Summary
Peroxisome proliferator-activated receptor γ (PPARγ) and its ligands are important regulators of lipid metabolism, inflammation, and diabetes. We previously demonstrated that anucleate human platelets express the transcription factor PPARγ and that PPARγ ligands blunt platelet activation. To further understand the nature of PPARγ in platelets, we determined the platelet PPARγ isoform(s) and investigated the fate of PPARγ following platelet activation. Our studies demonstrated that human platelets contain only the PPARγ1 isoform and after activation with thrombin, TRAP, ADP or collagen PPARγ is released from internal stores. PPARγ release was blocked by a cytoskeleton inhibitor, Latrunculin A. Platelet-released PPARγ was complexed with the retinoid X receptor (RXR) and retained its ability to bind DNA. Interestingly, the released PPARγ and RXR were microparticle associated and the released PPARγ/RXR complex retained DNA-binding ability. Additionally, a monocytic cell line, THP-1, is capable of internalizing PMPs. Further investigation following treatment of these cells with the PPARγ agonist, rosiglitazone and PMPs revealed a possible transcellular mechanism to attenuate THP-1 activation. These new findings are the first demonstrating transcription factor release from platelets, revealing the complex spectrum of proteins expressed and expelled from platelets, and suggest that platelet PPARγ has an undiscovered role in human biology.
Keywords: Microparticles, Peroxisome Proliferator-Activated Receptor γ(PPARγ), Platelets, Retinoic X Receptor (RXR), transcription factors
Introduction
Peroxisome proliferator-activated receptor γ (PPARγ), a ligand activated transcription factor, is one of three PPAR subtypes (also PPARα and PPARδ/β) [1]. PPARγ is widely expressed by adipose tissue [2] and immune system cells [3, 4]. Two PPARγ isoforms, PPARγ1 and PPARγ2, are products of the same gene, but result from differential promoter use, and alternative RNA splicing [5]. The PPARγ2 isoform is mainly expressed in adipose tissue [2], whereas PPARγ1 is more widely expressed [5]. Although PPARγ was originally described as a nuclear receptor, it has both cytoplasmic and nuclear distribution [3, 6]. As a transcription factor, PPARγ functions as a heterodimer with the Retinoid X Receptor (RXR) to initiate transcription of genes containing PPAR response elements (PPRE) [7].
PPARγ ligands include the synthetic clinically used thiazolidinediones [8] (rosiglitazone and pioglitazone) and naturally occurring ligands such as 15-deoxy-Δ12,14-prostaglandin J2 (15d-PGJ2) [9, 10] and lysophosphatidic acid (LPA) [11]. PPARγ and its ligands are widely studied because they are potent insulin sensitizers used to treat type 2 diabetes mellitus. Furthermore, rosiglitazone reduces the incidence of type 2 diabetes mellitus in at risk patients [12].
Recently, we reported PPARγ expression in human platelets [13], suggesting a new role for platelets in inflammation [14]. Upon activation, platelets release pro-inflammatory mediators including IL-1β, sCD40L, TGFβ, and TXA2, some of which activate endothelial cells and produce chemokines to recruit inflammatory cells [15–17]. Emerging evidence implicates inflammation in the development of type 2 diabetes mellitus and cardiovascular disease [18–20]. PPARγ ligands dampen platelet activation and studies in human patients with atherosclerosis have shown that TZD agonists of PPARγ reduce platelet activation, not only to inhibit plaque progression, but remarkably to promote regression of existing atherosclerotic plaques [21].
Platelet activation by physiological agonists or high shear stress leads to the highly regulated formation and release of their contents in soluble form or via membrane-bound vesicles [22]. One vesicle is the platelet microparticle (PMP) ranging in size from 0.1 to 1.0 µm [23] that surface-expose proteins to regulate inflammatory [24, 25] and hemostatic processes [26, 27]. PMPs are also involved in pathogenic processes and elevated in atherosclerosis [28], type 2 diabetes mellitus [29], and cancer [30].
Our laboratory recently discovered that human platelets express PPARγ and that PPARγ ligands attenuate platelet-release of the pro-inflammatory and pro-coagulant mediators sCD40L and TXA2, a cyclooxygenase product that enhances platelet activation [13]. Herein, we investigated the PPARγ isoform and fate in activated human platelets. Surprisingly, we found that PPARγ is complexed with RXR and is released from activated platelets. Some of the released PPARγ is associated with PMPs, which can be transferred to THP-1 cells. Moreover, in the presence of the PPARγ agonist, rosiglitazone, and PMP-containing PPARγ, THP-1 activation is dampened suggesting a novel transcellular mechanism of regulation.
Materials and Methods
Blood Collection and preparation of washed human platelets
Whole blood was obtained under IRB approval from male and female donors (21–55 years of age) that were NSAID-free for two weeks prior to donation with a body mass index (BMI) ≤25. Blood was collected by venipuncture into a citrate phosphate dextrose adenine solution containing collection bag (Baxter Fenwal, Round Lake, IL) or Vacutainer tubes containing buffered citrate sodium (BD Biosciences, Franklin Lakes, NJ). Platelet-rich plasma (PRP) was obtained by centrifugation (250xg/15 min/room temperature (RT)), diluted with an equal volume of Krebs-Ringer Bicarbonate Buffer (KRB) (Sigma, St. Louis, MO) pH 5.0 containing 15 mM sodium bicarbonate and 19 mM sodium citrate and centrifuged (200xg/10 min). The platelet pellet was washed in KRB pH 6.0, centrifuged (950xg/10 min), and resuspended in KRB pH 7.4. Platelets were counted on an Abbott Cell-Dyn 1700 (Abbott Park, IL). On average, 5.5×1010 platelets/unit of blood were obtained, and the platelet purity was 99 to 99.99% as described [13].
Platelet Activation
Nine x107 platelets in KRB pH 7.4 were incubated (37°C) with platelet activators: Thrombin 0.8 U/mL (Sigma), Thrombin Receptor Activator Peptide-6 (TRAP) 50 µM (Bachem Biosciences Inc., King of Prussia, PA), collagen 10 µg/mL (Chrono-log Corporation, Havertown, PA), adenosine diphosphate (ADP) 10 µM (Chrono-log Corporation), and phorbol 12-myristate 13-acetate (PMA) 0.2 µM (Sigma). After treatment, platelets were centrifuged (1200xg/1 min/RT), and supernatants and pellets analyzed. For some studies, platelets were incubated with the cytoskeletal inhibitor, Latrunculin A (Lat A) (Sigma), for 20 min prior to activation.
Culture of Megakaryocyte Cell Lines
Meg-01 [31] and M-07e cells [32] were cultured in RPMI 1640 medium (Gibco, Grand Island, NY) as previously described [13]. Meg-01 and M-07e are both human leukemia cell lines at the megakaryoblast stage of development.
Western blots for PPARγ and RXR
Total protein was isolated from platelets as described [13]. For platelet mediator release experiments, equal volumes of supernatant or lysate were used for Western blot analysis for PPARγ using a rabbit polyclonal anti-PPARγ (BIOMOL), a mouse monoclonal anti-PPARγ2 (Chemicon International, Temecula, CA) and for RXR using a rabbit polyclonal anti-RXR (Santa Cruz Biotechnology, Santa Cruz, CA) as described [10, 13, 33]. Human adipose tissue was used as a positive control for PPARγ and RXR.
Co-Immunoprecipitation (co-IP) of PPARγ and RXR
Lysates from unactivated platelet and supernatants from TRAP-activated platelets were immunoprecipitated for PPARγ using an anti-PPARγ antibody (Santa Cruz) or for RXR using a rabbit polyclonal antibody (Santa Cruz). Control samples were incubated with a mouse IgG1 isotype control antibody (for PPARγ control IP) or with rabbit serum (for RXR control IP) (both from Santa Cruz). Total protein (50 µg) was incubated in IP buffer (50 mM HEPES [pH 7], 0.1% NP-40, 250 mM NaCl, 5 mM EDTA, 10 mM NaF, 0.1 mM Na3VO4, 50 uM ZnCl2 supplemented with 0.1 mM phenylmethylsulfonyl fluoride, 1 mM dithiothreitol, and protease inhibitor cocktail) with antibody (5 µg) overnight at 4°C. Antibody complexes were precipitated using Protein G Plus Agarose (Santa Cruz) (2 hours/4°C). Complexes were pelleted and washed 5 times in IP buffer. The beads were denatured by boiling in Western buffer (Sigma). Western blots for PPARγ and RXR were performed as described above.
Real-Time RT-PCR for RXR
RNA was isolated from Meg-01 cells, platelets from 3 individual donors, and human adipose tissue as a positive control using an RNeasy Kit according to the manufacturer’s protocol (Qiagen, Valencia, CA). Reverse transcription reactions contained 0.5 µg of RNA and were performed as described [10]. A negative control without RT did not produce product. Quantitative real-time RT-PCR was performed for RXRα, RXRβ, and 7S rRNA as a control as published [34]. The cycle threshold values were normalized to 7S rRNA and compared to the normalized value for human adipose tissue.
Electrophoretic mobility shift assay (EMSA)
Gel shift assay of supernatants from unactivated or activated human platelets and PMP lysates was carried out as previously described [13]. The EMSA consensus sequence used for PPARγ was (5’CAAAACTAGGTCAAAGGTCA-3’).
PPARγ Activity Assay
Samples for the PPARγ activity assay were prepared as described for the EMSA. Specific PPARγ DNA-binding was assessed in platelet supernatants and PMP lysate (5 µg) using a TransAM PPARγ activity assay kit (Active Motif) as described [13]. PMP lysates were untreated or pretreated with PPARγ agonist, rosiglitazone −20 µM for 30 mins. at 37°C prior to DNA-binding.
Platelet Microparticle (PMP) Isolation
PMP isolation was previously described by Heijnen et. al. [35]. Final samples of washed platelet suspension contained 2–4×109 cells. The PMP pellet and PMP-poor supernatant were analyzed by Western blot or the pellet was resuspended in 1% paraformaldehyde (PFA) containing 5 mM EDTA for microscopic studies. Greater than 90% of the isolated particles were <1 µm in size determined by light microscopy.
Immunofluorescence
Platelets applied to poly-L-lysine coated polystyrene chamber slides (BD Biosciences) were fixed in 2% PFA and blocked with normal goat serum. Platelets were incubated with a PPARγ antibody (BIOMOL) in 0.005% Triton® X-100. PMPs were incubated in PBS containing either a chicken anti-PPARγ (Novus Biologicals) or anti-RXR antibodies. After washing, a goat anti-rabbit-FITC (Jackson ImmunoResearch)-donkey anti-chicken-FITC (Genway Biotech, Inc), or goat anti-rabbit allophycocyanin (APC) (Santa Cruz) were used. Normal rabbit serum served as a control for polyclonal antibodies or secondary antibody only for the chicken antibody. Following fixation, THP-1 cells were and labeled with a mouse anti-PKCa (BD Bioscience) (in 0.05% triton X-100, 1:250, RT, 1 hour). After washing, cells were labeled with anti-mouse IgG conjugated to biotin (1hr in triton/ RT) and then washed. Finally, strepavidin conjugated to APC was added (1 hr in triton/ RT). Slides were treated with Vectashield mounting medium (Vector Laboratories, Burlingame, CA). Images were acquired using an Olympus BX51 light microscope (Olympus, Melville, NY), photographed with a SPOT camera and analyzed with SPOT RT software (New Hyde Park, NY).
Live Imaging Microscopy
Live images were acquired using CytoViva technology (Aetos Technologies, Inc., Auburn, Alabama). This is a high-resolution, optical illumination microscopy that provides resolving power below 100nm and allows imaging of live samples in real time.
PPARγ Uptake Studies
A promonocytic cell line, THP-1, was cultured as described for Meg-01. 5×105 cells/0.5 mL were plated in 12-well culture plates with fresh medium. Isolated PMPs, derived from 1×109 platelets, were labeled with a PPARγ-FITC antibody, and added to THP-1 cell culture and incubated (37°C/1 hour). PMP labeled with secondary antibody only served as a negative control. THP-1 cells were centrifuged and washed twice in PBS. A fraction of the cells were fixed in 1% PFA and prepared for microscopy. Serial images were captured using a Leica TCS SP Spectral Confocal microscope (Leica, Heidelberg, Germany), photographed with a SPOT digital camera and analyzed with Image-Pro Plus v.3 software. Remaining THP-1 cells were analyzed by Western blot for PPARγ (chicken anti-PPARγ) followed by donkey anti-chicken-HRP (Jackson Immunologicals), and mouse monoclonal anti-actin followed by goat anti-mouse-HRP (both from Oncogene research products). Transcellular studies were carried out as described above for uptake. PPARγ agonists (rosiglitazone, 20 µM) and PMP were added to THP-1 cells and coincubated for an hour. The cells were harvested as described above.
Statistical Analysis
Experiments were repeated from a minimum of three individuals and as many as 20 individuals except where stated. Differences between means were evaluated by one-way ANOVA. A value of less than 0.05 was considered statistically significant.
Results
Human platelets contain PPARγ1, but not PPARγ2 protein
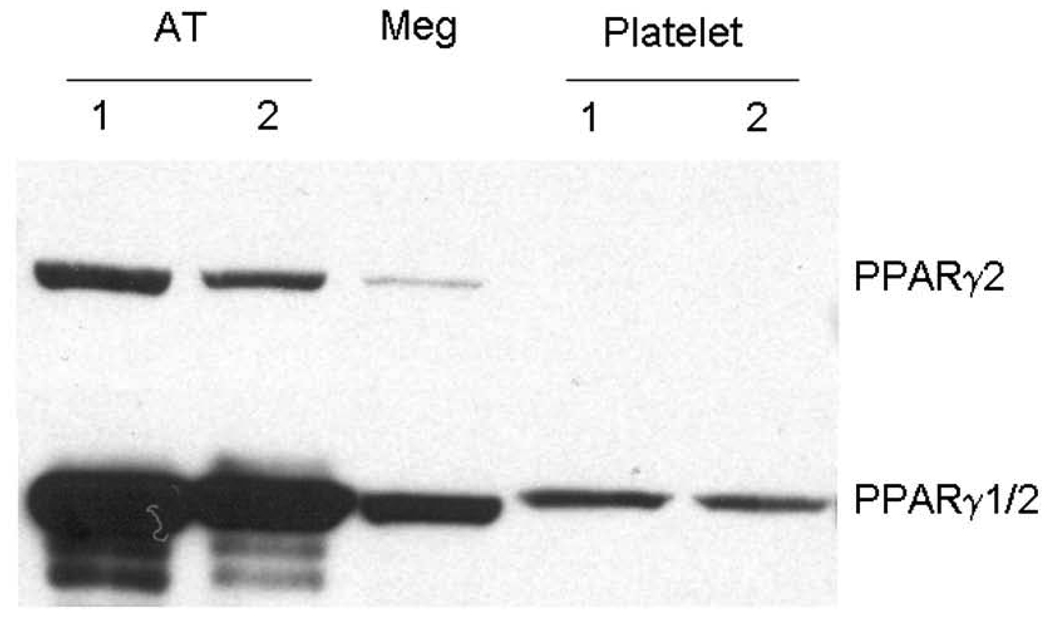
We previously demonstrated that human platelets and megakaryocytes express PPARγ [13]. To determine PPARγ isoform expression (γ1 and/or γ2) in platelets and Meg-01 cells, a Western blot was preformed using a monoclonal antibody specific for the unique amino-terminus of PPARγ2 (Figure 1). The human megakaryoblast cell line, Meg-01, expressed low levels of PPARγ2. In contrast, washed human platelets did not express detectable PPARγ2. All samples contained PPARγ as determined using an antibody that recognizes the common region of PPARγ1 and PPARγ2. Purified platelets from six individual donors were all negative for PPARγ2.
Figure 1. Human platelets contain only PPARγ1 protein.
A) Western blot analysis for PPARγ2 (top) reveals Meg-01 cells do contain PPARγ2 protein. Platelets from two individuals are negative for PPARγ2. Adipose tissue (AT) was used as a positive control. The bottom blot depicts total PPARγ (both PPARγ1 and 2) in the same samples. (10 µg protein/lane).
Human platelets release PPARγ upon activation
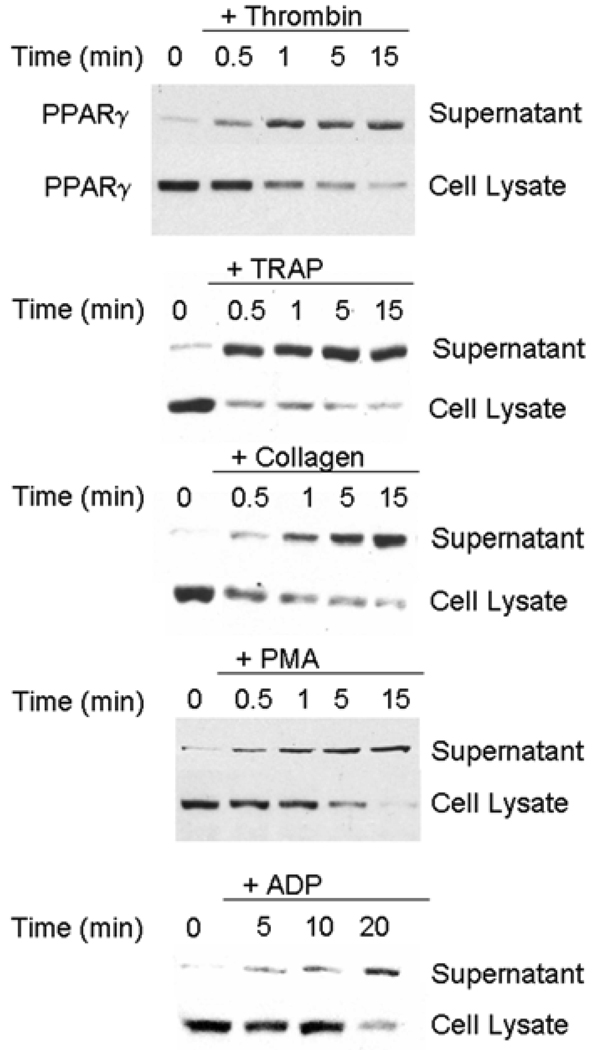
We next investigated PPARγ’s fate in activated platelets. Interestingly, platelet PPARγ levels decreased following thrombin activation (Figure 2), suggesting PPARγ was released. Indeed, abundant PPARγ protein was detectable in thrombin-activated platelet supernatants as early as 30 seconds after activation. Conversely, PPARγ was detected within unactivated platelets, while corresponding supernatants contained little PPARγ. In addition to thrombin activation, TRAP, collagen, and PMA all induced PPARγ release from platelets within 30 seconds (Figure 2). PPARγ release was seen with the weak platelet agonist ADP; however PPARγ was not detectable in supernatants until 5 minutes after exposure (Figure 2).
Figure 2. PPARγ is released from activated platelets.
Platelets were activated with thrombin, TRAP, collagen, PMA, or ADP for the indicated times and a Western blot for PPARγ was performed on supernatants and cell lysates (equal volume). Results are representative of at least 10 experiments.
Latrunculin A blocks PPARγ release
PPARγ release was blocked by Latrunculin A (Lat A), which interferes with actin cytoskeletal reorganization [36]. Flaumenhaft et. al recently demonstrated that exposure to Lat A inhibits α-granule release [36]. To ascertain PPARγ release kinetics, platelets were exposed to Lat A prior to PMA treatment, and cell-free supernatants examined for PPARγ. PMA treatment of platelets (Figure 3) strongly released PPARγ compared to untreated samples. Pre-treatment of platelets with Lat A (200 µM) substantially inhibited PPARγ release suggesting the protein may be localized to a-granules (Figure 3). Further morphologic and release rate data will be necessary to pinpoint the subcellular location of PPARγ.
Figure 3. Platelet PPARγ release is inhibited by Lat A.
Platelets were pre-incubated with the actin cytoskeleton inhibitor Lat A for 20 min and then activated with PMA. Cell-free supernatants were examined by Western blot for PPARγ (equal volume).
Human platelets express the PPARγ binding partner RXR
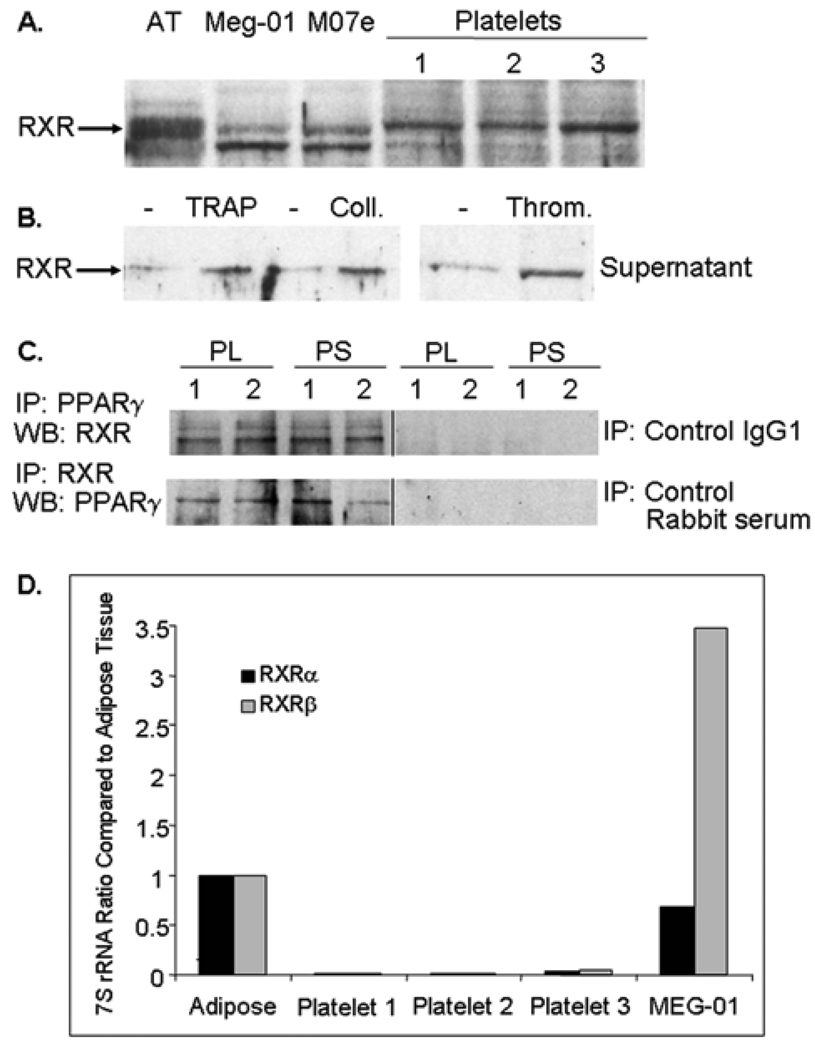
In nucleated cells, PPARγ forms a heterodimer with the Retinoid X Receptor (RXR) during transcription. We investigated in human platelets RXR expression, utilizing an antibody recognizing all forms of human RXR (RXRα, RXRβ, and RXRγ). Figure 4A shows that platelets purified from 3 individuals and 2 human megakaryocyte cell lines, Meg-01 and M-07e, express RXR protein. The two bands apparent in some lanes most likely represent different RXR forms. Platelets activated with TRAP, collagen, or thrombin all released RXR (Figure 4B). Co-IP studies were performed using an anti-PPARγ antibody and Western blotting for RXR and by the reciprocal experiment using an anti-RXR antibody for IP and Western blotting for PPARγ. Figure 4C demonstrates that PPARγ and RXR co-IP in both platelet lysates and TRAP-activated platelet supernatants. Since platelets contain some mRNAs [37], platelets and Meg-01 cells were evaluated for the presence of RXR mRNA. Real-time PCR was performed for RXRα and RXRβ, and levels were compared to adipose tissue which contains abundant RXR mRNA (Figure 4D). Platelets express 7S rRNA, which was used as a control for normalization; however, compared to adipose tissue and Meg-01 cells, which express RXRa and RXRβ RNA, platelets had little to no detectable product for either form of RXR.
Figure 4. Human platelets express RXR protein and release RXR upon activation.
(A) Platelets, and Meg-01 and M-07e human megakaryocyte cell lines were analyzed for RXR protein expression (10 µg/lane). (B) Platelets were unactivated or activated with TRAP, collagen, or thrombin (5 min) and cell-free supernatants analyzed for RXR protein. (C) PPARγ and RXR immunoprecipitations were performed using unactivated platelet lysates (PL), and supernatants (PS) from TRAP-activated platelets. IP samples contained 50 µg of total protein and an antibody to either PPARγ or RXR. The PPARγ IP was blotted for RXR and the RXR IP was blotted for PPARγ. Control antibody IPs were negative for RXR and PPARγ. (D) Real-time RT-PCR was performed for RXRα and RXRβ RNA isolated from platelets, Meg-01 cells, and human adipose tissue. The cycle threshold ratio of RXR to 7S rRNA was calculated and compared to adipose tissue (set at a ratio of 1).
PPARγ released from human platelets retains DNA-binding activity
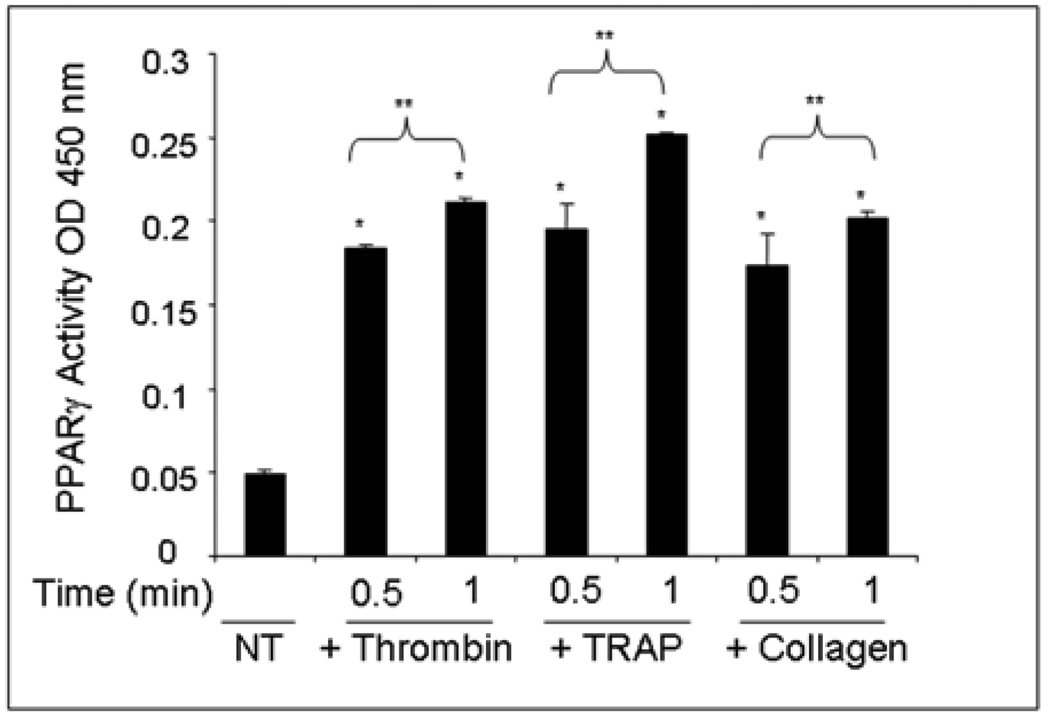
To examine PPARγ release further and to detect whether intact DNA-binding complexes of PPARγ were expelled, the TransAM PPARγ activity assay (Figure 5) and gel shift assay (EMSA) (data not shown) were used. Platelets were unactivated or activated with thrombin, TRAP, or collagen for 0.5 min and 1 min at which time cell-free supernatants were collected. The PPARγ activity assay utilizes a plate-bound PPRE-DNA oligonucleotide and a secondary incubation step with a PPARγ antibody. Increased binding to the DNA probe was observed in platelet supernatants with all three platelet activators as compared to the untreated control (Figure 5). Additionally, supernatants were incubated with a radiolabeled PPRE, and DNA-binding was assessed by EMSA. The EMSA results confirmed the PPARγ activity assay results, showing an increase in PPARγ DNA-binding in supernatants from activated platelets. A cold competitor (CC) control was performed for binding specificity using the 0.5 min TRAP sample and showed no shifted band.
Figure 5. The PPARγ released from human platelets retains DNA-binding activity.
Platelets were unactivated (NT = no treatment) or activated with thrombin, TRAP, or collagen (0.5 or 1 min). A PPARγ activity assay was performed using platelet supernatants. PPARγ was detected using a PPARγ antibody. The results are graphed as the OD value at 450 nm and are the average of duplicate samples. Error bars represent the standard deviation from the mean. The PPARγ activity released from the treated versus untreated platelets was significantly greater (*p<0.0001) for all three agonists at both 0.5 minute and 1.0 minute time points. The PPARγ activity released was significantly greater for each agonist at 1.0 minute than at 0.5 minute at (**p<0.05) for all three agonists.
PMPs from activated platelets contain PPARγ and RXR
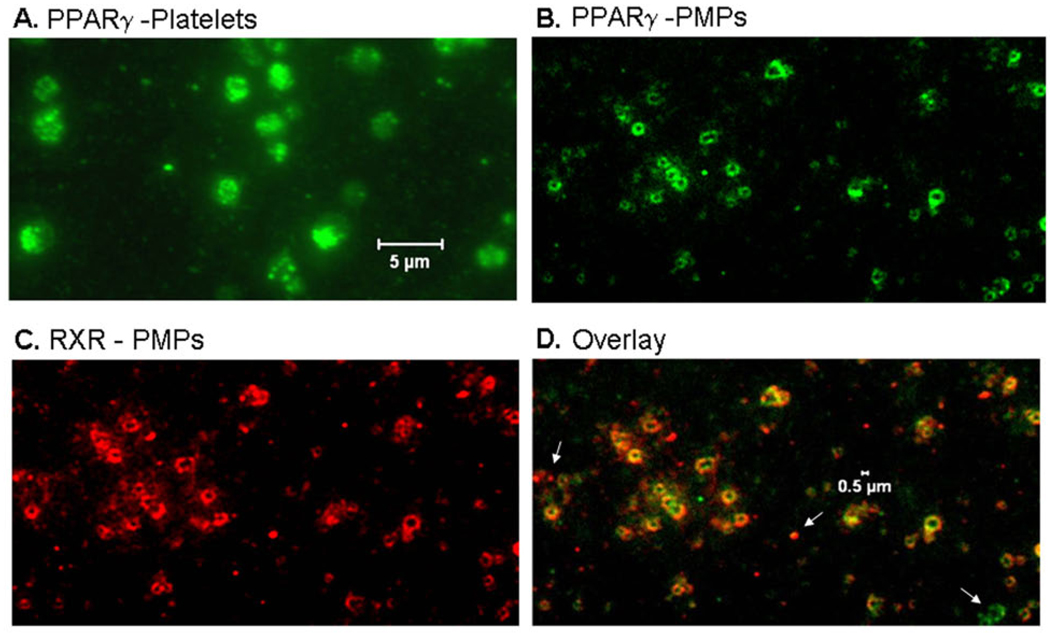
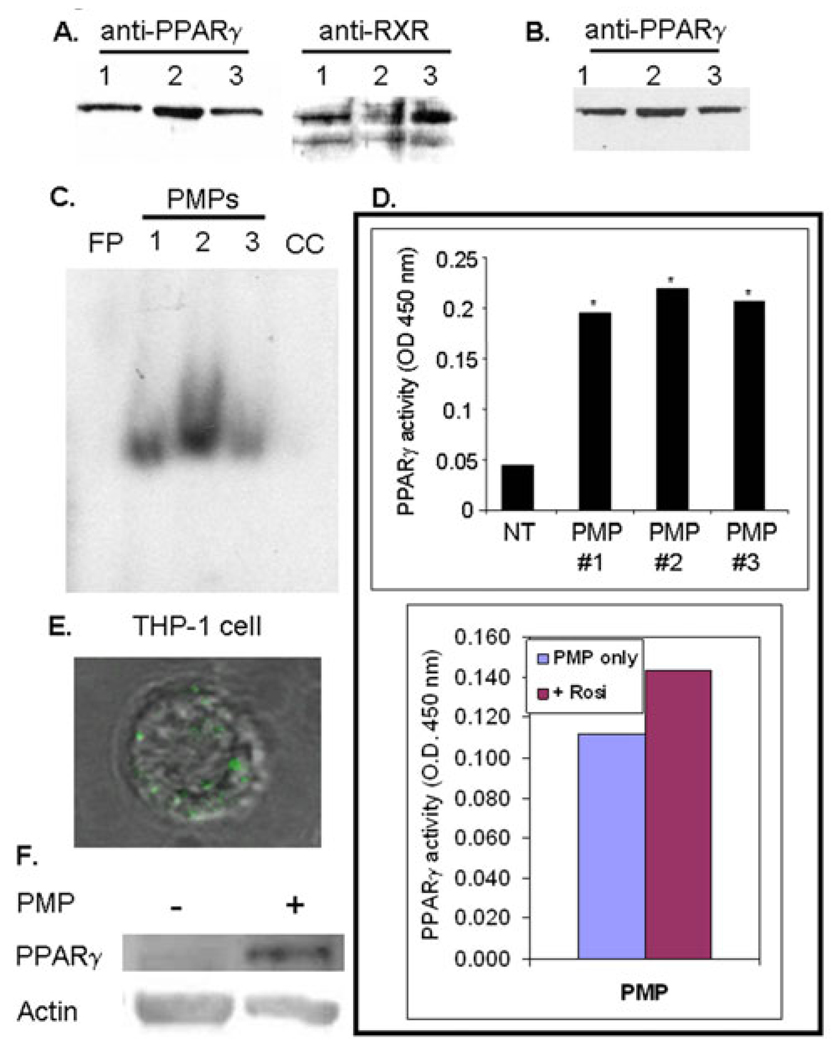
To determine whether platelet-released PPARγ was contained in PMPs, activated platelets and purified PMPs were labeled with a PPARγ antibody and visualized by fluorescence microscopy (Figure 6). A substantial portion of platelet PPARγ appeared to be compartmentalized in granules (Figure 6A). The staining clearly demonstrates that PMPs contain PPARγ protein (Figure 6B). In addition, PMPs contain RXR as depicted in the immunofluorescence staining in Figure 6C. An overlay of PPARγ and RXR staining in microparticles revealed most PMPs contain both PPARγ and RXR, although some PMPs are not double stained (Figure 6D), white arrows). Purified PMPs were also lysed and analyzed by Western blot for PPARγ and RXR (Figure 7A), and were positive for both proteins. The supernatant from the PMP isolation (PMP-poor fraction) was analyzed for PPARγ (Figure 7B) and showed there is PPARγ in PMP-poor fractions, suggesting that not all PPARγ is contained in PMPs. Since PMPs contained both PPARγ and RXR, an EMSA for PPARγ DNA-binding activity was performed on PMP lysates (Figure 7C). The PMP PPARγ bound to the PPRE DNA element as indicated by the shifted band. Additionally, PMPs isolated from TRAP-activated platelets were positive for PPARγ activity using the TransAM PPARγ assay (Figure 7D, top graph). These results argue that PMP PPARγ is already bound by an endogenous ligand. To assess the extent of PMP PPARγ DNA-binding, we used the TransAM PPARγ assay to measure whether DNA-binding could be augmented or inhibited following treatment with the PPARγ agonist rosiglitazone, or the antagonist, GW9662, respectively. Our results are representative of three individual experiments, but shown for one individual donor in Figure 7D (bottom graph) and demonstrates that in the presence of rosiglitazone, there is only a modest increase in binding activity in the PPARγ agonist treated PMP compared to untreated. We previously showed that PPARγ derived from platelet lysates also binds DNA without treatment with PPARγ agonists, but binds 3- to 4-fold more strongly in the presence of PPARγ agonists [13]. We hypothesize that the marginal increase in PMP PPARγ DNA-binding following treatment with rosiglitazone is consistent with the idea that PPARγ in PMPs is already bound to an endogenous ligand, or to a ligand and co-activator. Such associations would stabilize the PPARγ/RXR complex. Moreover, the addition of the antagonist GW9662 alone does not significantly block PMP PPARγ binding to DNA (data not shown). GW9662 acts by potently inhibiting PPARγ binding activity via covalent modification of a cysteine residue in the ligand-binding site of PPARγ and may destabilize PPARγ conformation [38]. Clearly, future studies are required to understand the mechanism(s) that governs PMP PPARγ DNA-binding and function.
Figure 6. Microparticles released by activated human platelets contain PPARγ and RXR.
Platelets were activated with TRAP and PMPs isolated. Both the platelet (A) and PMP (B) fractions were labeled with an anti-PPARγ antibody and visualized by fluorescence microscopy (final magnification 1000X). (B) Immunofluorescence labeling of PMPs with anti-PPARγ (green) or (C) anti-RXR (red) antibodies indicates that PMPs co-express PPARγ and RXR (overlay) (D) (final magnification 1000X). White arrows indicate PMPs that are single positive for either PPARγ or RXR.
Figure 7. PMP-derived PPARγ retains DNA-binding activity.
(A) Purified PMPs were analyzed for PPARγ and RXR protein expression (10 µg/lane). (B) PMPs were pelleted by centrifugation and the PMP-poor supernatant analyzed for PPARγ (5 µg protein/lane). Each lane in (A) and (B) represents a different donor (1–3). (C) An EMSA was performed using PMP lysate incubated with radiolabeled probe containing the PPAR DNA consensus sequence. FP-free probe, CC-PMP sample #3 incubated with cold competitor. (D) Top graph: A PPARγ activity assay was performed on isolated PMPs. DNA-bound PPARγ was detected using a PPARγ antibody. The results are graphed as the OD value at 450 nm. The platelet microparticle preparations from treated versus a no treatment control (NT) had PPARγ activity that was greater at *p < 0.0001 for each of the three experiments. Bottom graph: PPARγ activity was repeated on PMPs in the presence of the PPARγ agonist, rosiglitazone (Rosi, 20 µM), to detect augmentation of DNA-binding. (E) PMPs containing fluorescently labeled PPARγ were incubated with THP-1 cells. PPARγ-FITC labeled PMP are internalized as represented in an overlay of corresponding fluorescence and brightfield confocal images (final magnification 1000X). (F) THP-1 cells were incubated with or without PMP, and analyzed for PPARγ protein and actin.
Platelet-released PPARγ is transferred to THP-1 cells
Microparticles have been reported to be taken up by other cells such as macrophages [23]. Whether PPARγ-containing PMPs could be taken up by THP-1 cells was tested using PPARγ-FITC labeled PMPs. Our results show microparticles containing PPARγ are transferred to THP-1 cells as demonstrated by the appearance of FITC label in optical serial sections of THP-1 cells (Figure 7E). In support of the fluorescence data, PPARγ protein levels were measured by Western blot in THP-1 cells incubated with and without PMPs for 1 hour. Although THP-1 cells contain endogenous PPARγ, PPARγ expression in these cells is marginal until they are activated to induce upregulation of PPARγ protein [39]. The short 1 hour incubation was adequate for cells to take up PPARγ-containing PMPs, but not long enough for THP-1 cells to possibly be stimulated to make their own PPARγ. Figure 7F demonstrates PPARγ levels were increased in cells incubated with PMPs. Densitometry measurements (n=3) indicate an average 2.5 +/− 0.7 fold increase in PPARγ protein levels in PMP treated THP-1 cells compared to no treatment.
Transcellular Effects of PMP-derived PPARγ in THP-1 cells
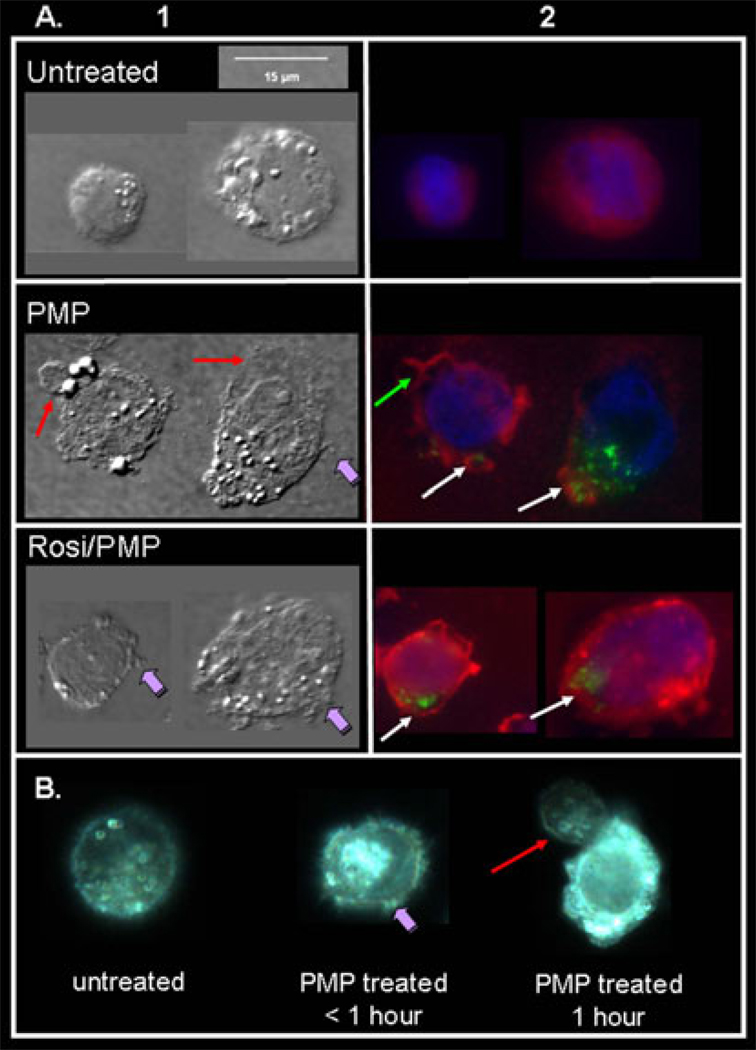
Recently, von Knethen et al reported that sequestration of PKCα in the cytosol leads to desensitization of monocytes/macrophages during sepsis [39]. During activation of the PPARγ expressing macrophage cell line, RAW 264.7, PKCα is translocated from the cytosol to the plasma membrane where it is subsequently depleted. In the presence of the synthetic PPARγ agonist, rosiglitazone, von Knethen and colleagues demonstrated that PKCα translocation was abrogated by direct protein/protein interaction with PPARγ1 [39]. As reported herein, platelets contain the PPARγ1 isoform only. To investigate whether internalized PMP PPARγ may elicit a similar function, we co-incubated THP-1 cells (a low PPARγ expressing cell line) for 1 hour with PMPs fluorescently labeled for PPARγ1 and rosiglitazone (Figure 8), and examined cell morphology and PMP PPARγ localization using both reflected light differential interference contrast (DIC) and fluorescence microscopy. Our data demonstrate that PMP treatment alone strongly activates THP-1 cells (Figure 8A; Column1, compare untreated and PMP), as evidenced by the formation of pseudopodia (purple arrow), lamellae (red arrows) and a general flattening of the cells when compared to untreated cells that remain more rounded. In the presence of rosiglitazone alone, the cells appeared similar to untreated cells (data not shown). Conversely, co-incubation of both rosiglitazone and PMPs demonstrates a dampening of THP-1 activation in cells that take up PPARγ-labeled PMPs (Figure 8; column 1, Rosi/PMP). These cells appear to have some features of both unactivated and activated cells, yet the observed cellular changes do not fully resemble that of PMP activated THP-1 cells. For example, the Rosi/PMP treated cells have some pseudopodia formation (purple arrows), but they are not as flat as the PMP treated cells and have not formed lamellae-type structures. Fixed and permeabilized THP-1 cells were also stained for PKCα and the nucleus counterstained to investigate PMP PPARγ and PKCα localization. Consistent with the changes in cellular morphology, cells activated with platelet-derived PMPs exhibit membrane localization of PKCα (Figure 8A; Column 2, green arrow) as well as reduced PKCα fluorescence suggesting depletion of PKCα protein in these cells (Figure 8A; Column 2, white arrows indicate PMP PPARγ uptake). Following co-incubation of both PPARγ-containing PMPs and rosiglitazone, THP-1 cells that contain fluorescent PMPs retain more PKCα fluorescence that is dispersed mainly throughout the cytosol, suggesting a PPARγ dependent transcellular attenuation of THP-1 activation (Figure 8A; Column 2, Rosi/PMP). Further investigation is required to determine the mechanism of this transcellular attenuation of THP-1 activation and to specifically localize PMP PPARγ.
Figure 8. PMP-derived PPARγ dampens THP-1 cell activation in the presence of Rosiglitazone.
THP-1 cells (1×106 cells) were incubated with and without purified PMPs from 1×109 platelets and labeled for PPARγ (green). Cells were thoroughly washed and applied to chamber slides. Fixed and permeabilized cells were stained with anti-PKCa (red). Cell nuclei were counterstained for DAPI (blue). (A) Reflected light differential interference contrast (DIC) microscopy was used to reveal changes in cellular morphology as shown in Column 1 following various treatments. THP-1 cells are activated by PMP treatment. The overlay in Column 2 demonstrates PMP PPARγ and PKCa localization with respect to the nucleus and cytosol. THP-1 cell activation is attenuated in the presence of both PMP PPARγ and rosiglitazone when compared to PMP alone as evidenced by cell morphology (Column 1) and cytosolic retention of PKCa in PMP containing cells (Column 2). The data is representative of three individual experiments.in the presence of rosiglitazone (Rosi, 20 µM) and PMPs. (B) Live images of THP-1 cells untreated or incubated with PMPs (< 1 hour or 1 hour) show changes in cell morphology following activation.
We are currently developing novel techniques via live imaging of cells in culture that allow us to study biological processes in real time (Figure 8B). Live imaging of THP-1 cells clearly show changes in the plasma membrane (PM) upon addition of PMPs (Figure 8B, compare untreated cell to PMP treated cells (<1 hour and 1 hour)), such as the dynamic formation of pseudopodia (purple arrow) and lamellae-type structures (red arrow), which are evident within the first hour of treatment. Additionally, within the lamellae-type structures, there is accelerated movement of cellular contents that is not witnessed in untreated cells. This activity is consistent with cytoskeletal rearrangement as well as other changes associated with cellular activation. This new high definition microscopic technology will be used in the future to further visualize PMP activation and real time uptake of fluorescent PMP PPARγ under various conditions including PPARγ agonist treatment of cells.
Discussion
The results presented herein are the first to demonstrate that PPARγ, in particular PPARγ1, is released from human platelets in response to platelet agonists. Interestingly, human platelets also contain the PPARγ binding partner RXR, and some PPARγ is released from activated platelets as a functional heterodimer (PPARγ/RXR). The expelled PPARγ was found in platelet releasate and associated with PMPs, and both forms of released PPARγ retain functional DNA-binding capability. To our knowledge, this is the first report demonstrating release of transcription factors from activated human platelets. Transcription factors thus constitute a new class of platelet-released molecules that could have a broad range of biological effects and possibly be used as biomarkers of platelet activation.
We found that PPARγ release could be prevented by pre-treatment with Lat A, an inhibitor of cytoskeletal reorganization. This suggests PPARγ release is in part dependent on cytoskeletal changes associated with platelet activation [36]. It is known that PMP formation and release involves cytoskeletal rearrangement [40] and is necessary for α-granule release [36], suggesting PPARγ may be a constituent of α-granules. Future studies will be necessary to clearly define the subcellular localization of PPARγ.
PPARγ from both unactivated and activated platelets binds the PPAR DNA consensus sequence in the absence of added PPARγ agonists [13]. This activity can be further enhanced by the addition of natural and synthetic PPARγ agonists. Our new data show that PMP PPARγ also retains substantial DNA-binding activity in the absence of added ligand, but in contrast to platelets, this activity is only modestly enhanced by agonist addition. In fact, PMPs are generated from activated platelets, which produce an endogenous ligand that promotes strong DNA-binding. Upon activation, platelets generate and release lysophosphatidic acid (LPA), a known PPARγ ligand [11]. Moreover, Johnson et al. have shown that PPARγ /RXR heterodimer stability is achieved by ligand binding and further strengthened by co-activator association [41]. Our results suggest that PMPs from activated platelets contain stable ligand bound to PPARγ /RXR heterodimers. Therefore, we were not surprised when GW9662 did not substantially inhibit PMP PPARγ DNA-binding activity, using an in vitro assay. GW9662 is an irreversible antagonist of PPARγ. It is hypothesized that binding of GW9662 does not stabilize PPARγ to the extent an agonist does and thus, dampens its activity. While the PMP PPARγ/RXR complex released from activated platelets does bind DNA, it may also function via a non-genomic mechanism.
PPARγ was originally characterized as a “transcription factor”, but it also has non-transcriptional functions. These include anti-inflammatory properties that are partially attributed to its transrepression ability. For example, PPARγ can physically bind to NF-κB, preventing nuclear translocation or enhancing transport of NF-κB out of the nucleus [6]. Furthermore, PPARγ can be SUMOylated and indirectly inhibit NF-κB by binding to the co-repressor complex on NF- κB DNA-binding sites [42]. Recent studies also provide evidence that cytosolic localization of PPARγ1, blocks translocation of PKCα to the membrane in monocytes and macrophages mediating cellular desensitization [39]. Thus, PPARγ may bind proteins within the platelet and prevent release of bioactive mediators. It was also recently shown that the nuclear hormone receptors, RXRα and RXRβ, are contained in human platelets and that RXR receptors can bind to the G-protein, Gq, and via a nongenomic mechanism inhibit platelet activation [43]. These data support the premise that nuclear receptors may be integral players in protein-protein interactions and cellular signaling separate from their transcriptional roles.
The release of PPARγ in association with PMPs is intriguing. PMPs are an important delivery and cell signaling system in inflammatory and hemostatic processes. PMPs can signal expression of adhesion molecules [28], modulate cell to cell interactions [44], and transfer functional receptors between cell types [45]. Our studies show that PPARγ labeled PMP can be internalized by the monocytic cell line, THP-1. In support of our findings, Koppler et. al demonstrated that microparticles released from three different human cell lines were transferred to monocytes and B cells [25]. Some released PPARγ is not associated with PMPs, and may be expelled as soluble protein or contained within platelet-released exosomes, a smaller class of microparticle (40–90 nm) without a pro-coagulant function. The function of platelet-released PPARγ is not yet elucidated, but it could serve as a biomarker of platelet activation or influence cells that incorporate a PMP. It is well established that PMP stimulation of THP-1 cells induces inflammatory cytokines and cell adhesion molecules [28]. Here we provide evidence that internalization of PPARγ-containing PMPs elicits a transcellular attenuation of THP-1 cell activation in the presence of a PPARγ agonist, rosiglitazone. The mechanism of this attenuation is currently under investigation. As stated above, PPARγ activation is known to educe effects in nucleated cells via non-genomic mechanisms that include protein modification and direct protein/protein binding with key signaling pathways such as NF-κB and PKCα. It is also documented that PPARγ agonists can influence cytoskeletal rearrangement in monocytes [46]. For example, the phospholipid mediator, platelet-activating factor (PAF), is a potent inflammatory molecule and functions in THP-1 macrophages to promote actin cytoskeletal rearrangement. It has been demonstrated that the PPARγ agonist, Pioglitazone, inhibits PAF mediated changes in the macrophage cytoskeleton to down regulate inflammation [47].
Platelet activation and release of their constituents plays a central role in hemostasis, immunomodulation and inflammation. Our exciting discovery that human platelets release PPARγ and RXR expands the spectrum of proteins contained within these cells. Additionally, we provide the foundation evidence that internalized platelet PPARγ protein appears capable of biologic activity in THP-1 cells. This discovery may represent a novel mechanism of transcellular regulation. Thus, it is important to further study the mechanism of PPARγ release and its regulation. Finally, released PPARγ and other proteins could serve as markers that platelets have been activated.
Acknowledgements
Grant support: This work was supported by T32-DE07165, DE011390, HL078603, HL086367, ES01247 and an EPA Center Grant (R827354).
The authors would like to thank Kelly Gettings for help with platelet preparation.
References
- 1.Daynes RA, Jones DC. Emerging roles of PPARs in inflammation and immunity. Nat Rev Immunol. 2002;2(10):748–759. doi: 10.1038/nri912. [DOI] [PubMed] [Google Scholar]
- 2.Yanase T, Yashiro T, Takitani K, et al. Differential expression of PPAR γ1 and γ2 isoforms in human adipose tissue. Biochem Biophys Res Commun. 1997;233(2):320–324. doi: 10.1006/bbrc.1997.6446. [DOI] [PubMed] [Google Scholar]
- 3.Padilla J, Leung E, Phipps RP. Human B lymphocytes and B lymphomas express PPAR-γ and are killed by PPAR-γ agonists. Clin Immunol. 2002;103(1):22–33. doi: 10.1006/clim.2001.5181. [DOI] [PubMed] [Google Scholar]
- 4.Ricote M, Li AC, Willson TM, et al. The peroxisome proliferator-activated receptor-γ is a negative regulator of macrophage activation. Nature. 1998;391(6662):79–82. doi: 10.1038/34178. [DOI] [PubMed] [Google Scholar]
- 5.Fajas L, Auboeuf D, Raspe E, et al. The organization, promoter analysis, and expression of the human PPARγ gene. J Biol Chem. 1997;272(30):18779–18789. doi: 10.1074/jbc.272.30.18779. [DOI] [PubMed] [Google Scholar]
- 6.Kelly D, Campbell JI, King TP, et al. Commensal anaerobic gut bacteria attenuate inflammation by regulating nuclear-cytoplasmic shuttling of PPAR-γ and RelA. Nat Immunol. 2004;5(1):104–112. doi: 10.1038/ni1018. [DOI] [PubMed] [Google Scholar]
- 7.Kliewer SA, Umesono K, Noonan DJ, et al. Convergence of 9-cis retinoic acid and peroxisome proliferator signalling pathways through heterodimer formation of their receptors. Nature. 1992;358(6389):771–774. doi: 10.1038/358771a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lehmann JM, Moore LB, Smith-Oliver TA, et al. An antidiabetic thiazolidinedione is a high affinity ligand for peroxisome proliferator-activated receptor γ (PPAR γ) J Biol Chem. 1995;270(22):12953–12956. doi: 10.1074/jbc.270.22.12953. [DOI] [PubMed] [Google Scholar]
- 9.Forman BM, Tontonoz P, Chen J, et al. 15-Deoxy-Δ12,14-prostaglandin J2 is a ligand for the adipocyte determination factor PPAR γ. Cell. 1995;83(5):803–812. doi: 10.1016/0092-8674(95)90193-0. [DOI] [PubMed] [Google Scholar]
- 10.Feldon SE, O'Loughlin CW, Ray DM, et al. Activated Human T Lymphocytes Express Cyclooxygenase-2 and Produce Proadipogenic Prostaglandins that Drive Human Orbital Fibroblast Differentiation to Adipocytes. Am J Pathol. 2006;169(4):1183–1193. doi: 10.2353/ajpath.2006.060434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McIntyre TM, Pontsler AV, Silva AR, et al. Identification of an intracellular receptor for lysophosphatidic acid (LPA): LPA is a transcellular PPARγ agonist. Proc Natl Acad Sci U S A. 2003;100(1):131–136. doi: 10.1073/pnas.0135855100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Investigators TDT. Effect of rosiglitazone on the frequency of diabetes in patients with impaired glucose tolerance or impaired fasting glucose: a randomized controlled trial. doi: 10.1016/S0140-6736(06)69420-8. The Lancet Prepublished on September 15, 2006;as. [DOI] [PubMed] [Google Scholar]
- 13.Akbiyik F, Ray DM, Gettings KF, et al. Human bone marrow megakaryocytes and platelets express PPARγ, and PPARγ agonists blunt platelet release of CD40 ligand and thromboxanes. Blood. 2004;104(5):1361–1368. doi: 10.1182/blood-2004-03-0926. [DOI] [PubMed] [Google Scholar]
- 14.Wagner DD, Burger PC. Platelets in inflammation and thrombosis. Arterioscler Thromb Vasc Biol. 2003;23(12):2131–2137. doi: 10.1161/01.ATV.0000095974.95122.EC. [DOI] [PubMed] [Google Scholar]
- 15.Lambert MP, Sachais BS, Kowalska MA. Chemokines and thrombogenicity. Thromb Haemost. 2007;97(5):722–729. doi: 10.1160/th07-01-0046. [DOI] [PubMed] [Google Scholar]
- 16.von Hundelshausen P, Petersen F, Brandt E. Platelet-derived chemokines in vascular biology. Thromb Haemost. 2007;97(5):704–713. doi: 10.1160/th07-01-0066. [DOI] [PubMed] [Google Scholar]
- 17.Henn V, Slupsky JR, Grafe M, et al. CD40 ligand on activated platelets triggers an inflammatory reaction of endothelial cells. Nature. 1998;391(6667):591–594. doi: 10.1038/35393. [DOI] [PubMed] [Google Scholar]
- 18.Tan KT, Lip GY. The potential role of platelet microparticles in atherosclerosis. Thromb Haemost. 2005;94(3):488–492. doi: 10.1160/TH05-03-0201. [DOI] [PubMed] [Google Scholar]
- 19.Blann AD, Tan KT, Tayebjee MH, et al. Soluble CD40L in peripheral artery disease. Relationship with disease severity, platelet markers and the effects of angioplasty. Thromb Haemost. 2005;93(3):578–583. doi: 10.1160/TH04-09-0586. [DOI] [PubMed] [Google Scholar]
- 20.Phipps RP. Atherosclerosis: the emerging role of inflammation and the CD40-CD40 ligand system. Proc Natl Acad Sci U S A. 2000;97(13):6930–6932. doi: 10.1073/pnas.97.13.6930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jackson SP, Schoenwaelder SM. Antiplatelet therapy: in search of the 'magic bullet'. Nat Rev Drug Discov. 2003;2(10):775–789. doi: 10.1038/nrd1198. [DOI] [PubMed] [Google Scholar]
- 22.Bode AP, Sandberg H, Dombrose FA, et al. Association of factor V activity with membranous vesicles released from human platelets: requirement for platelet stimulation. Thromb Res. 1985;39(1):49–61. doi: 10.1016/0049-3848(85)90121-5. [DOI] [PubMed] [Google Scholar]
- 23.Horstman LL, Jy W, Jimenez JJ, et al. New horizons in the analysis of circulating cell-derived microparticles. Keio J Med. 2004;53(4):210–230. doi: 10.2302/kjm.53.210. [DOI] [PubMed] [Google Scholar]
- 24.Weyrich AS, Lindemann S, Zimmerman GA. The evolving role of platelets in inflammation. J Thromb Haemost. 2003;1(9):1897–1905. doi: 10.1046/j.1538-7836.2003.00304.x. [DOI] [PubMed] [Google Scholar]
- 25.Koppler B, Cohen C, Schlondorff D, et al. Differential mechanisms of microparticle transfer to B cells and monocytes: anti-inflammatory properties of microparticles. Eur J Immunol. 2006;36(3):648–660. doi: 10.1002/eji.200535435. [DOI] [PubMed] [Google Scholar]
- 26.Sims PJ, Wiedmer T, Esmon CT, et al. Assembly of the platelet prothrombinase complex is linked to vesiculation of the platelet plasma membrane. Studies in Scott syndrome: an isolated defect in platelet procoagulant activity. J Biol Chem. 1991;264(29):17049–17057. [PubMed] [Google Scholar]
- 27.Tans G, Rosing J, Thomassen MC, et al. Comparison of anticoagulant and procoagulant activities of stimulated platelets and platelet-derived microparticles. Blood. 1991;77(12):2641–2648. [PubMed] [Google Scholar]
- 28.Nomura S, Tandon NN, Nakamura T, et al. High-shear-stress-induced activation of platelets and microparticles enhances expression of cell adhesion molecules in THP-1 and endothelial cells. Atherosclerosis. 2001;158(2):277–287. doi: 10.1016/s0021-9150(01)00433-6. [DOI] [PubMed] [Google Scholar]
- 29.Diamant M, Nieuwland R, Pablo RF, et al. Elevated numbers of tissue-factor exposing microparticles correlate with components of the metabolic syndrome in uncomplicated type 2 diabetes mellitus. Circulation. 2002;106(19):2442–2447. doi: 10.1161/01.cir.0000036596.59665.c6. [DOI] [PubMed] [Google Scholar]
- 30.Janowska-Wiexzorek AKJ, Marquez LA, Ratajczak J, Ratajczak MZ. Microvesicles derived from activated platelets: an under-appreciated modulator of the metastatic potential of tumor cells. Blood. 2003;102(11) 57b #3998. [Google Scholar]
- 31.Ogura M, Morishima Y, Ohno R, et al. Establishment of a novel human megakaryoblastic leukemia cell line, MEG-01, with positive Philadelphia chromosome. Blood. 1985;66(6):1384–1392. [PubMed] [Google Scholar]
- 32.Avanzi GC, Brizzi MF, Giannotti J, et al. M-07e human leukemic factor-dependent cell line provides a rapid and sensitive bioassay for the human cytokines GM-CSF and IL-3. J Cell Physiol. 1990;145(3):458–464. doi: 10.1002/jcp.1041450310. [DOI] [PubMed] [Google Scholar]
- 33.Ray DM, Bernstein SH, Phipps RP. Human multiple myeloma cells express peroxisome proliferator-activated receptor γ and undergo apoptosis upon exposure to PPARγ ligands. Clin Immunol. 2004;113(2):203–213. doi: 10.1016/j.clim.2004.06.011. [DOI] [PubMed] [Google Scholar]
- 34.Kaiser PC, Korner M, Kappeler A, et al. Retinoid receptors in ovarian cancer: expression and prognosis. Ann Oncol. 2005;16(9):1477–1487. doi: 10.1093/annonc/mdi265. [DOI] [PubMed] [Google Scholar]
- 35.Heijnen HF, Schiel AE, Fijnheer R, et al. Activated platelets release two types of membrane vesicles: microvesicles by surface shedding and exosomes derived from exocytosis of multivesicular bodies and alpha-granules. Blood. 1999;94(11):3791–3799. [PubMed] [Google Scholar]
- 36.Flaumenhaft R, Dilks JR, Rozenvayn N, et al. The actin cytoskeleton differentially regulates platelet alpha-granule and dense-granule secretion. Blood. 2005;105(10):3879–3887. doi: 10.1182/blood-2004-04-1392. [DOI] [PubMed] [Google Scholar]
- 37.Gnatenko DV, Dunn JJ, McCorkle SR, et al. Transcript profiling of human platelets using microarray and serial analysis of gene expression. Blood. 2003;101(6):2285–2293. doi: 10.1182/blood-2002-09-2797. [DOI] [PubMed] [Google Scholar]
- 38.Leesnitzer LM, Parks DJ, Bledsoe RK, et al. Functional consequences of cysteine modification in the ligand binding sites of peroxisome proliferator activated receptors by GW9662. Biochemistry. 2002;41(21):6640–6650. doi: 10.1021/bi0159581. [DOI] [PubMed] [Google Scholar]
- 39.von Knethen A, Soller M, Tzieply N, et al. PPARgamma1 attenuates cytosol to membrane translocation of PKCalpha to desensitize monocytes/macrophages. J Cell Biol. 2007;176(5):681–694. doi: 10.1083/jcb.200605038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fox JE, Austin CD, Boyles JK, et al. Role of the membrane skeleton in preventing the shedding of procoagulant-rich microvesicles from the platelet plasma membrane. J Cell Biol. 1990;111(2):483–493. doi: 10.1083/jcb.111.2.483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Johnson BA, Wilson EM, Li Y, et al. Ligand-induced stabilization of PPARgamma monitored by NMR spectroscopy: implications for nuclear receptor activation. J Mol Biol. 2000;298(2):187–194. doi: 10.1006/jmbi.2000.3636. [DOI] [PubMed] [Google Scholar]
- 42.Pascual G, Fong AL, Ogawa S, et al. A SUMOylation-dependent pathway mediates transrepression of inflammatory response genes by PPAR-γ. Nature. 2005;437(7059):759–763. doi: 10.1038/nature03988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Moraes LA, Swales KE, Wray JA, et al. Nongenomic signaling of the retinoid X receptor through binding and inhibiting Gq in human platelets. Blood. 2007;109(9):3741–3744. doi: 10.1182/blood-2006-05-022566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Scholz T, Temmler U, Krause S, et al. Transfer of tissue factor from platelets to monocytes: role of platelet-derived microvesicles and CD62P. Thromb Haemost. 2002;88(6):1033–1038. [PubMed] [Google Scholar]
- 45.Janowska-Wieczorek A, Majka M, Kijowski J, et al. Platelet-derived microparticles bind to hematopoietic stem/progenitor cells and enhance their engraftment. Blood. 2001;98(10):3143–3149. doi: 10.1182/blood.v98.10.3143. [DOI] [PubMed] [Google Scholar]
- 46.Singh N, Webb R, Adams R, et al. The PPAR-gamma activator, Rosiglitazone, inhibits actin polymerisation in monocytes: involvement of Akt and intracellular calcium. Biochem Biophys Res Commun. 2005;333(2):455–462. doi: 10.1016/j.bbrc.2005.05.127. [DOI] [PubMed] [Google Scholar]
- 47.Sumita C, Maeda M, Fujio Y, et al. Pioglitazone induces plasma platelet activating factor-acetylhydrolase and inhibits platelet activating factor-mediated cytoskeletal reorganization in macrophage. Biochim Biophys Acta. 2004;1673(3):115–121. doi: 10.1016/j.bbagen.2004.04.002. [DOI] [PubMed] [Google Scholar]