Abstract
Faced with the current wealth of genomic data, it is essential to have robust and reliable methods of converting DNA sequences into their functional gene products. We demonstrate here that when conditions are established that take advantage of the replication-associated virus amplification, the virus-induced shutdown of host protein synthesis as well as the activation of signalling pathways that normally occur during virus replication, adenovirus biology can be exploited to generate a potent kinase expression system. Residual virus in the protein production has always been a limitation for adenovirus systems and we describe a DNA intercalator/ultraviolet light treatment that eliminates residual adenovirus in protein preparations that has no deleterious effect on enzyme activity. The use of mammalian cells in combination with adenovirus generated a variety of active enzymes which could not be produced in Escherichia coli or baculovirus-infected insect cells. Thus, the utility of adenovirus-mediated enzyme expression as a versatile alternative to established protein production technologies is demonstrated.
INTRODUCTION
Amidst the current abundance of new genomic data, it becomes increasingly important to have reliable methods of converting these DNA sequences into their functional gene products. Only a small subset of the mammalian kinome, for example, (1) has been studied at the biochemical level and a full understanding of the biology of these enzymes will require methods to generate functional proteins. Currently, the two most popular methods of generating a protein product from a gene sequence are to introduce the sequence into the appropriate plasmid for production in bacteria or into a baculovirus vector for production in insect cells. However, as increasing numbers of the more exotic mammalian kinases are examined, it becomes clear that expression of these proteins in either prokaryotic or insect cells does not always result in soluble and functional enzymes. Some of these failures can be attributed to the requirement for mammalian specific chaperones, co-factors or post-translational modification (e.g. phosphorylation, glycosylation) that are missing in the bacterial or insect cells. Thus, methods to overexpress and purify mammalian enzymes using mammalian cells can be of substantial value.
The use of adenovirus as a protein expression system in mammalian cells is not a recent development. Recombinant forms of adenovirus were some of the first viruses used to direct exogenous protein expression in eukaryotic cells. Unfortunately, the initial experiments were performed in cells that (we now know) have limited amounts of the adenovirus receptor CAR, the expression levels were not strong (2) and early methods of recombinant adenovirus construction were cumbersome. In addition, although replication-defective adenoviruses pose no health risk, many regulatory agencies require that the viruses and any material containing residual virus (e.g. purified proteins derived from the viruses) must be handled with appropriate containment. Elimination of adenovirus is difficult because of the high titers of virus produced, and because, unlike enveloped virus such as baculovirus, adenovirus is stable to detergents, chloroform and most conditions used to purify native proteins; thus residual levels of adenovirus can be found in the purified protein. As a consequence, there has been only a limited use of adenovirus systems for recombinant enzyme production (see, for example, 3) and adenovirus use in this field has been largely overshadowed by bacterial and baculovirus systems.
A variety of simplified adenovirus construction techniques have now been developed that accelerate the construction of viruses. Because of the resulting wide availability of these vectors, as well as the problems that were being identified with Escherichia coli and baculovirus systems, it seemed appropriate to re-examine adenovirus function for the production of recombinant enzymes in mammalian cells. We describe here optimized expression conditions combined with a simple one-step affinity purification to generate 100 µg to 10 mg quantities of enzymatically active mammalian protein kinases that could not be produced by either bacterial or baculovirus systems. An essential problem to solve, if this method is to be generally useful for enzyme production, is the problem of residual adenovirus in the purified protein. Thus, we have identified a DNA intercalator/UV irradiation method that eliminates residual replicating adenovirus in the recombinant enzyme preparations without affecting recombinant kinase activity. This method can serve as a valuable alternative for producing enzymes that require a mammalian system for activity.
MATERIALS AND METHODS
Construction and purification of recombinant adenoviruses
Recombinant E1/E3 defective adenoviruses were constructed using homologous recombination in E.coli (4). All viruses contained an expression cassette comprising the CMV immediate early promoter driving the open reading frame of interest followed by a rabbit β globin intron/polyadenylation signal. The expression cassette is inserted into an adenovirus type 5 genome in place of nucleotides 342–3523, which removes the E1 region. In addition the genomes contain a deletion of adenovirus 5 E3 region of nucleotides 28592–30472, a destruction of the SpeI site at nucleotide 27082 and the addition of SpeI linkers at each terminal repeat. Recombinant viral genomes were released from the plasmid backbone by digestion with SpeI, purified by phenol/chloroform extraction and passage over a gel filtration column (Pharmacia Nick column). Virus cultures were initiated by transfecting the linearized genomes into 293 cells (5) using polyethylenimine (PEI) (6). After amplification, virus was purified by banding twice on CsCl gradients, transferred into 150 mM NaCl, 20 mM HEPES pH 7.5 (HBS)/40% glycerol by passage over a gel filtration column and stored at –80°C as previously described (7). Viral quantitation was based on protein content using the conversion 1 mg viral protein equals 3.4 × 1012 virus particles (8).
Open reading frames
All open reading frames were generated by PCR from commercial cDNA preparations and cloned into derivatives of pPM7 followed by assembly into adenovirus (see above). The following cDNAs were used.
The full-length wild type RICK open reading contained a cDNA identical to GenBank accession number AF027706 with a carboxy terminal haemagglutinin (HA) epitope tag. Kinase essential lysine residue 47 was mutated to arginine (K47R) to generate a kinase inactive form of the protein. RICK deleted of CARD domain (RICKdC) was generated from either the kinase wt or kinase dead (K47R) genes and contained the first 353 amino acid residues of RICK with either a carboxy terminal HA tag (RICKdC, wt or kr) or with a carboxy terminal strep-tag (RICKwtdCst, RICKkrdCst).
The RIP kinase domain construct (RIPkinwtST) contained a cDNA encoding residues 1–300 of the RIP protein identical to the sequence described in GenBank accession number NM_003804 plus a carboxy terminal strep-tag. The kinase dead version (RIPkinKRst) has the essential lysine 45 mutated to arginine (K45R) plus a carboxy terminal strep-tag.
The cdk9 construct contained a cDNA identical to GenBank accession number P50750, and NM_001261; a kinase dead version of the gene was prepared by mutating both kinase essental lysine 48 to arginine and the aspartic acid 167 to asparagine in cdk9KRDN. The cyclin T1 construct contained a cDNA identical to cyclin T1 (GenBank accession number XM_028819). Wild type cdk9, KRDN cdk9 and cyclin T1 were prepared with a carboxy terminal strep-tag.
The p38α construct contained a cDNA identical to GenBank accession number L35253; a kinase dead version of the gene was prepared by mutating lysine 53 to arginine.
The IKKbeta construct contained a cDNA identical to GenBank accession number AF080158 and AF031416.
Extraction of protein
Infected cells were harvested, washed twice with HBS and stored frozen at –20°C after flash freezing in dry ice. The frozen cell pellet was thawed on ice with 2 ml lysis buffer 4 per 3 × 107 cells (equal to one 185 cm2 flask equivalent), pipetting gently to disperse cells. The lysis and extraction were performed on ice for 45 min with vortexing every 5–10 min. The material was sonicated in a bath sonicator for 5 min, an aliquot of the material was saved as the total sample. The material was then centrifuged for 10 min at 1950 g and the supernatant retained as extract. For cdk9/cyclin T1, infected cells were extracted directly after harvest without freezing, using LB7 instead of LB4.
Buffers
Lysis buffer 4 (LB4). 20 mM HEPES NaOH pH 7.5, 150 mM NaCl, 20 mM 2-glycerol phosphate, 10 mM NaF, 0.5% NP-40, 2 mM MgCl2, 2 mM EGTA, 1 mM Na3VO4, 2 mM DTT, 1 mM PMSF, protease inhibitor cocktail (Roche) with the last three components added just before use.
Lysis buffer 5 (LB5). 20 mM Tris–HCl pH 7.0, 20 mM 2-glycerol phosphate, 10 mM NaF, 0.5% NP-40, 2 mM MgCl2, 2 mM EGTA, 1 mM Na3VO4, 2 mM DTT, 1 mM PMSF, protease inhibitor cocktail (Roche).
Lysis buffer 7 (LB7). Identical to Lysis buffer 4 with 400 mM NaCl.
New storage buffer II (NSBII). 20 mM HEPES NaOH pH 7.5, 150 mM NaCl, 10% glycerol, 10 mM NaF, 2 mM MgCl2, 0.1 mM EGTA, 2 mM DTT, protease inhibitor cocktail (Roche) with the last two components added just before use.
Affinity purification
Streptavidin–Sepharose (Amersham Pharmacia) or streptactin–macropore (IBA) were washed with the same lysis buffer (at least 2 × 10 resin vol). Washed resin was mixed with extract (100 µl of packed resin per 185 cm2 flask equivalent of extract) for 60 min on ice with frequent gentle agitation to keep the resin dispersed. The resin plus extract were then centrifuged at 115 g for 3 min and the supernatant was saved as the flowthrough (FT). The resin was washed with 2 × 7 resin vol of lysis buffer 4 and 2 × 7 resin vol NSBII and the resin was then eluted twice with 7 resin vol of NSBII+ 2.5 mM desthiobiotin (Eluate). The eluates were pooled and were stored in aliquots at –80°C after flash freezing in dry ice.
Chromatographic purification of RICKdC
Cell lysis was performed as above using LB5. Extracts were inactivated with psoralen (see below) and then fractionated over Red Sepharose (binding in LB5, elution with 300 mM NaCl). The salt content was diluted to 50 mM and the material was resolved on Q Sepharose using a NaCl gradient from 50 to 500 mM. Fractions were assayed for RICKdC content by western and for RICK autophosphorylation and histone 2b (H2b) or myelin basic protein (MBP) phosphorylation.
Psoralen inactivation
The method used conditions similar to those previously described (9). Lysates were transferred to Nunc 24-well cell culture plates (0.75–1.5 ml per well) and 1/200 vol of 33 mg/ml 8-methoxy psoralen (Sigma, M 3501, dissolved in DMSO) was added per well. The psoralen precipitate that formed was distributed in the sample by pipetting and the plate was irradiated with 365 nm UV light (plate on ice, lid on) for 30 min.
Analysis of virus content
Serial dilutions of test samples were performed in MEMalpha, 2% horse serum and added to 293 cells (40 000 cells/well, 24-well plate) for 2 h at 37°C. The medium was then changed to MEMalpha, 10% newborn calf serum. At 6–8 day post infection, cells were inspected microscopically for the presence of adenovirus (plaque formation, swollen, detached cells etc.) and then medium was removed, the cells were fixed for 5 min (1% formaldehyde, 150 mM NaCl), washed once with PBS and then stained for 10 min with crystal violet (3% w/v in 150 mM NaCl), washed twice with PBS, once with distilled water and then scanned.
Kinase assays: RICK autophosphorylation and Flashplate assay
Recombinant enzyme was incubated in 40 µl containing 50 mM HEPES, pH 7.5, 1 mM MgCl2, 1 mM MnCl2, 1.5 µM ATP, 1 µCi [γ-33P]ATP/probe. For autophosphorylation analysis, the samples were then incubated with 0.1% SDS at 99°C and resolved by SDS–PAGE (10% gel), fixed, stained and visualized by autoradiography. For Flashplate analysis, the samples included 20 ng/µl H2b. Reactions were performed in a Flashplate (basic, Perkin Elmer Life Sciences) well for 1 h at room temperature, the reaction was stopped by the addition of EDTA to 100 mM, washed four times with 100 mM EDTA, 100 mM sodium phosphate, pH 7.2, 0.01% Tween 20. Bound radioactivity was measured.
cdk9 assay
Kinase assay reactions were performed essentially as described in Grana et al. (10). Purified enzyme was prediluted with buffer A (20 mM HEPES, pH 7.4, 4 mM MgCl2, 1 mM DTT) the reaction was initiated by adding 25 µl diluted enzyme to an equal volume of buffer A containing substrate [1 mM cold ATP, 1 µCi/well, 1 µg/well GST-CTDII (GST fused to a triple repeat of YSPTSPS)]. After 1 h incubation at room temperature, the assay was stopped by adding 5 µl of 0.5 M EDTA, pH 8.0, half of the reaction volume was applied to a MAIP filter plate according to the manufacturer’s recommendations (Millipore). The dried plate was soaked with scintillation liquid, sealed with tape and incorporated radioactivity was measured employing a microbeta topcounter (Wallac Inc.).
p38 assay
Purified p38α samples were incubated in 25 mM HEPES, pH 7.5, 20 mM 2-glycerol phosphate, 20 mM MgCl2, 2 mM DTT, 0.1 mM sodium orthovanadate, 2 µg ATF-2 (Cell Signalling Technology) and 20 µM ATP for 30 min at 37°C. Reactions were then resolved by SDS–PAGE, transferred to nitrocellulose and probed with either anti-phospho-ATF-2 or anti-ATF-2 antisera (Cell Signalling) followed by the appropriate HRP conjugate and ECL.
Western analysis
Proteins were resolved on SDS–PAGE, and transferred to nitrocellulose. A total protein image was obtained after staining with Ponceau S. Blots were then blocked overnight in NET-gelatin, and either probed with the indicated primary antibody and was followed by the appropriate secondary antibody-horseradish peroxidase or blocked with 25 µg/ml avidin (Sigma) and then probed with streptactin–horseradish peroxidase (IBA). Finally, the images were developed using ECL reagents (Amersham).
RESULTS
Optimization of protein expression
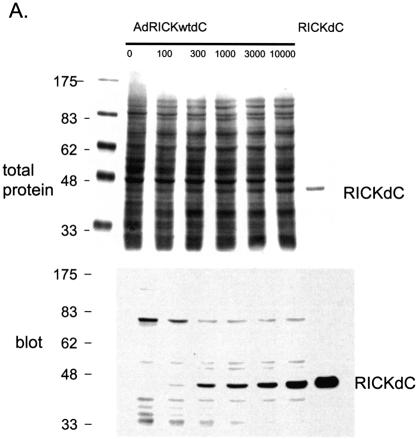
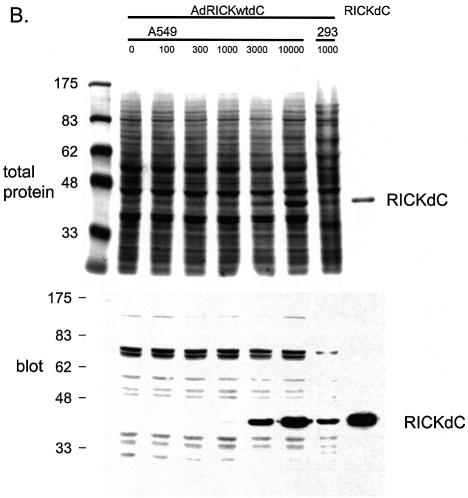
Most uses of recombinant adenovirus apply the virus to cells that do not support virus replication. However, for high level protein production, it may be useful to employ conditions that take advantage of both virus amplification and the inhibition of host translation that occurs during a productive adenovirus replication (11–13). Intuitively, if virus amplification occurs, one would use a low virus to cell ratio and indeed, if amplification and growth of the recombinant virus is the goal, then using more than 10–100 virus particles per cell (p/c) is wasteful, because no further amplification of virus occurs at higher multiplicities of infection (m.o.i.). However, for protein production, the parameters are indeed different. The expression of a transgene protein, the kinase domain from RICK [Rip2, CARDIAK, a kinase involved in innate immune response signalling (14–16)] was monitored as a function of various virus to cell ratios (Fig. 1). In 293 cells, in which the defective adenovirus can replicate due to E1 complementation (17), the yield of transgene product was found to increase using 100–10 000 p/c (Fig. 1A). In a replication system, one would normally use 10–100 p/c for maximum yields of virus, higher m.o.i. are not reflected in greater virus yields. However for protein production, best yields were obtained at 1000 p/c and higher (Fig. 1A, lanes 4–6). In contrast, transduction of A549 cells that do not support virus replication demonstrated that the best yields were obtained at 3000 p/c and higher (Fig. 1B, lanes 5 and 6), most likely because of the lack of amplification of the viral genome in these cells. A second important factor, host protein shut-off during replication, is apparent when comparing 293 and A549 cells. As an indication of this, prominent biotin-containing host proteins at ∼70 kDa are seen in both 293 and A549 uninfected cell extracts. In the extracts from 293 cells but not from A549 cells, the abundance of these proteins relative to the desired gene product declines dramatically at higher m.o.i., resulting in a higher product/total protein ratio. Thus the starting material for product purification is already significantly enriched by using the replicating system. A time course of expression demonstrated that maximum yield/cell was obtained at 48 h (results not shown). In conclusion, optimum transgene protein production is obtained using adenoviral replication conditions at relatively high m.o.i. (1000 p/c). Higher levels of transgene product are obtained at 10 000 p/c; however, the effort required to generate sufficient virus stocks becomes limiting at this m.o.i.
Figure 1.
Optimization of infection conditions. Recombinant adenovirus directing the expression of the RICKdC were used to infect either 293 (A) or A549 cells (B) at 100–10 000 virus particles/cell. At 48 h post-infection the cells were harvested, washed twice with HBS (150 mM NaCl, 20 mM HEPES, pH 7.5), lysed in SDS application buffer, heated at 99°C for 10 min, sonicated for 5 min in a sonicating water bath and 20 µl were loaded per well on a 10% acrylamide SDS gel. After resolution, the gel was transferred to nitrocellulose, probed with streptactin–HRP (IBA) and visualized by ECL. Also resolved was purified RICKdC (A and B, last lanes), non-infected cell lysates (A and B, first lanes).
Affinity purification
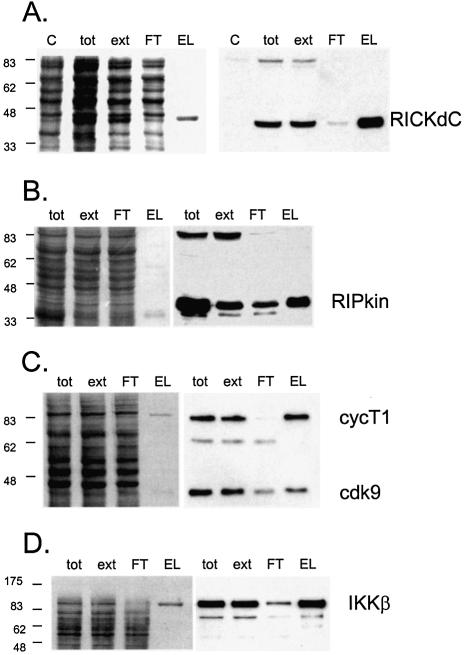
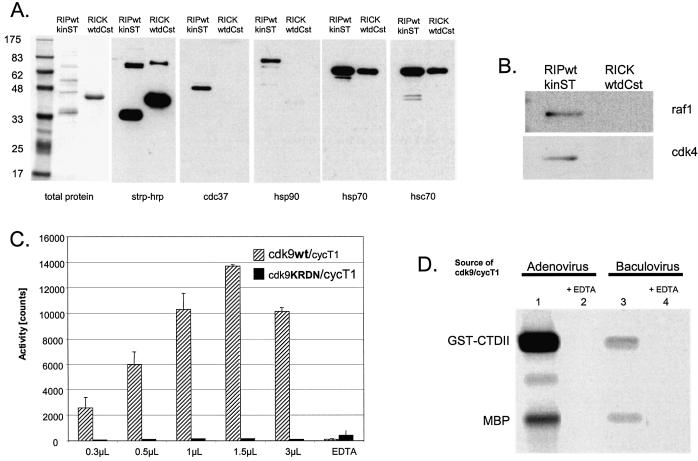
Initial results demonstrated that significant quantities of a recombinant protein can be generated. For purification of such a protein, an affinity purification tag was included in the system. Some years ago, a peptide was selected for its ability to bind streptavidin in a reversible fashion, and these studies led to the development of the strep-tag system (18,19, reviewed in 20). To test this system in the context of recombinant adenovirus, the RICKdC open reading frame was fitted with a C-terminal, nine amino acid strep-tag and cloned into an E1/E3 defective Ad5 genome. RICKdCst was expressed, extracted as described above, the extract was loaded on streptactin-macropore [an improved form of streptavidin (21,22)] washed extensively and then the strep-tagged protein was eluted with desthiobiotin. Both total protein staining (Fig. 2A, left panel) and staining with HRP-streptactin (Fig. 2A, right panel) revealed that a single protein band of the expected molecular mass (41 kDa) was present in the total (tot) and extracted (ext) fractions, depleted from the non-bound FT fraction and eluted from the resin with 2.5 mM desthiobiotin (Fig. 2A, EL). The eluted fraction demonstrated RICK kinase activity as measured by autophosphosphorylation (Fig. 3B) and a quantitative analysis of phosphorylation of H2b demonstrates that the kinase activity of the strep-tag purified material was substantially more active than the same molecule purified by ion exchange chromatography (Fig. 7B, compare RICKdC MQ purified by Red and MonoQ Sepharose to RICKdcST purified by streptactin–Sepharose).
Figure 2.
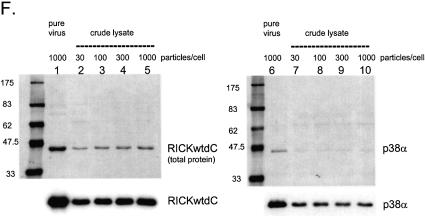
Expression and purification of several protein kinases. Recombinant adenoviruses directing the expression of the indicated protein kinases were used to infect 293 cells at 1000 virus particles/cell. At 48 h post-infection the cells were harvested, washed twice with HBS (150 mM NaCl, 20 mM HEPES, pH 7.5), lysed, extracted and purified by streptactin–macropore as described in Materials and Methods. Aliquots of the total material before extraction (tot), the extract (ext), unbound material after expsoure to resin (FT) and the material eluted with desthiobiotin (EL) were mixed with SDS application buffer, heated at 99°C for 10 min, sonicated for 5 min in a sonicating water bath and 20 µl was loaded per well on a 10% acrylamide SDS gel. After resolution, the gel was transferred to nitrocellulose, stained with Ponceau S (A–D, left panels) or blocked with NET/gelatin and avidin, probed with streptactin–horseradish peroxidase (IBA) and visualized by ECL (A–D, right panels). (E) Equal aliquots of purified proteins were resolved on a 10% acrylamide SDS gel and visualized by staining with colloidal Coomassie blue (Sigma). (F) Comparison of pure virus to crude lysate inocula. 293 cells were infected with either 1000 particles/cell of CsCl-purified adenovirus encoding RICKdCst or p38αST or with crude infected cell lysates of cells infected with the same adenoviruses, at 30, 100, 300 or 1000 particles/cell. At 48 h post-infection, cells were harvested, lysed and the strep-tagged protein purified as described above. Equal volumes of each of the eluate fractions were resolved on a 10% acrylamide SDS gel, transferred to nitrocellulose, stained for total protein with Ponceau S (upper panels) and then probed with streptactin–horseradish peroxidase (IBA) and visualized by ECL (lower panels).
Figure 3.
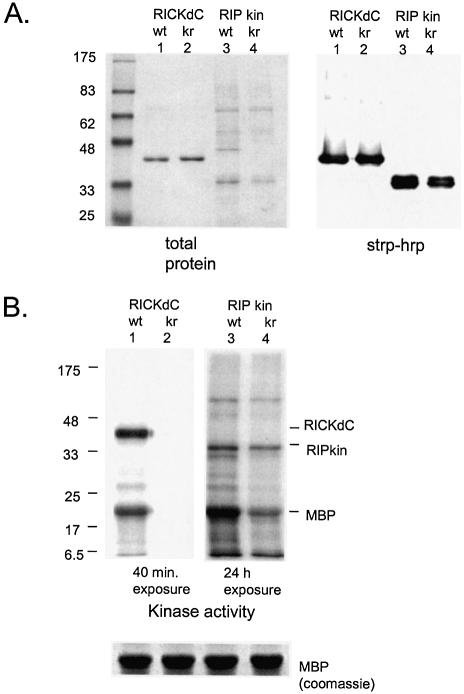
Analysis of contaminating kinase activity. (A) Protein content. RICKdC and RIPkin (both wild type and kinase inactive forms) were expressed in 293 cells, extracted and purified by streptavidin–Sepharose. Similar quantities of each protein were resolved by SDS–PAGE and stained for total protein (left side) or transferred to nitrocellulose, stained with streptavidin–HRP followed by ECL (right side). (B) Kinase activity. Equal quantities of RICKdC (wt or K47R) and RIP kin (wt or K45R) were incubated under RIP kinase conditions (see Materials and Methods) in the presence of [γ-33P]ATP and MBP as a substrate. The protein reactions were resolved by SDS–PAGE, and the radioactive pattern was revealed by exposure to X-ray film for either 40 min (RICK samples) or 24 h (RIP samples). The bottom panel shows equivalent MBP staining by Coomassie blue in four parallel reactions.
Figure 7.
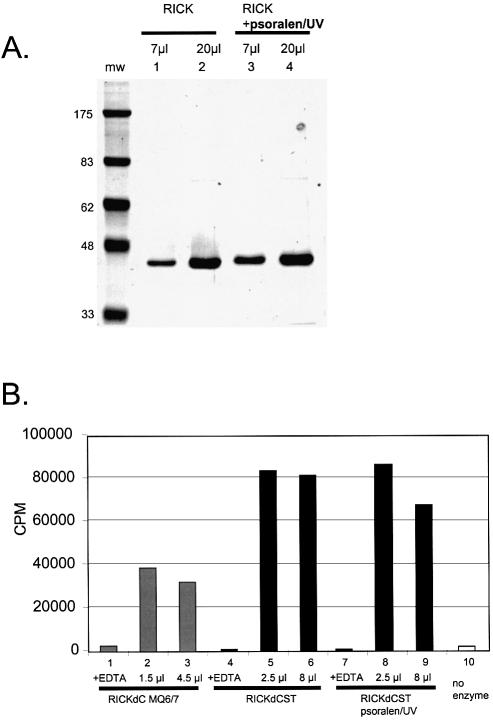
RICK kinase activity is not impaired by psoralen/UV treatment. RICKwtdCst was purified from control lysate or lysate treated for 30 min with psoralen/UV. Equal protein quantities were resolved on 10% acrylamide SDS gel and stained for total protein (Coomassie blue, A). In parallel, samples were incubated with [γ-33P]ATP under kinase appropriate conditions (see Materials and Methods) and the radioactivity incorporated into a histone H2b substrate was measured (B).
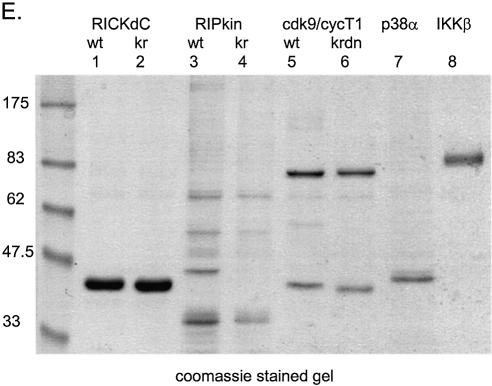
The broad utility of the method is demonstrated by obtaining similar expression and purification patterns with additional mammalian kinases including the kinase domain of the death signalling kinase RIP (Fig. 2B, but see below); the cyclinT1/cdk9 complex (Fig. 2C and below); NF-κB activating kinase IKKβ (Fig. 2D) and the MAP kinase p38α (Fig. 5A). A Coomassie blue-stained gel demonstrating the purity of a collection of these kinases is shown in Figure 2E.
Figure 5.
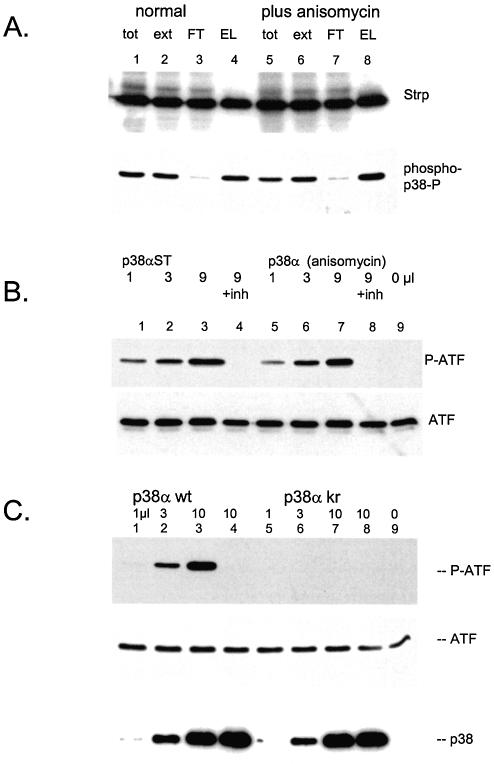
p38α is generated in an active form. (A) Adenovirus directing a strep-tagged p38α was used to infect 293 cells. One portion of cells was treated with anisomycin for 30 min before harvest. Lysate preparation and purification by streptactin–macropore was performed as described above; aliquots were resolved on 10% acrylamide SDS gels, transferrred to nitrocellulose, blocked with NET/gelatin and avidin, probed with streptactin–horseradish peroxidase (top panel) or probed with anti-phospho-p38 (NEB) (bottom panel). (B) The indicated quantities of p38α purified from either control cells (lanes 1–4) or anisomycin stimulated cells (lanes 5–8) were incubated with ATF2 as described in Materials and Methods. Samples for lane 4 and 8 included 5 µM SB203580. The reactions were resolved on 10% acrylamide SDS gels, transferred to nitrocellulose blocked with NET/gelatin probed with anti-phospho-ATF (top panel) or antisera against total ATF (bottom panel). (C) The indicated quantities of purified p38α wild type (lanes 1–4) or kinase inactive p38α (lanes 5–8) were incubated with ATF2 as described in Materials and Methods. The reactions for lane 4 and 8 included 5 µM SB203580. The reactions were resolved on 10% acrylamide SDS gels, transferred to nitrocellulose and probed with anti-phospho-ATF2 antisera (top panel), anti-ATF2 sera (middle panel) or streptactin–horseradish peroxidase to visualize p38α (bottom panel).
The generation of CsCl-purified adenovirus to be used as inoculum requires time and effort and as one reviewer pointed out, the procedure would be simplified if crude viral lysates could be used. We compared the yield of purified kinases obtained with pure virus to that obtained using comparable quantities of crude infected cell lysates. Because it is difficult to accurately measure viral concentrations in crude lysates, a broad range of inoculum volumes was tested calculated to span the 100–1000 particle/cell window. As shown in Figure 2F, purified RICKdC and p38α protein can be obtained using the unfractionated lysates as inoculum (RICK, lanes 2–5, p38α lanes 7–10); however, the yield of protein is slightly lower than that obtained with pure virus inoculum. (RICK lane 1; p38α, lane 6; per 185 cm2 flasks: RICKdC pure 36 µg; RICKdC crude lysate: 20 µg; p38α pure virus: 28 µg; p38α crude lysate: 20 µg). Furthermore, the resulting purified protein has a slightly different pattern of co-purified contaminants most likely due to the presence of high concentrations of viral capsid proteins added with the crude inoculum. Thus, the use of purified virus inoculum results in slightly higher yields and purity. However, for many proteins, the yields may still be sufficiently high to justify omitting the virus purification. That no increase in pure protein yield occurs with increasing crude lysate inoculum is likely to be due to limiting toxicity caused by free viral capsid components in the crude lysate.
The question of co-purifying eukaryotic kinases
For kinase production, a major consideration with the use of mammalian or insect systems is the issue of co-purifying cellular kinases. Because E.coli lacks classical serine/threonine and tyrosine kinases, there is little concern about copurifying a host kinase with the target kinase. However, with mammalian kinases overexpressed in eukaryotic cells, there is the possibility that either specific or non-specific interactions of the recombinant enzyme with cellular factors can lead to the copurification of proteins in addition to the target enyzme. This is especially important when producing kinases or other enzymes for drug screening efforts in which a contaminating enzyme activity can complicate the search for specific inhibitors. To demonstrate enzyme purity, a mutated version of the protein is prepared with the kinase activity genetically disrupted by mutating either the highly conserved lysine residue that forms part of the ATP binding pocket, or by mutating the aspartic acid residue in the kinase motif (23). The mutant protein is then expressed and subjected to the same level of purification. A measurement of residual kinase activity in such a preparation indicates the quantity of co-purifying cellular kinases.
Such an analysis was performed with the wild type RICK kinase domain and a kinase inactive RICK K47R mutant. Both proteins were expressed using adenovirus, purified (Fig. 3A) and equal protein quantities were tested for kinase activity (Fig. 3B). Substantial autophosphorylation and phosphorylation of a substrate (MBP) is observed with the wild type RICK kinase domain, no detectable kinase activity was found associated with the RICK K47R preparation (Fig. 3B, compare lanes 1 and 2) demonstrating the absence of non-RICK kinases in the preparations. The kinase domain from RIP was also prepared in both wild type and kinase inactive forms (Figs 2B and 3A). Although the wild type protein domain showed both autophosphorylation and kinase activity against MBP, (Fig. 3B, lane 3) substantial kinase activity was also found associated with the preparation of kinase inactive RIP (RIP K45R Fig. 3B, lane 4), demonstrating the presence of kinase activities in addition to the RIP molecule. Indeed, the total protein-staining pattern of the purified RIP preparations revealed a number of additional protein species (Figs 2E, lanes 3 and 4, and 3A, lanes 3 and 4) which were examined in more detail. The chaperone protein cdc37 interacts with a number of kinases and facilitates, in complexes with hsp90, the appropriate folding of molecules including Raf (24,25), Akt (26), the IKK complex (27), cdk9/cycT1 (28), cdk4 and cdk6 (29). Purified RIP contained quantities of 40 and 90 kDa proteins, western analysis demonstrated that indeed the purified RIP (but not similarly purified RICK) contained both cdc37 and hsp90 (Fig. 4A). Because both RICK and RIP preparations contained detectable quantities of the inducible (hsp70) and constitutive (hsc70) 70 kDa heat shock protein (Fig. 4A), this family of chaperones could not be the basis for the RIP contamination problem. However, cdc37/hsp90 can form multimeric complexes with a number of additional kinases; we tested for their presence in the RIP preparations. Indeed, cdk4 and raf1, two known cdc37 binding kinases (24,25,29), were present in the purified RIPwt and RIPK45R preparations but not in similarly purified RICK preparations (Fig. 4B). Thus, the presence of the kinases cdk4 and raf1 and perhaps additional cdc37 binding kinases could account for the kinase activity observed with purified, kinase-dead RIP K45R. Fortunately, this phenomenon of co-purifying kinases is rather the exception than the rule. To date, all other kinases that we have purified using these methods have shown negligible kinase activity associated with the kinase dead mutant form (see below for cdk9 and p38α).
Figure 4.
(A and B) Copurifying proteins. RICKdC and RIPkin (both as wild type kinase) were expressed in 293 cells, extracted and purified by streptavidin–Sepharose. Similar quantities of each protein were resolved by SDS–PAGE and stained for total protein (left side) or transferred to nitrocellulose, stained with either streptavidin-HRP or the indicated antisera followed by HRP-conjugated secondary antibody and then by ECL. (C) cdk9/cycT1 wt versus KRDN activity. Equal quantities of cdk9/cycT1 (wt or KRDN) were incubated under cdk9 kinase conditions in the presence of [γ-33P]ATP and an RNA polymerase II carboxy-terminal peptide as a substrate (see Materials and Methods). (D) Comparison of adenovirus and baculovirus-produced cdk9/cyclinT1. For baculovirus production, Sf9 cells were infected with baculoviruses directing the expression of His-tagged cdk9 and cyclin T1. Purification was performed essentially as described (30). Equal quantities of purified protein derived from adenovirus (lanes 1 and 2, see Materials and Methods) or baculovirus (lanes 3 and 4) were tested in the absence of EDTA (lanes 1 and 3) or presence of 10 mM EDTA (lanes 2 and 4) for their ability to phosphorylate GST-CTDII plus MBP as described in Materials and Methods.
The purification of enzyme complexes
For some eukaryotic kinases, it is essential to generate and purify multi-protein complexes. For example, the cellular kinase cdk9 requires the interaction with cyclin T1 for activity as the transcription elongation factor P-TEFb (30,31). Because maturation of cdk9 activity requires interaction of cdk9 with the cyclin T1 during synthesis and folding (28), the two components of the cdk9/cyclinT1 kinase complex were expressed as affinity tagged proteins by co-infection with adenoviruses and purified as described above (Fig. 2C). Both cdk9 and cyclinT bear affinity tags, thus it was important to demonstrate that other kinases reported to interact with cyclin T1 are not in the preparation. Immunoblot analysis demonstrated the absence of cdk2 and cdk4 and the purified cdk9/cycT1 kinase activity was not inhibited by established cdk2 and 4 inhibitors (results not shown). The kinase dead mutant of cdk9 with the essential lysine and aspartic acid residues mutated to arginine and asparagine (K48R, D167N), was expressed with cycT1, purified to the same extent as the wild type kinase/cycT1 complex and assayed for kinase activity. No detectable kinase activity was present in the cdk9K48RD167N mutant protein preparation (Fig. 4C) even when the sample was assayed at 10-fold higher concentration than the wild type cdk9 (compare 0.3 µl of wt with 3 µl of KRDN). Thus, although the potential exists for cyclin T1 to recruit and copurify other cyclin T1 binding kinases, the high level of target protein expression, the shut down of host protein expression by adenovirus and the purification method, results in a cdk9/cyclin T1 preparation that is free of other cyclinT1 binding kinases.
A comparison with baculovirus
cdk9/cyclin T1 can be generated using baculovirus infection of insect cells (30). To compare the adenovirus and baculovirus system, cdk9/cyclinT1 was expressed and purified from SF9 cells and the kinase activity was compared with the adenovirus-produced material. With the adenovirus system, we typically obtain 10 µg of pure cdk9/cyclinT1 per 3 × 107 cells. In our hands, to obtain a similar mass of the kinase with the baculovirus system required three times more cells, and the resulting purified material had one-fifth to one-tenth the kinase activity as the adenovirus-derived material (compare Fig. 4D, lanes 1 and 3).
Activation of kinases
For a number of kinases involved in signalling cascades, care must taken to ensure that the kinase is appropriately activated. This is especially well established for components of the MAP kinase cascade where the recombinant enzyme must be either genetically altered with an acidic amino acid residue, mimicking phosphorylation activation, or treated with an activated upstream kinase. For example, the map kinase p38α is normally produced in E.coli and activated using MKK3 or MKK6, which must also be prepared in an active form (32,33). The use of adenovirus in mammalian cells allows one to take advantage of endogenous signalling pathways to generate active forms of the enzyme. The p38 pathway is activated during adenovirus infection (34,35), thus the potential exists that the process of infection and replication may generate active enzyme without additional manipulations. To test this idea, p38α was expressed using this system. Initially, equal quantities of cells were used and one portion was stimulated with anisomycin before harvest in order to deliberately activate the p38 pathway (36,37). The protein was then purifed from the two extracts and examined for p38 kinase activity.
One measure of p38 activation is the hallmark activation phosphorylation on threonine 180 and tyrosine 182, (38,39) which can be readily measured using phospho-specific antibodies to these sites. The p38α generated using the adenovirus system was found to be already in a phosphorylated form (Fig. 5A, lane 4). Deliberate activation of p38 with anisomycin lead to no greater level of the p38 phosphorylation (Fig. 5A, lane 8). A more direct assay of p38 activation was performed by measuring the ability of the purified kinase to phosphorylate the transcription factor ATF-2. p38α purified from non-stimulated cells was already active in phosphorylating ATF-2 (Fig. 5B, lane 1–3) and no further increase in activity was obtained by treating the cells with anisomycin before harvest (Fig. 5B, lanes 5–7) consistent with the results obtained by examining the phosphorylation state of p38 (Fig. 5A, bottom panel). These results are consistent with the reports that p38 is activated during the course of adenovirus infection and replication (34,35); thus no further manipulation of this pathway is required to obtain active p38. The specificity of the p38α kinase activity observed here is demonstrated by the inhibition of the phosphorylation with the p38 inhibitor SB203580 (Fig. 5B, lane 4 and 8).
As a control to demonstrate the purity of the p38 preparation, a kinase inactive mutant p38α was generated (p38αKR) expressed and purified in the same manner (Fig. 5C, bottom panel). When tested for activity, the kinase inactive mutant indeed showed no activity [Fig. 5C, compare lanes 5–7 (kinase inactive) to lanes 1–3 (kinase active)] demonstrating that the activity observed with the wild type enzyme preparation can be attributed to p38α and not co-purifying mammalian kinases.
The question of contaminating adenovirus
Although the recombinant adenoviruses used here are replication-defective and pose no significant health threat, in most countries, recombinant adenovirus and all material derived from these viruses must be handled with appropriate containment. One can calculate that a typical lysate produced under the conditions described contains 1010 to 1012 virus particles per milliliter and even the extensive purification allowed by affinity chromatography is not sufficient to remove virus to <1 in 1012/ml. Thus, we sought a method of chemically inactivating the adenovirus present in the protein preparations.
It was previously demonstrated that 8-methoxy psoralen has the ability to enter the adenovirus capsid and upon irradiation with long wavelength UV, the compound form single- and double-stranded crosslinks of the adenoviral genome that block both viral transcription and replication (9,40,41). This provides a method of inactivating adenovirus that is accepted by clinical standards (see for example 42). Psoralen inactivation can be used to inactivate baculovirus in a recombinant protein preparation without altering the antigenic properties of the protein (43) and the treatment does not impair the entry function of the adenovirus capsid (9,40,41). However, a direct analysis of the consequences of psoralen/UV treatment on enzyme activity has not yet been performed.
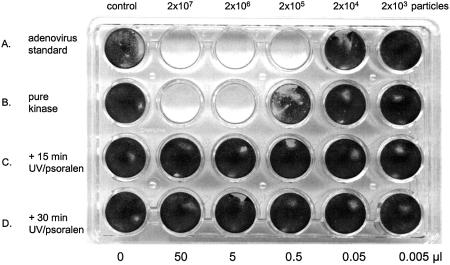
Cytopathic effect assays were performed to assess the quantity of adenovirus remaining in purified kinase preparations. In this assay, surviving (non-infected) cells stain with crystal violet, the presence of replicating adenovirus results in cell death and detachment from the substrate (referred to as cytopathic effect, CPE) and loss of crystal violet staining; clear areas in the monolayer indicate the presence of replicating adenovirus. An analysis of virus content of a purified kinase preparation shows complete killing of the cells with 50 and 5 µl of the material, (Fig. 6, row B) with substantial CPE observed microscopically at 0.05 and 0.005 µl. In comparison with a pure adenovirus standard (Fig. 6, row A) it can be calculated that the purified kinase preparation contains between 104 and 105 virus particles/µl. This is not surprising as the starting extract contains ∼109 viral particles/µl (results not shown). However, if the lysate used as the starting material for kinase preparation is treated with psoralen/UV light before purification, the replicating adenovirus content is eliminated. Treatment with 15 min of UV largely eliminates (row C, cytopathic effect only detectable microscopically) and exposure for 30 min of UV light combined with 8-methoxy psoralen completely eliminates all trace of virus replication (Fig. 4D). More sensitive plaque assays (not shown) and RT–PCR (9) have demonstrated the elimination of viral replication and transcription with this method.
Figure 6.
Assay of replicating adenovirus content before and after psoralen/UV treatment. 293 cells (40 000 cells/well, 24-well plate) were exposed to the indicated particles/cell m.o.i. of pure adenovirus (row A) or 50, 5, 0.5, 0.05 and 0.005 µl of pure RICKwtdCst kinase (El1 fraction from streptavidin–Sepharose fraction, (row B) or the same quantity of kinase purified from lysates exposed to 8-methoxy psoralen/265 nm UV light for 15 (row C) or 30 min (row D). At 7 days post-infection the cells were fixed with formaldehyde and stained with crystal violet. See Materials and Methods for details.
It is essential to demonstrate that this method does not impair the function of the recombinant enzyme. The 30 min exposure to psoralen/UV had no deleterious effect on the yield and purity of RICKdC with the similar enzyme yield and purity obtained from control extracts (Fig. 7A, lanes 1 and 2) and psoralen/UV treated extracts (Fig. 7A, lanes 3 and 4). The psoralen/UV had no detectable influence on RICK kinase activity (Fig. 7B, compare control, lanes 4–6, and psoralen/UV treated, lanes 7–9). Similar results were obtained with a variety of other mammalian kinases including cdk9/cyclinT1 and p38α (results not shown, but note that Fig. 4C uses psoralen-treated cdk9/cyclinT1).
DISCUSSION
We report here conditions that take advantage of the biology of a commonly available adenovirus vector for the production of recombinant kinases. The method uses mammalian cells for expression, and this may be important if post-translational modification, host-specific folding or activation of the enzyme by upstream kinases is required. A simple affinity purification method allows the enzyme to be purified in a single step, and an important part of this method is the chemical inactivation of residual adenovirus in the purified protein using a UV activated DNA intercalating agent, an important issue for the use of these proteins in other application. The method provides useful quantities of enzymatically active kinases when other expression systems give poor results (e.g. for RICK and cdk9/cyclin T1). Although the focus here has been on kinases, it is likely that the system will have applications with a variety of other mammalian enzymes.
Although the yields of purified protein will vary from protein to protein, some general comments on the yields obtained with this method can be made. The preparation is conveniently performed with material derived from 18, 185 cm2 tissue culture flasks of 293 cells (∼3 × 107 cells/flask), yielding 36 ml of extract and 12 ml of material (0.04–0.2 µg/µl) eluted from streptavidin–Sepharose. Thus a typical preparation would yield 0.5–2 mg of purified RICK. Perhaps a more critical parameter is the number of enzymatic assay points provided. In our applications, a 185 cm2 flask yields 0.2 ml (cdk9/cyclin T1) to 2 ml (RICK) of purified protein with 1 µl sufficient for a high throughput, radioactive assay point. Thus, 200–2000 assay points can be obtained from 185 cm2 flask.
A critical issue is the time involved in the construction and the purification of the adenovirus vectors. It currently takes 6–8 weeks to generate CsCl purified quantities of a novel adenovirus. This is substantially slower than E.coli systems, but comparable to baculovirus. The methods described here used CsCl-purified virus for the infections to generate kinase primarily because in establishing the methods it was essential to use known quantities of the adenovirus. It is also possible to use non-purified lysates for the kinase production infections (Fig. 2F) and this would save several weeks. One should point out that the same adenovirus preparations described here can also be used for biological experiments, thus providing additional justification for the construction and purification of these reagents.
In summary, the technique describe here provides a useful system for obtaining native, active, purified enzymes from mammalian cells.
REFERENCES
- 1.Manning G., Whyte,D.B., Martinez,R., Hunter,T. and Sudarsanam,S. (2002) The protein kinase complement of the human genome. Science, 298, 1912–1934. [DOI] [PubMed] [Google Scholar]
- 2.Karlsson S., Humphries,R.K., Gluzman,Y. and Nienhuis,A.W. (1985) Transfer of genes into hematopoietic cells using recombinant DNA viruses. Proc. Natl Acad. Sci. USA, 82, 158–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Massie B., Dionne,J., Lamarche,N., Fleurent,J. and Langelier,Y. (1995) Improved adenovirus vector provides herpes simplex virus ribonucleotide reductase R1 and R2 subunits very efficiently. Biotechnology, 13, 602–608. [DOI] [PubMed] [Google Scholar]
- 4.Chartier C., Degryse,E., Gantzer,M., Dieterle,A., Pavirani,A. and Mehtali,M. (1996) Efficient generation of recombinant adenovirus vectors by homologous recombination in Escherichia coli.J. Virol., 70, 4805–4810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Graham F.L., Smiley,J., Russell,W.C., Nairn,R. (1977) Characteristics of a human cell line transformed by DNA from human adenovirus type 5. J. Gen. Virol., 36, 59–74. [DOI] [PubMed] [Google Scholar]
- 6.Michou A.-I., Lehrmann,H., Saltik,M. and Cotten,M. (1999) Mutational analysis of the avian adenovirus CELO, which provides a basis for gene delivery vectors. J. Virol., 73, 1399–1410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cotten M., Baker,A., Birnstiel,M.L., Zatloukal,K. and Wagner,E. (1996) Adenovirus polylysine DNA conjugates. In Dracopoli,N.C., Haines,J.L., Korf,B.R., Moir,D.T., Morton,C.C., Seidman,C.E., Seidman,J.G. and Smith,D.R. (eds), Current Protocols in Human Genetics. John Wiley and Sons, Inc., New York, NY, pp. 12.3.1–12.3.33. [Google Scholar]
- 8.Lemay P., Boudin,M.L., Milleville,M. and Boulanger,P. (1980) Human adenovirus type 2 protein IIIa. I. Purification and characterization. Virology, 101, 131–143. [DOI] [PubMed] [Google Scholar]
- 9.Cotten M., Saltik,M., Kursa,M., Wagner,E., Maass,G. and Birnstiel,M.L. (1994) Psoralen treatment of adenovirus particles eliminates virus replication and transcription while maintaining the endosomolytic activity of the virus capsid. Virology, 205, 254–261. [DOI] [PubMed] [Google Scholar]
- 10.Grana X., De Luca,A., Sang,N., Fu,Y., Claudio,P.P., Rosenblatt,J., Morgan,D.O. and Giordano,A. (1994) PITALRE, a nuclear CDC2-related protein kinase that phosphorylates the retinoblastoma protein in vitro. Proc. Natl Acad. Sci. USA, 91, 3834–3838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yoder S.S., Robberson,B.L., Leys,E.J., Hook,A.G., Al-Ubaidi,M., Yeung,C.Y., Kellems,R.E. and Berget,S.M. (1983) Control of cellular gene expression during adenovirus infection: induction and shut-off of dihydrofolate reductase gene expression by adenovirus type 2. Mol. Cell. Biol., 3, 819–828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.O'Malley R.P., Duncan,R.F., Hershey,J.W. and Mathews,M.B. (1989) Modification of protein synthesis initiation factors and the shut-off of host protein synthesis in adenovirus-infected cells. Virology, 168, 112–118. [DOI] [PubMed] [Google Scholar]
- 13.Huang J.T. and Schneider,R.J. (1990) Adenovirus inhibition of cellular protein synthesis is prevented by the drug 2-aminopurine. Proc. Natl Acad. Sci. USA, 87, 7115–7119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Thome M., Hofmann,K., Burns,K., Martinon,F., Bodmer,J.L., Mattmann,C. and Tschopp,J. (1998) Identification of CARDIAK, a RIP-like kinase that associates with caspase-1. Curr. Biol., 8, 885–888. [DOI] [PubMed] [Google Scholar]
- 15.McCarthy J.V., Ni,J. and Dixit,V.M.(1998) RIP2 is a novel NF-kappaB-activating and cell death-inducing kinase. J. Biol. Chem., 273, 16968–16975. [DOI] [PubMed] [Google Scholar]
- 16.Inohara N., del Peso,L., Koseki,T., Chen,S. and Nunez,G. (1998) RICK, a novel protein kinase containing a caspase recruitment domain, interacts with CLARP and regulates CD95-mediated apoptosis. J. Biol. Chem., 273, 12296–12300. [DOI] [PubMed] [Google Scholar]
- 17.Jones N. and Shenk,T. (1979) Isolation of adenovirus type 5 host range deletion mutants defective for transformation of rat embryo cells. Cell, 17, 683–689. [DOI] [PubMed] [Google Scholar]
- 18.Schmidt T.G. and Skerra,A. (1994) One-step affinity purification of bacterially produced proteins by means of the Strep tag and immobilized recombinant core streptavidin. J. Chromatogr. A, 676, 337–345. [DOI] [PubMed] [Google Scholar]
- 19.Schmidt T.G., Koepke,J., Frank,R. and Skerra,A. (1996) Molecular interaction between the Strep-tag affinity peptide and its cognate target, streptavidin. J. Mol. Biol., 255, 753–766. [DOI] [PubMed] [Google Scholar]
- 20.Skerra A. and Schmidt,T.G. (2000) Use of the Strep-Tag and streptavidin for detection and purification of recombinant proteins. Methods Enzymol., 326, 271–304. [DOI] [PubMed] [Google Scholar]
- 21.Voss S. and Skerra,A. (1997) Mutagenesis of a flexible loop in streptavidin leads to higher affinity for the Strep-tag II peptide and improved performance in recombinant protein purification. Protein Eng., 10, 975–982. [DOI] [PubMed] [Google Scholar]
- 22.Korndorfer I.P. and Skerra,A. (2002) Improved affinity of engineered streptavidin for the Strep-tag II peptide is due to a fixed open conformation of the lid-like loop at the binding site. Protein Sci., 11, 883–893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hanks S.K. and Hunter,T. (1995) Protein kinases 6. The eukaryotic protein kinase superfamily: kinase (catalytic) domain structure and classification. FASEB J., 9, 576–596. [PubMed] [Google Scholar]
- 24.Silverstein A.M., Grammatikakis,N., Cochran,B.H., Chinkers,M. and Pratt,W.B. (1998) p50(cdc37) binds directly to the catalytic domain of Raf as well as to a site on hsp90 that is topologically adjacent to the tetratricopeptide repeat binding site. J. Biol. Chem., 273, 20090–20095. [DOI] [PubMed] [Google Scholar]
- 25.Grammatikakis N., Lin,J.H., Grammatikakis,A., Tsichlis,P.N. and Cochran,B.H. (1999) p50(cdc37) acting in concert with Hsp90 is required for Raf-1 function. Mol. Cell. Biol., 19, 1661–1672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Basso A.D., Solit,D.B., Chiosis,G., Giri,B., Tsichlis,P. and Rosen,N. (2002) Akt forms an intracellular complex with heat shock protein 90 (Hsp90) and Cdc37 and is destabilized by inhibitors of Hsp90 function. J. Biol. Chem., 277, 39858–39866. [DOI] [PubMed] [Google Scholar]
- 27.Chen G., Cao,P. and Goeddel,D.V. (2002) TNF-induced recruitment and activation of the IKK complex require Cdc37 and Hsp90. Mol. Cell, 9, 401–410. [DOI] [PubMed] [Google Scholar]
- 28.O'Keefe B., Fong,Y., Chen,D., Zhou,S. and Zhou,Q. (2000) Requirement for a kinase-specific chaperone pathway in the production of a Cdk9/cyclin T1 heterodimer responsible for P-TEFb-mediated tat stimulation of HIV-1 transcription. J. Biol. Chem., 275, 279–287. [DOI] [PubMed] [Google Scholar]
- 29.Lamphere L., Fiore,F., Xu,X., Brizuela,L., Keezer,S., Sardet,C., Draetta,G.F. and Gyuris,J. (1997) Interaction between Cdc37 and Cdk4 in human cells. Oncogene, 14, 1999–2004. [DOI] [PubMed] [Google Scholar]
- 30.Peng J., Zhu,Y., Milton,J.T. and Price,D.H. (1998) Identification of multiple cyclin subunits of human P-TEFb. Genes Dev., 12, 755–762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wei P., Garber,M.E., Fang,S.M., Fischer,W.H. and Jones,K.A. (1998) A novel CDK9-associated C-type cyclin interacts directly with HIV-1 Tat and mediates its high-affinity, loop-specific binding to TAR RNA. Cell, 92, 451–462. [DOI] [PubMed] [Google Scholar]
- 32.Khokhlatchev A., Xu,S., English,J., Wu,P., Schaefer,E. and Cobb,M.H. (1997) Reconstitution of mitogen-activated protein kinase phosphorylation cascades in bacteria. Efficient synthesis of active protein kinases. J. Biol. Chem., 272, 11057–11062. [DOI] [PubMed] [Google Scholar]
- 33.Davies S.P., Reddy,H., Caivano,M. and Cohen,P. (2000) Specificity and mechanism of action of some commonly used protein kinase inhibitors. Biochem. J., 351, 95–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tibbles L.A., Spurrell,J.C., Bowen,G.P., Liu,Q., Lam,M., Zaiss,A.K., Robbins,S.M., Hollenberg,M.D., Wickham,T.J. and Muruve,D.A. (2002) Activation of p38 and ERK signaling during adenovirus vector cell entry lead to expression of the C-X-C chemokine IP-10. J. Virol., 76, 1559–1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tamanini A., Rolfini,R., Nicolis,E., Melotti,P. and Cabrini,G. (2003) MAP kinases and NF-kappaB collaborate to induce ICAM-1 gene expression in the early phase of adenovirus infection. Virology, 307, 228–242. [DOI] [PubMed] [Google Scholar]
- 36.Gould G.W., Cuenda,A., Thomson,F.J. and Cohen,P. (1995) The activation of distinct mitogen-activated protein kinase cascades is required for the stimulation of 2-deoxyglucose uptake by interleukin-1 and insulin-like growth factor-1 in KB cells. Biochem. J., 311, 735–738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hazzalin C.A., Cano,E., Cuenda,A., Barratt,M.J., Cohen,P. and Mahadevan,L.C. (1996) p38/RK is essential for stress-induced nuclear responses: JNK/SAPKs and c-Jun/ATF-2 phosphorylation are insufficient. Curr. Biol., 6, 1028–1031. [DOI] [PubMed] [Google Scholar]
- 38.Han J., Lee,J.D., Bibbs,L. and Ulevitch,R.J.(1994) A MAP kinase targeted by endotoxin and hyperosmolarity in mammalian cells. Science, 265, 808–811. [DOI] [PubMed] [Google Scholar]
- 39.Derijard B., Raingeaud,J., Barrett,T., Wu,I.H., Han,J., Ulevitch,R.J. and Davis,R.J. (1995) Independent human MAP-kinase signal transduction pathways defined by MEK and MKK isoforms. Science, 267, 682–685. [DOI] [PubMed] [Google Scholar]
- 40.Cotten M., Wagner,E., Zatloukal,K., Phillips,S., Curiel,D.T. and Birnstiel,M.L. (1992) High-efficiency receptor-mediated delivery of small and large (48 kilobase gene constructs using the endosome-disruption activity of defective or chemically inactivated adenovirus particles. Proc. Natl Acad. Sci. USA, 89, 6094–6098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Baker A. and Cotten,M. (1997) Delivery of bacterial artificial chromosomes into mammalian cells with psoralen-inactivated adenovirus carrier. Nucleic Acids Res., 25, 1950–1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schreiber S., Kampgen,E., Wagner,E., Pirkhammer,D., Trcka,J., Korschan,H., Lindemann,A., Dorffner,R., Kittler,H., Kasteliz,F., Kupcu,Z., Sinski,A., Zatloukal,K., Buschle,M., Schmidt,W., Birnstiel,M., Kempe,R.E., Voigt,T., Weber,H.A., Pehamberger,H., Mertelsmann,R., Brocker,E.B., Wolff,K. and Stingl,G. (1999) Immunotherapy of metastatic malignant melanoma by a vaccine consisting of autologous interleukin 2-transfected cancer cells: outcome of a phase I study. Hum. Gene Ther., 10, 983–993. [DOI] [PubMed] [Google Scholar]
- 43.Weightman S.A. and Banks,M. (1999) Photochemical inactivation of recombinant baculovirus. J. Virol. Methods, 81, 179–182. [DOI] [PubMed] [Google Scholar]