Abstract
Somatic cells can be reprogrammed to a pluripotent state through the ectopic expression of defined transcription factors. Understanding the mechanism and kinetics of this transformation may shed light on the nature of developmental potency and suggest strategies with improved efficiency or safety. Here we report an integrative genomic analysis of reprogramming of mouse fibroblasts and B lymphocytes. Lineage-committed cells show a complex response to the ectopic expression involving induction of genes downstream of individual reprogramming factors. Fully reprogrammed cells show gene expression and epigenetic states that are highly similar to embryonic stem cells. In contrast, stable partially reprogrammed cell lines show reactivation of a distinctive subset of stem-cell-related genes, incomplete repression of lineage-specifying transcription factors, and DNA hypermethylation at pluripotency-related loci. These observations suggest that some cells may become trapped in partially reprogrammed states owing to incomplete repression of transcription factors, and that DNA de-methylation is an inefficient step in the transition to pluripotency. We demonstrate that RNA inhibition of transcription factors can facilitate reprogramming, and that treatment with DNA methyltransferase inhibitors can improve the overall efficiency of the reprogramming process.
Mouse and human cells can be reprogrammed to pluripotency through ectopic expression of defined transcription factors1–9 (‘direct reprogramming’). Generation of such induced pluripotent stem (iPS) cells may provide an attractive source of patient-specific stem cells (reviewed in refs 10, 11). However, the mechanism and nature of molecular changes underlying the process of direct reprogramming remain largely mysterious11. It is a slow and inefficient process that currently requires weeks, with most cells failing to repro-gramme2,9,12–14. A clearer understanding of the process would enable development of safer and more efficient reprogramming strategies, and might shed light on fundamental questions concerning the establishment of cellular identity.
To identify possible obstacles to reprogramming and to use this knowledge to devise ways to accelerate the transition to full pluripotency, we undertook a comprehensive genomic characterization of cells at various stages of the reprogramming process. The characterization involved gene expression profiling, chromatin state maps of key activating and repressive marks (histone H3 K4me3 and K27me3) and DNA methylation analysis.
Response to reprogramming factors
We first studied the response of lineage-committed cells to ectopic expression of the four reprogramming factors Oct4 (also known as Pou5f1), Sox2, Klf4 and c-Myc. Because most induced cells fail to achieve successful reprogramming, we reasoned that genomic characterization might yield insights into the basis of the low overall efficiency of the method.
To eliminate heterogeneity caused by differential viral integration, we studied mouse embryonic fibroblasts (MEFs) isolated from chimaeric mice that had been generated from an iPS cell line carrying integrated doxycycline (Dox)-inducible lentiviral vectors with the four reprogramming factors and a Nanog–GFP (green fluorescent protein) reporter gene13,15. We induced the expression of the repro-gramming factors and obtained gene expression profiles at days 4, 8, 12 and 16 (Supplementary Data). Fluorescence-activated cell sorting (FACS) analysis on day 16 showed that ~20% of the cells stained positive for the stem-cell marker SSEA1, but only ~1.2% had achieved complete reprogramming, as indicated by activation of the Nanog–GFP reporter (Supplementary Fig. 1) and consistent with previous reports13,14.
The immediate response to induction of the reprogramming factors (>3-fold change by day 4) is characterized by de-differentiation from the wild-type MEF state and upregulation ofproliferative genes. De-differentiation is evident in a significant decrease (5–40-fold) in expression levels of typical mesenchymal genes expressed in MEFs (for example, Snai1 and Snai2). The proliferative response is evident in upregulation of genes with functions such as DNA replication (Poli, Rfc4 and Mcm5) and cell cycle progression (Ccnd1 and Ccnd2); this response may be consistent with expression of reprogramming factor c-Myc10,16.
We also detected a strong increase in the expression of stress-induced and anti-proliferative genes. In particular, we detected a sustained 5–10-fold upregulation of Cdkn1a and Cdkn2a, which encode cyclin-dependent kinase (CDK) inhibitors that are key effectors of multiple differentiation and tumour suppressor pathways. Cdkn1a is a downstream target of the reprogramming factor Klf4 (ref. 17), whereas Cdkn2a is known to be activated by deregulated c-Myc expression18. This response was followed by gradual upregulation of genesassociated with differentiating MEFs (Pparg, Fabp4 and Mgp) on days 12–16.This suggests that induction of the reprogramming factors triggers normal ‘fail-safe’ mechanisms that act to prevent uncontrolled proliferation, which may prevent the majority of cells from reaching a stably de-differentiated state.
We also detected strong upregulation of lineage-specific genes from unrelated lineages. These include axon guidance factors (Epha7 and Ngef), epidermal proteins (Krt14, Krt16, Ivl and Sprr1a) and glomerular proteins (Podxl). We speculate that this gene activation reflects responses to the reprogramming factors Sox2 and Klf4, which, independent of their roles in embryonic stem cell regulation, function in neural, epidermal and kidney differentiation10,17.
Pluripotent cell lines
We next studied the changes to gene expression patterns and epigenetic states seen in successfully reprogrammed iPS cells. We analysed three cell lines: MEF-derived iPS cells carrying an Oct4–GFP reporter (MCV8.1; corresponding to subclone 8.1 in ref. 12); mature-B-lymphocyte-derived iPS cells carrying a Nanog–GFP reporter (B-iPS)15; and wild-type embryonic stem cells (V6.5)19.
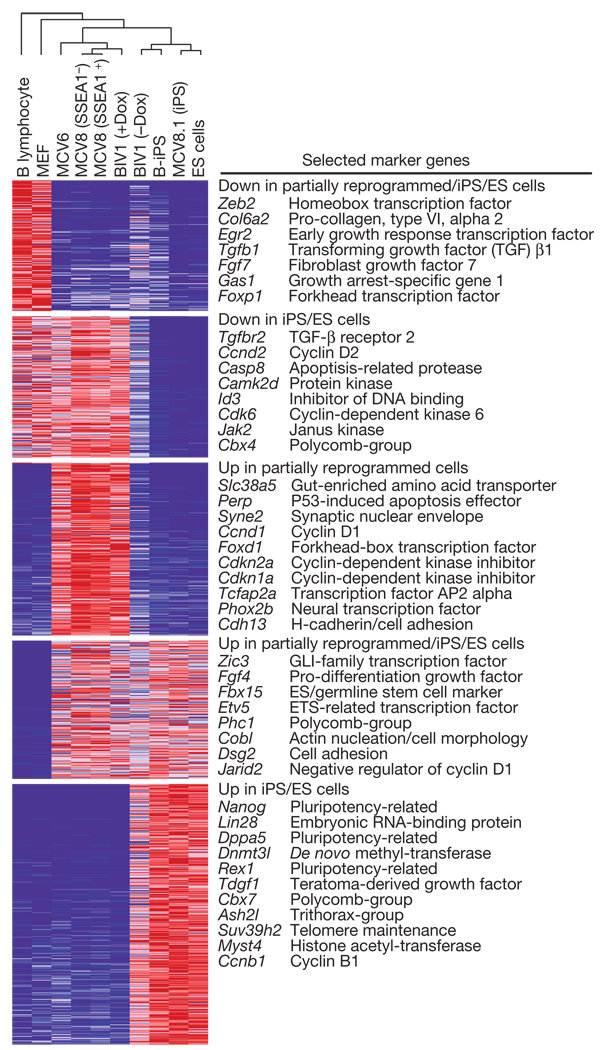
We found that the genome-wide expression profiles of Oct4- or Nanog-iPS cells derived from different cell types and systems are highly similar, but not identical, to wild-type embryonic stem cells (Fig. 1), consistent with recent studies of independent cell lines2,4,9,20. For example, the iPS and embryonic stem cell lines share high expression levels of genes related to maintenance of pluripotency and self-renewal such as Oct4, Sox2, Nanog, Lin28, Zic3, Fgf4, Tdgf1 and Rex1 (also known as Zfp42), and low expression levels for most lineage-specifying transcription factors and other developmental genes. Consistent with the characteristically short cell cycle of embryonic stem cells, the iPS cells show low expression of cyclin D (Ccnd1 and Ccnd2)21.
Figure 1. Gene expression profiling.
Relative expression levels across differentiated, partially reprogrammed and pluripotent cell populations. The dendrogram was generated by complete linkage hierarchical clustering using Pearson correlation on all measured genes. Only genes with at least twofold difference between any pair of samples from different classes are shown in the heat map. Red, white and blue indicate higher, identical and lower relative expression, respectively. ES cells, embryonic stem cells.
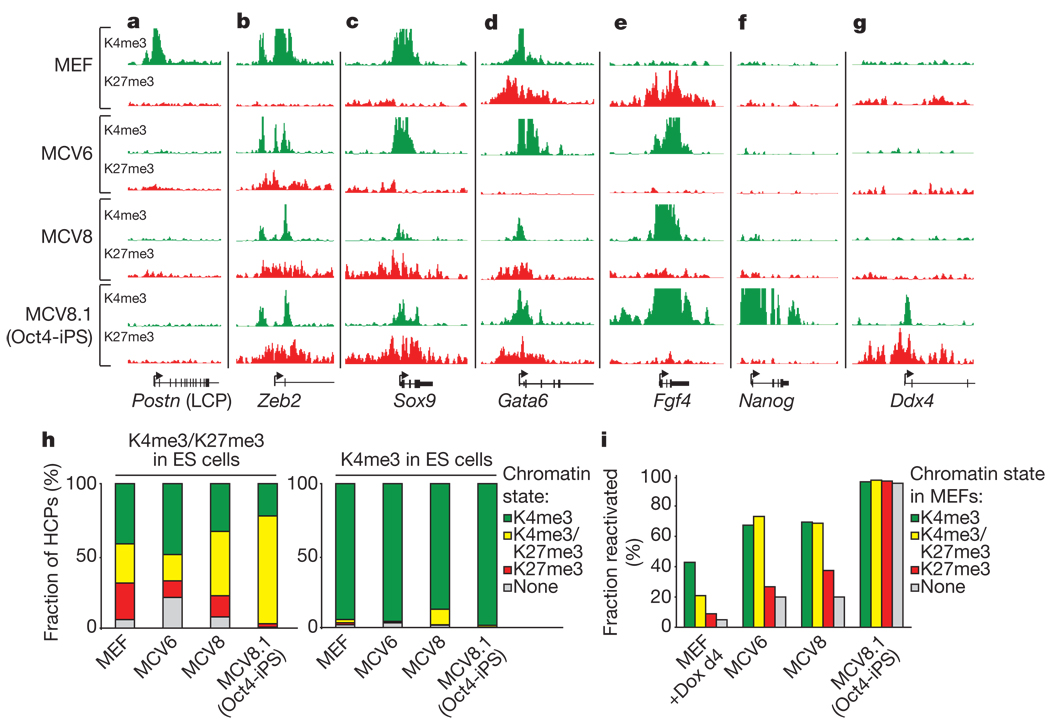
To determine whether iPS cells have also regained embryonic-stem-cell-like chromatin states, we generated genome-wide maps showing the location of H3K4me3 and H3K27me3 from the MEF-derived MCV8.1 cell line using ChIP-Seq. Previously we described the differences in these chromatin modifications between wild-type embryonic stem cells and MEFs22. In embryonic stem cells, virtually all high-CpG promoters (HCPs) are enriched with H3K4me3; a subset of these HCPs, associated with repressed developmental genes, are also enriched with H3K27me3 (‘bivalent’). In MEFs, most HCPs that are bivalent in embryonic stem cells resolve to become monovalent (H3K4me3- orH3K27me3-only). Some pluripotency- and germline-specific genes show loss of both H3K4me3 and H3K27me3 in somatic cells, and this correlates with DNA hypermethylation (ref. 23, and A.M. et al., unpublished observations).
The chromatin state maps of the iPS cell line MCV8.1 are markedly similar to those of embryonic stem cells both near promoters and in intergenic regions (Fig. 2 and Supplementary Figs 2–6). Most (>97%) HCPs that lack H3K4me3-enrichment in MEFs have regained this mark in MCV8.1 cells. At all pluripotency- and germ-line-specific genes examined, the promoters have regained H3K4me3-enrichment and show DNA hypomethylation (Fig. 3). At genes encoding lineage-specific transcription factors that are bivalent and transcriptionally silent in embryonic stem cells, the bivalent pattern is typically re-established (~80% of HCPs classified as bivalent in wild-type embryonic stem cells, and ~95% of loci encoding key developmental transcription factors; Fig. 2b–d, g).
Figure 2. Chromatin state maps.
a, Loss of H3K4me3 correlates with inactivation of MEF-specific low-CpG promoters (LCPs), such as that of Postn (periostin), during reprogramming. b, The transcription factor Zeb2 is marked by H3K4me3 and expressed in MEFs, but gains H3K27me3 and is silenced in partially and fully reprogrammed cells. c, The mesoderm/neural-crest transcription factor Sox9 is marked by H3K4me3 only and remains active in MCV6. d, The endodermal transcription factor Gata6 inappropriately lost H3K27me3 and is activated in MCV6 cells. e, The autocrine growth factor Fgf4 loses H3K27me3, gains H3K4me3 and becomes highly expressed in both partially and fully reprogrammed cells. f, The pluripotency gene Nanog gains H3K4me3 and is active only in iPS cells. g, The germline-specific gene Ddx4 gains H3K4me3 and H3K27me3 in iPS cells only, and remains poised for activation in germ cells. h, Chromatin states for high-CpG promoters (HCPs) in MEFs and reprogrammed cells, conditional on their state in embryonic stem cells. i, Fraction of genes with HCPs expressed in embryonic stem cells, but not wild-type MEFs, that have been re-activated in cells at various stages of reprogramming, conditional on their chromatin state inMEFs. Most HCPs markedbyH3K27me3 onlyorby neither mark are not re-actived in partially reprogrammed cells. d4, day 4.
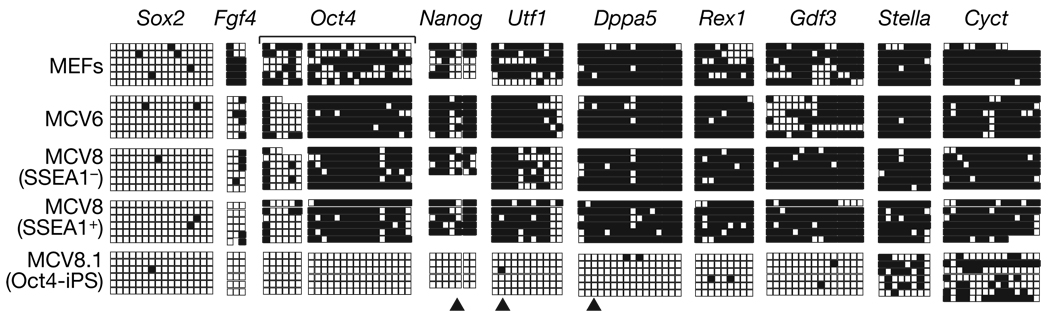
Figure 3. DNA methylation analysis.
Bisulphite sequencing of promoters or enhancers with Oct4/Sox2 binding sites near pluripotency-related and germ-cell-specific (Stella and Cyct) genes, as catalogued in ref. 23. Empty squares indicate unmethylated and filled squares methylated CpG dinucleotides. Most assayed sites are hypermethylated in differentiated and partially reprogrammed cells. Sox2 is enriched with H3K27me3 in non-pluripotent cells and accordingly hypomethylated in all cell types. Triangles show sites used for COBRA analysis (see text).
We conclude that direct reprogramming to a pluripotent state involves re-activation of endogeneous pluripotency-related genes, establishment of an ‘open’ chromatin state (as indicated by genome-wide H3K4me3 enrichment and DNA de-methylation), and comprehensive Polycomb-mediated repression of lineage-specifying genes (as indicated by bivalent chromatin states involving H3K27me3-enrichment).
Partially reprogrammed cell lines
Only a subset of the stably de-differentiated cells obtained in the absence of drug selection show evidence of complete reprogramming to a pluripotent state. Previously we derived clonal cell lines that can be maintained in relatively stable ‘partially reprogrammed’ states in the absence of drug selection12. We reasoned that characterizing such cells might help to identify key barriers in the late stages of the process. Accordingly, we studied three partially reprogrammed independent cell lines established during attempts to reprogramme MEFs or mature B lymphocytes (Fig 1–Fig 3).
MCV8
This cell line, which corresponds to subclone 8 from ref. 12, was established during our attempt to reprogramme MEFs carrying an Oct4–GFP reporter with constitutive retroviruses. It produces heterogeneous cultures of cells with mainly fibroblast-like morphology, with ~20–30% positive for the stem cell marker SSEA1 (Supplementary Figs 7 and 8) and occasional interspersed embryonic-stem-cell-like colonies at late passages. Multiple secondary subclones from these embryonic-stem-cell-like colonies have been shown to establish homogeneous GFP-positive iPS cell lines (including the MCV8.1 line characterized above12). Proviral integration patterns showed that the same parental cells in the MCV8 population gave rise to both GFP-positive and -negative cells, suggesting that complete reprogramming depends on stochastic epigenetic events11,12.
The gene expression patterns of MCV8 cells are clearly distinct from both MEFs and iPS cells (Fig. 1). MCV8 cultures show down-regulation of both structural genes (Col1a1 and Col1a2) and regulatory factors (Snai1, Snai2 and Zeb2) expressed in MEFs, upregulation of some lineage-specific genes with neural, epidermal or endodermal functions (presumably as a consequence of Sox2 and Klf4 expression), and particularly high expression of proliferative genes. Interestingly, high levels of expression can also be detected for several of the CDK inhibitors (Cdkn1a and Cdkn2a) induced by the reprogramming factors. It is unclear how the partially reprogrammed cells have escaped the presumed anti-proliferative effects of these genes, but possible explanations include compensation by overexpression of proliferative genes, repression of differentiation pathways (MCV8 is cultured in the presence of the differentiation inhibitor LIF and expresses the LIF receptor at 2–3-fold higher levels than embryonic stem cells) or transformation (but we note that MCV8 cells have not lost the ability to re-differentiate, see below).
The pattern of re-activation of genes expressed in embryonic stem cells in MCV8 is strongly correlated with chromatin state in MEFs (Fig. 2i). Several genes related to self-renewal and proliferation of embryonic and adult stem cells show re-activation, including the autocrine growth factor Fgf4 (ref. 24) and the transcription factor Zic3 (ref. 25), but genes directly related to pluripotency show low or undetectable expression. Of HCPs that are enriched with H3K4me3 in MEFs but are not expressed at detectable levels, most (~70%) are re-activated in MCV8. In contrast, transcriptionally silent HCPs that are enriched in MEFs for H3K27me3 only or for neither mark are significantly less likely to be re-activated (~35% and ~20%, respectively; PFisher < 10−6).
There are notable differences in the chromatin states of MCV8, MEFs and MCV8.1 iPS cells (Fig. 2). Examining HCPs that are bivalent in embryonic stem cells demonstrates that MCV8 cells show bivalent chromatin structures at 70% more of these loci (n=1,467) than seen in the MEFs (n=859), but at ~40% fewer than in MCV8.1 iPS cells (n = 2,360); this is consistent with partial de-differentiation (~88% of the bivalent loci in MCV8 are also biva-lentinMCV8.1). There are many more HCPs that lack H3K4me3 and H3K27me3 in MCV8 than in MCV8.1 (n =311 versus 31), and these genes include the majority of pluripotency- and germ-cell-specific loci. Using bisulphite sequencing, we confirmed that this chromatin state correlates with DNA hypermethylation (Fig. 3).
We initially sorted MCV8 cells into SSEA1-positive and -negative cells and analysed them separately. However, we found no major differences in expression levels or DNA methylation patterns between the two fractions (Fig 1 and Fig 3; Supplementary Data). Moreover, when the two subpopulations were cultured separately, both reverted to a heterogeneous state within 1–2 passages (Supplementary Fig. 9). Similar results were obtained from sorting by major histocompatibility complex surface expression, which decreases on reprogramming (Supplementary Fig. 10; Supplementary Data). Thus, although these surface markers may provide some enrichment for cells that are amenable to full reprogramming14, they do not seem to discriminate between significantly different cell states within MCV8 cultures.
MCV6
This cell line was also established during our attempt to reprogramme Oct4–GFP MEFs (subclone 6 from ref. 12).It produces homogeneous cultures with compact colonies and embryonic-stem-cell-like morphology (Supplementary Fig. 8). It differs from MCV8 in that it has different proviral integrations and has never spontaneously given rise to fully reprogrammed cells (Supplementary Fig. 7).
The gene expression profile and chromatin state maps from MCV6 are largely similar to those of MCV8, but we found two notable differences. First, MCV6 has fewer genes with bivalent chromatin signatures, and a disproportionately large fraction of HCPs with neither H3K4me3- nor H3K27me3-enrichment (7% versus ~2.5% in MEFs and MCV8). Second, MCV6 expresses high levels of several lineage-specifying transcription factors that are expressed at low or undetectable levels in MCV8 or iPS cells, including Sox9 (Fig. 2c) and Gata6 (Fig. 2d). The latter observation suggests that MCV6 may have become trapped in a more differentiated state than MCV8.
BIV1
This cell line was established during our attempt to reprogramme B lymphocytes with inducible lentiviral vectors15. It had lost surface expression of all common lymphoid markers and did not require any lymphoid cytokines for growth, but also showed no evidence of achieving complete reprogramming during 50days of continuous Dox-mediated viral expression (as judged by the absence of SSEA1- or GFP-positive cells). After Dox withdrawal and loss of any detectable viral expression (see below), the cells continued to proliferate with a more fibroblast-like morphology and, after more than ten additional days in culture, spontaneously gave rise to some GFP-positive embryonic-stem-cell-like colonies, but at a lower frequency than MCV8 (Supplementary Figs 8 and 11).
The gene expression profile and chromatin state maps from BIV1 cells grown with Dox show notable similarities to those of MCV8, including: downregulation of lineage-specific genes, such as the B lymphocyte master regulator Pax5; high expression of proliferative genes; activation of neural and epidermal genes; low levels of H3K4me3 and H3K27me3 enrichment relative to embryonic stem cells, consistent with DNA hypermethylation (see below); and incomplete activation of pluripotency-related loci (Fig. 1; Supplementary Figs 2–6 and 12). Notably, the expression profiles of BIV1, MCV8 and MCV6 are more similar to each other (r2 > 0.9 for any pair) than to the lineage-committed cell types from which they originated or to any of the pluripotent cell types (r2 < 0.8 for any pair; Fig. 1). This suggests that the three cell lines may represent relatively common intermediate states induced by the four reprogramming factors (Oct4, Sox2, Klf4 and c-Myc). (The three lines also show expression of Fbx15, suggesting that they may be similar to the Fbx15-selected cells obtained during initial attempts to generate iPS cells7.)
By comparing the expression profiles of BIV1 cultures before and after Dox withdrawal, we found that Dox withdrawal resulted in: upregulation of mesenchymal extracellular matrix genes (Col1a1 and Col2a1), consistent with the shift to a more fibroblast-like morphology; downregulation of most inappropriately expressed neural and epidermal genes, which is consistent with these genes being induced by overexpression of Sox2 or Klf4; and upregulation of some iPS and embryonic-stem-cell-specific genes (Dppa5 (also known as Dppa5a), Lin28 and Dnmt3l), which is consistent with the eventual emergence of rare GFP-positive colonies. Thus, continuous overexpression of the reprogramming factors may paradoxically have stabilized BIV1 cells in its partially reprogrammed state.
In summary, the three partially reprogrammed cell lines appear to represent similar (but distinct) cell states that emerge at an intermediate stage in the direct reprogramming process. The states are characterized by: re-activation of genes related to stem cell renewal and maintenance, but not pluripotency; incomplete repression of lineage-specific transcription factors; and incomplete epigenetic remodelling, including persistent DNA hypermethylation.
Inhibition of Dnmt1 accelerates reprogramming
Because the partially reprogrammed cell lines show DNA hypermethylation at pluripotency-related genes, we hypothesized that loss of DNA methylation (or a closely linked epigenetic mark, such as H3K9 methylation26) is a critical and inefficient step in the transition from a partially reprogrammed state to pluripotency.
Partially reprogrammed cell lines
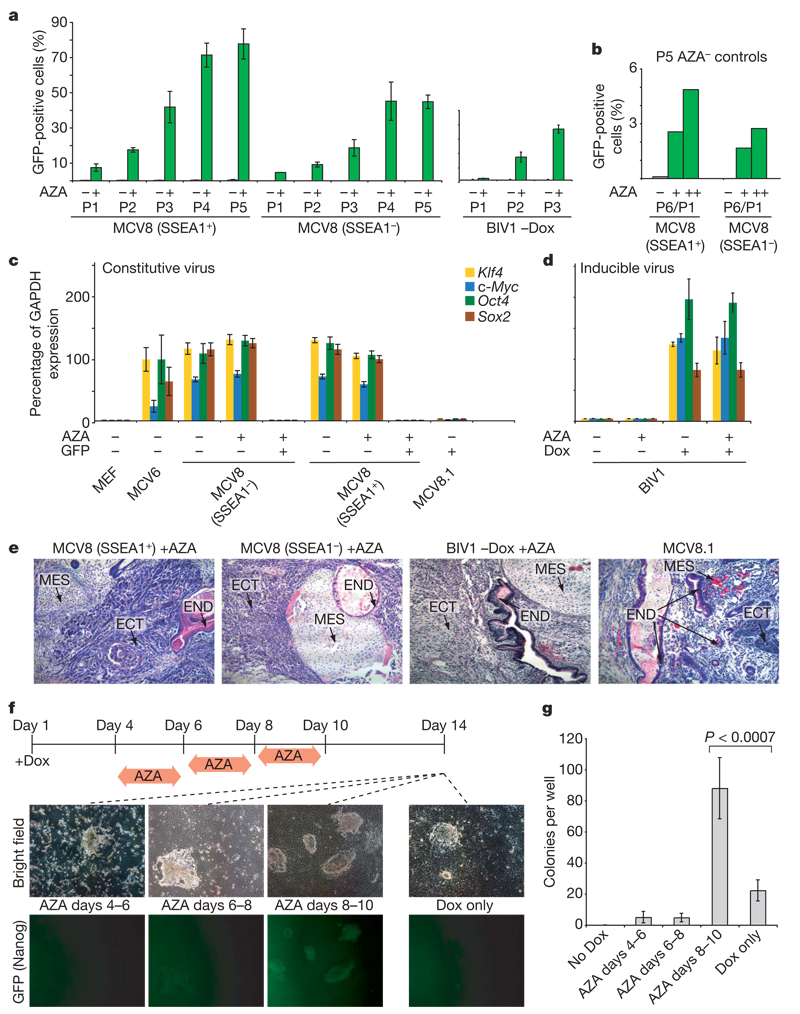
We tested this notion by treating cells with the DNA methyltransferase inhibitor 5-aza-cytidine (AZA) and found that it induced a rapid and stable transition to a fully reprogrammed iPS state. We initially studied SSEA1-positive MCV8 cells, treating them with AZA for 48 h and monitoring the subsequent appearance of GFP-positive cells (Fig. 4a and Supplementary Fig. 7). GFP-positive cells appeared at a frequency of 7.5% after one passage, comparedto0.25%inuntreated cells. After five passages, GFP-positive cells comprised 77.8% of the treated population, whereas the proportion in untreated cells remained stably low (0.41%). We obtained similar results when treating the SSEA1-negative fraction. (When untreated cells from the fifth passage were subsequently treated with AZA, GFP-positive cells appeared at a similar rate as in the initial treatment; Fig. 4b.) We also found robust induction of the GFP reporter after AZA treatment of BIV1 (–Dox) cells (Fig. 4a and Supplementary Fig. 13a).
Figure 4. Inhibition of Dnmt1 accelerates the transition to pluripotency.
a, MCV8 (sorted by FACS using a SSEA1-specific antibody) and BIV1 (−Dox) were either exposed to AZA for 48h (green) or kept in regular embryonic stem cell medium (grey). The number of Oct4–GFP-positive cells was analysed over multiple passages (P) by FACS. b, Untreated MCV8 control cells from passage 5 were subsequently subjected to AZA treatment for 48 h (+) or 120h (++), and resulting Oct4–GFP-positive cells were counted after one passage. P6/P1, total passage 6/passage 1 after AZA treatment. c, AZA treatment does not influence retroviral expression levels. d, AZA treatment has no influence on lentiviral expression in uninduced or induced BIV1 cells. e, Pluripotencyof all AZA-treated lines and MCV8.1 was demonstrated by teratoma formation. ECT, ectoderm; END, endoderm; MES, mesoderm. f, Overall efficiency of AZA treatment. Nanog–GFP MEFs were plated on 6-well plates (4 wells per time point with Dox, and 2 wells without). Cells were treated with AZA during one of the indicated intervals. On day 14, colony formation was analysed by fluorescence microscopy (representative panels are shown). g, Number of alkaline-phosphatase-positive, embryonic-stem-cell-like colonies obtained from each treatment. AZA treatment during days 8–10 resulted in a ~4-fold increase in efficiency over untreated controls. For a, c, d and g, error bars show standard deviations (n = 2, 2, 2 and 4, respectively).
We evaluated the cellular state and developmental potency of the GFP-positive MCV8 and BIV1 cells obtained after AZA treatment and FACS. Both populations stained positive for the stem-cell marker SSEA1. Combined bisulphite restriction analysis (COBRA) revealed significant de-methylation of CpGs near the pluripotency-related genes Dppa5, Nanog and Utf1 (Supplementary Fig. 14), implying that re-activation was not limited to the GFP-tagged reporters. The viral transgenes showed low or undetectable expression levels (Fig. 4c, d) indicating that AZA treatment did not interfere with viral silencing, which is required for full reprogramming9, and that the emergence of GFP-positive cells was not caused by viral re-activation. Finally, subcutaneous injection into severe combined immunodeficiency (SCID) mice led to teratoma formation in 3–4 weeks (Fig. 4e), demonstrating that the GFP-positive cells had undergone a stable transition to the pluripotent state. (Untreated MCV8 or BIV1 cells did not generate teratomas in the same time frame.)
To exclude nonspecific effects of AZA, we treated MCV8 cells with small interfering RNAs (siRNAs) or lentiviral short hairpin RNAs (shRNAs) against Dnmt1, which also led to the appearance GFP-positive cells within one passage (up to 1.7%; Supplementary Fig. 13b–d). We conclude that transient inhibition of Dnmt1 is sufficient to transition MCV8 and BIV1 cells rapidly from a partially repro-grammed state to a pluripotent state.
Populations of lineage-committed cells
We next used the chimaera-derived Nanog–GFP MEFs (described previously) to test whether AZA treatment could increase the overall reprogramming efficiency. The cells were grown in the presence of Dox from day 1, and AZA was administered for 48 h starting on day 4, 6 or 8. The reprogramming efficiency was determined by counting embryonic-stem-cell-like colonies at day 14 (Fig. 4f, g).
We found that starting AZA treatment on days 4 and 6 led to high cell death and no overall gain in efficiency. The cell death may reflect the fact that most cells are still in a differentiated state: genome-wide hypomethylation is known to induce apoptosis in differentiated cells, whereas embryonic stem cells are resistant27–29. In contrast, there was a consistent fourfold increase in the number of embryonic-stem-cell-like colonies in the cultures treated with AZA starting on day 8 (P <0.007; t-test). Moreover, most (>95%) embryonic-stem-cell-like colonies were GFP-positive in the treated cells, whereas only a minority (<25%) were GFP-positive in the untreated controls (a proportion consistent with refs 9, 12–14). Whereas early AZA treatment is counter-productive to reprogramming, there may be a sufficient number of partially reprogrammed cells in the population to outweigh its cytotoxic effect.
We conclude that de-methylation of one or more (unknown) loci is a critical step in the late stages of direct reprogramming, and that inhibition of Dnmt1 lowers this kinetic barrier, thereby facilitating the transition to pluripotency. A similar role for DNA demethylation has been reported recently during in vivo reprogramming in the germ line30.
Transcription-factor-knockdown
In contrast to the other partially reprogrammed cell lines, MCV6 did not respond to AZA treatment (Supplementary Fig. 7).We also noted previously that MCV6 cells never show spontaneous appearance of GFP-positive colonies. We hypothesized that expression of one or more lineage-specifying transcription factor may have stabilized these cells in a more differentiated state than MCV8 or BIV1.
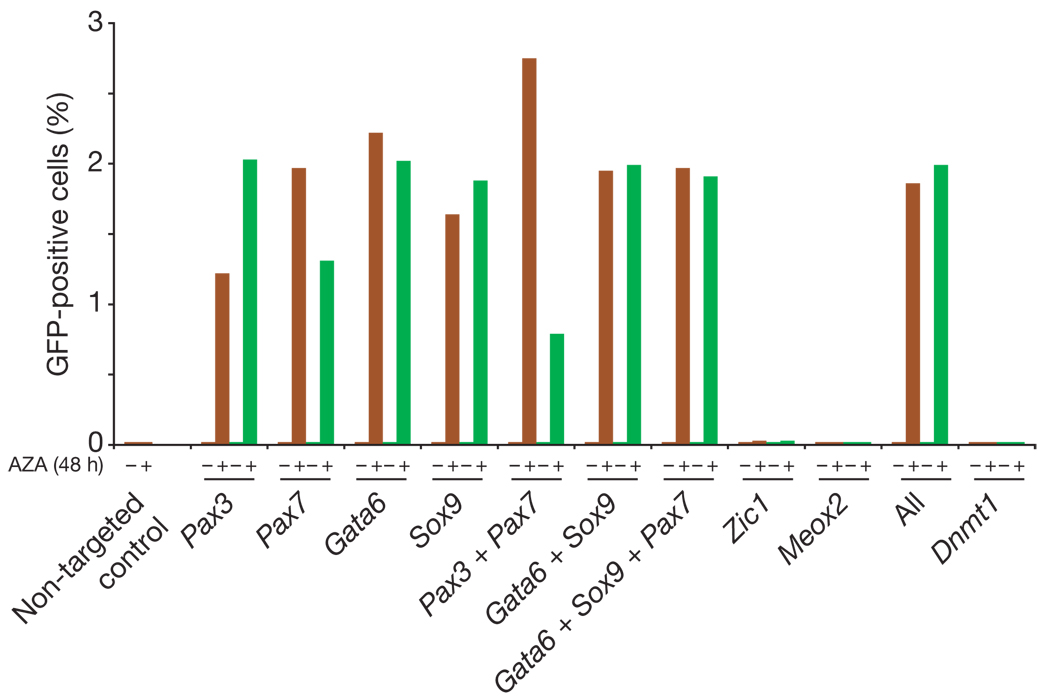
To test this hypothesis, we studied our genome-wide maps and identified lineage-specifying transcription factors that are expressed at low or undetectable levels in MCV8 or iPS cell populations. We transfected MCV6 cells with siRNAs against four transcription factors with >5-fold higher expression in MCV6 than in MCV8 (Gata6, Pax7, Pax3 and Sox9). This resulted in no significant response. However, when transfection ofsiRNA targeting any oneofthe factors was followed by treatment with AZA for 48 h, GFP-positive cells appeared at a significant frequency in all examined populations (16 independent transfections; Fig. 5 and Supplementary Fig. 15). For example, targeting the primitive endoderm marker Gata6 (ref. 31) generated ~2% GFP-positive cells within one passage of the subsequent AZA treatment. In contrast, no GFP-positive cells appeared in populations transfected with negative control siRNAs, or siRNAs targeted against transcription factors not expressed in MCV6 (Zic1 and Meox2) or against Dnmt1 (7 control populations; P< 4 × 10−4; Mann–Whitney U-test).
Figure 5. Transcription factor knockdown facilitates reprogramming.
MCV6 cells were plated onto 24-well dishes and transfected with siRNAs targeting expressed (Pax7, Pax3, Gata6, Sox9) or non-expressed (Zic1, Meox2) transcription factors. One plate was kept in embryonic stem cell medium and the second was exposed to AZA for 48h. Two independent siRNA sequences were used for duplicate experiments (red and green). FACS analysis was performed 48 h after AZA treatment (96 h after transfection) without passaging. The transfection efficiency was estimated as ~20% using Cy3-coupled GADPH control siRNA.
We conclude that re-activation or incomplete repression of lineage-specifying transcription factors during the reprogramming process blocks activation of the endogenous pluripotency regulatory network in MCV6. Transient silencing of one or more of these factors, combined with inhibition of Dnmt1, seems to shift the regulatory balance towards the pluripotent state, which may then be stabilized by autoregulatory feedback11.
Discussion
Several insights emerge from our integrative genomic analyses. First, the Oct4/Sox2/Klf4/c-Myc-based reprogramming process appears to be fairly general, with two independent strategies (constititutive retrovirus or inducible lentivirus) and two distinct cell types (MEFs and B lymphocytes) yielding similar immediate responses, partially reprogrammed states andasimilar mechanism for the final transition to pluripotency. Second, cells may fail to reprogramme successfully for several apparent reasons: the cells may induce anti-proliferative genes in response to proliferative stress; they may inappropriately activate or fail to repress endogenous or ectopic transcription factors, and become ‘trapped’ in differentiated states; and they may fail to reactivate hypermethylated pluripotency genes. Third, complete reprogramming can be facilitated by direct intervention against these failure modes, such as transient inhibition of Dnmt1 and expressed transcription factors.
We expect that further characterization of intermediate states and alternative small molecule treatments will yield critical insights that will help facilitate the desired transitions, making reprogramming efficient and safe for use in regenerative medicine. More generally, our data are consistent with a modelofdevelopment in which cellular states are defined by transcription factors and stabilized by epigenetic remodelling. Integrative gene expression and epigenomic profiling provides a powerful tool for defining and guiding directed transitions between these states.
Note added in proof: The work by A.M. et al. cited in the text as unpublished observations has now been accepted for publication32.
METHODS SUMMARY
Embryonic stem and iPS cells were cultivated on irradiated MEFs. MEFs were infected for 16–20h with the Moloney-based retroviral vector pLIB (Clontech) containing the complementary DNAs of Oct4, Sox2, Klf4 and c-Myc. Cell lines containing the inducible lentiviruses and a ROSA26-targeted M2rtTA were induced with 2 µg ml−1 of Dox. AZA treatment was performed for 48 h or as indicated at a concentration of 0.5 mM.
Bisulphite treatment was performed with the Qiagen EpiTect Kit. For chromatin immunoprecipiation, cells were harvested and cross-linked with formaldehyde (final concentration 1%) for 10 min at 37°C, were washed twice with cold PBS (plus protease inhibitors), frozen and kept at −80 °C. Chromatin immunoprecipiation, library construction, sequencing, identification of enriched intervals and chromatin state classification were performed as described previously22. RNA was isolated using Trizol followed by a second round of purification using RNeasy columns (Qiagen). RNA was then processed and analysed as described elsewhere22.
Reverse transfections were performed in 24-well dishes according to manufacturer’s instructions using the siPORT NeoFX transfection agent (Ambion) and Silencer Select (Ambion/ABI) siRNAs for the respective targets.
Fluorescently conjugated antibodies were used for FACS analysis and cell sorting. Cell sorting was performed by using FACS-Aria (BD-Biosciences), and consistently achieved cell sorting purity of >97%. For determining GFP-positive cell numbers by FACS, we counted >50,000 cells.
Full Methods and any associated references are available in the online version of the paper at www.nature.com/nature.
METHODS
Viral infections and cell lines
MEFs used to derive primary iPS cell lines by infections with inducible lentiviruses were harvested at 13.5 days post coitum from F1 matings between ROSA26–M2rtTA mice33 and Nanog–GFP mice13. Secondary Nanog–GFP MEFs were isolated using neomycin selection. Lentiviral preparation and infection with Dox-inducible lentiviruses encoding Oct4, Klf4, c-Myc and Sox2 cDNA driven by the tetracycline operator (TetO) and a minimal cytomegalovirus (CMV) promoter were described previously13. MCV6 and MCV8 were generated by retroviral infection of Oct4–GFP MEFs as described previously12.
Cell culture
Infected MEFs or secondary inducible MEFs15 were cultured and expanded in standard embryonic stem medium and conditions12. Culture and viral induction were performed as described13,15 and BIV1 was obtained as a stable line and grown under regular embryonic stem cell conditions in the presence or absence of 2 µg ml−1 Dox. AZA treatment was performed for 48h orasindicated at aconcentration of 0.5 mM. Higher doses showed similar effects but increased toxicity.
Expression profiling
RNA was isolated using Trizol followed by a ond round of purification using RNeasy Columns (Qiagen). RNA was then processed and analysed as described elsewhere22. Absolute expression values were Robust Multi-Array (RMA)-normalized, truncated to absolute intensity values ≥20, and visualized using GenePattern (http://www.broad.mit.edu/cancer/software/genepattern/).
Chromatin immunoprecipitation and Illumina/Solexa sequencing
Cells were harvested and cross-linked with formaldehyde (final concentration 1%) for 10 min at 37 °C. They were washed twice with cold PBS (plus protease inhibitors), frozen and kept at –80 °C. Chromatin immunoprecipitation, library construction, sequencing, identification of enriched intervals and chromatin state classification were performed as described previously22.
Bisulphite sequencing and COBRA
Genomic DNA was isolated and bisulphite conversion was performed in a thermocycler using the Qiagen EpiTect Kit according to manufacturer’s instructions with two additional cycles (5 min at 99 °C and 3h at 60°C) at the end. When using 2µg genomic DNA as starting material, converted DNA was eluted in 40µl elution buffer (Qiagen) and 2 µl were used and amplified with previously described primer sets23 and the following additional primer pairs (CyctF: GAAGGATTAAATAGATGTATAAGA AAATAT; CyctR: AAACCCTAATTATAAACAAATACAAC; Sox2F: GGTTTA GGAAAAGGTTGGGAATA; Sox2R: AACCAAAATAAAACAAAACCCATAA). PCR was performed in 25-µl reactions using EpiTect MSP Kit (Qiagen) mastermix according to the manufacturer’s instructions with a 45s annealing step at 50 °C (35 cycles). PCR products were gel-purified, TOPO-cloned (Invitrogen) and sequenced. COBRA for Dppa5, Nanog and Utf1 was performed using15µl of the gel-purified DNA. Dppa5 was digested for 4 h at 65 °C with Taq1 (TCGA). Nanog and Utf1 were digested with HpyCHIV (ACGT) for 4h at 37°C. Digested products were run on 2% agarose gels.
Knockdown of transcription factors and Dnmt1
Reverse transfections were performed in 24-well dishes according to the manufacturer’s instructions using the siPORT NeoFX transfection agent (Ambion). The following Silencer Select (Ambion/ABI) siRNAs were used (the sequence shown is the sense strand): negative control siRNA (4390843: sequence not provided), positive control Cy3 GAPDH siRNA (AM4649: sequence not provided), Pax3 siRNA (s71259, GCCCACGUCUAUUCCACAA; s71260, GCUCCGAUAUUGACUCUGA), Pax7 siRNA (s71271, CCCUCAGUGAGUUCGAUUA; s71272, CCACAUCC GUCACAAGAUA), Gata6 siRNA (s66489, CAAAAAUACUUCUCCUUCU; s66490, CCUCUGCACGCUUUCCCUA), Sox9 siRNA (s74192, AGACU CACAUCUCUCCUAA; s74193, AAGUUGAUCUGAAGCGAGA), Meox2 siRNA (s69792, GCAGUGAAUCUAGACCUCA; s69793, GCCCAUCAU AAUUAUCUGA), Zic1 siRNA (s76384, CAAAAAGUCGUGCAACAAA; s76385, GGGACUUUCUGUUCCGCA) and Dnmt1 siRNA (s65071, GGU AGAGAGUUACGACGAA; s65072, CAACGGAUCCUAUCACACU). Dnmt1 was stably knocked down using five independent shRNAs from the RNA interference consortium (TRC; http://www.broad.mit.edu/genome_bio/trc/). shRNA1 (TRCN0000039024; target: GCTGACACTAAGCTGTTTGTA), shRNA2 (TRCN0000039025; target: GCCTTTACTTTCAACATCAAA), shRNA3 (TRCN0000039026; target: CCGCACTTACTCCAAGTTCAA), shRNA4 (TRCN0000039027; target: CCCGAAGATCAACTCACCAAA) and shRNA5 (TRCN0000039028; target: GCAAAGAGTATGAGCCAATAT). MCV8 cells were infected overnight and selected in puromycin (final, 2µg ml−1) for 48h.
Quantitative RT–PCR
Total RNA was isolated using RNeasy Kit (Qiagen). Three micrograms of total RNA was treated with DNase I to remove potential contamination of genomic DNA using a DNA-Free RNA kit (Zymo Research). Retroviral expression levels were determined as described previously9. For inducible lentiviral expression, 1 µg of DNase I-treated RNA was reverse transcribed using a First Strand Synthesis kit (Invitrogen) and ultimately resuspended in 100 µl of water. Quantitative PCR analysis was performed in triplicate using 1/50 of the reverse transcription reaction in an ABI Prism 7000 (Applied Biosystems) with Platinum SYBR green qPCR SuperMix-UDG with ROX (Invitrogen). Primers used for amplification were as follows: c-Myc: F, 5'-ACCTAACTCGAGGAGGAGCTGG-3', and R, 5'-TCCACATAGCGTAAA AGGAGC-3'; Klf4: F, 5'-ACACTGTCTTCCCACGAGGG-3', and R, 5'-GGCATTAAAGCAGCGTATCCA-3'; Sox2: F, 5'-CATTAACGGCACACTG CCC-3', and R, 5'-GGCATTAAAGCAGCGTATCCA-3'; Oct4: F, 5'-AGCCTGGCCTGTCTGTCACTC-3', and R, 5'-GGCATTAAAGCAGC GTATCCA-3'. To ensure equal loading of cDNA into qRT–PCR reactions, GAPDH messenger RNA was amplified using the following primers: F, 5'-TTCACCACCATGGAGAAGGC-3', and R, 5'-CCCTTTTGGCTCCACCCT-3'. Data were extracted from the linear range of amplification. All graphs of qRT–PCR data shown represent samples of RNA that were DNase-treated, reverse transcribed, and amplified in parallel to avoid variation inherent in these procedures.
Flow cytometry analysis and cell sorting
The following fluorescently conjugated antibodies (PE, FITC, Cy-Chrome or APC-labelled) were used for FACS analysis and cell sorting: anti-SSEA1 (RnD Systems), anti-Igk, anti-Igλ1,2,3, anti-CD19, anti-B220, anti-sIgM and anti-sIgD (all obtained from BD-Biosciences). Cell sorting was performed by using FACS-Aria (BD-Biosciences), and consistently achieved cell sorting purity of >97%. For determining GFP-positive cell numbers by FACS, we counted >50,000 cells.
Supplementary Material
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
Acknowledgements
We thank the staff of the Broad Institute Genome Sequencing Platform, Genetic Analysis Platform and RNAi Platform for assistance with reagents and data generation. This research was supported by funds from the National Institutes of Health, the National Human Genome Research Institute, the National Cancer Institute, and the Broad Institute of MIT and Harvard.
Footnotes
Author Information All analysed data sets can be obtained from http://www.broad.mit.edu/seq_platform/chip/. Microarray and sequence data have been submitted to the NCBI GEO database under accession numbers GSE10871 and GSE11074, respectively. Reprints and permissions information is available at http://www.nature.com/reprints.
References
- 1.Aoi T, et al. Generation of pluripotent stem cells from adult mouse liver and stomach cells. Science. 2008 doi: 10.1126/science.1154884. [DOI] [PubMed] [Google Scholar]
- 2.Maherali N, et al. Directly reprogrammed fibroblasts show global epigenetic remodeling and widespread tissue contribution. Cell Stem Cells. 2007;1:55–77. doi: 10.1016/j.stem.2007.05.014. [DOI] [PubMed] [Google Scholar]
- 3.Nakagawa M, et al. Generation of induced pluripotent stem cells without Myc from mouse and human fibroblasts. Nature Biotechnol. 2008;26:101–106. doi: 10.1038/nbt1374. [DOI] [PubMed] [Google Scholar]
- 4.Okita K, Ichisaka T, Yamanaka S. Generation of germline-competent induced pluripotent stem cells. Nature. 2007;448:313–317. doi: 10.1038/nature05934. [DOI] [PubMed] [Google Scholar]
- 5.Park IH, et al. Reprogramming of human somatic cells to pluripotency with defined factors. Nature. 2008;451:141–146. doi: 10.1038/nature06534. [DOI] [PubMed] [Google Scholar]
- 6.Takahashi K, et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 7.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 8.Yu J, et al. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318:1917–1920. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- 9.Wernig M, et al. In vitro reprogramming of fibroblasts into a pluripotent ES-cell-like state. Nature. 2007;448:318–324. doi: 10.1038/nature05944. [DOI] [PubMed] [Google Scholar]
- 10.Yamanaka S. Strategies and new developments in the generation of patient-specific pluripotent stem cells. Cell Stem Cells. 2007;1:39–49. doi: 10.1016/j.stem.2007.05.012. [DOI] [PubMed] [Google Scholar]
- 11.Jaenisch R, Young R. Stem cells, the molecular circuitry of pluripotency and nuclear reprogramming. Cell. 2008;132:567–582. doi: 10.1016/j.cell.2008.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Meissner A, Wernig M, Jaenisch R. Direct reprogramming of genetically unmodified fibroblasts into pluripotent stem cells. Nature Biotechnol. 2007;25:1177–1181. doi: 10.1038/nbt1335. [DOI] [PubMed] [Google Scholar]
- 13.Brambrink T, et al. Sequential expression of pluripotency markers during direct reprogramming of mouse somatic cells. Cell Stem Cell. 2008;2:151–159. doi: 10.1016/j.stem.2008.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stadtfeld M, et al. Defining molecular cornerstones during fibroblast to iPS cell reprogramming in mouse. Cell Stem Cell. 2008;2:230–240. doi: 10.1016/j.stem.2008.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hanna J, et al. Direct reprogramming of terminally differentiated mature B lymphocytes to pluripotency. Cell. 2008;133:250–264. doi: 10.1016/j.cell.2008.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Adhikary S, Eilers M. Transcriptional regulation and transformation by Myc proteins. Nature Rev. Mol. Cell Biol. 2005;6:635–645. doi: 10.1038/nrm1703. [DOI] [PubMed] [Google Scholar]
- 17.Rowland BD, Peeper DS. KLF4, p21 and context-dependent opposing forces in cancer. Nature Rev. Cancer. 2006;6:11–23. doi: 10.1038/nrc1780. [DOI] [PubMed] [Google Scholar]
- 18.Gregory MA, Qi Y, Hann SR. The ARF tumor suppressor: keeping Myc on a leash. Cell Cycle. 2005;4:249–252. [PubMed] [Google Scholar]
- 19.Rideout WM, III, et al. Generation of mice from wild-type and targeted ES cells by nuclear cloning. Nature Genet. 2000;24:109–110. doi: 10.1038/72753. [DOI] [PubMed] [Google Scholar]
- 20.Lowry WE, et al. Generation of human induced pluripotent stem cells from dermal fibroblasts. Proc. Natl Acad. Sci. USA. 2008;105:2883–2888. doi: 10.1073/pnas.0711983105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Orford KW, Scadden DT. Deconstructing stem cell self-renewal: genetic insights into cell-cycle regulation. Nature Rev. Genet. 2008;9:115–128. doi: 10.1038/nrg2269. [DOI] [PubMed] [Google Scholar]
- 22.Mikkelsen TS, et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448:553–560. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Imamura M, et al. Transcriptional repression and DNA hypermethylation of a small set of ES cell marker genes in male germline stem cells. BMC Dev. Biol. 2006;6:34. doi: 10.1186/1471-213X-6-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Silva J, Smith A. Capturing pluripotency. Cell. 2008;132:532–536. doi: 10.1016/j.cell.2008.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lim LS, et al. Zic3 is required for maintenance of pluripotency in embryonic stem cells. Mol. Biol. Cell. 2007;18:1348–1358. doi: 10.1091/mbc.E06-07-0624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bernstein BE, Meissner A, Lander ES. The mammalian epigenome. Cell. 2007;128:669–681. doi: 10.1016/j.cell.2007.01.033. [DOI] [PubMed] [Google Scholar]
- 27.Jackson-Grusby L, et al. Loss of genomic methylation causes p53-dependent apoptosis and epigenetic deregulation. Nature Genet. 2001;27:31–39. doi: 10.1038/83730. [DOI] [PubMed] [Google Scholar]
- 28.Lei H, et al. De novo DNA cytosine methyltransferase activities in mouse embryonic stem cells. Development. 1996;122:3195–3205. doi: 10.1242/dev.122.10.3195. [DOI] [PubMed] [Google Scholar]
- 29.Meissner A, et al. Reduced representation bisulfite sequencing for comparative high-resolution DNA methylation analysis. Nucleic Acids Res. 2005;33:5868–5877. doi: 10.1093/nar/gki901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hajkova P, et al. Chromatin dynamics during epigenetic reprogramming in the mouse germ line. Nature. 2008;452:877–881. doi: 10.1038/nature06714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Singh AM, et al. A heterogeneous expression pattern for Nanog in embryonic stem cells. Stem Cells. 2007;25:2534–2542. doi: 10.1634/stemcells.2007-0126. [DOI] [PubMed] [Google Scholar]
- 32.Meissner A, et al. Genome-scale DNA methylation maps of pluripotent and differentiated cells. Nature. doi: 10.1038/nature07107. (in the press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Beard C, et al. Efficient method to generate single-copy transgenic mice by site-specific integration in embryonic stem cells. Genesis. 2006;44:23–28. doi: 10.1002/gene.20180. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.