Abstract
Whole genome amplification (WGA) procedures such as primer extension preamplification (PEP) or multiple displacement amplification (MDA) have the potential to provide an unlimited source of DNA for large-scale genetic studies. We have performed a quantitative evaluation of PEP and MDA for genotyping single nucleotide polymorphisms (SNPs) using multiplex, four-color fluorescent minisequencing in a microarray format. Forty-five SNPs were genotyped and the WGA methods were evaluated with respect to genotyping success, signal-to-noise ratios, power of genotype discrimination, yield and imbalanced amplification of alleles in the MDA product. Both PEP and MDA products provided genotyping results with a high concordance to genomic DNA. For PEP products the power of genotype discrimination was lower than for MDA due to a 2-fold lower signal-to-noise ratio. MDA products were indistinguishable from genomic DNA in all aspects studied. To obtain faithful representation of the SNP alleles at least 0.3 ng DNA should be used per MDA reaction. We conclude that the use of WGA, and MDA in particular, is a highly promising procedure for producing DNA in sufficient amounts even for genome wide SNP mapping studies.
INTRODUCTION
Single nucleotide polymorphisms (SNPs) that occur on the average once every kilobase pair in the human genome are the most abundant form of genetic variation (1). As a consequence of the Human Genome Project and other large SNP discovery efforts, information on more than four million SNPs is available in public databases. The great interest in SNPs originates in their potential use as markers for whole genome linkage disequilibrium mapping to elucidate genes underlying complex, multifactorial disorders (2). Recent studies on linkage disequilibrium patterns in the human genome indicate that very dense SNP maps with hundreds of thousands, or even millions of markers may be needed in genome wide association studies (3,4). Even if most of the currently used genotyping techniques rely on amplification of the genomic DNA by the polymerase chain reaction (PCR) prior to genotyping (5), the amount of DNA obtainable from patient or population samples would be an obstacle for SNP mapping studies on this scale. Given the large efforts involved in the collection of DNA samples from well characterized patient or population cohorts, it is desirable that the collected samples could serve as a long-lasting resource for future genetic studies. The amount of DNA is often limiting also in SNP genotyping studies on a more modest scale when the only available source of DNA are biobanked tumor or other tissue samples, buccal swabs or blood stains collected on filter paper.
One approach for creating an infinite source of DNA for current and future SNP studies is to immortalize the cell samples by transformation with Epstein–Barr virus (6). The transformation procedure is, however, labor intensive, and therefore expensive to apply on a large scale. Moreover, it is not applicable to already existing biobanked DNA sample collections. A technically more feasible approach for increasing the amount of DNA is to use a whole genome amplification (WGA) procedure such as primer extension preamplification with random femtomers (PEP) (7) or degenerate oligonucleotides (DOP–PCR) (8) as primers in PCR. The PEP and DOP–PCR procedures were originally designed for analysis of single cells or very small DNA samples. However, imbalanced amplification of microsatellite (9) and SNP alleles (10,11) as well as incomplete coverage of the genome in the amplification products (12) has been observed. A second concern related to these WGA methods is the possible introduction of artificial sequence variation into the amplification products via the degenerate PCR primers used.
An isothermal procedure for rolling circle amplification of DNA templates using the DNA polymerase from the Φ29 bacteriophage was first introduced as an amplification method for circularized DNA (13). The method has later been adapted for amplification of linear templates, and it is a promising alternative to PEP and DOP–PCR (12). This isothermal multiple displacement amplification (MDA) procedure uses random hexamers containing phosphorothioate-modified nucleotides as primers, and relies on the high processivity, high fidelity and strand displacement ability of the enzyme (14,15). According to quantitative real time PCR analysis, MDA using the Φ29 DNA polymerase provided a less biased representation of different genomic loci than PEP or DOP–PCR (12). When using WGA to increase the amount of DNA for SNP genotyping, a more critical requirement than balanced amplification of different genomic loci is balanced amplification of both alleles of each SNP at the same genomic loci. MDA products have been genotyped successfully at a few microsatellite and SNP loci (12,16), but only a few markers were analyzed and no attempt to evaluate the robustness of the genotyping results has been made.
We present here a systematic, quantitative evaluation of WGA with PEP and MDA for generating DNA templates for SNP genotyping. In this evaluation, we applied an improved protocol for PEP (17), in which a high fidelity PCR system is used in combination with thermo cycling conditions slightly modified from the original protocol. We genotyped a panel of 45 SNPs located in different genomic regions using multiplex, four-color fluorescent minisequencing in a microarray format. The results from PEP and MDA templates were compared to those obtained from genomic DNA. The minisequencing method is particularly useful for this evaluation because it is based on the high sequence-specificity of nucleotide incorporation by a DNA polymerase and therefore allows accurate, quantitative determination of the ratio between two SNP alleles (18). Thus, it also facilitates detection and determination of the magnitude of possible imbalanced amplification of the alleles of a SNP during the WGA procedures. The performance of the WGA methods were evaluated with respect to genotyping success, signal-to-noise ratios, power of discrimination between homozygous and heterozygous SNP genotypes, yield and authenticity of allele representation in the WGA product by the quantitative minisequencing method.
MATERIALS AND METHODS
SNP assay design
Forty-five SNPs distributed on all human chromosomes (except chromosome 10 and the Y-chromosome) were included in the panel. SNPs located in repetitive elements were excluded using the RepeatMasker program (http://ftp.genome.washington.edu/cgi-bin/RepeatMasker). PCR primers were designed using the Oligo Primer Analysis Software v.6.65 (Molecular Biology Insights Inc., Cascade, CO) or the Primer3 Software (http://www-genome.wi.mit.edu/cgi-bin/primer/primer3_www.cgi). The PCR products spanning the SNPs gave one unique hit in the genome according to BLAST analysis (http://www.ncbi.nlm.nih.gov/BLAST). Minisequencing primers annealing immediately adjacent to each SNP were designed for both DNA polarities. The 5′-end of the minisequencing primers contained a 20 bp ‘tag-sequence’ from the Affymetrix GeneChip® Tag collection (Affymetrix, Santa Clara, CA). Hairpin loop formation in the tagged minisequencing primers were evaluated by analyses with the NetPrimer software (http://www.premierbiosoft.com/netprimer/netprimer.html). The Swedish allele frequencies of the SNPs were determined using solid-phase minisequencing of pooled DNA samples as previously described (19), and the frequencies of the minor allele varied between 0.05 and 0.5. The capture oligonucleotides (cTags) complementary to the tag-sequences contained 15 T-residues and an NH2-group at their 3′-ends. The oligonucleotides were synthesized by Sigma Genosys (Cambridgeshire, UK) or Integrated DNA Technologies Inc. (Skokie, IL). Supplement 1 lists the dbSNP identification number, genomic location, variation, allele frequencies and the sequences of the PCR and minisequencing primers for all SNPs analyzed.
Preparation of arrays
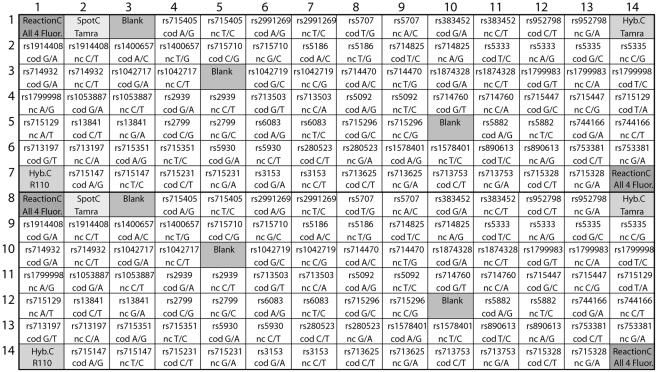
The cTags were covalently coupled via their NH2-groups to CodeLink™ Activated Slides (Amersham Biosciences, Uppsala, Sweden) according to the manufacturer’s instructions. The cTags were printed to form 80 subarrays per slide with spots of 130 µm in diameter and a spot-to-spot distance of 185 µm using a ProSys 5510A instrument (Cartesian Technologies Inc., Irvine, CA) with four Stealth Micro Spotting Pins (SMP3) (TeleChem International Inc., Sunnyvale, CA). In each subarray, a total of 98 cTags were printed as duplicate spots. The remaining amino-reactive groups on the slides were blocked with ethanolamine according to the manufacturer’s instructions. The subarray design is shown in detail in Figure 1.
Figure 1.
Schematic illustration of the array design. Each square represents one spot and the 14 × 14 spots constitutes one subarray as depicted in Figure 2. The capture oligonucleotides (cTags) arrayed in the horizontal rows 1–7 are duplicated in rows 8–14. SNPs are identified by their dbSNP number and cTags for both polarities, indicated by coding (cod) and non-coding (nc), have been arrayed. The nucleotide variations are given as corresponding to the interrogated DNA strand. A cTag (ReactionC) was included in the array to correspond to a minisequencing primer added in the minisequencing extension to serve as a reaction control together with four single stranded oligonucleotide templates, mimicking a four allelic SNP. Labeled cTags (SpotC) are arrayed to control the spotting procedure. Printed cTags without corresponding tagged minisequencing primers (Blank) were used to calculate the average background. To ensure that no leaking had occurred between subarrays, two cTags (Hyb.C) with corresponding differently labeled oligonucleotides added in an alternating pattern over the slide during the hybridization were included. The fluorophore expected to give signal is indicated below the control cTags.
DNA samples
Genomic DNA from 15 individuals of a Centre d’Étude du Polymorphisme Humain (CEPH) family (Utah pedigree no. 1362, individuals 10860, 10861, 11982–11987, 11989–11994 and 11996) was obtained from the Coriell Cell repositories (http://arginine.umdnj.edu). The family consisted of nine last generation children and their predecessors in two generations.
Multiple displacement amplification (MDA)
For MDA, reagents supplied with the GenomiPhi DNA Amplification Kit (Amersham Biosciencies, Uppsala, Sweden) were used. MDA reactions were performed with 3, 0.3, 0.03 or 0.003 ng genomic DNA to select the optimal amount for further use. In most cases 3 ng of genomic DNA in 1 µl was added to 9 µl of Sample Buffer (50 mM Tris–HCl pH 8.2, 0.5 mM EDTA containing random hexamer primers), and denatured at 95°C for 3 min. One microliter of Φ29 DNA polymerase mix including additional random hexamers was mixed on ice with 9 µl of Reaction Buffer containing dNTPs, and the mixture was added to the denatured sample. The MDA reaction was allowed to proceed for 18 h at 30°C. The enzyme was deactivated by heating to 65°C for 10 min. The success of the MDA reaction and the absence of product in negative control samples were assessed by agarose gel electrophoresis.
Primer-extension preamplification (PEP)
For PEP, enzymes and reagents supplied with the Expand High Fidelity (EHF) PCR System (Roche Diagnostics, Basel, Switzerland) were used. PEP reactions were performed with 3, 0.3, 0.03 or 0.003 ng genomic DNA to select the optimal amount. In most cases, 3 ng of genomic DNA was amplified using 120 µM random femtomer primers, 0.6 mM dNTPs, 0.15 mg/ml gelatine, 5 mM MgCl2 and 0.12 U/µl EHF Polymerase mix in 60 µl EHF Buffer without MgCl2. The mixture was heated to 94°C for 3 min and the PEP reactions were allowed to proceed by 50 cycles of 94°C for 40 s, 37°C for 2 min, ramping to 55°C with 0.10°C/s, 55°C for 4 min and 68°C for 30 s and a final step at 68°C for 7 min in a Thermal Cycler PTC-225 (MJ Research, Watertown, MA). The success of the PEP reaction and the absence of product in negative control samples were assessed by agarose gel electrophoresis.
Multiplex PCR
With the exception of initial titration experiments, 9 ng of genomic DNA or WGA product corresponding to 9 pg of original genomic DNA were subjected to multiplex PCR. This amount equals 6 µl of a 1:100 dilution of the MDA product and 9 µl of a 1:50 dilution of the PEP product. Ten optimized multiplex PCRs were performed in a volume of 20 µl under six different reaction conditions using two to six primer pairs at concentrations ranging from 0.2 to 0.3 µM for 35 or 40 cycles on a Thermal Cycler PTC-225 (MJ Research, Watertown, MA). The combinations of primers in the multiplex PCRs are indicated in Supplement 1.
Cyclic minisequencing
The multiplex PCR products from each individual were pooled and concentrated using a Microcon YM-30 device (Millipore Corporation, Bedford, MA). The volume of the eluate was adjusted to 35 µl by adding 3.5 µl of 10× PCR buffer containing 15 mM MgCl2 and H2O. To remove remaining dNTPs and primers, 7.5 µl aliquots of the concentrated PCR-products were treated with 0.1 U/µl shrimp alkaline phosphatase and 0.5 U/µl exonuclease I (USB Corporation, Cleveland, OH) in 11 µl of 4.4 mM MgCl2, 50 mM Tris–HCl pH 9.5 for 60 min at 37°C with subsequent deactivation of the enzymes at 95°C for 15 min. Cyclic minisequencing reactions were performed with 0.1 µM ddATP-Texas Red, ddCTP-Tamra and ddGTP-R110, 0.2 µM of ddUTP-Cy5 (Perkin-Elmer Life Sciences, Boston, MA), the 90 minisequencing primers at 10 nM each, and 0.064 U/µl Thermo Sequenase™ DNA Polymerase (Amersham Biosciences, Uppsala, Sweden) in 15.6 µl of 4.6 mM MgCl2 and 0.02% Triton-X100. The cyclic extension reaction was performed in a Thermal Cycler PTC-225 (MJ Research, Watertown, MA) with an initial 96°C for 3 min followed by 33 cycles at 95°C and 55°C for 30 s each.
Capture on microarrays
A custom made reaction rack holding the arrayed slides with a silicon grid to give 80 separate reaction chambers was preheated to 42°C. Twenty-two microliters of hybridization mixture, containing 15.6 µl of the minisequencing reaction product from above and 0.26 nM of Tamra- or R110-labeled hybridization control oligonucleotide in 900 mM NaCl, 90 mM sodium citrate, pH 7.0 (6× SSC), was pipetted into each reaction chamber and allowed to hybridize to the arrayed cTags under humid conditions at 42°C for 2.5 h. After hybridization, the slides were briefly rinsed at room temperature (22°C) with 4× SSC and subsequently washed with 2× SSC and 0.1% SDS twice for 5 min at 42°C and with 0.2× SSC for 1 min at room temperature. Finally the slide was spin dried in a centrifuge for 5 min at 900 r.p.m.
Signal detection
Fluorescence signals were measured using a ScanArray® 5000 instrument (Perkin-Elmer Life Science, Boston, MA) with the laser power kept constant at 80% and the photo-multiplier tube gain adjusted to obtain equal signal levels from the reaction control spots in all four laser channels. The excitation lasers were: Blue Argon 488 nm for R110; Green HeNe 543.8 nm for Tamra; Yellow HeNe 594 nm for Texas Red and Red HeNe 632.8 nm for Cy5. The signal intensities were determined with the QuantArray® analysis 3.1 software (Perkin-Elmer Life Sciences).
Genotype assignment
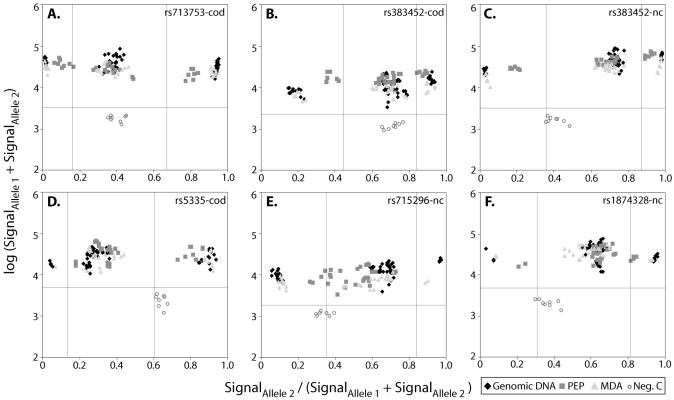
The QuantArray file was exported to the SNPSnapper v3.81beta software (http://www.bioinfo.helsinki.fi/SNPSnapper/) for genotype assignment. The three different subsets of data from standard PCR, PEP or MDA were analyzed both separately and simultaneously to allow comparison of the different clustering properties. Genotypes were assigned based on scatter plots with the logarithm of the sum of both fluorescence signals (SignalAllele1 + SignalAllele2) plotted against the fluorescence signal ratios [SignalAllele2/(SignalAllele1+SignalAllele2)], see Figure 4. For successful genotyping, the two duplicate spots and the two polarities were required to give concordant genotype calls.
Figure 4.
Examples (A–F) of scatter plots used to assign the genotypes of six SNPs. The logarithms of the sum of the fluorescence signals corresponding to the two alleles of a SNP in each sample are plotted on the y-axis. The signal ratio between the signal from one allele divided by the sum of the signals from both alleles are plotted on the x-axis. The closed diamonds show the results from genomic DNA, the medium-gray squares from the PEP and the light-gray triangles from the MDA products. The open circles are the results from negative controls included on the same array and used to set the cut-off levels for acceptable signals, illustrated by the horizontal lines. The vertical lines have been included to clarify the scatter plots and are positioned at equal distance from the homozygous and heterozygous clusters in genomic DNA. The SNPs are denoted by their dbSNP identification number, and the DNA polarities analyzed are indicated by cod or nc.
RESULTS AND DISCUSSION
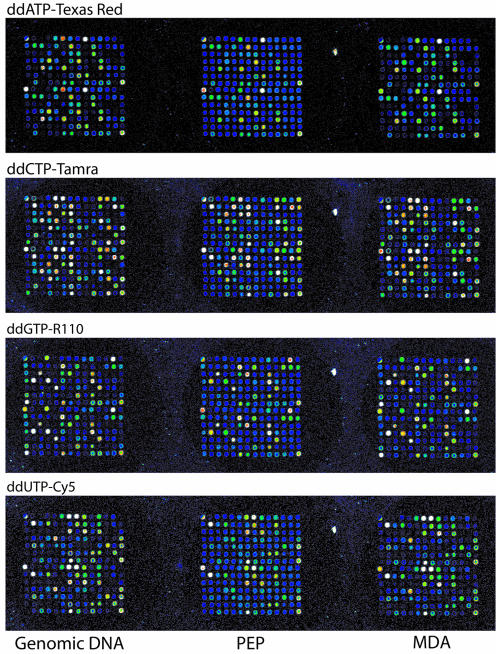
We used our in-house four-color fluorescence tag-array minisequencing system (18) for multiplex genotyping of a representative panel of SNPs for evaluation of WGA using MDA and PEP in comparison to genomic DNA. For this comparison we assembled a panel of 45 SNPs from different genomic locations, see Supplement 1. The SNPs were analyzed in DNA samples from 15 family members from the CEPH collection. The regions that span the SNPs were amplified by multiplex PCRs from the WGA products and from genomic DNA prior to genotyping. The ‘array of arrays’ format of our method (20) enabled us to analyze all SNPs in both DNA polarities in all 15 samples and in all three template types in the same experiment on a single microarray slide, facilitating accurate, quantitative comparison of the results. In total 15 000 fluorescent signals were generated per experiment. Figure 2 shows an example of a ‘subarray’ for one sample after scanning at four wavelengths to detect the dideoxynucleotides labeled with four different fluorophores incorporated in the minisequencing reactions.
Figure 2.
Images obtained by scanning a ‘subarray’ at four wavelengths. The results from one CEPH individual genotyped by tag-array minisequencing for the panel of 45 SNPs with primers in both DNA polarities after capture at duplicate positions in a subarray are shown. The images from genomic DNA as well as from PEP and MDA products are shown in three vertical rows of subarrays. Each microscope slide carries 80 subarrays. The fluorescent labels used for the four ddNTPs are indicated above the horizontal rows of subarrays. The obtained signals are reproduced with an artificial rainbow scale with blue as low and white as saturated signal.
Genotyping success
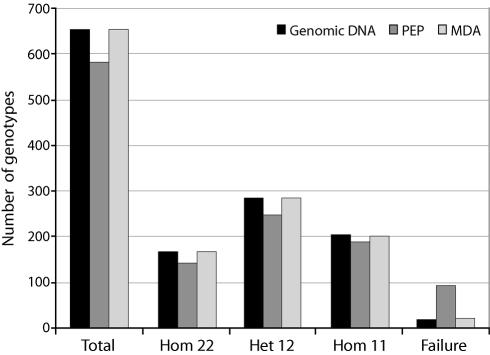
The genotypes determined from genomic DNA were considered as the correct results to which the genotypes for the 45 SNPs obtained with the two WGA methods were compared. Indirect evidence for the accuracy of the results from genomic DNA is provided by the fact that no Mendelian inheritance conflicts were detected in the three-generation CEPH family samples. The results were also reproduced in three independent experiments. For genomic DNA, 34 of the SNPs gave concordant genotypes for both DNA polarities, while for 10 SNPs acceptable genotype calls were made from one DNA polarity only. One SNP failed in all samples due to low signals, probably caused by PCR failure. In the PEP products, three additional SNPs failed, while the same SNPs that were called in genomic DNA were also called in MDA products. The overall success rate for the genomic DNA samples was 97.0%, i.e. 655 genotypes out of a maximum of 675 were called. The genotyping results from the PEP and MDA assays were concordant to the genomic result in 88.7 (581 out of 655 correct genotypes in genomic DNA) and 99.7% (653 out of 655) of the cases respectively (Fig. 3). The most frequent reasons for failure were low signals and poor clustering of signals (49%), or erroneous genotype calls (21%) from the PEP products.
Figure 3.
Genotyping success rates using genomic DNA (black), PEP (medium gray) and MDA (light gray) products as template for multiplex PCR when 15 DNA samples were analyzed for 45 SNP to generate 675 genotypes. The number of genotypes is given on the y-axis of the diagram. The total results as well as the results for homozygous (denoted Hom 22 and Hom 11) and heterozygous (Het 12) genotypes and failures are indicated.
Signal-to-noise
Although the four fluorescence signal patterns defining the genotypes in all three DNA templates are similar, the scanned array in Figure 2 reveals that the background fluorescence in most of the spots on the array is clearly higher after the PEP procedure. A frequently expressed concern regarding PEP is that sequence alterations could be introduced into the DNA templates via the fully degenerate femtomers used as primers in PEP. In our experiments the numeric fluorescence signals measured on the array from mis-incorporated dideoxynucleotides were consistently higher for the PEP products than when genomic DNA or MDA products served as templates (Table 1). The average background signals from control spots with printed capture oligonucleotides (cTags) without corresponding tagged minisequencing primers were also higher for the PEP products than for the two other DNA templates (Table 1). Thus our results do not provide evidence for introduction of sequence alterations by the random femtomer PEP primers at detectable levels. The signal intensities from correctly incorporated nucleotides do not differ between the methods. Therefore the signal-to-noise ratios for the PEP products are about half of those obtained in genomic and MDA templates. Table 1 show the average signal-to-noise ratios calculated for all SNP alleles in all samples. Also with respect to signal-to-noise ratios, genomic DNA and MDA products yielded strikingly similar results.
Table 1. Average minisequencing signals and signal-to-noise ratios for all SNPs in all samples.
| Fluorescence signals | Signal-to-noisea | |||
|---|---|---|---|---|
| Correctb | Mis-incorporatedc | Backgroundd | ||
| Genomic DNA | 30 020 | 2960 | 553 | 36 |
| PEP | 28 430 | 5310 | 3737 | 14 |
| MDA | 28 800 | 2970 | 508 | 34 |
aSignal-to-noise ratios obtained by dividing the fluorescence signals from the correct nucleotides by the fluorescence signal for the mis-incorporated nucleotides for a given SNP.
bSignals from correctly incorporated nucleotides i.e. one and two nucleotides for homozygous and heterozygous samples respectively.
cSignals from mis-incorporated nucleotides i.e. three and two nucleotides for homozygous and heterozygous samples respectively.
dAverage fluorescence signals from arrayed capture oligonucleotides (cTags) without corresponding tagged minisequencing primers, see Figure 1.
Occasionally, we observed high molecular weight DNA in negative controls when MDA and PEP products were analyzed on an ethidium bromide (EtBr)-stained agarose gel. Formation of these spurious high-molecular weight products has also been noticed by other users of the MDA system. The use of modified random hexamers with two 5′-terminal nitroindole residues has been reported to prevent the formation of primer-directed template independent DNA synthesis (21). In our study, the negative controls containing spurious high-molecular weight DNA did not result in PCR products or called genotypes.
Power of discrimination between genotypes
In our minisequencing system the genotypes are assigned based on the numeric fluorescence signals extracted from the microarrays using the SNPSnapper software (http://www.bioinfo.helsinki.fi/SNPSnapper). Figure 4 shows examples of scatter plots used for assignment of the genotypes for six SNPs. Tight clustering of signal ratios from different samples indicates robust genotyping, and the larger the distance between the clusters are, the better is the power of discrimination between homozygous and heterozygous genotypes. Differences between SNPs in the positions and tightness of the clusters are especially noticeable for the heterozygous genotypes. The reason for the differences in clustering properties between SNPs is that the flanking sequence as well as the fluorophores attached to the dideoxynucleotides affect the efficiency and sequence specificity of nucleotide incorporation by the DNA polymerase (18). The different properties of the fluorophores, such as molar extinction coefficients, emission spectra and quantum yield, as well as unspecific background also affect the obtained signal intensities and signal ratios.
The signal ratios from genomic DNA and MDA products constantly cluster identically, while the clusters from PEP products are less tight and positioned closer to each other than the corresponding clusters from genomic DNA and MDA products. In Figure 4E, less tight clusters from PEP products than from genomic DNA is particularly evident. Non-overlapping mean signal ratios were obtained from all three types of template and the overall average signal ratios for all genotyped SNPs are given in Table 2. With genomic DNA or MDA products as templates, the average signal ratios are surprisingly close to the theoretical values of 1.0 or 0 for homozygotes and 0.5 for heterozygotes.
Table 2. Average signal ratios and power of genotype discrimination for all SNPs in all samples.
| Signal ratiosa | Power of discriminationb | |||
|---|---|---|---|---|
| Hom 22 | Het 12 | Hom 11 | ||
| Genomic DNA | 0.94 ± 0.07 | 0.50 ± 0.22 | 0.07 ± 0.07 | 0.44 ± 0.18 |
| PEP | 0.84 ± 0.11 | 0.51 ± 0.17 | 0.15 ± 0.10 | 0.36 ± 0.14 |
| MDA | 0.94 ± 0.06 | 0.50 ± 0.20 | 0.07 ± 0.07 | 0.44 ± 0.17 |
aThe ratio between the signals from allele 2 and the sum of the signals from alleles 1 and 2.
bThe distance between the homozygous and heterozygous genotype clusters.
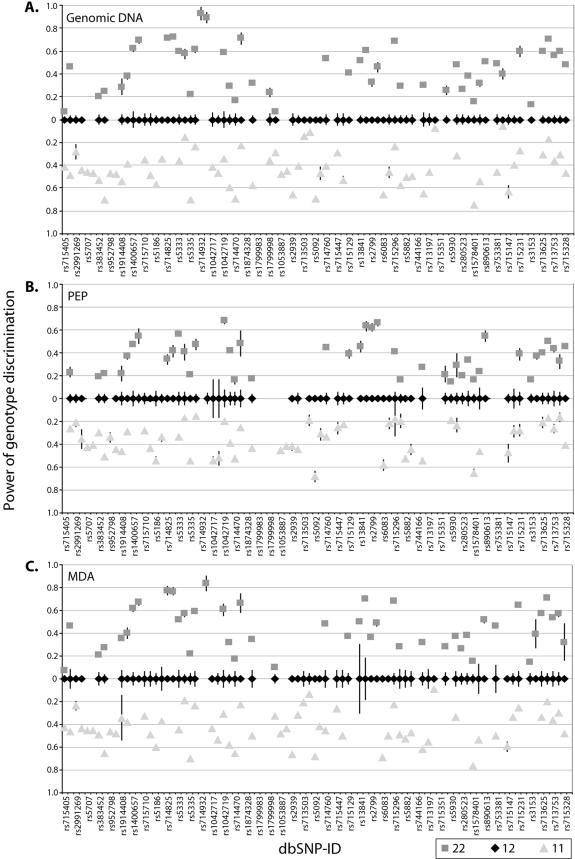
The power of genotype discrimination obtained in our study for each of the individual SNPs in both polarities for genomic DNA, PEP and MDA products are illustrated graphically in Figure 5. The corresponding average numerical values for all genotyped SNPs are given in Table 2, and show that the average distance between homozygous and heterozygous genotype clusters that reflect the power of genotype discrimination are essentially identical for the genomic and MDA products and somewhat lower for the PEP products. The previously discussed higher unspecific background of the PEP procedure contributes to the lower power of genotype discrimination.
Figure 5.
Power of genotype discrimination for all SNPs with genomic DNA (A), PEP product (B) or MDA product (C) as template. The dbSNP identification number is given below the panels and the results for the coding DNA polarity are given to the left of the non-coding. For each SNP, the power of genotype discrimination is defined as the distance between the homozygous genotype clusters for allele 1 (light-gray triangles) and allele 2 (medium-gray squares) and the heterozygote cluster (black diamonds). The distance is given by the absolute value when subtracting the average signal ratio for the homozygous cluster from the average signal ratio for the heterozygous cluster. The standard deviations for the ratios are indicated by vertical black bars.
Yield
The amount of genomic DNA subjected to WGA, and the amount of WGA product subjected to multiplex PCR in the experiments presented above were chosen based on initial titration experiments. Both the amount of genomic DNA and the level of dilution of the WGA products were varied 10-fold, and the products were visualized on EtBr-stained agarose gels. In the comparison of PEP and MDA to genomic DNA as templates for SNP genotyping, 3 ng of genomic DNA, which corresponds to 1000 genome equivalents were subjected to both PEP and WGA with a subsequent dilution to the equivalent of 9 pg of the original genomic DNA (three genome equivalents) for multiplex PCR followed by genotyping by minisequencing. The amount of genomic DNA consumed per SNP varied between 1.5 and 4.5 pg depending on the multiplexing level of the PCR. Thus in our study, up to a 2000-fold reduction in DNA consumption was achieved by the WGA procedures without compromising the genotyping accuracy. The dilution of the MDA product to an amount corresponding to 0.9 pg of genomic DNA also yielded visible EtBr-stained products in agarose gel electrophoresis, although in the comparison to PEP, 9 pg was used.
According to Dean et al. (12), a plateau of ∼20–30 µg of DNA (6 × 106–9 × 106 genome equivalents) will be reached in a 100 µl MDA reaction independently of the initial DNA amount subjected to the reaction, which would correspond to an almost 10 000-fold amplification when starting from 3 ng of genomic DNA. In the same study, PEP performed with 1 ng of human genomic DNA was reported to yield 360-fold amplification (12). In our previous study, we estimated a 1000-fold amplification of the genome in single cells using PEP (11).
In the MDA amplification, thiophospate modified random hexamers (5′-NpNpNpNpSNpSN-3′) are protected from degradation by the Φ29 DNA polymerase proofreading 3′–5′ exonuclease activity. This improvement was earlier shown to increase the yield 40-fold compared to standard random hexamers when applied to a double stranded circular M13 DNA template (22). A potential drawback of MDA amplification is that due to the dependence on hyperbranched amplification, the yield may decrease when using template with lower molecular weight, as has been predicted by mathematical modeling (21). PEP, however, which is based on exponential amplification, would not be affected.
Imbalanced amplification of alleles
Using quantitative real time PCR, an amplification bias of 102 to 104 between genomic loci was observed in PEP products, compared to 3-fold bias in MDA products (12). Using comparative genomic hybridization (CGH), a significant amplification bias between different genomic sequences was detected particularly at the ends of the chromosomes when using MDA (21). However, it has been argued that this may have been due to the low amount of Φ29 DNA polymerase used (16). For SNP genotyping a more important consideration than balanced amplification of different genomic loci is that the two alleles of the same SNP locus are amplified with equal efficiency during WGA. In our early study, in which we amplified genomic DNA from a single cell by PEP, and genotyped a panel of 10 SNPs, we observed significant imbalanced allelic representation at all heterozygous nucleotide positions (11). The imbalanced amplification of the two SNP alleles resulted in mistyping of four out of 52 analyzed heterozygote SNPs as homozygotes (8%) because the amount of one allele in the PCR products remained below the detection threshold.
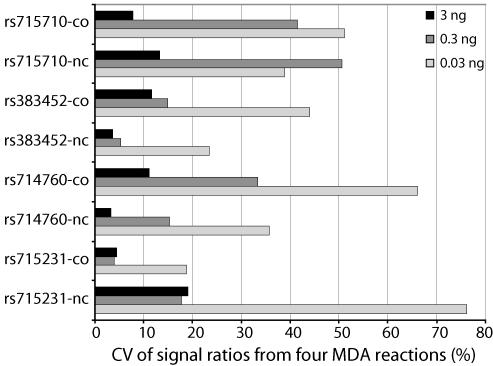
To investigate the relationship between the original amount of DNA used for MDA and the representation of the two SNP alleles in the PCR products, we genotyped a subset of heterozygous SNPs. Ten-fold dilutions containing between 3 ng and 3 pg of genomic DNA were subjected to MDA, and MDA product corresponding to 9, 0.9, 0.09 pg and 9 fg of the original genomic DNA were analyzed by PCR and minisequencing. The MDA reactions, in which 3 pg of genomic DNA (approximately one genome equivalent) was used, yielded fluorescence signals in eight out of 32 SNP genotyping reactions. Absence of signals was presumably caused by absence of DNA due to stochastic pipetting errors when DNA was added to the reaction mixtures. The observation that only a part of the SNPs yielded signals indicates imbalanced amplification between these SNP loci at this low amount of MDA template. With 0.03 ng of genomic DNA (∼10 genome equivalents), measurable signals were obtained for 22 out of 32 SNP genotyping reactions, but the signal intensity ratios varied so much between the parallel assays that assignment of the SNP genotypes would have been difficult. The variation in signal ratios is probably caused by imbalanced amplification of the two SNP alleles at the same locus. For five of the SNPs, 0.3 ng of genomic DNA (∼100 genome equivalents) gave fluorescent signal ratios that allowed assignment of the genotypes based on coefficient of variation (CV) values <20%. The most reproducible fluorescent signals, seen as low CV values for the signal ratios in Figure 6, was obtained with 3 ng of genomic DNA. The CV values in Figure 6 clearly show that the variation in the amplification of the two SNP alleles depends on the amount of genomic DNA subjected to MDA. This result is concordant with a study by Stenman et al. (23), who observed increased stochastic variation in signal ratios after competitive PCR with less than 1000 molecules of mRNA (23).
Figure 6.
Coefficient of variation (CV) for signal ratios obtained by analyzing both polarities of four SNPs in a heterozygous sample. The SNPs are indicated on the left of the diagram. Each of the products from four parallel MDA reactions was analyzed in triplicate multiplex PCR and minisequencing reactions. The CV values for the signal ratios are given on the horizontal axis. MDA reactions were performed with 3 (black), 0.3 (medium-gray) or 0.03 ng (light-gray) of genomic DNA.
Conclusion
In the present study we found that WGA using MDA allows robust and accurate genotyping of SNPs. The MDA procedure with Φ29 DNA polymerase produced genotyping results that were indistinguishable from those obtained with genomic DNA with respect to genotyping success, signal-to-noise ratios and power of discrimination between genotypes. The major determinant for successful genotyping is the amount of DNA subjected to MDA, and according to our results, about 1000 genome equivalents (3 ng) of DNA should be used. The MDA product obtained from 3 ng of genomic DNA is sufficient for hundreds (or even thousands) of multiplex PCRs. Assuming an average multiplexing level of 10 SNPs per PCR, a genome wide association study using 300 000 SNP markers would require <1 µg of genomic DNA. This opens up new possibilities for large clinical or epidemiological studies for elucidation of the genetic background to complex disorders using small tissue or cell samples.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at NAR Online.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Elin Cederholm, Kristina Larsson, Raul Figueroa, Ann-Christin Wiman and David Fange for their excellent technical assistance. The GenomiPhi DNA Amplification Kits were made available to us by Amersham Biosciences. Financial support for the study was provided by the Swedish Research Council, the K & A Wallenberg foundation and by the European Commission (contracts QLRT-2001-00004 and QLRT-2001-01325).
REFERENCES
- 1.Sachidanandam R., Weissman,D., Schmidt,S.C., Kakol,J.M., Stein,L.D., Marth,G., Sherry,S., Mullikin,J.C., Mortimore,B.J., Willey,D.L. et al. (2001) A map of human genome sequence variation containing 1.42 million single nucleotide polymorphisms. Nature, 409, 928–933. [DOI] [PubMed] [Google Scholar]
- 2.Risch N. and Merikangas,K. (1996) The future of genetic studies of complex human diseases. Science, 273, 1516–1517. [DOI] [PubMed] [Google Scholar]
- 3.Shifman S., Kuypers,J., Kokoris,M., Yakir,B. and Darvasi,A. (2003) Linkage disequilibrium patterns of the human genome across populations. Hum. Mol. Genet., 12, 771–776. [DOI] [PubMed] [Google Scholar]
- 4.Phillips M.S., Lawrence,R., Sachidanandam,R., Morris,A.P., Balding,D.J., Donaldson,M.A., Studebaker,J.F., Ankener,W.M., Alfisi,S.V., Kuo,F.S. et al. (2003) Chromosome-wide distribution of haplotype blocks and the role of recombination hot spots. Nature Genet., 33, 382–387. [DOI] [PubMed] [Google Scholar]
- 5.Syvanen A.C. (2001) Accessing genetic variation: genotyping single nucleotide polymorphisms. Nat. Rev. Genet., 2, 930–942. [DOI] [PubMed] [Google Scholar]
- 6.Katakura Y., Alam,S. and Shirahata,S. (1998) Immortalization by gene transfection. Methods Cell. Biol., 57, 69–91. [DOI] [PubMed] [Google Scholar]
- 7.Zhang L., Cui,X., Schmitt,K., Hubert,R., Navidi,W. and Arnheim,N. (1992) Whole genome amplification from a single cell: implications for genetic analysis. Proc. Natl Acad. Sci. USA, 89, 5847–5851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Telenius H., Carter,N.P., Bebb,C.E., Nordenskjold,M., Ponder,B.A. and Tunnacliffe,A. (1992) Degenerate oligonucleotide-primed PCR: general amplification of target DNA by a single degenerate primer. Genomics, 13, 718–725. [DOI] [PubMed] [Google Scholar]
- 9.Cheung V.G. and Nelson,S.F. (1996) Whole genome amplification using a degenerate oligonucleotide primer allows hundreds of genotypes to be performed on less than one nanogram of genomic DNA. Proc. Natl Acad. Sci. USA, 93, 14676–14679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Grant S.F., Steinlicht,S., Nentwich,U., Kern,R., Burwinkel,B. and Tolle,R. (2002) SNP genotyping on a genome-wide amplified DOP-PCR template. Nucleic Acids Res., 30, e125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Paunio T., Reima,I. and Syvanen,A.C. (1996) Preimplantation diagnosis by whole-genome amplification, PCR amplification and solid-phase minisequencing of blastomere DNA. Clin. Chem., 42, 1382–1390. [PubMed] [Google Scholar]
- 12.Dean F.B., Hosono,S., Fang,L., Wu,X., Faruqi,A.F., Bray-Ward,P., Sun,Z., Zong,Q., Du,Y., Du,J. et al. (2002) Comprehensive human genome amplification using multiple displacement amplification. Proc. Natl Acad. Sci. USA, 99, 5261–5266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lizardi P.M., Huang,X., Zhu,Z., Bray-Ward,P., Thomas,D.C. and Ward,D.C. (1998) Mutation detection and single-molecule counting using isothermal rolling-circle amplification. Nature Genet., 19, 225–232. [DOI] [PubMed] [Google Scholar]
- 14.Esteban J.A., Salas,M. and Blanco,L. (1993) Fidelity of phi 29 DNA polymerase. Comparison between protein-primed initiation and DNA polymerization. J. Biol. Chem., 268, 2719–2726. [PubMed] [Google Scholar]
- 15.Blanco L., Bernad,A., Lazaro,J.M., Martin,G., Garmendia,C. and Salas,M. (1989) Highly efficient DNA synthesis by the phage phi 29 DNA polymerase. Symmetrical mode of DNA replication. J. Biol. Chem., 264, 8935–8940. [PubMed] [Google Scholar]
- 16.Hosono S., Faruqi,A.F., Dean,F.B., Du,Y., Sun,Z., Wu,X., Du,J., Kingsmore,S.F., Egholm,M. and Lasken,R.S. (2003) Unbiased whole-genome amplification directly from clinical samples. Genome Res., 13, 954–964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dietmaier W., Hartmann,A., Wallinger,S., Heinmoller,E., Kerner,T., Endl,E., Jauch,K.W., Hofstadter,F. and Ruschoff,J. (1999) Multiple mutation analyses in single tumor cells with improved whole genome amplification. Am. J. Pathol., 154, 83–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lindroos K., Sigurdsson,S., Johansson,K., Ronnblom,L. and Syvanen,A.C. (2002) Multiplex SNP genotyping in pooled DNA samples by a four-colour microarray system. Nucleic Acids Res., 30, e70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Syvanen A.C., Sajantila,A. and Lukka,M. (1993) Identification of individuals by analysis of biallelic DNA markers, using PCR and solid-phase minisequencing. Am. J. Hum. Genet., 52, 46–59. [PMC free article] [PubMed] [Google Scholar]
- 20.Pastinen T., Raitio,M., Lindroos,K., Tainola,P., Peltonen,L. and Syvanen,A.C. (2000) A system for specific, high-throughput genotyping by allele-specific primer extension on microarrays. Genome Res., 10, 1031–1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lage J.M., Leamon,J.H., Pejovic,T., Hamann,S., Lacey,M., Dillon,D., Segraves,R., Vossbrinck,B., Gonzalez,A., Pinkel,D. et al. (2003) Whole genome analysis of genetic alterations in small DNA samples using hyperbranched strand displacement amplification and array-CGH. Genome Res., 13, 294–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dean F.B., Nelson,J.R., Giesler,T.L. and Lasken,R.S. (2001) Rapid amplification of plasmid and phage DNA using Phi 29 DNA polymerase and multiply-primed rolling circle amplification. Genome Res., 11, 1095–1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stenman J., Lintula,S., Rissanen,O., Finne,P., Hedstrom,J., Palotie,A. and Orpana,A. (2003) Quantitative detection of low-copy-number mRNAs differing at single nucleotide positions. Biotechniques, 34, 172–177. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.