Abstract
Amelogenesis imperfecta (AI) is caused by AMEL, ENAM, MMP20 and KLK4 gene mutations. Mice lacking expression of the AmelX, Enam and Mmp20 genes have been generated. These mouse models provide tools for understanding enamel formation and AI pathogenesis. This study describes the AI phenotypes and relates them to their mouse model counterparts. Human AI phenotypes were determined in a clinical population of AI families and published cases. Human and murine teeth were evaluated using light and electron microscopy. A total of 463 individuals from 54 families were evaluated and mutations in the AMEL, ENAM and KLK4 genes were identified. The majority of human mutations for genes coding enamel nonproteinase proteins (AMEL and ENAM) resulted in variable hypoplasia ranging from local pitting to a marked, generalized enamel thinning. Specific AMEL mutations were associated with abnormal mineralization and maturation defects. Amel and Enam null murine models displayed marked enamel hypoplasia and a complete loss of prism structure. Human mutations in genes coding for the enamel proteinases (MMP20 and KLK4) cause variable degrees of hypomineralization. The murine Mmp20 null mouse exhibits both hypoplastic and hypomineralized defects. The currently available Amel and Enam mouse models for AI exhibit enamel phenotypes (hypoplastic) that are generally similar to those seen in humans. Mmp20 null mice have a greater degree of hypoplasia than humans with MMP20 mutations. Mice lacking expression of the currently known genes associated with the human AI conditions provide useful models for understanding the pathogenesis of these conditions.
Key Words: Enamel, Amelogenesis imperfecta, Mouse, Human, Gene, Mutation
Introduction
Amelogenesis imperfectas (AI) are a clinically and genetically diverse group of conditions caused by mutations in genes critical for normal enamel formation. Mutations in the AMELX, ENAM, MMP20 and KLK4 genes are associated with specific AI types having X-linked, autosomal dominant and autosomal recessive modes of inheritance [Wright, 2006]. The pathogenesis of these conditions and the developmental mechanisms leading to the specific phenotypes remain poorly understood. This is largely due to the lack of adequate cell models for studying the complexities of enamel formation. Given the uniqueness of enamel development and the cell-specific expression of many of the genes involved in tooth development in general, as well as the enamel in particular, identification and functional characterization of enamel-forming genes in humans is significantly limited. The generation of animal models provides an important resource to study normal and abnormal enamel development. Generation of mice lacking expression of genes associated with enamel formation provides a potentially useful tool for understanding biomineralization of enamel and the pathogenesis of the different AI types [Gibson et al., 2001; Caterina et al., 2002].
Amelogenin is the most abundant extracellular matrix (ECM) protein in developing enamel. Amelogenins are encoded by 2 single copy genes on chromosome Xp22.3–p22.1 and on chromosome Yp11 [Salido et al., 1992; Fincham and Simmer, 1997]. Mutations in the X chromosome amelogenin gene (AMELX) cause a variety of changes in the amelogenin protein and are associated with AI phenotypes ranging from hypoplastic to hypomineralized enamel [Wright et al., 2003]. Enamelin is a relatively low-abundance matrix protein in developing enamel and is encoded by the ENAM gene which is located within a cluster of genes critical to biomineralization on chromosome 4q21 [Hu and Yamakoshi, 2003]. Mutations in ENAM cause AI types characterized by localized pitted enamel or generalized thin enamel [Rajpar et al., 2001; Mardh et al., 2002; Hart et al., 2003a]. MMP20 and KLK4 are proteinases critical for processing the enamel matrix, thereby allowing the enamel crystallites to grow into space previously occupied by the ECM [Simmer and Hu, 2002]. The genes coding for these proteins are located on chromosomes 11q23 and 19q13, respectively, and are associated with autosomal recessive forms of AI. Abnormal proteinase activities result in hypomaturation AI that is characterized by enamel that is deficient in mineral content but is of normal enamel thickness [Hart et al., 2004; Kim et al., 2005]. Despite extensive studies as to how these proteins orchestrate the biomineralization of enamel, our knowledge of the complex processes that result in the structure and composition of enamel remains lacking. The purpose of this study was to compare and contrast the phenotypes of human enamel from individuals affected with AI to those mouse enamels that have been genetically modified by deletion of specific enamel matrix protein genes.
Methods
Human genotypes and phenotypes for AI were determined from a large clinical cohort that has been recruited to evaluate the etiology and pathogenesis of these conditions. This study was approved by the Institutional Review Board and all study participants provided informed consent prior to participation. DNA was collected from blood or saliva and candidate genes were sequenced using previously published techniques and primer sets for the AMELX, ENAM, MMP20 and KLK4 genes [Hart et al., 2002, 2003b, 2004; Kim et al., 2005]. All individuals were clinically evaluated by 1 of 2 examiners and the dentition photographed and dental radiographs taken whenever possible. Exfoliated primary or permanent teeth slated for therapeutic extraction were collected for histological analysis. The teeth were evaluated with light microscopy by cutting thin sections using a diamond blade. Samples were also cut, polished and etched or fractured for evaluation using scanning electron microscopy.
Mice lacking expression of Amelx, Enam and Mmp20 have been generated in the laboratories of several authors and have been described previously [Gibson et al., 2001; Caterina et al., 2002; Hu et al., 2008]. The teeth from these animals were examined using light and scanning electron microscopy. Sample preparation was similar to that for the human samples. The gross and histological enamel phenotypes of mouse and human teeth were compared.
Results
Families segregating for the AI trait were recruited and a total of 463 individuals from 54 families were enrolled. The families had a variety of AI types, with 18 autosomal dominant, 26 autosomal recessive and 10 X-linked traits. Coding regions including intron-exon junctions of candidate genes (AMELX, ENAM, AMTN, AMBN, MMP20 and KLK4) were priority sequenced based initially on reported phenotype-genotype relationships and if no mutation was identified, remaining candidate genes were sequenced. Mutations were identified in the AMELX, ENAM and KLK4 genes and no mutations were identified in MMP20, AMTN (amelotin) or AMBN (ameloblastin) genes.
The majority of human mutations for genes coding nonproteinase enamel proteins (AMELX and ENAM) resulted in variable degrees of hypomineralization and hypoplasia that ranged from pitting and grooves to a marked, generalized thinning of the enamel. Three different AMELX mutations were identified. Two mutations (AMELX g.3458delC and AMELX g.4046delC) were associated with enamel that was of normal or nearly normal thickness with a prismatic structure and decreased mineral content apparently due to maturation defects. The third AMELX mutation resulted in loss of the C terminus and a hypoplastic phenotype (AMELX g.4046delC; fig. 1). In addition to being markedly decreased in thickness, the enamel lacked a prismatic architecture. Females having these mutations showed a mosaic phenotype with areas of more normal enamel adjacent to regions of more affected (thin/hypomineralized) enamel. Two different ENAM mutations were identified (ENAM g.8344delG and ENAM g.13185_13186insAG, the later mutation having been reported previously [Ozdemir et al., 2005a]) and both were associated with generalized thin enamel that had a complete lack of prism structure (fig. 1). The thin layer of enamel had a rough surface and lacked any evidence of a prismatic structure. Backscatter analysis showed a laminated type of enamel pattern. Human mutations in genes coding for the enamel proteinases (MMP20 and KLK4) cause variable degrees of hypomineralization, but appear to have a normal enamel thickness based on radiographic assessment [Hart et al., 2004; Kim et al., 2005; Ozdemir et al., 2005b]. No MMP20 mutations were identified in this study population and there have been no histological evaluations reported of MMP20 enamel.
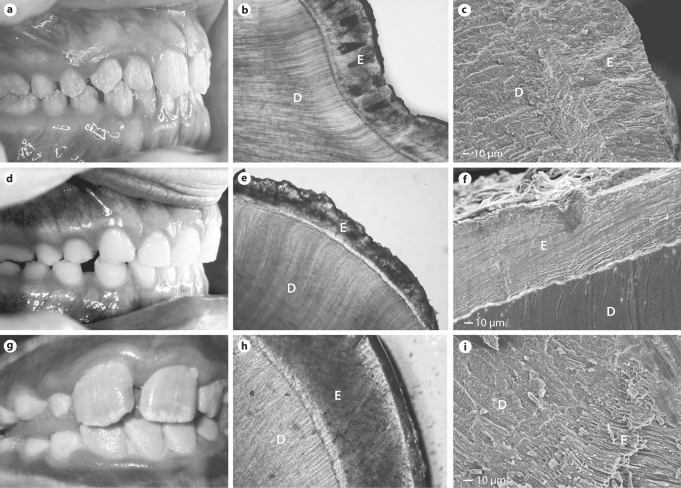
Fig. 1.
Highly diverse clinical appearances of the human dentition result from AMELX g.4046delC (a, female dentition), ENAM g.8344delG (d) and KLK4 g.2142G>A mutations (g). The dentin (D) appears normal, while the enamel (E) affected by these different mutations shows varying degrees of opacity and hypoplasia as seen with light microscopy (b, AMELX;e, ENAM;h, KLK4). These different mutations also have markedly different effects that frequently disrupt the normal prismatic structure in the AMELX (c) and ENAM (f) enamel but not in the KLK4 AI enamel (i) as seen with scanning electron microscopy.
Murine KO models involving Amel and Enam genes are both associated with marked enamel hypoplasia and a complete loss of prism structure. The mandibular incisors in both these models show a loss of the typical yellow brown coloration seen in the wild-type mice (fig. 2). The enamel surface in both the AmelX and Enam null mice is rough compared with the wild-type enamel. The AmelX null mouse enamel is reduced from approximately 100 to 10 μm and shows no prismatic structure. The Enam null mouse enamel is only a few micrometers thick and shows no organization into prism structure (fig. 3). The Mmp20 null mouse exhibits both hypoplastic and hypomineralized defects and shows areas of enamel loss [Caterina et al., 2002].
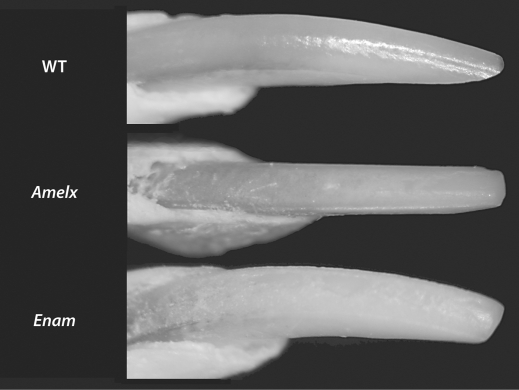
Fig. 2.
Wild-type mice (WT) typically have smooth enamel with a yellowish-brown coloration. Mice null for Amelx and Enam show a roughened surface, abnormal wear on the incisal edge and a white opaque appearance.
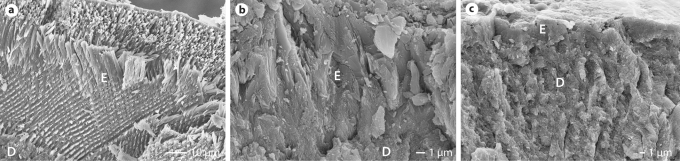
Fig. 3.
Scanning electron microscopy reveals the typical prismatic enamel (E) architecture in the wild-type mouse (a) and the complete loss of prisms in both the Amelx null (b) and Enam null (c) mice. Similar to humans, the dentin (D) appears structurally normal.
Discussion
The enamel phenotypes associated with AI in humans are diverse and range from a decrease in the amount of enamel to enamel that is of normal thickness but lacks the high level of mineralization seen in normal enamel. Both phenotypes have been reproduced in mouse models that have been generated to perturb the normal expression of essential ECM proteins involved in amelogenesis [Gibson et al., 2001; Seedorf et al., 2007]. The AI-associated enamel phenotypes in humans and mice appear to differ depending on whether the mutation/knockout involves genes encoding for an ECM protein (for example, amelogenin or enamelin) versus the ECM proteinases (MMP20 and KLK4). The currently available Amelx and Enam mouse models for AI exhibit enamel phenotypes (hypoplastic) that are generally similar to those seen in humans. There is a loss of discernable prismatic architecture in both humans having mutations that causes a loss of amelogenin (signal peptide mutations) or of the C terminus [Wright et al., 2003]. Similarly, the mice lacking amelogenin show no evidence of a prism structure [Gibson et al., 2001]. Humans having ENAM mutations in the present study showed generalized thin enamel with a roughened surface similar to the Enam null mouse. The developmental mechanism in humans with ENAM mutations that cause localized pitted hypoplastic defects likely differ from that causing generalized hypoplasia in the Enam null mice [Mardh et al., 2002; Hart et al., 2003b; Ozdemir et al., 2005a]. Humans having ENAM mutations that cause haploinsufficiency are associated with localized hypoplasia, whereas those having generalized hypoplasia are thought to represent a dominant negative effect. The Enam null mouse more closely resembles the latter defect, but the lack of a laminated enamel appearance could be due to the differences in developmental mechanisms between the complete loss of protein (Enam null mouse) and the dominant negative mechanism in humans. Mice lacking expression of Mmp20 have a greater degree of hypoplasia coupled with hypomineralization, while humans with MMP20 mutations tend to exhibit primarily hypomineralization defects. However, it is possible that humans with MMP20 mutations could have varying levels of enamel hypoplasia, as to our knowledge there have been no published histological studies of teeth from individuals with a known MMP20 mutation. While reasonably detailed analysis of human enamel affected by a KLK4 mutation has been reported, there is no mouse model to study the specific mechanism of how this autosomal recessive trait results in the enamel phenotype observed [Hart et al., 2004].
The number of known human mutations in genes coding for the enamel ECM and proteinases continues to grow. There are well over 20 mutations in 4 genes and the present and other studies suggest that at least several additional AI-associated genes will be identified in the near future. Of the 463 study participants studied from 54 families, mutations were detected in only 9 kindreds. Linkage in 2 large families with autosomal dominant hypocalcified AI was identified to the 8q24.3 locus as has been reported by others [Mendoza et al., 2007]. The present study and many recent investigations excluding known candidate genes in the AI families tested suggest that the current candidate genes account for less than 25% of the AI cases [Kim et al., 2006]. Continued clinical investigations are clearly indicated to identify new genetic loci and genes associated with AI and to characterize the variability and diversity of resulting phenotypes, so we may better understand the role of these enamel-related genes in amelogenesis.
Comparison of mouse and human pathogenesis caused by different perturbations in the genes responsible for generating the ECM of enamel shows marked similarities and some subtle differences. This may arise for several reasons. It is well known that animal models of human diseases do not always recapitulate the human disease phenotype. While many human mutations alter function of the normal protein product, most mouse models for AI candidate genes are designed to knock out the gene. Such null models cannot fully recapitulate the functional range of AI-associated human gene mutations. Additionally, the effect of possible epigenetic interactions is limited in current murine models, compared to the more diverse genetic background that exists in the human population studied to date. Development of mice that have gene changes similar to those in humans (such as point mutations and missense mutations) may provide even better models for understanding the specific mechanisms involved in particular AI subtypes [Gibson et al., 2007]. However, the current mouse models provide a powerful tool for helping understand the role of the genes and their ECM products in amelogenesis and biomineralization. This is especially true given the lack of cellular models for investigating amelogenesis and mineralization of this highly mineralized tissue.
Acknowledgements
Supported in part by NIDCR grants DE12879 (J.T.W.), DE011089 (C.W.G.), DE01627 (J.D.B.) and DE11301 (J.H.).
Abbreviations used in this paper
- AI
amelogenesis imperfecta
- ECM
extracellular matrix
References
- Caterina J.J., Skobe Z., Yanli Ding J.S., Simmer J.P., Birkedal-Hansen H., J.D. Bartlett. Enamelysin (matrix metalloproteinase 20)-deficient mice display an amelogenesis imperfecta phenotype. J Biol Chem. 2002;277:49598–49604. doi: 10.1074/jbc.M209100200. [DOI] [PubMed] [Google Scholar]
- Fincham A.G., Simmer J.P. Dental Enamel. Chichester: John Wiley & Sons; 1997. Amelogenin proteins of developing dental enamel; pp. 118–134. [DOI] [PubMed] [Google Scholar]
- Gibson C.W., Yuan Z.A., Hall B., Longenecker G., Chen E., Thyagarajan T., Sreenath T., Wright J.T., Decker S., Piddington R., Harrison G., Kulkarni A.B. Amelogenin-deficient mice display an amelogenesis imperfecta phenotype. J Biol Chem. 2001;276:31871–31875. doi: 10.1074/jbc.M104624200. [DOI] [PubMed] [Google Scholar]
- Gibson C.W., Yuan Z.A., Li Y., Daly B., Suggs C., Aragon M.A., Alawi F., Kulkarni A.B., Wright J.T. Transgenic mice that express normal and mutated amelogenins. J Dent Res. 2007;86:331–335. doi: 10.1177/154405910708600406. [DOI] [PubMed] [Google Scholar]
- Hart P.S., Aldred M.A., Crawford P.J.M., Wright N.J., Hart T.C., Wright J.T. Two amelogenin gene mutations cause different amelogenesis imperfecta phenotypes. Arch Oral Biol. 2002;47:255–260. doi: 10.1016/s0003-9969(02)00003-1. [DOI] [PubMed] [Google Scholar]
- Hart T.C., Hart P.S., Gorry M.C., Michalec M.D., Ryu O.H., Uygur C., Ozdemir D., Firatli S., Aren G., Firatli E. Novel ENAM mutation responsible for autosomal recessive amelogenesis imperfecta and localised enamel defects. J Med Genet. 2003a;40:900–906. doi: 10.1136/jmg.40.12.900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hart P.S., Michalec M.D., Seow W.K., Hart T.C., Wright J.T. Identification of the enamelin (g.8344delG) mutation in a new kindred and presentation of a standardized ENAM nomenclature. Arch Oral Biol. 2003b;48:589–596. doi: 10.1016/s0003-9969(03)00114-6. [DOI] [PubMed] [Google Scholar]
- Hart P.S., Hart T.C., Michalec M.D., Ryu O.H., Simmons D.G., Hong S.P., Wright J.T. Mutation in kallikrein 4 causes autosomal recessive hypomaturation amelogenesis imperfecta. J Med Genet. 2004;41:545–549. doi: 10.1136/jmg.2003.017657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J.C., Yamakoshi Y. Enamelin and autosomal-dominant amelogenesis imperfecta. Crit Rev Oral Biol Med. 2003;14:387–398. doi: 10.1177/154411130301400602. [DOI] [PubMed] [Google Scholar]
- Hu J.C., Hu Y., Smith C.E., McKee M.D., Wright J.T., Yamakoshi Y., Papagerakis P., Hunter G.K., J.Q. Feng, F. Yamakoshi, J.P. Simmer. Enamel defects and ameloblast-specific expression in Enam knock-out/lacZ knock-in mice. J Biol Chem. 2008;283:10858–10871. doi: 10.1074/jbc.M710565200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J.W., Simmer J.P., Hart T.C., Hart P.S., Ramaswami M.D., Bartlett J.D., Hu J.C. MMP-20 mutation in autosomal recessive pigmented hypomaturation amelogenesis imperfecta. J Med Genet. 2005;42:271–275. doi: 10.1136/jmg.2004.024505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J.W., Simmer J.P., Lin B.P., Seymen F., Bartlett J.D., Hu J.C. Mutational analysis of candidate genes in 24 amelogenesis imperfecta families. Eur J Oral Sci. 2006;114(suppl 1):3–12. doi: 10.1111/j.1600-0722.2006.00278.x. discussion 39–41, 379. [DOI] [PubMed] [Google Scholar]
- Mardh K.C., Backman B., Holmgren G., Hu J.C., Simmer J.P., K. Forsman-Semb. A nonsense mutation in the enamelin gene causes local hypoplastic autosomal dominant amelogenesis imperfecta (AIH2) Hum Mol Genet. 2002;11:1069–1074. doi: 10.1093/hmg/11.9.1069. [DOI] [PubMed] [Google Scholar]
- Mendoza G., Pemberton T.J., Lee K., Scarel-Caminaga R., Mehrian-Shai R., Gonzalez-Quevedo C., Ninis V., Hartiala J., H. Allayee, M.L. Snead, S.M. Leal, S.R. Line, P.I. Patel. A new locus for autosomal dominant amelogenesis imperfecta on chromosome 8q24.3. Hum Genet. 2007;120:653–662. doi: 10.1007/s00439-006-0246-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozdemir D., Hart P.S., Firatli E., Aren G., Ryu O.H., Hart T.C. Phenotype of ENAM mutations is dosage-dependent. J Dent Res. 2005a;84:1036–1041. doi: 10.1177/154405910508401113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozdemir D., Hart P.S., Ryu O.H., Choi S.J., Ozdemir-Karatas M., Firatli E., Piesco N., Hart T.C. MMP20 active-site mutation in hypomaturation amelogenesis imperfecta. J Dent Res. 2005b;84:1031–1035. doi: 10.1177/154405910508401112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajpar M.H., Harley K., Laing C., Davies D.M., Dixon M.J. Mutation of the gene encoding the enamel-specific protein, enamelin, causes autosomal-dominant amelogenesis imperfecta. Hum Mol Genet. 2001;10:1673–1677. doi: 10.1093/hmg/10.16.1673. [DOI] [PubMed] [Google Scholar]
- Salido E., Yen P., Koprivnikar K., Yu L.C., Shapiro L. The human enamel protein gene amelogenin is expressed from both the X and the Y chromosomes. Am J Hum Genet. 1992;50:303–316. [PMC free article] [PubMed] [Google Scholar]
- Seedorf H., Klaften M., Eke F., Fuchs H., Seedorf U., Hrabe de Angelis M. A mutation in the enamelin gene in a mouse model. J Dent Res. 2007;86:764–768. doi: 10.1177/154405910708600815. [DOI] [PubMed] [Google Scholar]
- Simmer J.P., Hu J.C. Expression, structure, and function of enamel proteinases. Connect Tissue Res. 2002;43:441–449. doi: 10.1080/03008200290001159. [DOI] [PubMed] [Google Scholar]
- Wright J.T. The molecular etiologies and associated phenotypes of amelogenesis imperfecta. Am J Med Genet A. 2006;140:2547–2555. doi: 10.1002/ajmg.a.31358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright J.T., Hart P.S., Aldred M.J., Seow W.K., Crawford P.J.M., Hong S.P., C. Gibson, T.C. Hart. Relationship of phenotype and genotype in X-linked amelogenesis imperfecta. Connect Tissue Res. 2003;44(suppl):72–78. [PubMed] [Google Scholar]