Abstract
There is growing recognition that mammalian cells produce many thousands of large intergenic transcripts1–4. However, the functional significance of these transcripts has been particularly controversial. Although there are some well-characterized examples, most (>95%) show little evidence of evolutionary conservation and have been suggested to represent transcriptional noise5,6. Here we report a new approach to identifying large non-coding RNAs using chromatin-state maps to discover discrete transcriptional units intervening known protein-coding loci. Our approach identified ~1,600 large multi-exonic RNAs across four mouse cell types. In sharp contrast to previous collections, these large intervening non-coding RNAs (lincRNAs) show strong purifying selection in their genomic loci, exonic sequences and promoter regions, with greater than 95% showing clear evolutionary conservation. We also developed a functional genomics approach that assigns putative functions to each lincRNA, demonstrating a diverse range of roles for lincRNAs in processes from embryonic stem cell pluripotency to cell proliferation. We obtained independent functional validation for the predictions for over 100 lincRNAs, using cell-based assays. In particular, we demonstrate that specific lincRNAs are transcriptionally regulated by key transcription factors in these processes such as p53, NFκB, Sox2, Oct4 (also known as Pou5f1) and Nanog. Together, these results define a unique collection of functional lincRNAs that are highly conserved and implicated in diverse biological processes.
There are at present only about a dozen well-characterized lincRNAs in mammals, with transcript sizes ranging from 2.3 to 17.2kilobases (kb)7,8. These lincRNAs have distinctive biological roles through diverse molecular mechanisms, including functioning in X-chromosome inactivation (Xist, Tsix)8,9, imprinting (H19, Air)7,10, trans-acting gene regulation (HOTAIR)11 and regulation of nuclear import (Nron)12. Importantly, these well-characterized lincRNAs show clear evolutionary conservation confirming that they are functional.
Genomic projects over the past decade have used shotgun sequencing and microarray hybridization1–4 to obtain evidence for many thousands of additional non-coding transcripts in mammals. Although the number of transcripts has grown, so too have the doubts as to whether most are biologically functional5,6,13. The main concern was raised by the observation that most of the intergenic transcripts show little to no evolutionary conservation5,13. Strictly speaking, the absence of evolutionary conservation cannot prove the absence of function. But, the markedly low rate of conservation seen in the current catalogues of large non-coding transcripts (<5% of cases) is unprecedented and would require that each mammalian clade evolves its own distinct repertoire of non-coding transcripts. Instead, the data suggest that the current catalogues may consist largely of transcriptional noise, with a minority of bona fide functional lincRNAs hidden amid this background. Thus, to expand our understanding of functional lincRNAs, we are faced with two important challenges: (1) identifying lincRNAs that are most likely to be functional; and (2) inferring putative functions for these lincRNAs that can be tested in hypothesis-driven experiments.
To address the first challenge, we took an entirely different approach to discovering functional lincRNAs on the basis of exploiting chromatin structure. We recently developedan efficient method14 to create genome-wide chromatin-state maps, using chromatin immunoprecipitation followed by massively parallel sequencing (ChIP-Seq). We observed that genes actively transcribed by RNA polymerase II (Pol II) are marked by trimethylation of lysine4 of histoneH3 (H3K4me3) at their promoter and trimethylation of lysine36 of histone H3 (H3K36me3) along the length of the transcribed region14. We will refer to this distinctive structure as a ‘K4–K36 domain’. We proposed that, by identifying K4–K36 structures that reside outside known protein-coding gene loci, we could systematically discover lincRNAs.
To test this hypothesis, we searched for K4–K36 domains in genome-wide chromatin-state maps of four mouse cell types: mouse embryonic stem cells (ESCs), mouse embryonic fibroblasts (MEF), mouse lung fibroblasts (MLF) and neural precursor cells (NPC). We identified K4–K36 domains of at least 5 kb in size that did not overlap regions containing protein-coding genes as well as known microRNAs15 and endogenous short interfering RNAs (siRNAs)16,17. This analysis revealed 1,675 K4–K36 (1,250 conservatively defined) domains that do not overlap with known annotations; examples are shown in Fig. 1 (Supplementary Table 1).
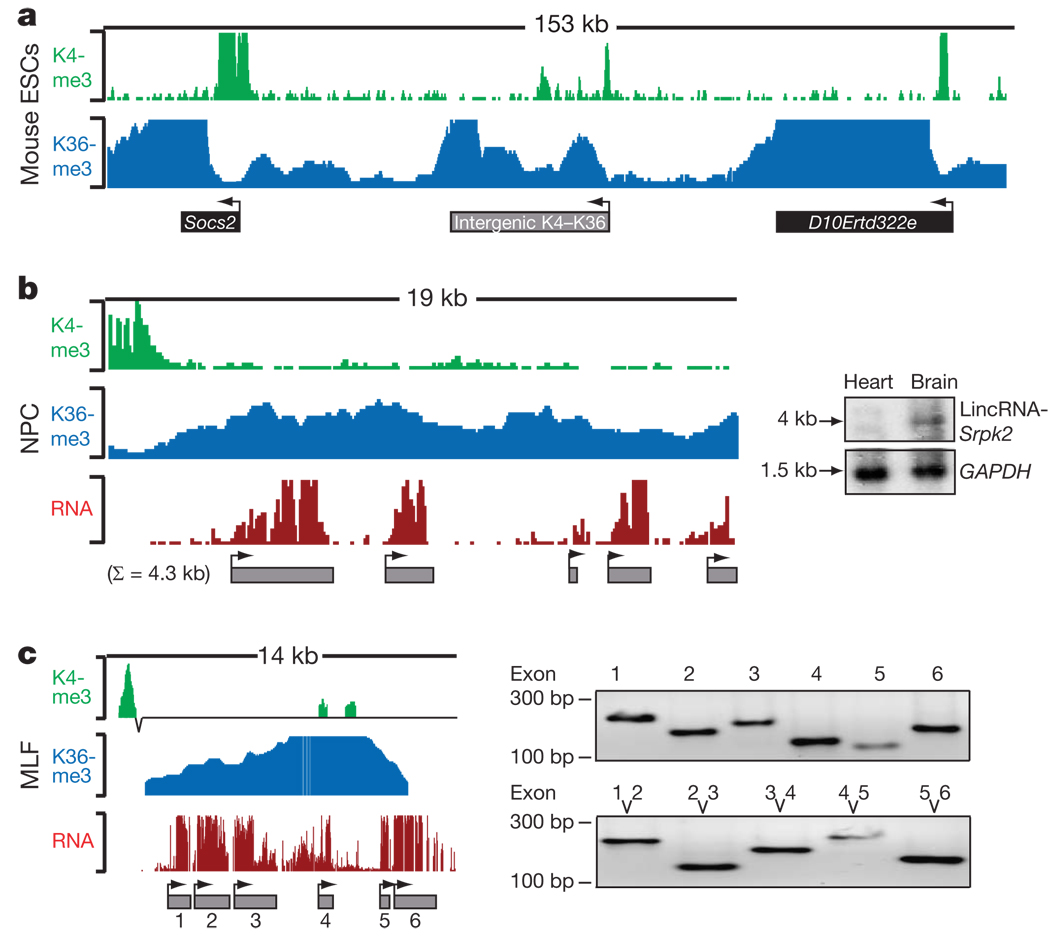
Figure 1. Intergenic K4–K36 domains produce multi-exonic RNAs.
a, Example of an intergenic K4–K36 domain and the K4–K36 domains of two flanking protein-coding genes. Each histone modification is plotted as the number of DNA fragments obtained by ChIP-Seq at each position. Black boxes indicate known protein-coding regions and grey boxes are intergenic K4–K36 domains. Arrowheads indicate the orientation of transcription. b, Intergenic K4–K36 domains were interrogated for presence of transcription by hybridizing RNA to DNA tiling arrays. The RNA hybridization intensity is plotted in red. RNA peaks were determined and are represented by grey boxes. The presence of a spliced transcript was validated by hybridization to a northern blot (right). c, Connectivity between the inferred exons was validated by PCR with reverse transcription (RT–PCR). Right top shows RT–PCR validation of each exon, right bottom shows RT–PCR across each consecutive exon.
Having identified K4–K36 loci with no previous annotation, we addressed: (1) whether these gene loci produce large multi-exonic RNA molecules; (2) whether the RNA molecules encode proteins or are non-coding transcripts; and (3) whether the RNA molecules, their promoters and their chromatin structure show conservation across mammals.
To test whether the intergenic K4–K36 domains produce RNA transcripts, we selected a random sample of 350 regions and designed DNA microarrays containing oligonucleotides that tile across the regions (Methods) as well as various control regions. We hybridized poly(A)+-selected RNA from each of the four cell types to the arrays. We developed an algorithm (Methods) to identify regions of significant hybridization and used it to define putative exons of transcripts detected at the loci. For ~70% of the intergenic loci with K4–K36 domains present in a cell type, we found clear evidence of RNA transcription in that cell type (Fig. 1 and Supplementary Table 2 and Table 3). The proportion is similar to that for the protein-coding genes: ~72% of K4–K36 domains corresponding to known protein-coding genes show significant hybridization (P < 0.05, Wilcoxon test). In addition, we confirmed the presence of 93 out of 107 (87%) randomly selected exons, representing at least one exon from 19 out of 20 K4–K36 domains tested. We also confirmed the connectivity of consecutive exons in 52 out of 67 (78%) cases, including one from each of 16 K4–K36 domains tested (Fig. 1c and Supplementary Table 4). Furthermore, we validated the presence of discrete transcripts by hybridization to RNA northern blots in 15 of 17 tested loci (Fig. 1b, Supplementary Fig. 1, Supplementary Table 5 and Methods).
To determine whether the transcripts encode previously unknown protein-coding genes or non-coding RNAs, we used an established metric (the codon substitution frequency, CSF18,19) to assess characteristic evolutionary signatures of protein-coding domains. Analysing both the overall genomic locus (Fig. 2a and Supplementary Table 6) and the exons themselves (Supplementary Fig. 2 and Methods), we found that >90% of the intergenic K4–K36 domains fall well below the threshold of known protein-coding genes and resemble known lincRNAs (Fig. 2a). The result indicates that most of the loci do not encode protein-coding genes. Consistent with this, fewer than 2.5% of the exons show any similarity to known protein-coding genes, using the BLASTX program (Methods).
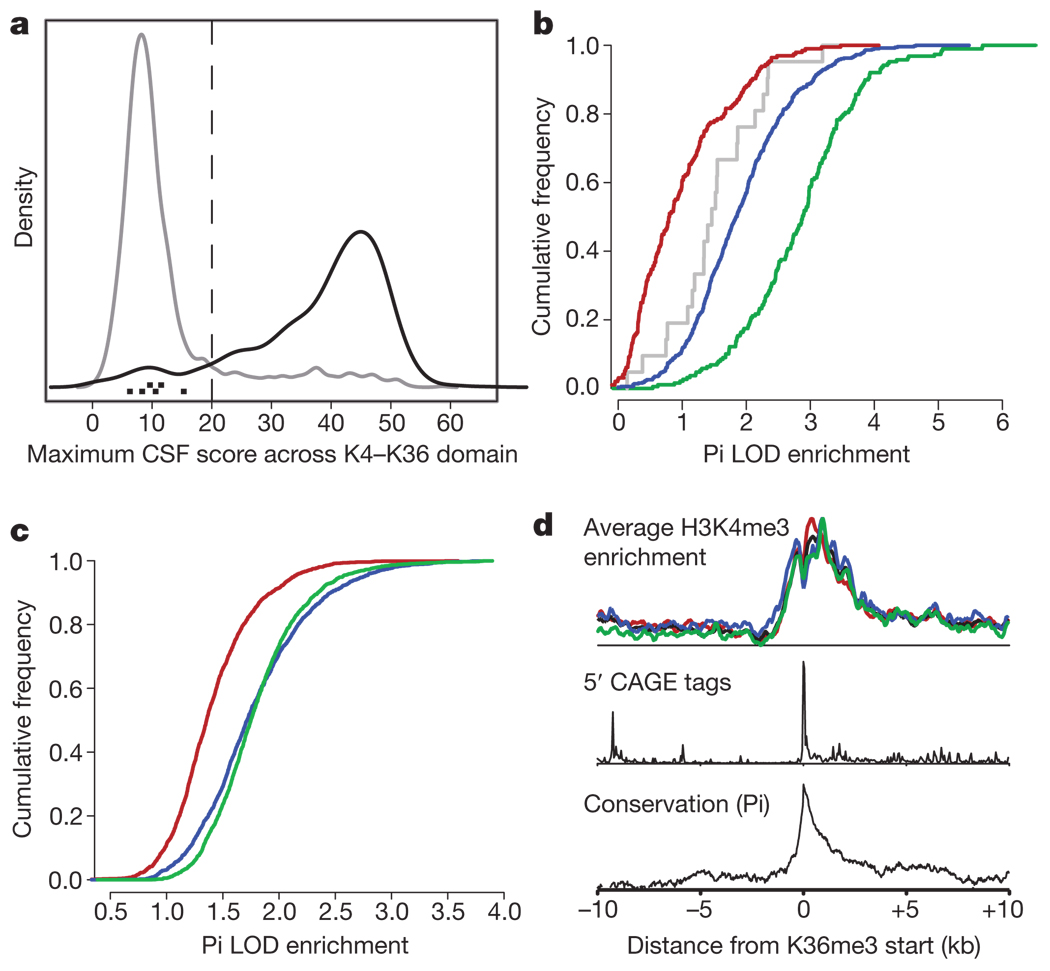
Figure 2. lincRNA K4–K36 domains do not encode proteins and are conserved in their exons and promoters.
a, Density plot of the maximum CSF score (Methods) across intergenic K4–K36 domains (grey) and known protein-coding genes (black). The maximum CSF scores for known lincRNAs are indicated as black points at the bottom. b, Cumulative distribution of sequence conservation across mammals for lincRNA exons (blue), protein-coding exons (green), introns (red) and known non-coding RNA exons (grey). c, Cumulative distribution of sequence conservation for lincRNA promoters (blue), random intergenic regions (red), and protein-coding promoters (green). LOD, logarithm of the odds ratio; Pi is the conservation metric (see Supplementary Methods). d, Enrichment of various promoter features plotted as the distance from the start of the K36me3 region averaged across all lincRNAs. Enrichment in each cell type of K4me3 domains across mouse ESCs (red), MEF (black), MLF (blue) and NPC (green) is shown (top panel). Enrichment of 5′ CAGE-tag density representing the 5′ end of RNA molecules (middle panel) and conservation scores in the K4me3 region are shown (bottom panel).
To assess the extent of nucleotide sequence conservation in the RNA transcripts, we used a method that explicitly models the underlying substitution rate (Methods) across 21 mammalian genomes (M.G. and X. Xie, submitted, Methods). We found that the lincRNA exons show clear sequence conservation when compared to other intergenic regions (Fig. 2b, Supplementary Fig. 3 and Supplementary Tables 2 and 7). Furthermore, the transcribed regions are highly enriched for conserved elements (defined by the PhastCons program20) compared to other intergenic regions (P < 0.0001, permutation test). The conservation level is similar to that seen for known lincRNAs, although it is lower than that seen for protein-coding exons, probably reflecting a lower degree of constraint on RNA structures than on amino-acid codons. The presence of strong purifying selection provides firm evidence that most K4–K36-defined lincRNAs must be biologically functional in mammals.
We used the same method to assess the conservation of the lincRNAs promoters (marked by the K4 domain). The lincRNA promoter regions show strong conservation, being essentially indistinguishable from known protein-coding genes (Fig. 2c and Supplementary Table 8). Furthermore, the lincRNA promoters show a notable enrichment of ‘CAGE tags’ (obtained by capturing the 7-methylguanosine cap at the 5′-end of Pol II transcripts) that mark transcriptional start sites21 (Fig. 2d). Most of the lincRNA promoters regions (85%) contain a significant cluster of CAGE tags, with the density tightly localized around the promoter. In addition, the lincRNA promoters show strong enrichment for binding of RNA PolII in mouse ESCs (P < 2 × 10−16; Supplementary Fig. 4).
To investigate whether the K4–K36 chromatin structures observed at the loci are conserved across species, we constructed chromatin-state maps in human lung fibroblasts and MLF. Notably, ~70% of the K4–K36 domains in human also had a K4–K36 domain in the orthologous region of the mouse genome (Supplementary Table 9). The proportion is similar to that seen for protein-coding genes (~80%).
Together, the results show that most of the K4–K36 domains encode multi-exonic, non-protein-coding transcripts and the loci show clear conservation of nucleotide sequence and chromatin structure. Moreover, transcription and processing of these lincRNAs appears to be similar to that for protein-coding genes—including Pol II transcription, 5′-capping and poly-adenylation.
Having identified a large set of conserved lincRNAs, the next important challenge is to develop a method to infer putative functions that can be tested experimentally. To this end, we began by creating an RNA expression compendium of both lincRNAs and protein-coding genes across a wide range of tissues. We hybridized poly-adenylated RNA from 16 mouse samples to a custom lincRNA array. The samples included the original four cell types (mouse ESCs, NPC, MEF and MLF), a time course of embryonic development (whole embryo, hindlimb and forelimb at embryonic days 9.5, 10.5 and 13.5), and four normal adult tissues (brain, lung, ovary and testis) (Supplementary Fig. 5 and Supplementary Table 10).
The expression data contains a wealth of information about the lincRNAs. As an example, we searched for lincRNAs with an expression pattern opposite to the known lincRNA HOTAIR. Notably, we found that the most highly anti-correlated lincRNA in the genome lies in the HOXC cluster, in the same euchromatic domain as HOTAIR; we call this lincRNA Frigidair (Fig. 3c). This suggests that Frigidair may repress HOTAIR or perhaps activate genes in the HOXD cluster.
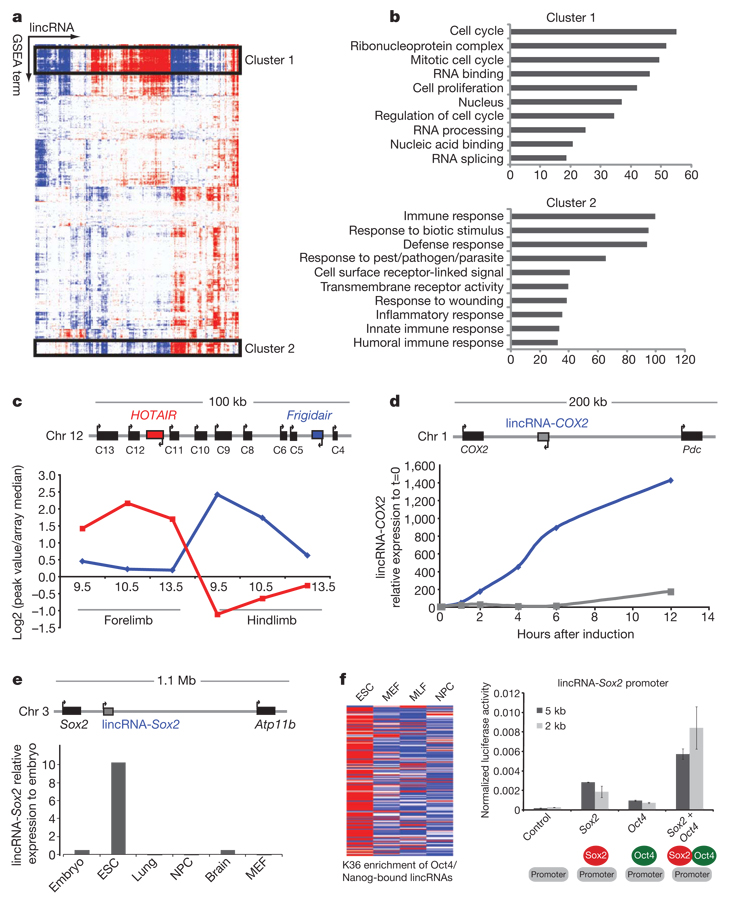
Figure 3. lincRNAs show strong associations with other lincRNAs and with several biological processes.
a, Association matrix of lincRNA and functional gene sets. Functional gene sets (columns) and lincRNAs (rows) are shown as positively (red), negatively (blue) or not associated (white) with lincRNA expression profiles. The black boxes highlight two significant biclusters in the matrix. b, Gene ontology of the protein-coding genes in these clusters is shown and plotted as the −log(P value) for the enrichment of each Gene Ontology term. c, Map of mouse genomic locus (Hoxc) containing HOTAIR. HOTAIR (red) and Frigidair (blue) show diametrically opposed expression patterns between mouse forelimb (anterior) and mouse hindlimb (posterior). d, Map of genomic locus containing COX2 along with the location of lincRNA-COX2. Quantitative RT–PCR shows that lincRNA-COX2 is upregulated in TLR4-stimulated cells (blue) but not TLR3-stimulated cells (grey). e, A map of the genomic locus containing Sox2 shows a lincRNA ~50 kb upstream that is expressed specifically in ESCs. f, K36me3 enrichment across four cell types for lincRNAs bound by Oct4 or Nanog (left). Red indicates high enrichment, blue denotes low enrichment. The lincRNA-Sox2 promoter was cloned into a luciferase reporter construct and assayed for transcriptional activity with Sox2 and Oct4 alone, together and controls (right). The y-axis represents the transcriptional activity of this promoter relative to a Renilla construct. Error bars are ± s.d. of three replicate transfections.
To take a more systematic approach, we also analysed RNA expression data for protein-coding genes from published sources14,22 and generated further data for the embryonic development time course. We clustered the lincRNA and protein-coding genes into sets with correlated expression patterns (Supplementary Fig. 6a). We used Gene Set Enrichment Analysis (GSEA) to construct a matrix of the association of each lincRNA with each of ~1,700 functional gene sets (Fig. 3a and Supplementary Table 10 and Table 11)23. We next performed biclustering on the gene set matrix to identify sets of lincRNAs that are associated with distinct sets of functional categories24. This analysis revealed numerous sets of lincRNAs associated with distinct and diverse biological processes (Fig. 3a). These include cell proliferation, RNA binding complexes, immune surveillance, ESC pluripotency, neuronal processes, morphogenesis, gametogenesis and muscle development (Fig. 3b and Supplementary Fig. 7).
To assess the validity of the inferred functional associations, we examined the gene sets associated with HOTAIR. HOTAIR showed negative association with HOXD genes (false discovery rate (FDR) <0.018) and positive association with ‘Chang Serum Response’ (FDR < 0.001), a known predictor of breast cancer meta-stasis25. Both results are consistent with the known properties of HOTAIR, including a role in breast cancer metastasis11,26.
We then sought to obtain independent experimental validation of the inferred biological functions for many of the lincRNAs. We focused on three large clusters of lincRNAs associated with the p53-mediated DNA damage response in MEF, NFκB signalling in dendritic cells, and ESC pluripotency, on the basis of their expression pattern across tissues.
We exposed p53+/+ and p53−/− MEF to a DNA damaging agent and profiled the resulting expression changes on our lincRNA micro-array (Methods)27. We found 39 lincRNAs that were significantly induced in p53+/+ but not in p53−/− cells (Methods, Supplementary Fig. 8 and Supplementary Table 12). Approximately half of these lincRNAs resided in the cluster associated with p53-mediated DNA damage response, confirming the validity of the functional inference (P<10−7). Notably, we found that the promoters of these 39 lincRNAs were significant enriched for the p53 cis-regulatory binding element (versus all lincRNA promoters, P< 0.01, Wilcoxon test; Supplementary Fig. 8 and Supplementary Table 13). This suggests that p53 directly binds and regulates the expression of at least some of these lincRNA genes.
We stimulated CD11C+ bone-marrow-derived dendritic cells with a specific agonist of the Toll-like receptor Tlr4, which signals through NFκB. We found that 20lincRNAs showed marked upregulation after Tlr4-stimulation (Supplementary Table 14). Consistent with the inferences described earlier, 80% of these induced lincRNAs resided in the cluster associated with NFκB signalling. The greatest change in expression was observed in a lincRNA that is located ~51kbupstream of the protein-coding gene COX2 (also known as Ptgs2), a critical inflammation mediator that is directly induced by NFκB on Tlr4 stimulation; we refer to this as lincRNA-COX2. We found that lincRNA-COX2 is induced ~1,000-fold over the course of 12h after Tlr4 stimulation (Fig. 3d). In contrast, stimulation of Tlr3, which signals through IRF3, led to only weak induction of lincRNA-COX2 (Fig. 3d).
Using published data from mouse ESCs, we identified 118 lincRNAs in which the promoter loci were bound by the core transcription factors Oct4 and Nanog28 (Supplementary Table 15). Of those represented on our expression array 72% resided in the cluster associated with pluripotency, again supporting the validity of the functional inference. We noticed that one of these lincRNAs, which is only expressed in ESCs, is located ~100 kb from the Sox2 locus, which encodes another key transcription factor associated with pluripotency (Fig. 3e).We cloned the promoter of this locus(which we will refer to as lincRNA-Sox2) upstream of a luciferase reporter gene and transfected the construct into mouse cells transiently expressing Oct4, Sox2, or both, as well as several controls. We found that Sox2 and Oct4 were each sufficient to drive expression of this lincRNA promoter, and the expression of both Oct4 and Sox2 together caused synergistic increases in expression (Fig. 3f). To our knowledge, this is the first experimental validation of a lincRNA promoter being directly regulated by key transcription factors such as Sox2 and Oct4.
The ultimate proof-of-function will be to demonstrate that RNA-interference-mediated knockout of each lincRNAs has the predicted phenotypic consequences. Towards this end, we examined a recently published short hairpin RNA screen of (presumed) protein-coding genes to identify genes that regulate cell proliferation rates in mouse ESCs29. The screen involved genes and some unidentified transcripts that had been identified as expressed in ESCs and showing rapid decrease in expression after retinoic acid treatment. Of the top ten hits in the screen, one corresponded to a gene of unknown function. We discovered that this gene corresponds to one of our lincRNAs (located ~181 kb from Enc1) contained in both the ‘cell cycle and cell proliferation’ cluster (FDR < 0.001) and the ‘ESC’ cluster (FDR < 0.001; Supplementary Fig. 9 and Supplementary Table 16). This provides functional confirmation that this lincRNA has a direct role in cell proliferation in ESCs, consistent with the analysis above.
Our results address the two key issues in the study of lincRNAs. We show that chromatin structure can identify sets of lincRNAs that show a high degree of evolutionary conservation, indicating that they are biologically functional. (We do not exclude the possibility that lincRNAs identified by shotgun sequencing that fail to show conservation are nonetheless functional, but other evidence will be required to establish this point.) We also provide a functional genomics pipeline for inferring putative roles for lincRNAs. The approach suggested functional roles for 150 lincRNAs that we studied on microarrays, and the independent experiments provided support for the predicted pathways for ~85 lincRNAs. The pipeline thus provides a useful guide for hypothesis-driven functional studies.
A fundamental issue will now be to determine the biological functions and the mechanisms by which lincRNAs act. One clue may come from the distribution of lincRNAs across the genome. We noted that several of the lincRNAs were located near genes encoding transcription factors (such as Sox2, Klf4, Myc and Brn1). Analysing the set of lincRNAs, we found that the genes neighbouring lincRNAs were strongly biased towards those encoding transcription factors (P < 0.001, permutation test; Supplementary Fig. 10 and Supplementary Table 17) and other proteins factors related to transcription. A second clue may come from our previous observation that HOTAIR11 represses gene expression and is associated with chromatin remodelling proteins, together with recent similar observations for XIST30. On the basis of these observations, we speculate that many lincRNAs may be involved in transcriptional control—perhaps by guiding chromatin remodelling proteins to target loci—and that some transcription factors and lincRNAs may act together, with the transcription factor activating a transcriptional program and the lincRNA repressing a previous transcriptional program. Testing these speculations will require biochemical and genetic studies, including gene knockdown in appropriate settings. Whatever their functions, the highly conserved lincRNAs represent an important new contingent in the growing population of the modern ‘RNA world’.
METHODS SUMMARY
Identifying intergenic K4–K36 domains and RNA
Enriched K4–K36 domains were identified using a sliding window approach across the genome and assessing the significance of each window. We filtered the list of K4–K36 enriched domains to eliminate known annotations. DNA tiling arrays (Nimblegen) were designed to tile intergenic K4–K36 domains. Transcribed regions were defined using a sliding window approach.
Conservation and coding potential
To detect sequence constraint we used a method that explicitly modelled the rate of mutation and level of constraint. We took the maximum 12-base-pair window score for each exonic region. We normalized for the size differences between exons by computing a size matched random genomic score (Supplementary Methods).
We tested the protein-coding potential of K4–K36 domains by determining the maximum CSF score observed across the entire genomic locus. We computed the CSF scores across sliding windows of 90 base pairs and scanned all six possible reading frames in each window.
Protein-coding gene expression profiles
We generated a correlation matrix between lincRNAs and between lincRNAs and protein-coding genes by computing the Pearson correlation for all pairwise combinations. This matrix was clustered and visualized using the Gene Pattern platform for integrative genomics (http://genepattern.broad.mit.edu/). Functional associations were computed using Gene Set Enrichment Analysis (GSEA) (Supplementary Methods).Inbrief, we used each lincRNA as a profile, computed the Pearson correlation for each protein-coding gene and then ranked the protein-coding genes by their correlation coefficient. Gene sets were filtered by an FDR < 0.05 and an association matrix was generated.
Supplementary Material
is linked to the online version of the paper at www.nature.com/nature.
Acknowledgements
We would like to thank our colleagues at the Broad Institute, especially J. P. Mesirov for discussions and statistical insights, X. Xie for statistical help with conservation analyses, J. Robinson for visualization help, M. Ku, E. Mendenhall and X. Zhang for help generating ChIP samples, and N. Novershtern and A. Levy for providing transcription factor lists. M. Guttman is a Vertex scholar, I.A. acknowledges the support of the Human Frontier Science Program Organization. This work was funded by Beth Israel Deaconess Medical Center, National Human Genome Research Institute, and the Broad Institute of MIT and Harvard.
Footnotes
Microarray data have been deposited in the Gene Expression Omnibus (GEO) under accession number GSE13765.
Reprints and permissions information is available at www.nature.com/reprints
References
- 1.Bertone P, et al. Global identification of human transcribed sequences with genome tiling arrays. Science. 2004;306:2242–2246. doi: 10.1126/science.1103388. [DOI] [PubMed] [Google Scholar]
- 2.Carninci P, et al. The transcriptional landscape of the mammalian genome. Science. 2005;309:1559–1563. doi: 10.1126/science.1112014. [DOI] [PubMed] [Google Scholar]
- 3.Kapranov P, et al. Large-scale transcriptional activity in chromosomes 21 and 22. Science. 2002;296:916–919. doi: 10.1126/science.1068597. [DOI] [PubMed] [Google Scholar]
- 4.Rinn JL, et al. The transcriptional activity of human chromosome 22. Genes Dev. 2003;17:529–540. doi: 10.1101/gad.1055203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ponjavic J, Ponting CP, Lunter G. Functionality or transcriptional noise? Evidence for selection within long noncoding RNAs. Genome Res. 2007;17:556–565. doi: 10.1101/gr.6036807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Struhl K. Transcriptional noise and the fidelity of initiation by RNA polymerase II. Nature Struct. Mol. Biol. 2007;14:103–105. doi: 10.1038/nsmb0207-103. [DOI] [PubMed] [Google Scholar]
- 7.Brannan CI, Dees EC, Ingram RS, Tilghman SM. The product of the H19 gene may function as an RNA. Mol. Cell. Biol. 1990;10:28–36. doi: 10.1128/mcb.10.1.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brown CJ, et al. A gene from the region of the human X inactivation centre is expressed exclusively from the inactive X chromosome. Nature. 1991;349:38–44. doi: 10.1038/349038a0. [DOI] [PubMed] [Google Scholar]
- 9.Lee JT, Davidow LS, Warshawsky D. Tsix, a gene antisense to Xist at the X-inactivation centre. Nature Genet. 1999;21:400–404. doi: 10.1038/7734. [DOI] [PubMed] [Google Scholar]
- 10.Sotomaru Y, et al. Unregulated expression of the imprinted genes H19 and Igf2r in mouse uniparental fetuses. J. Biol. Chem. 2002;277:12474–12478. doi: 10.1074/jbc.M109212200. [DOI] [PubMed] [Google Scholar]
- 11.Rinn JL, et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129:1311–1323. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Willingham AT, et al. A strategy for probing the function of noncoding RNAs finds a repressor of NFAT. Science. 2005;309:1570–1573. doi: 10.1126/science.1115901. [DOI] [PubMed] [Google Scholar]
- 13.Wang J, et al. Mouse transcriptome: neutral evolution of ‘non-coding’ complementary DNAs. Nature. 2004;431:1–2. doi: 10.1038/nature03016. [PubMed] [Google Scholar]
- 14.Mikkelsen TS, et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448:553–560. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Griffiths-Jones S, Grocock RJ, van Dongen S, Bateman A, Enright AJ. miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res. 2006;34:D140–D144. doi: 10.1093/nar/gkj112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tam OH, et al. Pseudogene-derived small interfering RNAs regulate gene expression in mouse oocytes. Nature. 2008;453:534–538. doi: 10.1038/nature06904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Watanabe T, et al. Endogenous siRNAs from naturally formed dsRNAs regulate transcripts in mouse oocytes. Nature. 2008;453:539–543. doi: 10.1038/nature06908. [DOI] [PubMed] [Google Scholar]
- 18.Clamp M, et al. Distinguishing protein-coding and noncoding genes in the human genome. Proc. Natl Acad. Sci. USA. 2007;104:19428–19433. doi: 10.1073/pnas.0709013104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lin MF, et al. Revisiting the protein-coding gene catalog of Drosophila melanogaster using 12 fly genomes. Genome Res. 2007;17:1823–1836. doi: 10.1101/gr.6679507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Siepel A, et al. Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res. 2005;15:1034–1050. doi: 10.1101/gr.3715005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Carninci P, et al. Genome-wide analysis of mammalian promoter architecture and evolution. Nature Genet. 2006;38:626–635. doi: 10.1038/ng1789. [DOI] [PubMed] [Google Scholar]
- 22.Su AI, et al. Large-scale analysis of the human and mouse transcriptomes. Proc. Natl Acad. Sci. USA. 2002;99:4465–4470. doi: 10.1073/pnas.012025199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Subramanian A, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl Acad. Sci. USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tanay A, Sharan R, Shamir R. Discovering statistically significant biclusters in gene expression data. Bioinformatics. 2002;18 Suppl 1:S136–S144. doi: 10.1093/bioinformatics/18.suppl_1.s136. [DOI] [PubMed] [Google Scholar]
- 25.Chang HY, et al. Robustness, scalability, and integration of a wound-response gene expression signature in predicting breast cancer survival. Proc. Natl Acad. Sci. USA. 2005;102:3738–3743. doi: 10.1073/pnas.0409462102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Carrio M, Arderiu G, Myers C, Boudreau NJ. Homeobox D10 induces phenotypic reversion of breast tumor cells in a three-dimensional culture model. Cancer Res. 2005;65:7177–7185. doi: 10.1158/0008-5472.CAN-04-1717. [DOI] [PubMed] [Google Scholar]
- 27.Ventura A, et al. Cre-lox-regulated conditional RNA interference from transgenes. Proc. Natl Acad. Sci. USA. 2004;101:10380–10385. doi: 10.1073/pnas.0403954101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Loh YH, et al. The Oct4 and Nanog transcription network regulates pluripotency in mouse embryonic stem cells. Nature Genet. 2006;38:431–440. doi: 10.1038/ng1760. [DOI] [PubMed] [Google Scholar]
- 29.Ivanova N, et al. Dissecting self-renewal in stem cells with RNA interference. Nature. 2006;442:533–538. doi: 10.1038/nature04915. [DOI] [PubMed] [Google Scholar]
- 30.Zhao J, Sun BK, Erwin JA, Song JJ, Lee JT. Polycomb proteins targeted by a short repeat RNA to the mouse X chromosome. Science. 2008;322:750–756. doi: 10.1126/science.1163045. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
is linked to the online version of the paper at www.nature.com/nature.