by the QM/MM calculations using the small subsystem as a function of the reaction coordinate for Thr or Ser phosphorylation, respectively. Changes in relevant bond distances as a function of the reaction coordinate for Thr (B) and Ser (C) phosphorylation, respectively.
Table 1.
Calculated activation energies (kcal/mol), Mulliken charges (e), selected optimized distances (Å) for the selected model systems.
| Small Thr | Small Ser | Small 1Mg2+ | Big | Big K52A | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Activation Energy |
17.1 | 14.1 | 5.0 | 13.8 | 19.5 | ||||||||||
| R† | TS | P* | R | TS | P | R | TS | P | R | TS | P | R | TS | P | |
| D(O1-P2) | 1.80 | 2.43 | 3.41 | 1.81 | 2.48 | 3.31 | 1.87 | 2.56 | 3.43 | 1.77 | 2.39 | 3.30 | 1.79 | 2.41 | 3.32 |
| D(P2-O3) | 3.60 | 2.43 | 1.83 | 3.15 | 2.46 | 1.81 | 3.50 | 2.61 | 1.84 | 3.54 | 2.39 | 1.79 | 3.66 | 2.41 | 1.80 |
| D(O3-H4) | 1.01 | 1.05 | 1.46 | 1.03 | 1.04 | 1.58 | 1.00 | 1.02 | 1.49 | 1.02 | 1.06 | 1.60 | 1.02 | 1.15 | 1.59 |
| Δq(ADP)Ψ | 0.00 | −0.38 | −0.46 | 0.00 | −0.41 | −0.51 | 0.00 | −0.27 | 0.47 | 0.00 | −0.49 | −0.55 | - | - | - |
| Δq(Asp147)Ψ | 0.00 | 0.01 | 0.361 | 0.00 | 0.04 | 0.541 | 0.00 | 0.05 | 0.61 | 0.00 | 0.20 | 0.651 | - | - | - |
Reactive state.
Product state.
Difference in net Mulliken charges for the selected moiety.
The product includes the charge of the transferred H4
A key step in the reaction is the proton transfer from Thr to Asp147. Interestingly, we find that the H+ is only transferred when the P2-O3 bond is almost fully formed (Figure 4B). For ERK2, our results thus argue against an alternative scenario in which a low barrier hydrogen transfer to the nearby Asp occurs at early stages of the phospho transfer process46. During the reaction, Lys149 rotates and remains in contact with the γ phosphate as it is transferred to Thr.
The small-QM system clearly point toward a specific phosphorylation mechanism. However, the energy difference of 14.3 kcal/mol between the reactant and product states is unrealistic (Figure 4A). A likely cause is that the phosphate is being transferred to a relatively small QM moiety, CH3CH2OH. To explore this effect, and possible changes in the mechanism, we performed additional calculations with a larger QM system that encompassed the small subsystem plus the entire target tripeptide for a total of 84 atoms (Figure 5A). We used the same reaction coordinate as in the small subsystem, and found no significant differences in the reaction mechanism. However, we obtained a significantly lower activation barrier of 13.8 kcal/mol (Figure 5B and Table 1), a stabilization 3.3 kcal/mol with respect to the small-QM system. The main factor for this difference appears to be the stabilization of the phosphorylated product state. The shape of the potential energy curve shows a remarkable stabilization of the phosphorylated peptide in comparison to the small QM subsystem. The difference in potential energy between reactants and products is 3.2 kcal/mol, showing that the larger subsystem including all of the peptide atoms stabilizes the formation of the phospho-Thr product.
Figure 5.
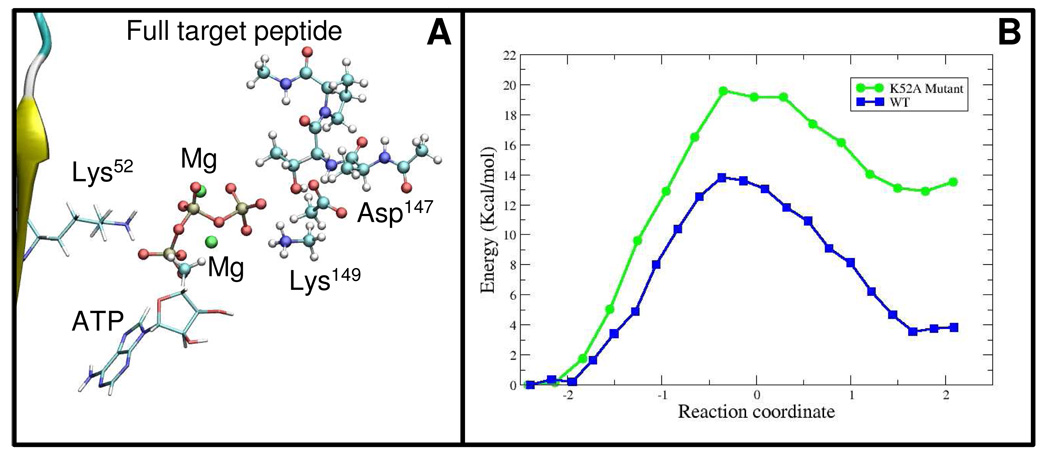
(A) The large quantum subsystem used in the QM/MM calculations (ball and stick rendering) showing the ATP moiety and the position of Lys52 (stick rendering). (B) Change in potential energy as function of the reaction coordinate for the large subsystem using the Wild Type enzyme (WT) and the Lys52 mutant (K52A).
To further explore the role of the different groups in the reaction, we performed a qualitative analysis of the Mulliken charge distribution for reactants, transition states, and products for the two models that we used. We also analyze the potential factors lowering the barrier for the phosphotransfer and stabilizing the ERK2-bound product state in the larger subsystem. Presumably, the γ-phosphate oxygen atoms could increase the nucleophilicity of the substrate hydroxyl group to attack P3. In agreement with this idea, our results show that upon formation of the complex the Thr target receives an extra charge from the enzyme of ~0.2e. At the transition state the leaving phosphate retains part of its negative charge; however there is a significant decrease of around ~0.2e, with a marked change in P3 of 0.1e, that is opposed to the extra charge of ~0.4e and ~0.5e gained by the ADP moiety in the small and big QM subsystems, respectively (Table 1). The charge of Lys149 does not significantly change in the reaction, which suggests that its main role is to position the ATP for catalysis, besides a possible stabilization of the leaving γ-phosphate along the reaction path. The most striking difference observed between the two models is the charge of Asp147; a significant increase in its charge upon formation of the transition state is obtained only in the large subsystem (~0.2e), suggesting that even though the hydrogen transfer has not begun at this point there is a charge transfer from the Thr to Asp147 at an early stage of the reaction. This implies that the conserved Asp in the kinase superfamily may have a role in the activation barrier by accepting charge from the target.
Detailed kinetic analyses of the phosphoryl transfer reaction catalyzed by ERK2 allowed us to relate our results to available experimental information17–19. The suggested ERK2 reaction pathway is shown in Scheme 1; based on solvent viscometric techniques, it has been proposed that the rate limiting step of the reaction is k3, the phosphotransfer step18. However a recent study proposed that both phosphoryl transfer, k3, and substrate release, k4, are the rate limiting steps17. Reported values for k3 are between 10 and 40 s−1 17, 18. An estimate of the reaction barrier obtained by transition state theory, is around 15.0 kcal/mol, which is in reasonable agreement with our results, assuming a prefactor of ~1 ps−1.
Scheme 1.
In summary, we propose a reaction mechanism for ERK2 that involves the dissociation of the γ-phosphate from ATP with the concomitant formation of the phosphopeptide. The proton transfer to the conserved Asp occurs late in the reaction process, but a partial charge transfer to Asp has occurred already at the transition state. Our study provides a detailed description of the phosphorylation catalyzed by MAPKs and therefore broadens our understanding of their function. Is important to remark that the mechanism proposed here is in agreement with previous experimental results17, 18.
Ser vs. Thr phosphorylation
We performed a comparative study by replacing the threonine residue in the target peptide by serine using the small system as reference. Significant differences were observed in the activation energy for Ser phosphorylation, 14.1 kcal/mol, a lowering of 3.0 kcal/mol over Thr (Figure 4A). However, the reaction mechanism, as judged by the changes in the most relevant reaction coordinates (Figure 4C), is very similar. The most striking difference is that the OH group of the smaller serine is closer to the ATP than that of the Thr peptide, as indicated by a reactant O3-P2 distance of ~3.15 Å compared to ~3.60 Å for Thr (Figure 4C). These results points out the importance of Pro in being able to direct the target for efficient catalytic activity. Another important difference is the remarkable stabilization of phospho-Ser with respect to phospho-Thr (~6.4 kcal/mol). Analysis of the changes in Mulliken charges along the reaction path may explain some of the observed differences. Most of the charge changes are similar to those observed before for Thr. However, the charge of the protonated Asp147 in the product state is less negative by ~0.2e than in the Thr case, which may explain the gain in relative stability of the products. The combination of both a decrease in the O3-P2 reactant distance and a stabilization of the product state may explain the reduced activation energy observed for Ser.
MAPKs can phosphorylate either Ser or a Thr residues in their different targets40. However, it is not known if there are differences in the reaction steps (k2, k−2, k3, and k4 in Scheme 1) between Ser and Thr. In a study with model peptides in which Thr was mutated to Ser, no significant changes in substrate production were observed but a detailed analysis of the different kinetic steps was not performed11. The differences observed in our model indicate that k3 should be higher for Ser, suggesting that this residue is a preferred target for ERK2. However, the final selectivity for Ser or Thr will also depend on the different binding affinities for the target peptides and/or dissociation constants of the phosphorylated products. Kinetic studies with target peptides that differ only in the phospho acceptor, Ser or Thr, may help elucidate this aspect of MAPKs selectivity.
Role of Lys52 in catalysis
Mutation of a highly conserved Lys in the kinase superfamily has been reported to reduce or totally abolish kinase activity47. Lys52 of ERK2 interacts with the α and β oxygen atoms of ATP (Figure 5A). Based on crystallographic evidence a possible role for the conserved Lys in the positioning of the ATP molecule was suggested21. It was shown that mutation of Lys52 to Arg distorts the ATP conformation and leads to a kinase with approximately 5% activity of the WT21. However, the Arg side chain overlaps with the ATP binding site and therefore interferes with the binding of the nucleotide21, making it difficult to clearly assess the Lys52 role.
We have analyzed the role of Lys52 in the reaction by generating a neutral Lys52. This model allows us to asses the relevance of Lys52 for catalysis without affecting the conformation of the active site. We conducted constrained optimizations along the previously determined reaction path. Only for the larger QM system we were able to obtain stable products, with a large energy difference between reactants and products, 12.9 kcal/mol in comparison to 3.2 kcal/mol for the wild type (Figure 5B). Moreover, the activation barrier was higher than in the wild type case, 19.5 kcal/mol, a difference of 5.7 kcal/mol. The increase in the activation barrier is consistent with experiments in which an ERK2 K52A mutant had less than 0.5% activity of the wild type protein21. Indeed, Robinson et al. showed that Lys52 has a small effect on Km for ATP but has an important effect on kcat 21. Taking these results into account, it appears that Lys52 affects kinase activity primarily by the stabilization of the transition and product states through electrostatic interactions. As we showed before, the Mulliken charge of the ADP moiety increases as the reaction proceeds. The positive Lys52 may thus be important for stabilization of the ADP product during the reaction.
Phosphorylation and Mg2+ cations
Kinases require Mg2+ cations as cofactors for efficient catalytic activity. Despite the abundant crystal structures and the kinetic characterization of the reaction mechanism the actual role that these cations play is still unclear. In our initial model two Mg2+ cations occupying the M1 and M2 positions were considered. In our simulations the two Mg2+ cations retain their positions during the reaction process, and the coordination remains the same except for the binding to the γ-phosphate. The distance between the M1 Mg2+ and the binding γ-phosphate oxygen is the same along the reaction pathway. In contrast, the distance between the M2 Mg2+ and the γ-phosphate oxygen increases from 2.16 Å to 3.26 Å. According to the calculated Mulliken charges in our models both Mg2+ ions receive charge from the system, ~0.4e for the M1 Mg2+ and ~0.3e for the M2 Mg2+, and these charges do not significantly change during the reaction.
Kinases have been shown to phosphorylate their substrates with only one Mg2+ bound20. In our initial model the Mg in the M1 site is coordinated by two oxygen atoms from the β and γ phosphates of ATP, the acidic oxygen atoms of Asp165, and two water molecules. The Mg bound in the M2 site is coordinated by two oxygen atoms from the α and γ phosphates of ATP, one oxygen from Asp165, one oxygen from Asn152 and a water molecule. To explore the possibility of catalysis with only one Mg2+ ion in the active site, we performed additional calculations. The first question that arises is which of the two binding sites, M1 and M2, is preferentially occupied. Crystallographic studies on PKA, which have been used as the model for analyzing the Ser/Thr kinase family, showed that M1 is the high-affinity Mg2+ site sufficient for catalysis, while the lower affinity M2 site becomes occupied when the concentration of Mg2+ exceeds that for ATP43. However, a careful examination of several other Ser/Thr kinases crystal structures showed that a single bound Mg2+ can be found also in the M2 site (e.g. in structures with PDB codes 1HCK, 1J1C, 1JKK, 1S9I). We noted that the reported Ser/Thr structures have been crystallized with ADP, ATP or non-hydrolyzable analogs; with or without inhibitors; in active or inactive states, and with or without substrates, which precludes a generalization for the whole Ser/Thr protein kinase family. Moreover, the binding site conformations were significantly different among the different structures. Nevertheless, most of the crystal structures of kinases that have high sequence identity with ERK2, and were crystallized with only one Mg2+, had the M2 site occupied instead of the M1 site. Among them, the mutant ERK2 structure 1GOL21, where Lys52 was mutated to arginine (Arg), has ATP and one Mg2+ bound in the M2 site. In agreement with these observations, when we generated one-Mg2+ models with the M1 or M2 Mg2+ moved from the catalytic site to the solvent, we found that only when the M2 Mg2+ was left in the binding site did the phospho moiety of ATP retain a conformation suitable for catalysis. The one-Mg2+ M2 site differed from the two-Mg2+ model mainly in the positioning of one oxygen of the β-phosphate that was binding the M1 site; this oxygen rotated and bonded the M2 Mg2+, in agreement with crystal structures of kinases with only one Mg2+ (Figure 6A)48. Interestingly, this result may explain why the kcat towards ELK1 of the ERK2 Asn152 to Ala mutant is reduced,49 as mutation of Asn152 may change the binding of Mg2+ to the M2 site..
We then used the model with one Mg2+ bound at M2 to analyze the reaction pathway. We followed the same reaction coordinate that we determined for the two-Mg2+ model. Interestingly, the reaction mechanism remained similar to the two-Mg2+ case (Table 1 and Figure S1). The O1-P2 distance was slightly increased, which could lead to a weakening of the O1-P2 bond. We also analyzed the differences in Mulliken charges along the reaction, and found that the M2 Mg2+ charge does not change from the two-Mg2+ model and is constant during the phospho transfer process. The extra charge, previously transferred to the M1 Mg2+ was distributed on the ATP, Asp147 and Lys149 almost equally. Surprisingly, we found an activation barrier of only 5.0 kcal/mol from the change of potential energy along the reaction coordinate. These effects could be caused by a destabilization of the reactant state, as the second Mg2+ strongly interacts with ATP. In summary, our results show that ERK2 is active with one Mg2+ bound to the M2 site and that the reaction mechanism seems to be similar to the two-Mg2+ case. A combination of NMR and QM methods, as used to understand Ca2+ binding to calmodulin50, may help to clarify these points further.
Conclusions
MAPKs regulate gene expression primarily through the phosphorylation of transcription factors51. However, how MAPKs bind and phosphorylate their targets is not yet fully understood. Structural and biophysical aspects of MAPKs have been studied in great detail, with more than 80 structures deposited in the PDB to date52. Even though these studies have resulted in a general understanding of the canonical kinase catalytic domain structure, structures of MAPKs with a target bound in the catalytic site have not been determined. A factor probably interfering with crystallization is that the binding sites of MAPKs for their peptide substrates are mostly outside of the catalytic domain53. Recent studies have provided compelling evidence that these binding sites for docking motifs play a key role in determining the substrate specificity of MAPKs54. Binding to this docking site and the active site usually involves unstructured regions in protein targets which may be another factor precluding crystallization. Here we constructed a model of the complex between ERK2 and a target peptide based on an extensive set of related kinase structures, and tested the model by conducting a QM/MM study of the reaction catalyzed by ERK2. Based on the structural model, we propose that the unique characteristics of Pro are the main reasons for MAPKs specificity towards Ser/Thr-Pro motifs. In particular, Pro has a medium size hydrophobic face, lacks an NH group, and is intrinsically rigid. The capability of positioning the target tightly at the right distance suggests also a role of Pro in catalysis. In this regard, the model that we presented here complements previous structural studies, thus providing further insight into functional aspects of this key family of protein kinases. Another important characteristic of Ser/Thr-Pro motifs in transcription factors is that the undergo Pro cis-trans isomerization, which can be regulated by the prolyl-isomerase Pin1 after phosphorylation by MAPKs55. The resulting enhancement in transcriptional response to growth factors can be attributed to a modified binding mechanism to their co-transcription factors, indicating that the Ser/Thr-Pro selectivity is used to regulate precisely the MAPK signaling routes.
We used the modeled ERK2-peptide complex to determine the reaction pathway catalyzed by this MAPK. The main findings are that the reaction occurs in one step with the Ser/Thr H+ being transferred to a conserved Asp at the end of the reaction. Based on previous QM/MM calculations on PKA16, 45 and CDK2,46 two different mechanisms have been proposed. In the PKA calculations,16, 45 the proton from the alcoholic amino acid was found to be transferred to a conserved Asp, and in CDK246 to a phosphate of the ATP. Our results align with the previous studies on PKA16, 45 and contrast to the results on CDK2.46 An activation barrier of 10 kcal/mol and a symmetrical transition state was found in the original work of Cheng and colleagues16 by performing constraint optimizations along a proposed reaction pathway. Later Valiev et al45 conducted a study on PKA using a QM/MM implementation of the nudged elastic band (NEB) method where no implicit assumption of the reaction coordinates was used. In this case an activation barrier of 15 kcal/mol and an overall similar transition state was determined. In agreement with the two studies on PKA, we find that the proton transfer occurs late in the reaction pathway. As stated above, the main differences arise with a Car-Parrinello QM/MM study46 that proposed an associative phosphoryl transfer with a 24 kcal/mol activation barrier. The position of two key residues that are conserved in the kinase superfamily, Asp147 and Lys149 in ERK2, could account for the observed differences. In the crystal structure of PKA with ADP, aluminum fluoride and a substrate peptide (1L3R56) and in the crystal structure of CDK2 with ATP and a substrate peptide (1QMZ 33) the target serine is located between the Asp and Lys residues, thus precluding the formation of a salt bridge between them. However, in the CDK2 study,46 a salt bridge formed upon molecular dynamics relaxation of the complex. The interaction with Lys likely reduces the proton affinity of Asp, thus eliminating it as the catalytic base. In contrast, here and in the PKA studies16, 45, the salt bridge is not formed and the Asp is free to accept the proton from the Ser/Thr residue. Interestingly, in a structure of PKA similar to 1L3R56, but with an inhibitor bound that lacks the Ser residue (1ATP43), the distance between the conserved Asp and Lys residues is 3.4 Å, almost 1Å shorter than in the 1L3R structure.56 Based on these results from simulation and experiment, we propose that the presence or absence of the salt bridge between Asp and Lys likely determines the proton acceptor and thus an important aspect of the kinase reaction mechanism.
The QM/MM methodology allows us to explore the energetic feasibility of proposed reaction mechanisms, and to analyze the role of different groups within the active site. We determined that a similar mechanism is employed for both Ser and Thr, but Ser phosphorylation has a lower activation barrier than Thr. This difference may be a regulating factor in targets with similar binding affinities. Our study also sheds light on the role of Lys52, a conserved residue in subdomain II whose mutation is often used to confirm kinase activity of novel gene products. We propose that this conserved Lys stabilizes the ADP moiety being formed in the transition and product states, besides positioning the ATP in a conformation conducive for the phosphoryl transfer. We showed that ERK2 is able to phosphorylate its targets even in the presence of only one Mg2+ in the M2 position, in agreement with previous experiments20, and that the reaction mechanism remains essentially unchanged, which helps explain why many kinases are active in the presence of low magnesium concentrations.
QM/MM methods are typically used in structures that have detailed information of the active site. In this study we were able to reconstruct the whole active site due to the wealth of structural information of the kinase superfamily. Good correlation with experiments suggests that this is a reasonable approach. Modeling techniques aiming at understanding protein reaction mechanisms in detail can be extended to cases where the structure of the complex has not been resolved if enough structural and biochemical data are available.
In summary, we constructed a model of the complex of ERK2, ATP, and a target peptide bound to the active site, and used it to analyze in detail the reaction catalyzed by the MAPK ERK2. As kinases from different families share a similar catalytic site, a consensus mechanism could be expected; however, differences may arise by slight conformational changes, specific restraints or substitution of the intervening groups not necessarily directly involved in catalysis. Thus, similar studies on other kinase families may shed additional light on the mechanisms whereby kinases transfer the phospho group from ATP to their specific protein targets.
Supplementary Material
Acknowledgments
Adrian Turjanski was supported by a PEW Latin American Fellow. The authors acknowledge support by the Intramural Research Programs of NIDCR, NIH (A.G.T and J.S.G.) and NIDDK, NIH (G.H.).
Footnotes
Supporting Information Available
S1. The structure of the 1Mg complex and the change in the most relevant distances during the reaction. This information is available free of charge via the Internet at http://pubs.acs.org/.
S2. Coordinates in PDB format of the ERK2 complex used for the QM/MM calculations
References
- 1.Raman M, Chen W, Cobb MH. Oncogene. 2007;26(22):3100–3112. doi: 10.1038/sj.onc.1210392. [DOI] [PubMed] [Google Scholar]
- 2.Chang L, Karin M. Nature. 2001;410(6824):37–40. doi: 10.1038/35065000. [DOI] [PubMed] [Google Scholar]
- 3.Weston CR, Davis RJ. Curr. Opin. Cell. Biol. 2007;19(2):142–149. doi: 10.1016/j.ceb.2007.02.001. [DOI] [PubMed] [Google Scholar]
- 4.Kyriakis JM, Avruch J. Physiol. Rev. 2001;81(2):807–869. doi: 10.1152/physrev.2001.81.2.807. [DOI] [PubMed] [Google Scholar]
- 5.Gioeli D, Mandell JW, Petroni GR, Frierson HF, Jr, Weber MJ. Cancer Res. 1999;59(2):279–284. [PubMed] [Google Scholar]
- 6.Hoshino R, Chatani Y, Yamori T, Tsuruo T, Oka H, Yoshida O, Shimada Y, Ari-i S, Wada H, Fujimoto J, Kohno M. Oncogene. 1999;18(3):813–822. doi: 10.1038/sj.onc.1202367. [DOI] [PubMed] [Google Scholar]
- 7.Lawrence MC, Jivan A, Shao C, Duan L, Goad D, Zaganjor E, Osborne J, McGlynn K, Stippec S, Earnest S, Chen W, Cobb MH. Cell Res. 2008;18(4):436–442. doi: 10.1038/cr.2008.37. [DOI] [PubMed] [Google Scholar]
- 8.Payne DM, Rossomando AJ, Martino P, Erickson AK, Her JH, Shabanowitz J, Hunt DF, Weber MJ, Sturgill TW. Embo J. 1991;10(4):885–892. doi: 10.1002/j.1460-2075.1991.tb08021.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Robinson MJ, Cheng M, Khokhlatchev A, Ebert D, Ahn N, Guan KL, Stein B, Goldsmith E, Cobb MH. J. Biol. Chem. 1996;271(47):29734–29739. doi: 10.1074/jbc.271.47.29734. [DOI] [PubMed] [Google Scholar]
- 10.Canagarajah BJ, Khokhlatchev A, Cobb MH, Goldsmith EJ. Cell. 1997;90(5):859–869. doi: 10.1016/s0092-8674(00)80351-7. [DOI] [PubMed] [Google Scholar]
- 11.Clark-Lewis I, Sanghera JS, Pelech SL. J. Biol. Chem. 1991;266(23):15180–15184. [PubMed] [Google Scholar]
- 12.Remenyi A, Good MC, Lim WA. Curr. Opin. Struct. Biol. 2006;16(6):676–685. doi: 10.1016/j.sbi.2006.10.008. [DOI] [PubMed] [Google Scholar]
- 13.Friesner RA, Guallar V. Annu. Rev. Phys. Chem. 2005;56:389–427. doi: 10.1146/annurev.physchem.55.091602.094410. [DOI] [PubMed] [Google Scholar]
- 14.Mulholland AJ, Grant GH, Richards WG. Protein Eng. 1993;6(2):133–147. doi: 10.1093/protein/6.2.133. [DOI] [PubMed] [Google Scholar]
- 15.Sanchez VM, Crespo A, Gutkind JS, Turjanski AG. J. Phys. Chem. B. 2006;110(36):18052–18057. doi: 10.1021/jp063099q. [DOI] [PubMed] [Google Scholar]
- 16.Cheng Y, Zhang Y, McCammon JA. J. Am. Chem. Soc. 2005;127(5):1553–1562. doi: 10.1021/ja0464084. [DOI] [PubMed] [Google Scholar]
- 17.Wang ZX, Wu JW. J. Biol. Chem. 2007;282(38):27678–27684. doi: 10.1074/jbc.M703161200. [DOI] [PubMed] [Google Scholar]
- 18.Prowse CN, Hagopian JC, Cobb MH, Ahn NG, Lew J. Biochemistry. 2000;39(20):6258–6266. doi: 10.1021/bi000277b. [DOI] [PubMed] [Google Scholar]
- 19.Waas WF, Rainey MA, Szafranska AE, Dalby KN. Biochemistry. 2003;42(42):12273–12286. doi: 10.1021/bi0348617. [DOI] [PubMed] [Google Scholar]
- 20.Waas WF, Dalby KN. Biochemistry. 2003;42(10):2960–2970. doi: 10.1021/bi027171w. [DOI] [PubMed] [Google Scholar]
- 21.Robinson MJ, Harkins PC, Zhang J, Baer R, Haycock JW, Cobb MH, Goldsmith EJ. Biochemistry. 1996;35(18):5641–5646. doi: 10.1021/bi952723e. [DOI] [PubMed] [Google Scholar]
- 22.Berman HM, Battistuz T, Bhat TN, Bluhm WF, Bourne PE, Burkhardt K, Feng Z, Gilliland GL, Iype L, Jain S, Fagan P, Marvin J, Padilla D, Ravichandran V, Schneider B, Thanki N, Weissig H, Westbrook JD, Zardecki C. Acta Crystallogr. D. Biol. Crystallogr. 2002;58(Pt 6 No 1):899–907. doi: 10.1107/s0907444902003451. [DOI] [PubMed] [Google Scholar]
- 23.Bellon S, Fitzgibbon MJ, Fox T, Hsiao HM, Wilson KP. Structure. 1999;7(9):1057–1065. doi: 10.1016/s0969-2126(99)80173-7. [DOI] [PubMed] [Google Scholar]
- 24.Xie X, Gu Y, Fox T, Coll JT, Fleming MA, Markland W, Caron PR, Wilson KP, Su MS. Structure. 1998;6(8):983–991. doi: 10.1016/s0969-2126(98)00100-2. [DOI] [PubMed] [Google Scholar]
- 25.Wang JM, Cieplak P, Kollman PA. J. Comput. Chem. 2000;21(12):1049–1074. [Google Scholar]
- 26.Case A, Pearlman DA, Caldwell JW, Cheatham TE, Wang JM, Ross WS, Simmerling C, Darden T, Merz KM, Stanton RV, Cheng A, Vincent JJ, Crowley M, Tsui V, Gohlke H, Radmer R, Duan Y, Pitera J, Massova I, Seibel GL, Singh UC, Weiner PA. Amber 7. San Francisco: University of California; 2002. [Google Scholar]
- 27.Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML. J. Chem. Phys. 1983;79(2):926–935. [Google Scholar]
- 28.Craft JW, Jr, Legge GB. J. Biomol. NMR. 2005;33(1):15–24. doi: 10.1007/s10858-005-1199-0. [DOI] [PubMed] [Google Scholar]
- 29.Meagher KL, Redman LT, Carlson HA. J. Comput. Chem. 2003;24(9):1016–1025. doi: 10.1002/jcc.10262. [DOI] [PubMed] [Google Scholar]
- 30.Essmann U, Perera L, Berkowitz ML, Darden T, Lee H, Pedersen LG. J. Chem. Phys. 1995;103(19):8577–8593. [Google Scholar]
- 31.Berendsen HJC, Postma JPM, Vangunsteren WF, Dinola A, Haak JR. J. Chem.Phys. 1984;81(8):3684–3690. [Google Scholar]
- 32.Ryckaert JP, Ciccotti G, Berendsen HJC. J. Comput. Phys. 1977;23(3):327–341. [Google Scholar]
- 33.Brown NR, Noble ME, Endicott JA, Johnson LN. Nat. Cell. Biol. 1999;1(7):438–443. doi: 10.1038/15674. [DOI] [PubMed] [Google Scholar]
- 34.Soler JM, Artacho E, Gale JD, Garcia A, Junquera J, Ordejon P, Sanchez-Portal D. J.Phys.:Condens. Matter. 2002;14(11):2745–2779. doi: 10.1088/0953-8984/20/6/064208. [DOI] [PubMed] [Google Scholar]
- 35.Perdew JP, Burke K, Ernzerhof M. Phys. Rev. Lett. 1996;77(18):3865–3868. doi: 10.1103/PhysRevLett.77.3865. [DOI] [PubMed] [Google Scholar]
- 36.Luo R, Wang JM, Kollman PA. Abstr. Paper Am. Chem. Soc. 2002;224:U470–U471. [Google Scholar]
- 37.Eichinger M, Tavan P, Hutter J, Parrinello M. J. Chem. Phys. 1999;110(21):10452–10467. [Google Scholar]
- 38.Rovira C, Schulze B, Eichinger M, Evanseck JD, Parrinello M. Biophys. J. 2001;81(1):435–445. doi: 10.1016/S0006-3495(01)75711-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Crespo A, Scherlis DA, Marti MA, Ordejon P, Roitberg AE, Estrin DA. J. Phys. Chem. B. 2003;107(49):13728–13736. [Google Scholar]
- 40.Chen Z, Gibson TB, Robinson F, Silvestro L, Pearson G, Xu B, Wright A, Vanderbilt C, Cobb MH. Chem. Rev. 2001;101(8):2449–2476. doi: 10.1021/cr000241p. [DOI] [PubMed] [Google Scholar]
- 41.Hamelberg D, Shen T, McCammon JA. J. Am. Chem. Soc. 2005;127(6):1969–1974. doi: 10.1021/ja0446707. [DOI] [PubMed] [Google Scholar]
- 42.Rainey MA, Callaway K, Barnes R, Wilson B, Dalby KN. J. Am. Chem. Soc. 2005;127(30):10494–10495. doi: 10.1021/ja052915p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zheng J, Knighton DR, ten Eyck LF, Karlsson R, Xuong N, Taylor SS, Sowadski JM. Biochemistry. 1993;32(9):2154–2161. doi: 10.1021/bi00060a005. [DOI] [PubMed] [Google Scholar]
- 44.Mildvan AS. Proteins. 1997;29(4):401–416. [PubMed] [Google Scholar]
- 45.Valiev M, Yang J, Adams JA, Taylor SS, Weare JH. J. Phys. Chem. B. 2007;111(47):13455–13464. doi: 10.1021/jp074853q. [DOI] [PubMed] [Google Scholar]
- 46.De Vivo M, Cavalli A, Carloni P, Recanatini M. Chemistry. 2007;13(30):8437–8444. doi: 10.1002/chem.200700044. [DOI] [PubMed] [Google Scholar]
- 47.Kamps MP, Sefton BM. Mol. Cell. Biol. 1986;6(3):751–757. doi: 10.1128/mcb.6.3.751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schulze-Gahmen U, De Bondt HL, Kim SH. J. Med. Chem. 1996;39(23):4540–4546. doi: 10.1021/jm960402a. [DOI] [PubMed] [Google Scholar]
- 49.Zhang J, Zhou B, Zheng CF, Zhang ZY. J. Biol. Chem. 2003;278(32):29901–29912. doi: 10.1074/jbc.M303909200. [DOI] [PubMed] [Google Scholar]
- 50.Biekofsky RR, Turjanski AG, Estrin DA, Feeney J, Pastore A. Biochemistry. 2004;43(21):6554–6564. doi: 10.1021/bi0497852. [DOI] [PubMed] [Google Scholar]
- 51.Turjanski AG, Vaque JP, Gutkind JS. Oncogene. 2007;26(22):3240–3253. doi: 10.1038/sj.onc.1210415. [DOI] [PubMed] [Google Scholar]
- 52.Lee SJ, Zhou T, Goldsmith EJ. Methods. 2006;40(3):224–233. doi: 10.1016/j.ymeth.2006.05.003. [DOI] [PubMed] [Google Scholar]
- 53.Lee T, Hoofnagle AN, Kabuyama Y, Stroud J, Min X, Goldsmith EJ, Chen L, Resing KA, Ahn NG. Mol. Cell. 2004;14(1):43–55. doi: 10.1016/s1097-2765(04)00161-3. [DOI] [PubMed] [Google Scholar]
- 54.Dimitri CA, Dowdle W, MacKeigan JP, Blenis J, Murphy LO. Curr. Biol. 2005;15(14):1319–1324. doi: 10.1016/j.cub.2005.06.037. [DOI] [PubMed] [Google Scholar]
- 55.Monje P, Hernandez-Losa J, Lyons RJ, Castellone MD, Gutkind JS. J. Biol. Chem. 2005;280(42):35081–35084. doi: 10.1074/jbc.C500353200. [DOI] [PubMed] [Google Scholar]
- 56.Madhusudan, Akamine P, Xuong NH, Taylor SS. Nat. Struct. Biol. 2002;9(4):273–277. doi: 10.1038/nsb780. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.