Summary
In many species, reducing nutrient intake without causing malnutrition extends lifespan [1-3]. Like DR (dietary restriction), modulation of genes in the insulin-signaling pathway, known to alter nutrient sensing, has been shown to extend lifespan in various species [1-4]. In Drosophila, the target of rapamycin (TOR) and the insulin pathways have emerged as major regulators of growth and size. Hence we examined the role of TOR pathway genes in regulating lifespan by using Drosophila. We show that inhibition of TOR signaling pathway by alteration of the expression of genes in this nutrient-sensing pathway, which is conserved from yeast to human, extends lifespan in a manner that may overlap with known effects of dietary restriction on longevity. In Drosophila , TSC1 and TSC2 (tuberous sclerosis complex genes 1 and 2) act together to inhibit TOR (target of rapamycin), which mediates a signaling pathway that couples amino acid availability to S6 kinase, translation initiation, and growth [5]. We find that overexpression of dTsc1, dTsc2, or dominant-negative forms of dTOR or dS6K all cause lifespan extension. Modulation of expression in the fat is sufficient for the lifespan-extension effects. The lifespan extensions are dependent on nutritional condition, suggesting a possible link between the TOR pathway and dietary restriction.
Introduction
A reduction in nutrient intake, also known as dietary restriction (DR), extends lifespan in various species including worm (C. elegans), fly (D. melanogaster), yeast (S. cerevisiae), and mouse (Mus musculus) [1, 2]. Like DR, modulation of genes in the insulin-signaling pathway, known to alter nutrient sensing, has been shown to extend lifespan in various species [1-4]. Evolutionary theory suggests that DR regulates certain genetic pathways that shift an organism's metabolic investment from reproduction and growth toward somatic maintenance [3, 6, 7], allowing the organism to survive under harsh environments until there are suitable reproductive conditions. In C. elegans, in response to crowding and starvation, the insulin/insulin-like growth factor (IGF-1) signaling pathway controls the formation of the dauer larva, an alternative developmental state that is nonreproducing, stress resistant, and long lived [8, 9]. Mutations in genes of the same pathway, daf-2 (insulin receptor) and age-1 (Phosphatidyl inositol-3 kinase), lead to a doubling of lifespan, and render the animals more sensitive to dauer formation [10]. Both the dauer-formation and lifespan-extension phenotypes are suppressed by mutations in daf-16, a forkhead family transcription factor [10, 11]. Drosophila mutations in Inr (insulin-like receptor) and chico (insulin-receptor substrate), both components of the insulin-signaling pathway, also regulate lifespan [12, 13]. A reduction of nutrients in the fly diet has been shown to extend lifespan in the fly [14-16]. Reduced availability of protein or yeast in the fly diet can reduce signaling through the InR/PI3K pathway, as measured by using a pleckstrin homology domain green fluorescent protein (PH-GFP), which is localized to the cell membrane when the PI3K pathway is active [17]. Furthermore, the lifespan extension that is observed by chico mutant flies is dependent on the nutritional content of fly food, suggesting an overlap between the two mechanisms of lifespan extension [14].
In Drosophila, nutrient availability and insulin signaling have been shown to be critical factors in regulating not only lifespan but also growth and size [18]. Inhibition of InR/PI3K signaling, by using mutations in the pathway, phenocopies the cellular and organismal effects of starvation, leading to a reduction in the size of the fly [17, 19, 20]. Tsc1, Tsc2, TOR, and S6 kinase have emerged as important regulators of growth and size in Drosophila in a pathway that is parallel to, but also interactive with, the insulin pathway (Figure S1) [21]. Recent evidence suggests that the fat body in Drosophila acts as a nutrient sensor, which uses TOR signaling to generate a humoral signal that modulates insulin signaling and growth in peripheral tissues [22]. TOR deficiency in the nematode C. elegans was recently shown to extend lifespan [23]. We therefore tested whether modulating the activities of genes in the TOR pathway might regulate lifespan.
Results and Discussion
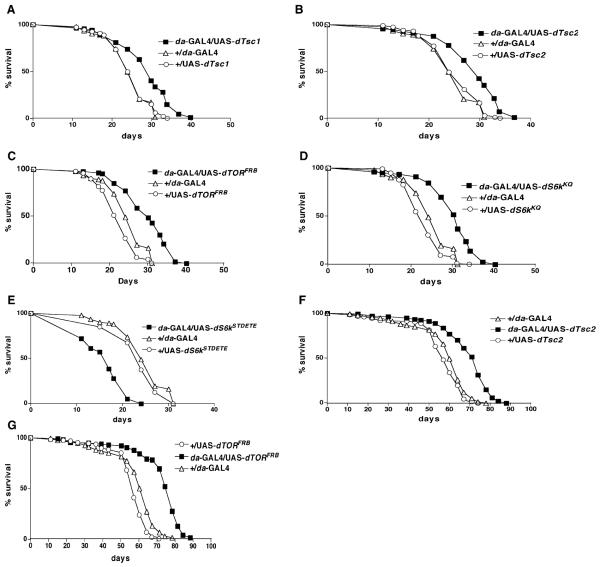
The Drosophila homologs of human Tsc1 (Hamartin) and Tsc2 (tuberin) function in vivo as a complex that controls growth and size in a cell-autonomous manner [24-27]. To examine their role in regulating lifespan, we overex-pressed dTsc1 and dTsc2 by using constructs provided by the Duojia Pan lab [5] through the ubiquitously expressed driver, daughterless (da)-GAL4. We found that overexpression in transgenic flies carrying UAS constructs containing dTsc1 or dTsc2 extended mean lifespan at 29°C by 14% and 12%, respectively (Figures 1A and 1B). Since GAL4 enhancer traps generally yield stronger effects at 29°C, we chose to perform most of the experiments at that temperature [28].
Figure 1. Regulation of Lifespan by Modulating Genes in the TOR Signaling Pathway.
Survival curves on standard laboratory food for male flies obtained after crossing the ubiquitously expressed da-GAL4 enhancer trap with various UAS constructs carrying genes in the TOR signaling pathway ([A]–[E] at 29°C, [F] and [G] at 25°C). All logrank test p values are based on comparison of the overexpressor strain with the driver alone. The plus symbol indicates the parental strain w1118. The mean lifespan (days) of the various genotypes were as follows. (A) da-GAL4/UAS-dTsc1 (closed squares), 29 days (n=136); +/da-GAL4 (driver alone) (open triangles), 26 days (n = 92); +/UAS-dTsc1 (open circles), 25 days (n = 109) (p < 0.0001). (B) da-GAL4/UAS-dTsc2 (closed squares), 29 days (n = 112); +/da-GAL4 (open triangles), 26 days (n = 92); +/UAS-dTsc2 (open circles), 25 days (n = 118) (p < 0.0001). (C) da-GAL4/UAS-dTORFRB (closed squares), 30 days (n = 126); +/da-GAL4 (open triangles), 26 days (n = 92); +/UAS-dTORFRB (open circles), 22 days (n = 98) (p < 0.0001). (D) da-GAL4/UAS-dS6kKQ (closed squares), 30 days (n = 122); +/da-GAL4 (open triangles), 26 days (n = 92); +/UAS-dS6kKQ (open circles), 23 days (n = 115) (p < 0.0001). (E) da-GAL4/UAS-dS6kSTDETE (closed squares), 16 days (n = 75); +/da-GAL4 (open triangles), 26 days (n = 92); +/UAS-dS6kSTDETE (open circles), 24 days (n = 40) (p < 0.0001). (F) da-GAL4/UAS-dTsc2 (closed squares), 69 days (n = 134); +/da-GAL4 (open triangles), 58 days (n = 133); +/UAS-dTsc2 (open circles), 56 days (n = 126) (p < 0.0001). (G) da-GAL4/UAS-dTORFRB (closed squares), 72 days (n = 112); +/da-GAL4 (open triangles), 58 days (n = 133); +/UAS-dTORFRB (open circles), 55 days (n = 128) (p < 0.0001).
dTsc1 and dTsc2 physically interact with dTOR [5], which is conserved from yeast to human as a nutrient sensor [18]. Loss of dTsc1 in Drosophila eye leads to an increase in cell size, provided that dTOR is present [5]. Surprisingly, however, dTOR overexpression causes a reduction in cell size, a phenotype similar to dTOR loss-of-function mutations, perhaps due to titration of cofactors required for TOR signaling [29]. We examined the effect of dTOR on lifespan by using three UAS constructs generously provided by the Thomas Neufeld lab [29]. One carries the full-length wild-type TOR gene. The second carries FRB, the 11 kDa FKBP12-rapamycin binding domain, which has been shown to prevent S phase entry when injected into human osteosarcoma cells [30]. The third carries TED (toxic effector domain), containing the 754 amino acid central region, which inhibits cell growth and arrests cells in G1 when overexpressed in yeast [29]. Ubiquitous overexpression with the da-GAL4 driver of UAS-dTORFRB led to a mean life-span increase at 29°C of 24% (Figure 1C). However, overexpression of UAS -dTORWT or UAS-dTORTED prevented eclosion to adulthood (data not shown).
S6 kinase activation upon phosphorylation has been implicated in mediating the downstream effects of TOR on translation initiation in flies and mammals [21]. S6 kinase phosphorylation of ribosomal protein S6 is accompanied by upregulation of a class of mRNAs containing an oligopyrimidine tract at their transcriptional start site termed 5′TOP [31]. Some 200 genes, most of which encode components of the translational apparatus including ribosomal proteins and elongation factors, have this sequence and can account for about 20% of total cellular mRNA [31]. Flies carrying homozygous mutations in dS6K show a developmental delay and a reduction in body size [32]. The stimulation of dS6K phosphorylation by dTOR is abrogated when dTsc1 and dTsc2 are overexpressed [5]. Furthermore, flies with reduced dTSC1 show increased dS6 kinase activation, and genetic reduction of S6 kinase level can rescue the lethality caused by loss of function of dTsc1 [33].
We examined the role of S6 kinase in regulating life-span by using dominant-negative and constitutively active constructs generously provided by the Mary Stewart lab [34]. The dominant-negative effect was achieved by replacing the conserved lysine in the ATP binding site by glutamine (UAS-dS6KKQ), which causes cell-size reduction. The constitutively active form was generated by replacing the phosphorylation sites of S6 kinase by acidic amino acids (UAS-dS6KSTDETE), causing an autonomous cell size increase [34]. By using da-GAL4 to drive ubiquitous overexpression of the dominant-negative form, we observed a mean lifespan increase of 22% at 29°C (Figure 1D). Conversely, overexpression of the constutively active form of S6 kinase caused a mean lifespan decrease of 34% at 29°C (Figure 1E). Overexpression of dTsc2 and dTORFRB was also tested at 25°C and led to a 20% and 26% increase in mean lifespan increase, respectively (Figures 1F and 1G).
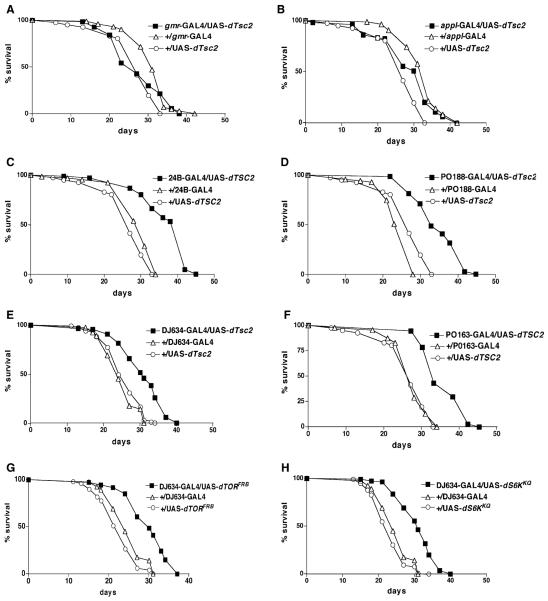
To determine which tissues are responsible for the lifespan extension, we employed various GAL4 drivers with specific GAL4 expression pattern (Figure S2) to overexpress dTsc2 via a UAS promoter. Overexpression in the eye by using the driver gmr-GAL4 or in the nervous system by using appl-GAL4 did not extend lifespan (Figures 2A and 2B). On the other hand, by using the drivers 24B-GAL4 and PO188-GAL4, enhancer traps that are predominantly expressed in the muscle and fat resulted in mean lifespan extensions of 27% and 37%, respectively, at 29°C (Figures 2C and 2D). The fat-specific drivers DJ634-GAL4 and PO163-GAL4, when used to over-express dTsc2, also led to a mean lifespan extension of 22% and 31%, respectively, at 29°C (Figures 2E and 2F). Using DJ634-GAL4 to overexpress the dominant-negative form of TOR (UAS-dTORFRB) or of S6 kinase (UAS-UAS-dS6KKQ) also led to mean lifespan increases of 30% and 29%, respectively, at 29°C (Figures 2G and 2H). These results indicate that manipulation of the TSC, TOR, and S6 kinase genes in the fat tissue is sufficient for their lifespan extension effects in Drosophila.
Figure 2. Dependence of Lifespan Extension on Overexpression in Specific Tissues.
Survival curves, on standard laboratory food, for male flies obtained by crossing various genes in the TOR pathway by using different tissue specific drivers along with appropriate control strains. gmr-GAL4 is expressed in the eye, appl-GAL4 in the nervous system, 24B-GAL4 and PO188-GAL4 in the muscle and fat tissues, and DJ634-GAL4 and PO163-GAL4 in the fat tissue. All logrank test p values were based on comparison of the overexpressor strain with the driver alone. The plus symbol indicates the parental strain w1118. The mean lifespan at 29°C of the various genotypes were as follows. (A) gmr-GAL4/ UAS-dTsc2 (closed squares), 27 days (n = 69); +/gmr-GAL4 (open triangles), 31 days (n = 102); +/UAS-dTsc2 (open circles), 27 days (n = 81) (p < 0.01). (B) appl-GAL4/ UAS-dTsc2 (closed squares), 33 days (n = 108); +/appl-GAL4 (open triangles), 32 days (n = 85); +/UAS-dTsc2 (open circles), 27 days (n = 81) (p < 0.001). (C) 24B- GAL4/UAS-dTsc2 (closed squares), 37 days (n = 107); +/24B-GAL4 (open triangles), 31 days (n = 67); +/UAS-dTsc2 (open circles), 27 days (n = 81) (p < 0.0001). (D) PO188-GAL4/UAS-dTsc2 (closed squares), 35 days (n = 76); +/PO188-GAL4 (open triangles), 24 days (n = 71); +/ UAS-dTsc2 (open circles), 27 days (n = 81) (p < 0.0001). (E) DJ634-GAL4/UAS-dTsc2 (closed squares), 30.6 days (n = 120); +/DJ634-GAL4 (open triangles), 24.6 days (n = 120); +/UAS-dTsc2 (open circles), 25.3 days (n = 118) (p < 0.0001). (F) PO163-GAL4/UAS-dTsc2 (closed squares), 36 days (n = 126); +/PO163-GAL4 (open triangles), 28 days (n = 87); +/UAS-dTsc2 (open circles), 27 days (n = 81) (p < 0.0001). (G) DJ634-GAL4/UAS-dTORFRB (closed squares), 31 days (n = 107); +/DJ634-GAL4 (open triangles), 25 days (n = 120); +/UAS-dTORFRB (open circles), 22 days (n = 98) (p < 0.0001). (H) DJ634-GAL4/UAS-dS6kKQ (closed squares), 31 days (n = 110); +/DJ634-GAL4 (open triangles), 25 days (n = 120); +/UAS-dS6kKQ (open circles), 23 days (n = 115) (p < 0.0001).
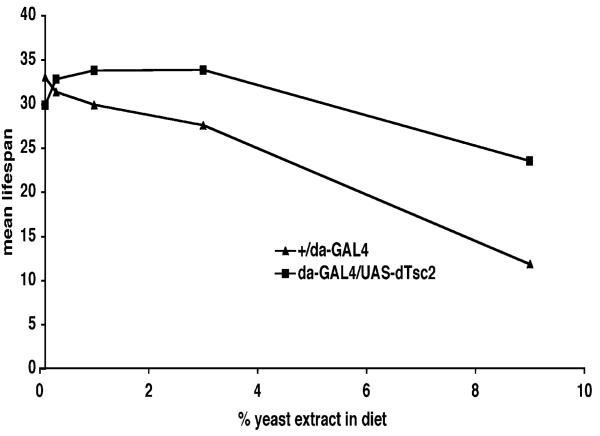
Amino acids have been shown to activate dS6k via TOR, an effect that can be abrogated in the presence of increased levels of dTsc1 and dTsc2 [5]. As nutrients in the diet can modulate lifespan and because the TOR pathway is a critical mediator of nutrient signaling, we asked whether the observed lifespan-extension effects are dependent on nutrient conditions. We tested this with overexpression of dTsc2 by using the ubiquitously expressing da-GAL4 driver. Flies were allowed to develop to adulthood under standard laboratory food and then maintained on specially prepared food containing various concentrations of yeast extract. At high concentrations of yeast extract, which may be regarded as the opposite of dietary restriction, the lifespan of control flies (da-GAL4/+) was severely reduced (Figure 3). However, overexpression of dTsc2 protects the fly from the deleterious effects of rich food, as if mimicking the effect of dietary restriction (Figure 3). Similar results were observed by overexpression of the dominant-negative form of S6 kinase (data not shown).
Figure 3. Lifespan Extension by dTsc2 Overexpression Is Dependent on the Concentration of Yeast Extract in the Diet.
Logrank test was used to calculate the p values. The plus symbol indicates the parental strain w1118. Mean lifespan at 29°C of adult male da-Gal4/+ (closed triangles) and da-GAL4/UAS-dTSC2 (closed squares) flies were: on 0.1% yeast extract, da-Gal4/+, 33 days (n = 125) and da-GAL4/UAS-dTSC2, 30 days (n = 164); on 0.3% yeast extract, da-Gal4/+, 31 days (n = 139) and da-GAL4/UAS-dTSC2, 33 days (n = 138); on 1% yeast extract, da-Gal4/+, 30 days (n = 182) and da-GAL4/UAS-dTSC2, 34 days (n = 164); on 3% yeast extract, da-Gal4/+, 28 days (n = 181) and da-GAL4/UAS-dTSC2, 34 days (n = 154); on 9% yeast extract, da-Gal4/+, 12 days (n = 150) and da-GAL4/UAS-dTSC2, 24 days (n = 156). Mean lifespan differences between flies overexpressing dTsc2 and controls were −9% (p < 0.3), +5% (p < 0.03), +13% (p < 0.0001), +23% (p < 0.0001), and 95% (p < 0.0001) when lifespan was measured on fly food containing 0.1%, 0.3%, 1%, 3%, and 9% yeast extract, respectively.
Recent evidence from Drosophila suggests that signaling through TSC is both parallel to and interacting with the insulin pathway [21, 25]. This is supported by the finding that heterozygosity of dTsc1 or dTsc2 is sufficient to rescue the lethality of loss-of-function dInR mutants [25]. However, the finding that loss-of-function mutations of dTsc1 and dPTEN, a phosphatase that negatively regulates the insulin-signaling pathway, cause cell autonomous and additive increases in cell size suggests that they may be in parallel pathways [25]. Furthermore, in Drosophila, dPTEN loss of function, which leads to an increase in cell size, is only slightly suppressible by loss of function of dFOXO, a fly homolog of C. elegans daf-16 [35]. However, the increase in cell size resulting from dTsc1 is enhanced by dFOXO loss of function [35]. Interestingly, unlike long-lived daf-2 mutants, the lifespan extension due to TOR deficiency in C. elegans is not suppressible by a daf-16 mutation [23]. However, the TOR mutant animals do not further extend lifespan in a daf-2 background, leading to the possibility that TOR may be acting downstream or separately from daf-16 to exert its lifespan effects [23].
Lifespan extension has been linked with other phenotypes, including stress resistance, metabolic rate, lipid level, reproductive capacity, and body size [16, 36, 37]. We compared the long-lived strains described above with their respective controls for resistance to starvation but found no significant differences (Table S1). Similarly, no significant differences were observed for weight and lipid content among these strains (Table S1). It may be that lifespan extension can be produced by mild modulation of these genes, whereas effects on other phenotypes require severe perturbations. While lifespan extension is observed by using the da-GAL4 driver to overexpress dTsc1 or dTsc2 alone, simultaneous overexpression of dTsc1 and dTsc2 prevented eclosion to adulthood (data not shown). Similarly, no change in size is observed if dTsc1 or dTsc2 alone are overexpressed in the eye, but a cell-autonomous decrease in size is seen when both are overexpressed simultaneously [25, 26]. Lifespan extension by chico is semidominant, but its effect on body size is recessive [13]. Dominant effects on lifespan are observed with the genes Inr, EcR, Indy, and Rpd3, but their effects on lifespan can be uncoupled from other phenotypes such as fecundity, stress resistance, or lipid accumulation.
In humans, mutations in TSC1 and TSC2 lead to tuberous sclerosis, a common disorder characterized by the presence of benign tumors in various tissues, with some having large cells. DR in mice has been shown to protect against age-related tumorigenesis [38, 39]. Our results suggest a link between lifespan extension by DR and the activities of genes in the TOR pathway. Hence, it is conceivable that the protective effects of DR on tumori-genesis and age-related decline might come from inhibition of such nutrient-responsive pathways.
Our results show that upregulation of dTsc2 in the fat is sufficient for lifespan extension effects in Drosophila. Reduction of daf-2 levels in the C. elegans nervous system has been shown to be sufficient for lifespan extension [40, 41]. However, the lifespan extensions due to mutations in the insulin pathway or germline ablation in C. elegans are dependent on daf-16 activity in the intestine, the fat storage tissue in C. elegans [41]. In Drosophila, the fat body has been proposed to modulate insulin signaling in peripheral tissues by secretion of dALS (acid-labile subunit) [22], which, in mammals, forms a ternary complex with insulin-like growth factor, leading to an extension of the half-life of its ligand [42]. Recently, mice with FIRKO (fat-specific insulin receptor knockout) have been shown to live 18% longer than controls [43]. Hence, it is possible that secondary endocrine signals downstream of the insulin and TOR signaling pathways are released from the fat, and these affect the rate of aging in other tissues. Juvenile hormone and ecdysone are two such endocrine signals that have been implicated in regulating lifespan in conjunction with the insulin pathway in Drosophila [4].
Supplementary Material
Acknowledgments
We thank members of the Benzer laboratory, including G. Carvalho, D. Walker, E. Fabrikant, T. Brummel, D. Tracey, and H. Dar Wang for helpful discussions and comments on the manuscript. We also thank D. Pan, T. Neufeld, and M. Stewart for generously providing us with fly strains. This work was supported by a grant from the American Federation for Aging Research and a postdoctoral fellowship from the John Douglas French Alzheimer's Foundation Research to P.K., a California Institute of Technology SURF scholarship to D.K., and grants to S.B. from the National Institutes of Health (AG16630), the National Science Foundation (MCB-9907939), and the Ellison foundation.
Footnotes
Supplemental Data
Supplemental data including a description of the Experimental Procedures and two figures are available at http://www.current-biology.com/cgi/content/full/14/10/885/DC1/.
References
- 1.Guarente L, Kenyon C. Genetic pathways that regulate ageing in model organisms. Nature. 2000;408:255–262. doi: 10.1038/35041700. [DOI] [PubMed] [Google Scholar]
- 2.Partridge L, Gems D. Mechanisms of ageing: public or private? Nat. Rev. Genet. 2002;3:165–175. doi: 10.1038/nrg753. [DOI] [PubMed] [Google Scholar]
- 3.Martin GM, Austad SN, Johnson TE. Genetic analysis of ageing: role of oxidative damage and environmental stresses. Nat. Genet. 1996;13:25–34. doi: 10.1038/ng0596-25. [DOI] [PubMed] [Google Scholar]
- 4.Tatar M, Bartke A, Antebi A. The endocrine regulation of aging by insulin-like signals. Science. 2003;299:1346–1351. doi: 10.1126/science.1081447. [DOI] [PubMed] [Google Scholar]
- 5.Gao X, Zhang Y, Arrazola P, Hino O, Kobayashi T, Yeung RS, Ru B, Pan D. Tsc tumour suppressor proteins antagonize amino-acid-TOR signalling. Nat. Cell Biol. 2002;4:699–704. doi: 10.1038/ncb847. [DOI] [PubMed] [Google Scholar]
- 6.Holliday R. Food, reproduction and longevity: is the extended lifespan of calorie-restricted animals an evolutionary adaptation? Bioessays. 1989;10:125–127. doi: 10.1002/bies.950100408. [DOI] [PubMed] [Google Scholar]
- 7.Kirkwood TL, Kapahi P, Shanley DP. Evolution, stress, and longevity. J. Anat. 2000;197:587–590. doi: 10.1046/j.1469-7580.2000.19740587.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Riddle DL, Swanson MM, Albert PS. Interacting genes in nematode dauer larva formation. Nature. 1981;290:668–671. doi: 10.1038/290668a0. [DOI] [PubMed] [Google Scholar]
- 9.Klass M, Hirsh D. Non-ageing developmental variant of Caenorhabditis elegans. Nature. 1976;260:523–525. doi: 10.1038/260523a0. [DOI] [PubMed] [Google Scholar]
- 10.Kenyon C, Chang J, Gensch E, Rudner A, Tabtiang R. A C. elegans mutant that lives twice as long as wild type. Nature. 1993;366:461–464. doi: 10.1038/366461a0. [DOI] [PubMed] [Google Scholar]
- 11.Vowels JJ, Thomas JH. Genetic analysis of chemosensory control of dauer formation in Caenorhabditis elegans. Genetics. 1992;130:105–123. doi: 10.1093/genetics/130.1.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tatar M, Kopelman A, Epstein D, Tu MP, Yin CM, Garofalo RS. A mutant Drosophila insulin receptor homolog that extends life-span and impairs neuroendocrine function. Science. 2001;292:107–110. doi: 10.1126/science.1057987. [DOI] [PubMed] [Google Scholar]
- 13.Clancy DJ, Gems D, Harshman LG, Oldham S, Stocker H, Hafen E, Leevers SJ, Partridge L. Extension of life-span by loss of CHICO, a Drosophila insulin receptor substrate protein. Science. 2001;292:104–106. doi: 10.1126/science.1057991. [DOI] [PubMed] [Google Scholar]
- 14.Clancy DJ, Gems D, Hafen E, Leevers SJ, Partridge L. Dietary restriction in long-lived dwarf flies. Science. 2002;296:319. doi: 10.1126/science.1069366. [DOI] [PubMed] [Google Scholar]
- 15.Rogina B, Helfand SL, Frankel S. Longevity regulation by Drosophila Rpd3 deacetylase and caloric restriction. Science. 2002;298:1745. doi: 10.1126/science.1078986. [DOI] [PubMed] [Google Scholar]
- 16.Nusbaum TJ, Rose MR. The effects of nutritional manipulation and laboratory selection on lifespan in Drosophila melanogaster. J. Gerontol. A Biol. Sci. Med. Sci. 1999;54:B192–B198. doi: 10.1093/gerona/54.5.b192. [DOI] [PubMed] [Google Scholar]
- 17.Britton JS, Lockwood WK, Li L, Cohen SM, Edgar BA. Drosophila's insulin/PI3-kinase pathway coordinates cellular metabolism with nutritional conditions. Dev. Cell. 2002;2:239–249. doi: 10.1016/s1534-5807(02)00117-x. [DOI] [PubMed] [Google Scholar]
- 18.Oldham S, Hafen E. Insulin/IGF and target of rapamycin signaling: a TOR de force in growth control. Trends Cell Biol. 2003;13:79–85. doi: 10.1016/s0962-8924(02)00042-9. [DOI] [PubMed] [Google Scholar]
- 19.Bohni R, Riesgo-Escovar J, Oldham S, Brogiolo W, Stocker H, Andruss BF, Beckingham K, Hafen E. Autonomous control of cell and organ size by CHICO, a Drosophila homolog of vertebrate IRS1–4. Cell. 1999;97:865–875. doi: 10.1016/s0092-8674(00)80799-0. [DOI] [PubMed] [Google Scholar]
- 20.Leevers SJ, Weinkove D, MacDougall LK, Hafen E, Waterfield MD. The Drosophila phosphoinositide 3-kinase Dp110 promotes cell growth. EMBO J. 1996;15:6584–6594. [PMC free article] [PubMed] [Google Scholar]
- 21.Marygold SJ, Leevers SJ. Growth signaling: TSC takes its place. Curr. Biol. 2002;12:R785–R787. doi: 10.1016/s0960-9822(02)01294-0. [DOI] [PubMed] [Google Scholar]
- 22.Colombani J, Raisin S, Pantalacci S, Radimerski T, Montagne J, Leopold P. A nutrient sensor mechanism controls Drosophila growth. Cell. 2003;114:739–749. doi: 10.1016/s0092-8674(03)00713-x. [DOI] [PubMed] [Google Scholar]
- 23.Vellai T, Takacs-Vellai K, Zhang Y, Kovacs AL, Orosz L, Muller F. Genetics: influence of TOR kinase on lifespan in C. elegans. Nature. 2003;426:620. doi: 10.1038/426620a. [DOI] [PubMed] [Google Scholar]
- 24.Ito N, Rubin GM. gigas, a Drosophila homolog of tuberous sclerosis gene product-2, regulates the cell cycle. Cell. 1999;96:529–539. doi: 10.1016/s0092-8674(00)80657-1. [DOI] [PubMed] [Google Scholar]
- 25.Gao X, Pan D. TSC1 and TSC2 tumor suppressors antagonize insulin signaling in cell growth. Genes Dev. 2001;15:1383–1392. doi: 10.1101/gad.901101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Potter CJ, Huang H, Xu T. Drosophila Tsc1 functions with Tsc2 to antagonize insulin signaling in regulating cell growth, cell proliferation, and organ size. Cell. 2001;105:357–368. doi: 10.1016/s0092-8674(01)00333-6. [DOI] [PubMed] [Google Scholar]
- 27.Tapon N, Ito N, Dickson BJ, Treisman JE, Hariharan IK. The Drosophila tuberous sclerosis complex gene homologs restrict cell growth and cell proliferation. Cell. 2001;105:345–355. doi: 10.1016/s0092-8674(01)00332-4. [DOI] [PubMed] [Google Scholar]
- 28.Seroude L, Brummel T, Kapahi P, Benzer S. Spatio-temporal analysis of gene expression during aging in Drosophila melanogaster. Aging Cell. 2002;1:47–56. doi: 10.1046/j.1474-9728.2002.00007.x. [DOI] [PubMed] [Google Scholar]
- 29.Hennig KM, Neufeld TP. Inhibition of cellular growth and proliferation by dTOR overexpression in Drosophila. Genesis. 2002;34:107–110. doi: 10.1002/gene.10139. [DOI] [PubMed] [Google Scholar]
- 30.Alarcon CM, Heitman J, Cardenas ME. Protein kinase activity and identification of a toxic effector domain of the target of rapamycin TOR proteins in yeast. Mol. Biol. Cell. 1999;10:2531–2546. doi: 10.1091/mbc.10.8.2531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Thomas G. The S6 kinase signaling pathway in the control of development and growth. Biol. Res. 2002;35:305–313. doi: 10.4067/s0716-97602002000200022. [DOI] [PubMed] [Google Scholar]
- 32.Montagne J, Stewart MJ, Stocker H, Hafen E, Kozma SC, Thomas G. Drosophila S6 kinase: a regulator of cell size. Science. 1999;285:2126–2129. doi: 10.1126/science.285.5436.2126. [DOI] [PubMed] [Google Scholar]
- 33.Radimerski T, Montagne J, Hemmings-Mieszczak M, Thomas G. Lethality of Drosophila lacking TSC tumor suppressor function rescued by reducing dS6K signaling. Genes Dev. 2002;16:2627–2632. doi: 10.1101/gad.239102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Barcelo H, Stewart MJ. Altering Drosophila S6 kinase activity is consistent with a role for S6 kinase in growth. Genesis. 2002;34:83–85. doi: 10.1002/gene.10132. [DOI] [PubMed] [Google Scholar]
- 35.Junger MA, Rintelen F, Stocker H, Wasserman JD, Vegh M, Radimerski T, Greenberg ME, Hafen E. The Drosophila Forkhead transcription factor FOXO mediates the reduction in cell number associated with reduced insulin signaling. J. Biol. 2003;2:20.1–20.17. doi: 10.1186/1475-4924-2-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lithgow GJ, Kirkwood TB. Mechanisms and evolution of aging. Science. 1996;273:80. doi: 10.1126/science.273.5271.80. [DOI] [PubMed] [Google Scholar]
- 37.Arking R, Force AG, Dudas SP, Buck S, Baker GT., 3rd. Factors contributing to the plasticity of the extended longevity phenotypes of Drosophila. Exp. Gerontol. 1996;31:623–643. doi: 10.1016/s0531-5565(96)00096-4. [DOI] [PubMed] [Google Scholar]
- 38.Hursting SD, Lavigne JA, Berrigan D, Perkins SN, Barrett JC. Calorie restriction, aging, and cancer prevention: mechanisms of action and applicability to humans. Annu. Rev. Med. 2003;54:131–152. doi: 10.1146/annurev.med.54.101601.152156. [DOI] [PubMed] [Google Scholar]
- 39.Weindruch R. Effect of caloric restriction on age-associated cancers. Exp. Gerontol. 1992;27:575–581. doi: 10.1016/0531-5565(92)90012-o. [DOI] [PubMed] [Google Scholar]
- 40.Wolkow CA, Kimura KD, Lee MS, Ruvkun G. Regulation of C. elegans life-span by insulinlike signaling in the nervous system. Science. 2000;290:147–150. doi: 10.1126/science.290.5489.147. [DOI] [PubMed] [Google Scholar]
- 41.Libina N, Berman JR, Kenyon C. Tissue-specific activities of C. elegans DAF-16 in the regulation of lifespan. Cell. 2003;115:489–502. doi: 10.1016/s0092-8674(03)00889-4. [DOI] [PubMed] [Google Scholar]
- 42.Boisclair YR, Rhoads RP, Ueki I, Wang J, Ooi GT. The acid-labile subunit (ALS) of the 150 kDa IGF-binding protein complex: an important but forgotten component of the circulating IGF system. J. Endocrinol. 2001;170:63–70. doi: 10.1677/joe.0.1700063. [DOI] [PubMed] [Google Scholar]
- 43.Bluher M, Kahn BB, Kahn CR. Extended longevity in mice lacking the insulin receptor in adipose tissue. Science. 2003;299:572–574. doi: 10.1126/science.1078223. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.