Introduction
Dietary restriction (DR) is a robust method of increasing life span in a variety of species (1, 2)(see Masoro Subfield History and Kristal Perspective). The shift of an organism's metabolic investment from reproduction and growth toward somatic maintenance, which allows the organism to survive under harsh environments until there are suitable reproductive conditions, has been proposed to be responsible for the increase in life span caused by DR (3). However, the molecular mechanism by which this shift in metabolism manifests as a change in life span remains to be elucidated.
Recently, the nutrient-sensing TOR (target of rapamycin) pathway (Fig. 1), which is conserved in organisms ranging from fungi to humans, has been implicated in the regulation of life span (4-6). TOR itself is a kinase that phosphorylates a number of different proteins and thus modulates their behavior. In this Perspective, we discuss the responsiveness of the TOR pathway to nutrients, as well as its downstream effectors that couple nutrients to growth, in relation to aging. We also discuss connections between the TOR pathway and the insulin-like signaling (ILS) pathway, which is also implicated in the control of life span (see “One for All”).
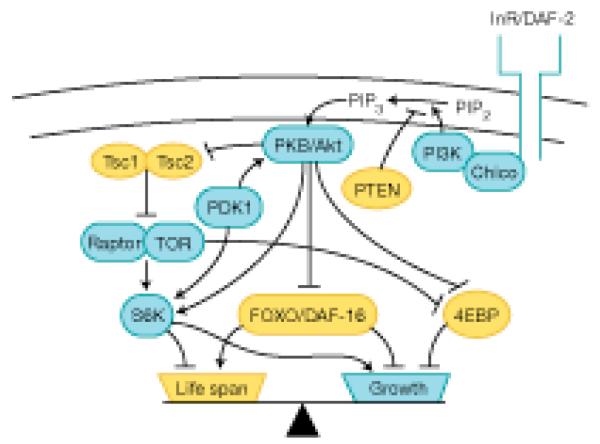
Fig. 1.
Balance between life span and growth is maintained by the activities of the TOR and insulin/IGF-1 signaling pathways. Arrows represent molecular interactions based on studies in various model organisms. Growth is slowed by increased activity of proteins shown in yellow and by decreased activity of proteins shown in blue. PIP2, phosphatidylinositol 4,5-bisphosphate; PIP3, phosphatidylinositol-3,4,5-trisphosphate; PKB, protein kinase B; PKD1, phosphoinositide-dependent kinase 1.
TOR and Life Span
Experiments published in 2003 by Vellai et al. revealed that down-regulation of TOR expression by RNA inhibition (RNAi) in Caenorhabditis elegans during adulthood results in life-span extension (4). Whether the ILS pathway is required for this phenotype was addressed by determining whether it was suppressible by mutation in the gene encoding DAF-16 ,a FOXO forkhead transcription factor that functions in the ILS pathway. If this pathway is active, DAF-16 becomes phosphorylated and is localized in the cytoplasm; if the pathway is inactive, DAF-16 enters the nucleus, where it positively regulates the expression of genes that are believed to promote life-span extension. Whereas down-regulation of the expression of DAF-2 [an insulin/insulin-like growth factor-1 (IGF-1)-like receptor] by RNAi causes life-span extension, a phenotype that is suppressed by daf-16 mutations, Vellai et al. showed that daf-16 mutations do not suppress the effects of TOR down-regulation, indicating that the ILS pathway is not involved.
More recently, Jia et al. (5) provided additional evidence for connections between the TOR pathway and life span by showing that heterozygous C. elegans daf-15 mutants display an extended life span as compared to the wild type. DAF-15 is the worm ortholog of the mammalian protein raptor (regulatory associated protein of mammalian TOR), which forms a stoichiometric complex with TOR. However, this life-span extension was not observable in a daf-16 mutant background, indicating, in contrast to the results from Vellai et al., that DAF-16 is required for these effects (5). This apparent discrepancy between the results of the two groups is explained by the negative regulation of DAF-15 by DAF-16 (5). In a daf-15 heterozygous mutant background, loss of DAF-16 may increase the abundance of DAF-15, negating the life-span extension effect caused by reduction of DAF-15 by the heterozygous mutation (5).
Life-span extension by mutations that affect the TOR pathway also occurs in flies; modulation of various genes that encode components of the TOR signaling pathway, including the products of the tuberous sclerosis complex genes (Tsc1 and Tsc2), TOR, and S6 kinase (S6K), extend life span (6)(see“More Without TOR”). The life-span extension displayed by mutant flies as compared to controls was found to be greater when they were supplied with rich nutrition as compared to poor nutrition, an observation that connects TOR's effects on life span with its previously characterized role in nutrient sensing.
TOR Links Nutrients to Growth
Mutations that affect components of the TOR pathway in a variety of organisms cause growth defects that point toward its role in nutrient sensing. In Saccharomyces cerevisiae, disruption of TOR1 leads to a phenotype similar to that seen in yeast treated with rapamycin, an antibiotic that inhibits TOR (7, 8). The cells arrest at the G1 phase of the cell cycle and display reduced protein synthesis, increased levels of autophagy (see below), and decreased amino acid transport (7). In Drosophila, larvae lacking TOR show similarities to amino acid-deprived animals, such as reduced nucleolar size and lipid vesicle aggregation in the larval fat body (a structure that acts as both the liver and the adipose tissue of the fly) (9). The role of TOR has also been examined in anautogenous mosquitoes (10). These mosquitoes use the reproductive strategy of anautogeny, requiring a blood meal to initiate egg maturation. Production of egg yolk protein precursors requires both the steroid hormone 20 hydroxyecdysone and the TOR pathway (10), indicating that the TOR pathway couples blood ingestion with egg production.
In C. elegans, deletion of the gene encoding TOR leads to developmental arrest at the L3 larval stage; these larvae exhibit an intestinal phenotype characterized by an increase in gut lumen size and a decreased ability to absorb and digest nutrients (11). A similar phenotype was observed for homozygous daf-15 mutants (5). TOR pathway mutants also show an increase in fat accumulation like that observed in daf-2 mutants (5, 11). Interestingly, neither the developmental arrest nor the fat accumulation of homozygous daf-15 mutants is suppressible by daf-16 mutations (5). Thus, although mutations that affect the TOR pathway can result in phenotypes similar to those caused by mutations that affect the ILS pathway, these data suggest that the TOR pathway is either acting downstream or parallel to the ILS pathway in the worm.
Interaction Between the TOR and ILS Pathways
Recent evidence from Drosophila suggests that signaling through TOR is both parallel to and interactive with signaling in the ILS pathway (12, 13). This idea is supported by the finding that heterozygosity of dTsc1 or dTsc2, which encode products that function upstream of TOR, is sufficient to suppress the lethality caused by loss of function of dInR, which, like daf-2, encodes an insulin/IGF-1-like receptor (13). However, the finding that loss-of-function mutations of dTsc1 and dPTEN, which encodes a phosphatase that negatively regulates the ILS pathway, cause cell-autonomous and additive increases in cell size, suggests that the products of these genes might function in parallel pathways (13). Furthermore, the increase in cell size resulting from mutations in dTsc1 is enhanced by the loss of function of dFOXO, which encodes a forkhead transcription factor that is the ortholog of C. elegans DAF-16 (14). Cross talk between the TOR pathway and the ILS pathway is apparent, because both activate the downstream effectors 4EBP (which can act as a repressor of translation) and S6K (which phosphorylates ribosomal protein S6) (7, 15). Further interactions between these pathways are suggested by transcriptional regulation of the gene encoding 4EBP by dFOXO in flies (16), and DAF-15 regulation by DAF-16 in worms (discussed above) (5).
The Need for Multiple Growth Pathways
Unicellular organisms sense and directly respond to nutrients, such as amino acids, to control growth. In multicellular organisms, the requirement for coordination of growth in different tissues creates a need for intercellular communication, which is achieved by diffusible growth factors. In order to gauge the appropriateness of these growth signals, multicellular organisms have also retained the ability to sense nutrients autonomously, as checkpoints for growth control. The TOR pathway has been implicated in this coordination of nutritional status and growth (17). Colombani et al. (17) showed that down-regulation of TOR signaling in the Drosophila fat body can activate an amino acid sensor in the fat body that might play a role in modulating the growth of peripheral tissues. A decrease in signaling by InR and phosphatidylinositol 3-kinase (PI3K, another component of the ILS pathway) in the peripheral tissues is observed upon activation of this amino acid sensor in the fat body. The fat body produces a Drosophila insulin-like peptide cofactor, dALS (acid labile subunit), and this activity has been suggested as a possible mechanism of humoral control of growth in peripheral tissues (17).
Downstream Effectors of TOR
TOR coordinates the activity of various targets that all seem to have a unifying role in regulating cellular growth in response to nutrients.
Autophagy
Autophagy, a process triggered in many organisms upon nutrient starvation that involves TOR (18-20), provides an attractive mechanism to explain the effects of the TOR pathway on life span. During autophagy, various cytoplasmic components are enclosed within a double-membrane structure (the autophagosome) and delivered to the vacuole for degradation. Recently, in C. elegans autophagy has been shown to be required for the life-span extension that occurs in daf-2 mutants as well as for dauer morphogenesis (an alternative developmental pathway that occurs under unfavorable growth conditions) (21). TOR regulates autophagy by modulating the behavior of APG13 and APG1, two proteins known to play a role in autophagy. TOR maintains APG13 in a phosphorylated state, a form that has a low affinity for APG1. Nutrient deprivation or inactivation of TOR by rapamycin leads to APG13 dephosphorylation and hence to interaction with and activation of APG1 to stimulate autophagy (20).
Protein synthesis
Protein synthesis is another process mediated by the TOR pathway. S6K phosphorylation and thus activation have been implicated in mediating the downstream effects of TOR on translation initiation in both flies and mammals. This idea is suggested by the observation that the TOR knockout phenotype can be suppressed by overexpressing S6K in flies (22). Consistent with the effect of S6K on growth, flies carrying homozygous mutations in dS6K show a developmental delay and a reduction in body size (23). Phosphorylation of ribosomal protein S6 by S6K is accompanied by up-regulation of a class of mRNAs that contain an oligopyrimidine tract at their transcriptional start site, termed 5′TOP (24). Most of these mRNAs encode components of the translational apparatus, including ribosomal proteins and elongation factors (24). Hence, increasing the translation of TOP messages amplifies the stimulatory effect of the TOR pathway on protein synthesis.
4EBP, a protein that is phosphorylated upon insulin stimulation, is also targeted by TOR to regulate translation and growth (7, 15). When inactive, the hypophosphorylated form acts as a translational repressor by binding the protein synthesis initiation factor eIF4E and thus blocking the binding of eIf4E with initiation factor eIF4G. This activity represses cap-dependent translation (during which the assembly of initiation factors on mRNA is stimulated by a 5′ cap and a polyadenylated tail on the mRNA) (7, 15). Overexpression of eIF4E in cultured mammalian cells causes transformation of fibroblasts and increased cell size, which can be reversed by increasing the abundance of 4EBP (15, 25).
Ribosome production
Ribosome production and protein synthesis are energetically costly processes that are co-regulated to meet the metabolic demands of the cell. In yeast, TOR signaling regulates ribosome biogenesis at both the transcriptional and translational levels. It is known to affect the transcription of ribosomal protein mRNAs by RNA polymerase II, as well as transcription of ribosomal RNA and transfer RNA by RNA polymerase I and RNA polymerase III (26, 27). The control of ribosome biogenesis by the TOR pathway, though not well understood, is thought to involve protein phosphatase 2a, mutations in which inhibit polyribosome formation (26, 27).
Amino acid transport
Amino acids are essential to cellular growth because they serve as building blocks for protein synthesis. The TOR pathway plays an important role in regulating the activity of amino acid permeases, proteins that transport amino acids across the plasma membrane, in S. cerevisiae (28). Inhibition of TOR function by rapamycin or nitrogen limitation induces ubiquitination and degradation of tryptophan permease (28).
Although a role for the TOR pathway in the control of amino acid transport has not been shown in worms, deletion of the gene encoding the intestinal peptide transporter shows some interesting interactions with the TOR and ILS pathways. Deletion of the C. elegans pep-2 gene (which is normally expressed in the intestine and encodes the ortholog of mammalian intestinal peptide transporter) abolishes the uptake of intact peptides from the gut lumen (29). As a result, there is a decrease in the rate of development, body size, and progeny number and an increase in tolerance to stress. Interestingly, the life-span extension caused by daf-2 mutations or down-regulation of TOR by RNAi is further enhanced by the pep-2 deletion. Furthermore, loss of TOR in the pep-2 mutant background enhances the enlargement of the gut lumen and the slow development that occurs in this mutant (29). These data suggest an important role for amino acid transport upstream of TOR signaling in C. elegans.
Conclusion
The evidence that TOR regulates life span provides a framework for exploring the link between nutrient limitation and life-span extension. It will be an interesting challenge to dissect the downstream effectors of the TOR and ILS pathways that regulate life span. Further analysis of these pathways might also provide information about connections between diet and cancer. Growth signaling pathways are frequently misregulated in cancer, and the TOR and ILS growth signaling pathways play an important role in tumor formation in some types of cancer [reviewed in (15)]. Additionally, DR reduces the incidence of a variety of tumors in rodents (30). Thus, the TOR and ILS pathways may be a key link that provides insight into the protective effects of DR on tumor formation.
Contributor Information
Pankaj Kapahi, Buck Institute for Age Research, Novato, CA 94945, USA. Pkapahi@buckinstitute.org (P.K.).
Brian Zid, Division of Biology at the California Institute of Technology, Pasadena, CA 91125, USA..
References
- 1.Guarente L, Kenyon C. Genetic pathways that regulate ageing in model organisms. Nature. 2000;408:255–262. doi: 10.1038/35041700. [DOI] [PubMed] [Google Scholar]
- 2.Partridge L, Gems D. Mechanisms of ageing: Public or private? Nat. Rev. Genet. 2002;3:165–175. doi: 10.1038/nrg753. [DOI] [PubMed] [Google Scholar]
- 3.Holliday R. Food, reproduction and longevity: Is the extended lifespan of calorie-restricted animals an evolutionary adaptation? Bioessays. 1989;10:125–127. doi: 10.1002/bies.950100408. [DOI] [PubMed] [Google Scholar]
- 4.Vellai T, Takacs-Vellai K, Zhang Y, Kovacs AL, Orosz L, Muller F. Genetics: Influence of TOR kinase on lifespan in C. elegans. Nature. 2003;426:620. doi: 10.1038/426620a. [DOI] [PubMed] [Google Scholar]
- 5.Jia K, Chen D, Riddle DL. The TOR pathway interacts with the insulin signaling pathway to regulate C. elegans larval development, metabolism and life span. Development. 2004;131:3897–3906. doi: 10.1242/dev.01255. [DOI] [PubMed] [Google Scholar]
- 6.Kapahi P, Zid BM, Harper T, Koslover D, Sapin V, Benzer S. Regulation of lifespan in Drosophila by modulation of genes in the TOR signaling pathway. Curr. Biol. 2004;14:885–890. doi: 10.1016/j.cub.2004.03.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Harris TE, Lawrence JC., Jr. TOR signaling. Sci. STKE. 2003;2003:re15. doi: 10.1126/stke.2122003re15. [DOI] [PubMed] [Google Scholar]
- 8.Heitman J, Movva NR, Hall MN. Targets for cell cycle arrest by the immunosuppressant rapamycin in yeast. Science. 1991;253:905–909. doi: 10.1126/science.1715094. [DOI] [PubMed] [Google Scholar]
- 9.Zhang H, Stallock JP, Ng JC, Reinhard C, Neufeld TP. Regulation of cellular growth by the Drosophila target of rapamycin dTOR. Genes Dev. 2000;14:2712–2724. doi: 10.1101/gad.835000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hansen IA, Attardo GM, Park JH, Peng Q, Raikhel AS. Target of rapamycin-mediated amino acid signaling in mosquito anautogeny. Proc. Natl. Acad. Sci. U.S.A. 2004;101:10626–10631. doi: 10.1073/pnas.0403460101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Long X, Spycher C, Han ZS, Rose AM, Muller F, Avruch J. TOR deficiency in C. elegans causes developmental arrest and intestinal atrophy by inhibition of mRNA translation. Curr. Biol. 2002;12:1448–1461. doi: 10.1016/s0960-9822(02)01091-6. [DOI] [PubMed] [Google Scholar]
- 12.Marygold SJ, Leevers SJ. Growth signaling: TSC takes its place. Curr. Biol. 2002;12:R785–R787. doi: 10.1016/s0960-9822(02)01294-0. [DOI] [PubMed] [Google Scholar]
- 13.Gao X, Pan D. TSC1 and TSC2 tumor suppressors antagonize insulin signaling in cell growth. Genes Dev. 2001;15:1383–1392. doi: 10.1101/gad.901101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Junger MA, Rintelen F, Stocker H, Wasserman JD, Vegh M, Radimerski T, Greenberg ME, Hafen E. The Drosophila Forkhead transcription factor FOXO mediates the reduction in cell number associated with reduced insulin signaling. J. Biol. 2003;2:20. doi: 10.1186/1475-4924-2-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shamji AF, Nghiem P, Schreiber SL. Integration of growth factor and nutrient signaling: Implications for cancer biology. Mol. Cell. 2003;12:271–280. doi: 10.1016/j.molcel.2003.08.016. [DOI] [PubMed] [Google Scholar]
- 16.Puig O, Marr MT, Ruhf ML, Tjian R. Control of cell number by Drosophila FOXO: Downstream and feedback regulation of the insulin receptor pathway. Genes Dev. 2003;17:2006–2020. doi: 10.1101/gad.1098703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Colombani J, Raisin S, Pantalacci S, Radimerski T, Montagne J, Leopold P. A nutrient sensor mechanism controls Drosophila growth. Cell. 2003;114:739–749. doi: 10.1016/s0092-8674(03)00713-x. [DOI] [PubMed] [Google Scholar]
- 18.Scott RC, Schuldiner O, Neufeld TP. Role and regulation of starvation-induced autophagy in the Drosophila fat body. Dev. Cell. 2004;7:167–178. doi: 10.1016/j.devcel.2004.07.009. [DOI] [PubMed] [Google Scholar]
- 19.Rusten TE, Lindmo K, Juhasz G, Sass M, Seglen PO, Brech A, Stenmark H. Programmed autophagy in the Drosophila fat body is induced by ecdysone through regulation of the PI3K pathway. Dev. Cell. 2004;7:179–192. doi: 10.1016/j.devcel.2004.07.005. [DOI] [PubMed] [Google Scholar]
- 20.Kamada Y, Funakoshi T, Shintani T, Nagano K, Ohsumi M, Ohsumi Y. Tor-mediated induction of autophagy via an Apg1 protein kinase complex. J. Cell Biol. 2000;150:1507–1513. doi: 10.1083/jcb.150.6.1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Melendez A, Talloczy Z, Seaman M, Eskelinen EL, Hall DH, Levine B. Autophagy genes are essential for dauer development and life-span extension in C. elegans. Science. 2003;301:1387–1391. doi: 10.1126/science.1087782. [DOI] [PubMed] [Google Scholar]
- 22.Radimerski T, Montagne J, Hemmings-Mieszczak M, Thomas G. Lethality of Drosophila lacking TSC tumor suppressor function rescued by reducing dS6K signaling. Genes Dev. 2002;16:2627–2632. doi: 10.1101/gad.239102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Montagne J, Stewart MJ, Stocker H, Hafen E, Kozma SC, Thomas G. Drosophila S6 kinase: a regulator of cell size. Science. 1999;285:2126–2129. doi: 10.1126/science.285.5436.2126. [DOI] [PubMed] [Google Scholar]
- 24.Thomas G. The S6 kinase signaling pathway in the control of development and growth. Biol. Res. 2002;35:305–313. doi: 10.4067/s0716-97602002000200022. [DOI] [PubMed] [Google Scholar]
- 25.Lazaris-Karatzas A, Montine KS, Sonenberg N. Malignant transformation by a eukaryotic initiation factor subunit that binds to mRNA 5′ cap. Nature. 1990;345:544–547. doi: 10.1038/345544a0. [DOI] [PubMed] [Google Scholar]
- 26.Zaragoza D, Ghavidel A, Heitman J, Schultz MC. Rapamycin induces the G0 program of transcriptional repression in yeast by interfering with the TOR signaling pathway. Mol. Cell. Biol. 1998;18:4463–4470. doi: 10.1128/mcb.18.8.4463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rohde J, Heitman J, Cardenas ME. The TOR kinases link nutrient sensing to cell growth. J. Biol. Chem. 2001;276:9583–9586. doi: 10.1074/jbc.R000034200. [DOI] [PubMed] [Google Scholar]
- 28.Schmidt A, Beck T, Koller A, Kunz J, Hall MN. The TOR nutrient signalling pathway phosphorylates NPR1 and inhibits turnover of the tryptophan permease. EMBO J. 1998;17:6924–6931. doi: 10.1093/emboj/17.23.6924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Meissner B, Boll M, Daniel H, Baumeister R. Deletion of the intestinal peptide transporter affects insulin and TOR signaling in Caenorhabditis elegans. J. Biol. Chem. 2004;279:36739–36745. doi: 10.1074/jbc.M403415200. [DOI] [PubMed] [Google Scholar]
- 30.Weindruch R. Effect of caloric restriction on age-associated cancers. Exp. Gerontol. 1992;27:575–581. doi: 10.1016/0531-5565(92)90012-o. [DOI] [PubMed] [Google Scholar]