Abstract
SCRATCHY is a methodology for the construction of libraries of chimeras between genes that display low sequence homology. We have developed a strategy for library creation termed enhanced crossover SCRATCHY, that significantly increases the number of clones containing multiple crossovers. Complementary chimeric gene libraries generated by incremental truncation (ITCHY) of two distinct parental sequences are created, and are then divided into arbitrarily defined sections. The respective sections are amplified by skewed sets of primers (i.e. a combination of gene A specific forward primer and gene B specific reverse primer, etc.) allowing DNA fragments containing non-homologous crossover points to be amplified. The amplified chimeric sections are then subjected to a DNA shuffling process generating an enhanced crossover SCRATCHY library. We have constructed such a library using the rat theta 2 glutathione transferase (rGSTT2) and the human theta 1 glutathione transferase (hGSTT1) genes (63% DNA sequence identity). DNA sequencing analysis of unselected library members revealed a greater diversity than that obtained by canonical family shuffling or with conventional SCRATCHY. Expression and high-throughput flow cytometric screening of the chimeric GST library identified several chimeric progeny that retained rat-like parental substrate specificity.
INTRODUCTION
Diversification of parental sequence(s) is a critical component in the directed evolution of enzymes and other proteins. In addition to conventional random mutagenesis using error-prone PCR, a variety of methods including family shuffling by sexual PCR (1,2) have been developed for the construction of highly diversified mutant libraries.
Family shuffling by sexual PCR is an efficient method for the generation of chimeric genes based on homologous recombination between two or more closely related parental sequences (1,2). Briefly, the parental genes are randomly fragmented by DNase I followed by reassembly under conditions allowing homology-dependent annealing and extension of the gene fragments. During the process, loci of the parental genes are allocated to the reassembled fragments forming chimeric progeny with various combinations of parental sequences. Consequently, the progeny generated by shuffling are scattered between and around the parental genes in sequence space resulting in a chimeric library that covers a wider area of functional sequence space compared with that covered by random mutagenesis of a single parental gene (2). The wider coverage of sequence space improves the odds of identifying sequences with a desired function. In fact, family shuffling by sexual PCR and its variants has been successfully applied to a variety of enzymes including cephalosporinases (2), biphenyl dioxygenases (3), thymidine kinases (4), catechol 2,3-dioxygenases (5) and triazine hydrolases (6).
The primary drawback of family shuffling is that recombination preferentially occurs in regions of high homology (7,8). Thus, the quantity and positioning of crossover points in the individual progeny are constrained, particularly when attempting chimeragenesis between distantly related sequences. In such cases, the result is typically poor diversity in the progeny sequence repertoire in addition to a high frequency of unshuffled parental sequences.
SCRATCHY (9) is an alternative method that enables combinatorial engineering of target proteins independent of sequence identity. SCRATCHY consists of a combination of two distinct methods for chimeric gene library construction: Incremental Truncation for the Creation of HYbrid enzymes [ITCHY (10,11)] and DNA shuffling (1). In addition to the re-distribution of non-homologous crossovers generated in ITCHY library construction (a single crossover per gene), homology-dependent crossovers are spontaneously generated during the shuffling process when the two parental sequences are sufficiently (>79%) similar (9). However, when attempting chimeragenesis between two distantly related parents, low sequence identity generally prevents the formation of homology-dependent crossovers during the shuffling step. In the crossover probability prediction, the average frequency of crossovers in SCRATCHY progeny is typically close to 1.0 (9). This is because of the absence of homologous recombination and the fact that allocation of non-homologous crossovers derived from ITCHY clones, which contain one crossover per gene, essentially follows a Poisson distribution centered on this crossover number. A further undesired consequence is that SCRATCHY libraries contain revertant wild-type parental sequences (i.e. progeny without crossovers) as a significant (>10–20%) population. The low complexity of the resulting chimeric sequences caused by the poor crossover numbers only allows coverage of a small and restricted area in the huge sequence space spread between and around the parental sequences.
Herein, we report the development of a simple and efficient method to improve sequence diversity in SCRATCHY libraries by increasing the frequency of non-homologous crossover events. This method allowed generation of a complex chimeric gene library of two distantly related proteins from which efficient chimeragenesis could not be achieved by other methods based on homologous recombination. Unlike other gene shuffling techniques, this methodology allows for the regulation of the average crossover number per progeny gene. Additionally, bias in the position of crossover points can be controlled using this method.
MATERIALS AND METHODS
Materials
The DNA polymerases rTaq and Pfu-turbo were purchased from TaKaRa (Japan) and Stratagene (USA), respectively. DNA nucleases and ligases were obtained from New England Biolabs (USA) and Roche (Germany), respectively, unless specifically noted. Primers were from Integrated DNA Technologies (USA). Plasmids bearing rat GSTT2.2 and human GSTT1.1 were a kind gift from Prof. Bengt Mannervik. The pET-28a expression vector was purchased from Novagen (USA). Plasmid isolation and agarose gel purification kits were obtained from Qiagen (USA). Terrific broth (TB) growth media was from Difco (UK). Monochlorobimane and 7-amino-4-chloromethyl coumarin were obtained from Molecular Probes (USA). All other reagents were purchased from Sigma-Aldrich (USA) unless specifically noted.
ITCHY library construction
The complementary rGST-hGST (r-hGSTT) and hGST-rGST (h-rGSTT) ITCHY libraries with non-homologous crossover points were generated using: (i) full-length rGSTT2 (DNA encoding amino acids 1–244) and the region encoding the C-terminal domain of hGSTT1 (DNA encoding amino acids 79–240). (ii) Full-length hGSTT1 (DNA encoding amino acids 1–240) and the region encoding the C-terminal domain of rGSTT2 (DNA encoding amino acids 79–244), respectively. The r-hGSTT ITCHY library was prepared according to Ostermeier et al. (10). The h-rGSTT library was prepared according to Lutz et al. (12). The resulting h-rGSTT and r-hGSTT chimeric clones were subjected to ORF selection by fusing a Neomycin resistance gene to the C-terminus of the hybrids. In-frame hybrids were then selected by plating the libraries on kanamycin plates (M.Ostermeier and S.J.Benkovic, unpublished results).
Chimeric section-specific amplification using skewed primers
The mammalian GSTT chimeric sequences constructed using ITCHY were arbitrarily divided into three overlapping sections: section 1 (1–266 nt), section 2 (240–456 nt) and section 3 (438-termination codon, where the A of the ATG initiation codon was defined as +1). DNA fragments having chimeric junctions in sections 2 or 3 were amplified by the following combinations of skewed primers; h2F (5′-CTG ACTACTGGTACCCTCAG-3′) and r2R (5′-CTGAGGAAC TTGTCCTCCAGACG-3′) for the second section of h-rGSTT, h3F (5′-CGAGGACAAGTTCCTCCAGA-3′) and r3R (5′- TACGCTGACGTCGAGTCTAAGCTTAGGGAATCCTG GCAATTC-3′) for the third section h-rGSTT, r2F (5′-CAGACCACTGGTACCCGGC-3′) and h2R (5′-TCTGGAG GAACTTGTCCTCGAGCAA) for the second section of r-hGSTT and r3F (5′-CTGGAGGACAAGTTCCTCAGGG-3′) and h3R (5′-TACGCTGACGTCGAGTCTAAGCTTAAC GGATCATGGCCAGC-3′) for the third section of r-hGSTT. The chimeric section-specific amplification was carried out using the following reaction conditions: 94°C for 60 s; 24 cycles of 94°C for 30 s, 50°C for 30 s, 72°C for 30 s. The amplified fragments from sections 2 and 3 were gel-purified and subjected to a DNA shuffling reaction with the parental fragments for section 1 as described below.
DNA shuffling of the amplified chimeric sections
DNA shuffling of the chimeric fragments was carried out as described previously (1,2). Briefly, the gel-purified PCR products were pooled and randomly digested with an appropriate amount of DNase I (Roche, Germany). The DNase I digests were extracted by phenol/chloroform followed by precipitation with ethanol. Partial denaturation of the purified DNA (65°C for 3 min) affected dissociation of small digested fragments, which remained associated via base pairing. Fragments of 50–100 bp were then gel-purified and subjected to a PCR reassembly reaction using libF (5′-TCACAC AGGAACAGAATCCAT-3′) and libR (5′-TTCGGATCCTACGCTGACGTCGAGTCTAAG-3′) primers. The underlined sequences indicate (a part of) NcoI and BamHI restriction sites. The bold letters in the libF primer indicate the initiation codon of the chimeric gene. The DNA fragments of 600–850 bp in lengths thus obtained were gel-purified, ligated with T-vector (pGEM-T, Promega) and constitute the enhanced crossover SCRATCHY library.
Construction of expression library
Escherichia coli Jude-1 cells [DH10B harboring the F’ factor derived from XL1-blue, exhibiting high electrocompetency (≥1 × 1010c.f.u./µg plasmid DNA), Hayhurst,A., Iverson,B.L. and Georgiou,G., in preparation] harboring the enhanced crossover SCRATCHY library (105 independent clones) were pooled, and the plasmids were purified. The NcoI and BamHI digested fragments (600–850 bp) from the plasmids were gel-purified, then subcloned into the expression vector pET-28a (Novagen). Finally, E.coli BL21(DE3) cells were transformed with the resulting plasmids to construct an expression library of chimeric GSTTs.
High-throughput screening of chimeric GSTTs
Cells were grown overnight at 30°C in TB media supplemented with 50 µg/ml kanamycin (TB/Kan) then subcultured (1:100 dilution) into fresh media and grown at 37°C with vigorous shaking. When the absorbance (OD600) reached 0.5–0.7, the cells were transferred to a 25°C shaker and allowed to equilibrate for 30 min. Subsequently, IPTG was added to a final concentration of 100 µM, and the cultures were incubated for an additional 3–4 h prior to harvesting. Aliquots (1 ml) were centrifuged at 2040 g for 5 min, the cell pellet was resuspended in 1 ml of phosphate buffered saline (PBS, pH7.4) and the suspensions were kept on ice prior to flow cytometric (FC) analysis. Ten to twenty microliter aliquots of the PBS cell suspensions were diluted to 1 ml in 1× PBS such that the resulting FC event rate of the suspension was 15–20 000 cells/s. The probe was added to a final concentration of 10 µM from a 10 mM dimethyl sulfoxide stock. The cells were labeled at 25°C for 2 min prior to the analysis.
FC analysis and sorting were carried out on a Cytomation MoFlo cell sorter with a standard L configuration on path 1 and a single photomultiplier on path 2. Events were triggered on side scatter (SSC) using a 488 nm argon ion laser (100 mW), and a 488/10 band pass filter on path 1. Forward scatter (FSC) 488 nm light was collected with a photodiode equipped with a 488/10 band pass filter. Rat-like parental GST activity was detected using monochlorobimane (MCB) or 7-amino-4-chloromethyl coumarin (CMAC). The glutathione (GSH) conjugates of these probes were excited with a broad band ultraviolet argon ion laser (75 mW), and the resulting fluorescence (FL) was detected through a 450/65 band pass filter. Both FSC × SSC and FL gates were set based on analysis of the parental control cells. The FL gate generally contained 1–60% of the parental rGSTT2-2 population while encompassing <0.1% of the events from the hGSTT1-1 negative control. Libraries were sorted directly to agar plates with TB media and 50 µg/ml kanamycin. Individual colonies were inoculated into TB/Kan media and grown overnight at 30°C, the cells were subcultured as above, and the synthesis of GST chimeras was induced by IPTG. Cellular fluorescence of monoclonal populations was analyzed by flow cytometry, as described above. Plasmid was isolated from clones that were confirmed to have rat-like parental activity using a QIAprep Spin Miniprep Kit. Purified plasmid was used as template in sequencing reactions using the T7 Prom (5′-TAATACGACTCACTATAGG-3′) and T7 Term (5′-GCTAGTTATTGCTCAGCGG-3′) primers.
RESULTS AND DISCUSSION
Overview of the construction of SCRATCHY library with enhanced crossovers
The distribution of the number of non-homologous crossovers within SCRATCHY libraries essentially follows a Poisson distribution. In conventional SCRATCHY (9), the chimeras are generated by the ITCHY process and therefore contain a single non-homologous crossover per gene. SCRATCHY libraries are formed by homologous shuffling of ITCHY libraries, and when the homology is low the resulting progeny often contain an average of only one non-homologous crossover event per gene (originating from the parental ITCHY clones).
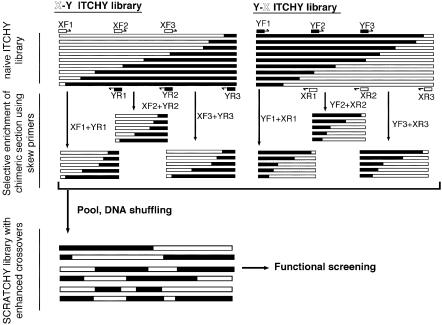
Figure 1 shows a schematic overview of the enhanced crossover SCRATCHY methodology. First, two ITCHY libraries, generated from two genes X and Y, are arbitrarily divided into a number of defined sections with short (>25 bp) overlapping sequences. Secondly, the defined sections are individually amplified by PCR using a skewed set of primers (e.g. gene X-specific forward primer F1 and gene Y-specific reverse primer R1 for the section X-Y1 amplification). The skewed sets of primers allow chimeric gene fragments having crossover points in the template section to be exponentially amplified. The enriched chimeric sections are subsequently assembled by overlap extension, or subjected to DNase I digestion followed by a reassembling PCR reaction to form SCRATCHY progeny with multiple non-homologous crossovers.
Figure 1.
Schematic overview of the construction of enhanced crossover SCRATCHY library. Initially, individual truncation libraries of the two complementary constructs (X-Y and Y-X ITCHY library) were created. After the ORF selection, the libraries are divided into arbitrarily defined sections. The sections are individually amplified by a skewed set of primers (e.g. a combination of XF1 and YR1 for the amplification of the first section of X-Y ITCHY library) to enrich the number of crossovers per gene. The amplified chimeric fragments from respective sections are pooled, and subjected to a shuffling reaction to generate the enhanced crossover SCRATCHY library with stochastic distribution of the number of crossovers per gene. Alternatively, the pooled chimeric fragments are allowed to assemble by overlapping extension to construct a library of chimeric genes with the defined number of crossovers.
For the example outlined in Figure 1, the number of non-homologous crossovers per progeny gene is increased to three by the selective enrichment of chimeric sections. It should be emphasized that the average number of crossovers can be controlled simply by increasing or decreasing the number of defined chimeric sections. Additionally, the positions of the chimeric sections can be subjectively defined on the basis of local sequence identity, substrate binding motifs, predicted secondary structures deduced from the parental amino acid sequences, etc. Thus, the crossover density in specific regions of interest can be controlled by increasing the number of defined sections in that region.
Construction of SCRATCHY library from mammalian GSTTs
We constructed and screened an enhanced crossover SCRATCHY library of two glutathione transferase genes (GSTs) belonging to the theta class of mammalian enzymes. Mammalian cytosolic GSTs are homodimeric enzymes capable of catalyzing the conjugation of glutathione to a wide variety of electrophilic compounds, and play a key role in cellular detoxification systems (reviewed in 13). The T1-1 and T2-2 subclasses of mammalian theta GSTs have been shown to have significant differences in their substrate specificities (14). For example, dichloromethane is known to be a characteristic substrate for the GSTT1-1 subclass. In contrast, it has been demonstrated that 1-menaphythyl sulfate is a preferred substrate for GSTT2-2 enzymes (15–17).
Human GSTT1-1 (hGSTT1-1, 240 aa in length) and rat GSTT2-2 (rGSTT2-2, 244 aa in length) share 54.5% amino acid identity. At the nucleotide sequence level, the genes show significant identity (74%) in the 5′-region encoding the glutathione binding site (N-terminal 1/3), whereas the nucleotide identity decreases to ∼60% in the remainder of the genes (C-terminal 2/3). In particular, the region encoding the C-terminal helix, which has been proposed to play an important role in substrate access to the active site (18), shows the least degree of sequence identity (50%).
Construction of a chimeric library from these two parental genes using conventional family shuffling by sexual PCR presents a significant challenge due to the low sequence identity between the two genes. In particular, the 3′-regions show a low frequency of continuously identical sequences, which are required for homologous recombination by family shuffling. Consequently, chimeric GSTT library construction using conventional family shuffling resulted in clones that had few if any crossovers, none of which occurred within the 3′-region. In fact, a restriction fragment length polymorphism analysis of PCR-amplified clones showed that >95% of the clones tested (n = 48) had fragment patterns identical to one or the other of the parental GSTT genes (data not shown). This result indicates that the generation of GSTT chimeric genes rarely occurs in a typical family shuffling reaction. PCR amplification of the shuffled library using hGSTT1-1 specific forward primer and rGSTT2-2 specific reverse primer allowed for chimeric gene specific enrichment. However, sequence analysis of 20 randomly selected enriched chimeric clones revealed that 95% of the homologous crossovers were found in the 5′-region, 5% were in just downstream of the 5′-region, and no crossovers were located in the 3′-region (data not shown). Recently, Broo et al. also reported that homologous recombination between these two mammalian GSTTs occurs exclusively in the 5′ region, and also noticed a large number of spontaneous point mutations generated by the family shuffling process (17). These data suggest that chimeras of theta class 1 and theta class 2 GSTs with crossovers in the 3′-region are rarely generated by homologous recombination.
In order to introduce crossovers at different locations within these genes, we first prepared ITCHY libraries from hGSTT1 and rGSTT2 in both orientations. ORF selection was then carried out to exclude frame-shifted crossovers from the ITCHY libraries. The ORF-selected ITCHY libraries were divided into three arbitrary sections on the basis of sequence identity between the two parental sequences. The parental sequences for section one (corresponding to amino acids 1–87, encompassing the entire N-terminal region) can be potentially recombined during the shuffling process by homologous recombination (described above), albeit at a very low frequency. Therefore, the parental sequences were used for this section in the shuffling reaction. Section 2 (216 bp, 59% nt identity, encoding amino acids 80–153) and section 3 (285 or 300 bp, 60% nt identity, encoding amino acid residues 147-stop codon) were amplified by skewed sets of primers as shown in Figure 1. The amplified chimeric sections (200– 300 bp in length for section 2, and 300–400 bp for the third section) were subsequently employed in DNA shuffling together with the first section.
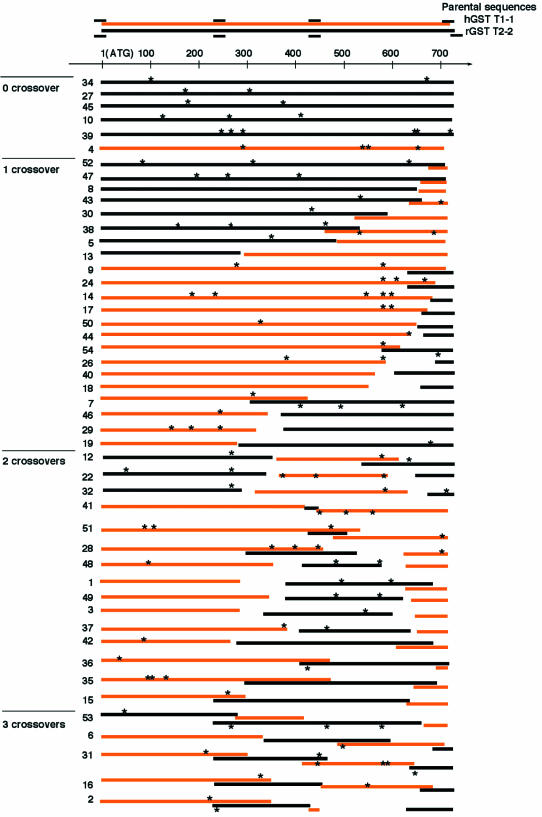
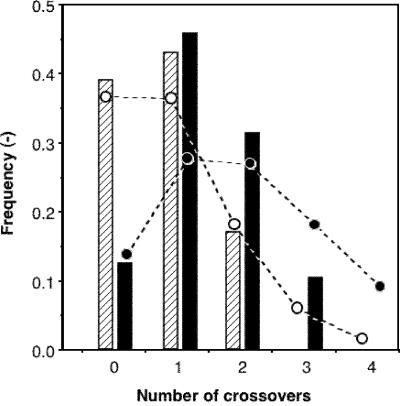
Figure 2 shows graphical representations of DNA sequences from 48 randomly selected enhanced crossover SCRATCHY clones. The population was comprised of six revertant clones (parent-like sequences), 22 clones with a single crossover, 15 clones with two crossovers, and five clones with three crossovers. The average number of crossovers per gene was 1.4. The distribution of crossover numbers in the sequenced clones is summarized and compared with that of 19 chimeric clones generated by conventional SCRATCHY (Fig. 3). These data demonstrate that >40% of the chimeras generated by enhanced crossover SCRATCHY have two or more non-homologous crossovers, whereas the corresponding fraction of chimeras from conventional SCRATCHY was <20%. No chimeras with three crossovers were observed in the latter library. Additionally, the frequency of revertant sequences with enhanced crossover SCRATCHY (12.5%) was substantially lower than that obtained in the conventional SCRATCHY library (38%). These results suggest that non-homologous crossovers enriched by the chimeric section-selective amplification were successfully propagated to the progeny genes. The positions of the crossovers are evenly distributed throughout sections 2 and 3, whereas no crossovers were found in section 1. Owing to the ORF-selection of ITCHY-derived chimeric sequences, most of the enhanced SCRATCHY progeny possess long ORFs (>600 bp), comparable in size with the parental genes.
Figure 2.
Schematic drawing of the DNA sequences of randomly selected SCRATCHY clones. The DNA sequences of individual chimeric progeny are depicted by sequential bars. Orange and black bars represent the hGSTT1-1 and rGSTT2-2-derived sequences, respectively. The positioning of the bars corresponds to their respective parental sequences (depicted on the top). Deletion and redundancy of the parental sequences at non-homologous crossover points are depicted as bars with a gap and overlapping bars, respectively. The small bars on the parental sequences indicate the annealing sites of skewed primers. The numerals on the left of progeny indicate clone numbers. Asterisks on the progeny sequences indicate point mutations spontaneously introduced during the shuffling process.
Figure 3.
Crossover frequency distribution. Bars represent the frequency of the clones having denoted number of crossover points. Hatched and closed bars indicate mammalian GSTT chimeric genes generated by conventional SCRATCHY (19 clones) and enhanced crossover SCRATCHY (48 clones), respectively. The dotted lines with white or black circles represent the theoretical distribution of the crossovers (1.0 and 2.0 crossovers per gene on average, respectively) calculated on the basis of a Poisson distribution.
Interestingly, no identifiable homologous crossover events were found among the 48 clones analyzed. Thus, it appears that crossovers were derived only from non-homologous recombination in the ITCHY-derived genes. The low frequency of homologous recombination in SCRATCHY clones is consistent with the results of the GSTT chimeric gene library construction using conventional family shuffling (discussed above). Thus, it appears that homologous recombination primarily occurs between cognate DNA fragments to form the respective predominantly parental sequences corresponding to the two starting GST genes. Most clones with non-homologous crossovers showed gapped or redundant sequences at their crossover points (Fig. 2). Such deletions and duplications are introduced in the incremental truncation and ligation reactions during ITCHY library construction. This intrinsic property of ITCHY chimeragenesis allows for the generation of diversity not only by the random positioning of crossover points, but also by varying gene length via the deletion or insertion of parental sequences. Although a large percentage of the redundant or gapped parental sequences may interfere with correct folding of the chimeric proteins, recent reports show that redundancy or gaps at crossover points can play an important role in enzyme evolution to novel substrate specificities (19), and can result in a dramatic improvement in protein solubility (20). These reports are notable examples in which a significant change in function required a drastic change in polypeptide sequence, which could not be generated by classical chimeragenesis based on homologous recombination. However, because ITCHY (10) or the other methods based on non-homologous recombination (19,20) yields exclusively single crossover chimeras, it fails to access sequence diversity generated by combinations of non-homologous crossovers. Likewise, SCRATCHY (9) as described above, is not able to generate chimeras with interchanged internal sequences or domains in high frequency. Therefore, methods able to generate multiple crossovers with a stochastic distribution of crossover numbers would be invaluable tools for the construction of initial libraries, despite the increased risk of disorder in protein folding caused by redundant or gapped parental sequences at the crossover points.
High-throughput screening of the GSTT enhanced crossover SCRATCHY library
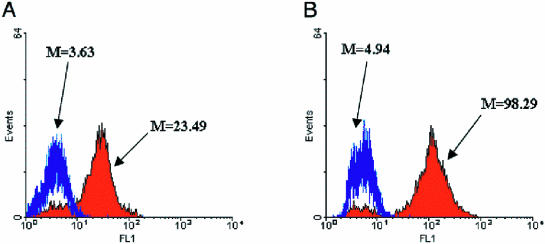
MCB can function as a substrate for the rGSTT2-2 enzyme while showing little to no reactivity with GSH in the presence of hGSTT1-1 (21). The utility of this molecule as a GST substrate is derived from the fact that MCB alone is effectively non-fluorescent when irradiated with UV light. However, upon conjugation to GSH, the conjugate becomes highly fluorescent. We have found that CMAC shows a similar reactivity profile to that of MCB in whole cell based assays and likewise shows an exceptional increase in fluorescence when conjugated to free thiols including GSH. Importantly, CMAC generally yields a 3-fold increase in signal-to-noise compared with MCB in FC analysis, and therefore is much better suited for library screening applications (Fig. 4).
Figure 4.
Fluorescence histograms of BL21(DE3) cells expressing parental hGSTT1-1 (blue) and rGSTT2-2 (red). Mean fluorescence intensities of individual populations are noted on the histograms. (A) Cells labeled for 1 min with 10 µM MCB. (B) Cells labeled for 1 min with 10 µM CMAC.
An enhanced crossover SCRATCHY GSTT library consisting of 105 independent clones was subcloned downstream from the T7 promoter in pET-28a and proteins were expressed in E.coli BL21 (DE3) cells. A total of 106 cells were analyzed at a rate of 15 000 cells/s, and 10 000 of these clones were sorted directly onto solid agar growth media based on their ability to catalyze the parental rat-like conjugation of endogenous GSH to CMAC. Monoclonal populations (20 clones) were isolated, re-grown and rat-like parental activity was confirmed by FC. Plasmid was isolated from mutants of interest for DNA sequencing and analysis.
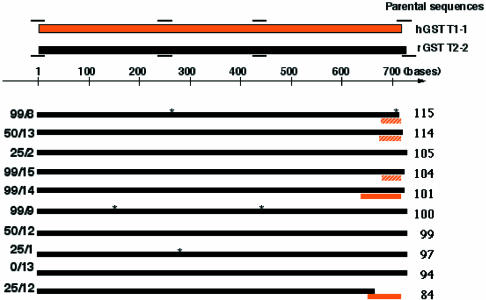
Figure 5 shows the DNA sequences of the representative positive clones retaining cellular GSTT2-2 activity comparable with parental rGSTT2-2. The majority of selected positive clones were comprised of sequences highly similar to the wild-type rGSTT2-2. This is not surprising considering that rat parental genes are represented in the library at a frequency of 10.4% (Figs 2 and 3). Nonetheless, sequence diversity was observed in the C-terminal helix (rGSTT2-2: P230-P244) of many of the selected positive clones. Some of the enzymatically active clones, such as 99/8 and 50/13, possess a truncated C-terminal helix from the rat enzyme with an out-of-frame junction to the adjacent hGSTT1-1-derived sequence. Presumably, these out-of-frame junctions were spontaneously generated by ‘Hybrid-duplex crossover’ recombination (9) during the shuffling process. More importantly, clone 25/12, which retained cellular activity comparable with wild-type rGSTT2-2, had an in-frame fusion to the human C-terminal helix at K223 of rGSTT2-2 (Fig. 5). As a result, this clone possesses the entire putative C-terminal helix of hGSTT1-1 instead of the C-terminal helix of rGSTT2-2. These results indicate that the C-terminal helix of rGSTT2-2 can be replaced by that of hGSTT1-1 without abolishing the substrate specificity of the rGSTT2-2 like chimera. Thus, it appears that despite the low evolutionary conservation of the C-terminal helix, it is not the predominant feature responsible for differences in the substrate specificities of the hGSTT1-1 and rGSTT2-2 enzymes. Because the sequence identity of the parental genes is as low as 50% in the region of the C-terminal helix, it would have been impossible to generate chimeric genes having crossover points in this region merely by homologous recombination. Consequently, the isolation of clone 25/12 illustrates the utility of non-homologous recombination methods as a tool for the elucidation of structure–function relationships as well as for directed evolution of proteins.
Figure 5.
DNA sequences of typical functionally selected chimeric progeny. The DNA sequences of individual active progeny are represented as in Figure 2. The numbers to the right of the progeny denote their normalized mean cellular fluorescence values with respect to wild-type rGSTT2-2 expressing cells (mean fluorescence = 100).
The generation of chimeric libraries from two or more parental genes (2–6) often provides benefits that are unattainable by more conventional mutagenic techniques such as error-prone PCR (2). Chimeragenesis allows efficient exploration of the functional DNA sequence space between parental genes. However, as was discussed above, canonical shuffling is of utility only for the generation of chimeras between genes with high homology. The development of homology-independent techniques such as ITCHY (10,11), SCRATCHY (9) and SHIPREC (20) has addressed some of the fundamental limitations of canonical gene shuffling technologies. Similarly, the concepts outlined herein should expand the scope and utility of SCRATCHY library construction protocols. In particular, the use of the enhanced crossover SCRATCHY technique for construction of mammalian GSTT libraries has augmented the average crossover number per progeny gene while significantly reducing the number of revertant parental sequences in the unselected population. Because this enrichment of crossover points is achieved by PCR based amplification, this technique should be easily adaptable to other enzymes and proteins.
Acknowledgments
ACKNOWLEDGEMENTS
This work was supported by a grant from the Foundation for Research. Karl Griswold was supported in part by an NIH Biotechnology Training graduate fellowship T32 GM08474.
REFERENCES
- 1.Stemmer W.P.C. (1994) DNA shuffling by random fragmentation and reassembly: in vitro recombination for molecular evolution. Proc. Natl Acad. Sci. USA, 91, 10747–10751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Crameri A., Raillard,S.-A., Bermudez,E. and Stemmer,W.P.C. (1998) DNA shuffling of genes from diverse species accelerates directed evolution. Nature, 391, 288–291. [DOI] [PubMed] [Google Scholar]
- 3.Kumamaru T., Suenaga,H., Mitsuoka,M., Watanabe,T. and Furukawa,K. (1998) Enhanced degradation of polychlorinated biphenyls by directed evolution of biphenyl dioxygenase. Nat. Biotechnol., 16, 663–666. [DOI] [PubMed] [Google Scholar]
- 4.Christians F.C., Scapozza,L., Crameri,A., Folkers,G. and Stemmer,W.P.C. (1999) Directed evolution of thymidine kinase for AZT phosphorylation using DNA family shuffling. Nat. Biotechnol., 17, 259–264. [DOI] [PubMed] [Google Scholar]
- 5.Kikuchi M., Ohnishi,K. and Harayama,S. (1999) Novel family shuffling methods for the in vitro evolution of enzymes. Gene, 236, 159–167. [DOI] [PubMed] [Google Scholar]
- 6.Raillard S.A., Krebber,A., Chen,Y., Ness,J.E., Bermudez,E., Trinidad,R., Fullem,R., Davis,C., Welch,M., Seffernick,J., Wackett,L.P., Stemmer,W.P.C. and Minshull,J. (2001) Novel enzyme activities and functional plasticity revealed by recombining highly homologous enzymes. Chem. Biol., 8, 891–898. [DOI] [PubMed] [Google Scholar]
- 7.Moore G., Maranas,C.D., Lutz,S. and Benkovic,S.J. (2001) Predicting crossover generation in DNA shuffling. Proc. Natl Acad. Sci. USA, 98, 3226–3231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Joern J.M., Meinhold,P. and Arnold,F.H. (2002) Analysis of shuffled gene libraries. J. Mol. Biol., 316, 643–656. [DOI] [PubMed] [Google Scholar]
- 9.Lutz S., Ostermeier,M., Moore,G.L., Maranas,C.D. and Benkovic,S.J. (2001) Creating multiple crossover DNA libraries independent of sequence identity. Proc. Natl Acad. Sci. USA, 98, 11248–11253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ostermeier M., Shim,J.H. and Benkovic,S.J. (1999) A combinatorial approach to hybrid enzymes independent of DNA homology. Nat. Biotechnol., 17, 1205–1209. [DOI] [PubMed] [Google Scholar]
- 11.Lutz S. and Benkovic,S.J. (2000) Homology-independent protein engineering. Curr. Opin. Biotechnol., 11, 319–324. [DOI] [PubMed] [Google Scholar]
- 12.Lutz S., Ostermeier,M. and Benkovic,S.J. (2001) Rapid generation of incremental truncation libraries for protein engineering using α-phosphothioate nucleotides. Nucleic Acids Res., 29, e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Armstrong R.N. (1997) Structure, catalytic mechanism and evolution of the glutathione transferase. Chem. Res. Toxicol., 10, 2–18. [DOI] [PubMed] [Google Scholar]
- 14.Landi S. (2000) Mammalian class theta GST and differential susceptibility to carcinogens. Mutat. Res., 463, 247–283. [DOI] [PubMed] [Google Scholar]
- 15.Jemth P. and Mannervik,B. (1997) Kinetic characterization of recombinant human glutathione transferase T1-1, a polymorphic detoxification enzyme. Arch. Biochem. Biophys., 348, 247–254. [DOI] [PubMed] [Google Scholar]
- 16.Jemth P., Stenberg,G., Chaga,G. and Mannervik,B. (1996) Heterologous expression, purification and characterization of rat class theta glutathione transferase T2-2. Biochem. J., 316, 131–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Broo K., Larsson,A.-K., Jemth,P. and Mannervik,B. (2002) An ensemble of theta class glutathione transferases with novel catalytic properties generated by stochastic recombination of fragments of two mammalian enzymes. J. Mol. Biol., 318, 59–70. [DOI] [PubMed] [Google Scholar]
- 18.Chelvanayagam G., Wilce,M.C.J., Parker,M.W., Tan,K.L. and Board,P.G. (1997) Homology model for the human GSTT2 theta class glutathione transferase. Proteins, 27, 118–130. [DOI] [PubMed] [Google Scholar]
- 19.Pikkemaat M.G. and Janssen,D.B. (2002) Generating segmental mutations in haloalkane dehalogenase: a novel part in the directed evolution toolbox. Nucleic Acids Res., 30, e85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sieber V., Martinez,C.A. and Arnold,F.H. (2001) Libraries of hybrid proteins from distantly related sequences. Nat. Biotechnol., 19, 456–460. [DOI] [PubMed] [Google Scholar]
- 21.Eklund B.I., Edalat,M., Stenberg,G. and Mannervik,B. (2002) Screening for recombinant glutathione transferases active with monochlorobimane. Anal. Biochem., 309, 102–108. [DOI] [PubMed] [Google Scholar]