Abstract
Brown adipose cells are specialized to dissipate chemical energy in the form of heat, as a physiological defense against cold and obesity1. PRDM16 (PRD1-BF1-RIZ1 homologous domain containing 16) is a 140 kDa zinc finger protein that robustly induces brown fat determination and differentiation2. Recent data suggests that brown fat cells arise in vivo from a myf5-positive, myoblastic lineage through the action of PRDM163; however, the molecular mechanisms responsible for this developmental switch is unclear. Here we show that PRDM16 forms a transcriptional complex with the active form of C/EBP-β (LAP), serving as a critical molecular unit that controls the cell fate switch from myoblastic precursors to brown fat cells. Forced expression of PRDM16 and C/EBP-β is sufficient to induce a fully functional brown fat program in naïve fibroblastic cells, including skin fibroblasts from mouse and man. Transplantation of fibroblasts expressing these two factors into mice gives rise to an ectopic fat pad with the morphological and biochemical characteristics of brown fat. As with endogenous brown fat, this synthetic brown fat tissue serves as a sink for glucose uptake, as determined by 18FDG-PET scanning. These data indicate that the PRDM16-C/EBP-β complex initiates brown fat development from myoblastic precursors, and may provide opportunities for the development of novel therapeutics for obesity and type-2 diabetes.
Because of the importance of brown adipose tissue (BAT) as a natural defense against hypothermia and obesity1, and its demonstrated presence in adult humans4–7, understanding the formation of this cell type in mechanistic detail may open new avenues to the development of novel classes of therapeutics for metabolic diseases such as obesity and type-2 diabetes. Several transcriptional regulators have been identified that positively or negatively control BAT development including Rb8, p1079, RIP14010 and FoxC211. Most recently, we have shown that PRDM16, a 140 kDa zinc finger protein, functions as a bi-directional switch in brown fat cell fate by stimulating the development of brown fat cells from white preadipocytes12,13 and from myf5-positive myoblastic precursors3 in vitro and in vivo. At a molecular level, PRDM16 works as a transcriptional coregulatory protein by coactivating PPAR-γ (peroxisome-proliferator-activated receptor-γ), which is considered the “master” gene of fat cell differentiation14,15, and this is almost certainly an important event in the myoblast to brown adipocyte conversion3. However, both isoforms of PPAR-γ are expressed at very low levels in both primary and immortalized myoblasts, while they are abundantly expressed in white and brown preadipocytes (Supplementary Fig. 1). Hence, it is very likely that PRDM16 initiates the process of myoblast to brown fat conversion by complexing with other DNA-binding factors, well before the coactivation of PPAR-γ.
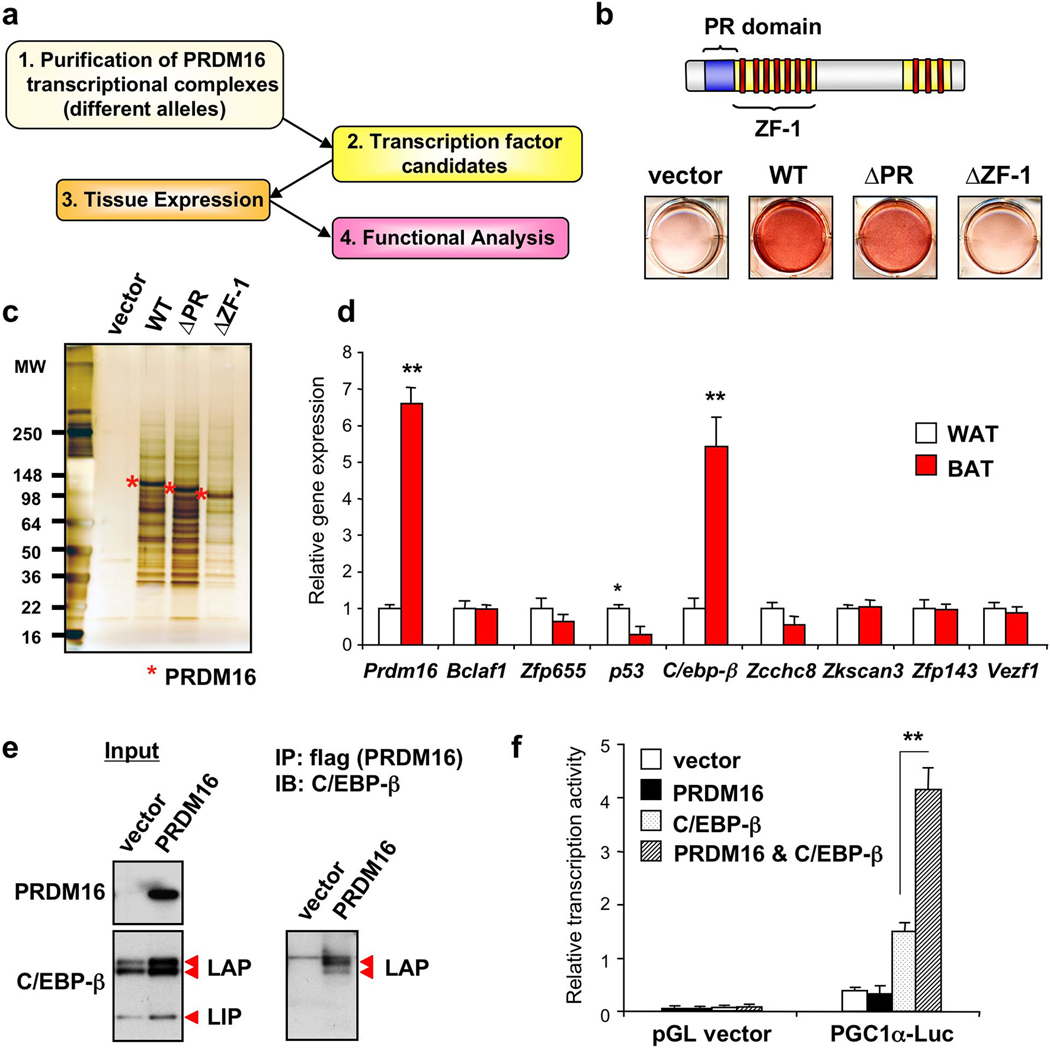
We therefore devised a strategy to address this, as illustrated in Fig. 1a. In brief, we performed proteomic analyses of transcriptional complexes formed with wild type PRDM16 or different mutant alleles that were differentiation-competent or -incompetent. Transcription factors that co-purified preferentially with differentiation-competent PRDM16 proteins were identified; their expression in white and brown fat was then analyzed and compared to that of PRDM16. Subsequently, we examined their function in the process of myoblast-brown fat conversion through PRDM16.
Figure 1. Identification of C/EBP-β as a critical binding partner in the PRDM16 transcriptional complex.
a, Strategy to identify key PRDM16 binding partners. b, C2C12 myoblasts expressing indicated viral vectors were stained with Oil-Red-O 6 days after inducing adipocyte differentiation. c, PRDM16 transcriptional complex was immunopurified from brown fat cells expressing full-length or deletion mutants of PRDM16. d, Gene expression of known or predicted transcription factors identified in the PRDM16 complex in BAT and WAT. n=6. e, Endogenous C/EBP-β was detected in the PRDM16 complex by Western blotting. Input was shown in left. f, Transcriptional activity of PGC-1α promoter in response to PRDM16 and/or C/EBP-β. n=3; error bars are s.e.m.; *P<0.05, **P<0.01.
As shown in Fig. 1b, wild type (WT) and a mutant protein lacking the PR (PRD1-BF-1-RIZ1 homologous)-domain (ΔPR; a.a.91–223), sharing high homology with a SET domain16 17, induced brown fat cell differentiation from myoblasts. In contrast, a mutant allele lacking zinc finger domain-1 (ΔZF-1; a.a. 224–447) completely lost its adipogenic function. The brown fat gene program was also induced by both WT and ΔPR, but not by ΔZF-1 (Supplementary Fig. 2). To avoid comparing proteomic analyses of complexes from cells of very different phenotypes, we expressed all three PRDM16 forms in bona-fide brown fat cells. PRDM16 complexes were then immunopurified to apparent homogeneity (Fig. 1c), and subjected to high-resolution “shotgun” sequencing by liquid chromatography with tandem mass spectrometry (LC-MS/MS)18. In total, 49 proteins were identified in differentiation-competent PRDM16 complexes, but only 8 of these (Bclaf1, Zfp655, p53, C/ebp-β, Zcchc8, Zkscan3, Zfp143 and Vezf1) are known or predicted transcription factors (Supplementary Table 1). Since we have assumed that the expression of a key initiating transcription factor would not be extinguished during the brown fat cell adipogenesis, and PRDM16 is highly enriched in BAT relative to white adipose tissue (WAT)13, we asked whether any of these factors were similarly enriched in BAT. As shown in Fig. 1d, the expression of only C/EBP-β was co-enriched with PRDM16 in BAT versus WAT. In addition, C/EBP-β protein was enriched in BAT, and further induced by cold exposure (Supplementary Fig. 3). Importantly, both primary and immortalized myoblasts express C/EBP-β at similar levels to those seen in preadipocytes (Supplementary Fig. 4), where this factor is thought to play a very important role in adipogenesis19,20. Our analyses have therefore been focused on C/EBP-β and its function in complex with PRDM16.
Brown fat cells express three forms of C/EBP-β, two active forms, named LAP (liver-enriched transcriptional activator protein) and a dominant-negative form, LIP (liver-enriched transcriptional inhibitory protein)21 (Fig. 1e, left). Interestingly, PRDM16 preferentially bound to LAP, but not to LIP (Fig. 1e, right and Supplementary Fig. 5). Independent co-expression assays in HEK293 cells confirmed the physical binding of PRDM16 and C/EBP-β. In addition, PRDM16 interacts with other C/EBP family members, C/EBP-α and -δ (Supplementary Fig. 6). This interaction is likely to be direct through the two zinc finger domains, because the zinc finger domains of the purified glutathione S-transferase (GST)-fused PRDM16 bound to in vitro translated C/EBP-β (Supplementary Fig. 7). Lastly, we asked if PRDM16 could affect the transcriptional activity of C/EBP-β. Since C/EBP-β is known to induce PGC-1α gene expression22, we performed a luciferase reporter assay using the -2kb PGC-1α promoter where the C/EBP binding sites have been characterized22. As shown in Fig. 1f, the expression of PRDM16 and C/EBP-β synergistically stimulated PGC-1α promoter activity. These data suggest that PRDM16 forms a transcriptional complex with active forms of C/EBP-β through direct interaction, and regulates its transcriptional activity.
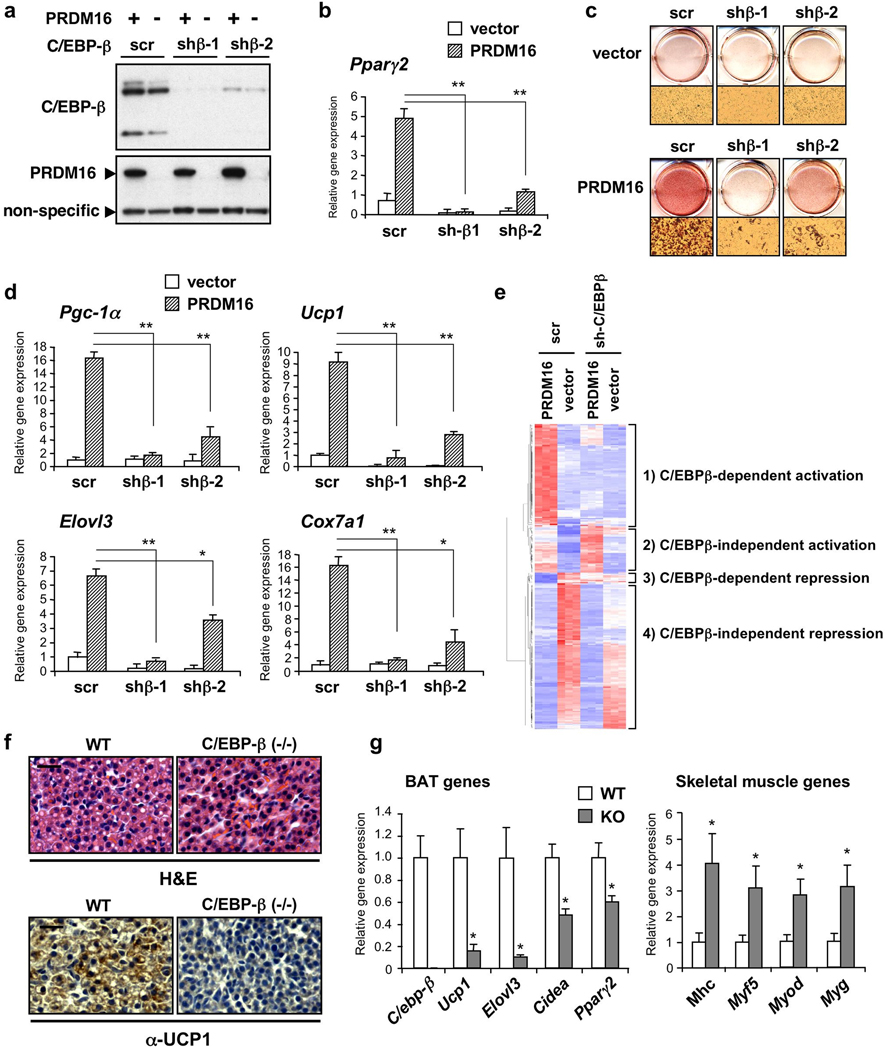
To examine the functional role of the interaction between PRDM16 and C/EBP-β in the myoblast-brown fat conversion, retroviruses expressing a short hairpin (sh) scrambled control RNA (scr) or shRNAs targeting C/EBP-β (shβ-1 and -2) were transduced together with PRDM16 or an empty vector to C2C12 myoblasts (Fig. 2a). As shown in Fig. 2b, knock-down of C/EBP-β significantly blunted the induction of Pparγ2 expression by PRDM16 in undifferentiated C2C12 myoblasts. Consistent with this result, Oil-Red-O staining showed that depletion of C/EBP-β blunted the adipogenesis induced by PRDM16 (Fig. 2c). Furthermore, induction of brown fat selective genes including Pgc-1α, Ucp1, Elovl3 and Cox7a1 were completely or partially blocked by knock-down of C/EBP-β, correlating with the knock-down efficacy (Fig. 2d). In addition, we ectopically expressed LIP, a dominant-negative form of C/EBP-β and this also significantly blunted PRDM16-induced adipogenesis and brown fat-selective gene expression (Supplementary Fig. 8).
Figure 2. C/EBP-β is required for the initiation of the myoblast to brown fat conversion through PRDM16.
a, Western blot analysis for C/EBP-β and PRDM16 in C2C12 myoblasts expressing scr or sh-C/EBP-β with PRDM16 or vector. b, Pparg2 gene expression. n=3. c, These cells were stained with Oil-Red-O 6 days after inducing adipocyte differentiation. d, BAT-selective gene expression. n=4. e, Microarray analysis of undifferentiated C2C12 myoblasts expressing scr or sh-C/EBP-β with PRDM16 or vector. n=3. f, Top: H&E staining of BAT from WT and C/EBP-β KO mice. Bottom: Immunohistochemistry to detect UCP1 expression. Scale bar, 20 µm. g, mRNA expression of BAT and skeletal-muscle-selective genes in BAT from E17.5 embryos. n=5–8; error bars are s.e.m.; *P<0.05, **P<0.01.
Next, we took a systematic approach to ask what fraction of the PRDM16-regulated genes require C/EBP-β at the initiating step of the myoblast-brown fat conversion. To this end, RNAs from undifferentiated C2C12 myoblasts expressing PRDM16 or control together with scr or sh-C/EBP-β, maintained under conditions non-permissive for differentiation, were subjected to Affymetrix microarray analysis. As shown in Fig. 2e, 316 genes were significantly elevated or reduced by PRDM16 (>2-fold, P<0.05), which were clustered into 4 groups: 1) genes elevated by PRDM16 in a C/EBP-β dependent manner; 2) genes elevated by PRDM16 in a C/EBP-β independent manner; 3) genes repressed by PRDM16 in a C/EBP-β dependent manner; and 4) genes repressed by PRDM16 in a C/EBP-β independent manner. The expression of a subset of genes identified by microarray analyses was validated by RT-PCR (Supplementary Fig. 9). Strikingly, the majority of genes activated by PRDM16 before differentiation (62/95, 65.3%) indeed required C/EBP-β, whereas most of the repressed genes (210/221, 95.0%) were not grossly altered by C/EBP-β depletion.
We further explored the genetic requirement for C/EBP-β in brown fat development by analyzing C/EBP-β deficient (KO) embryos. Defects in BAT of C/EBP-β null newborn or adult mice have been described, although the reported phenotype was inconsistent23,24. Since a large number of these embryos died within first 24 h after birth23,25, we have re-examined this issue in late gestation (stage E18.5). This should permit a clearer separation of developmental changes in the BAT, as opposed to those that might occur secondary to abnormalities in other tissues after birth. As shown in Fig. 2f, haematoxylin and eosin (H&E) staining showed that brown fat cells in KO embryos contained significantly less lipid droplets compared with those in WT, suggesting a defects in brown fat development per se (Fig. 2f, top). Moreover, UCP1 expression was severely reduced in KO embryos (Fig. 2f, bottom), consistent with the results of Tanaka et al23. We also conducted a definitive molecular characterization of the BAT from WT and KO embryos. Remarkably, BAT from C/EBP-β KO mice nearly phenocopied that from PRDM16 KO mice at the gene expression level; i.e. a broad reduction of BAT-selective gene expression, and a broad induction of the skeletal muscle gene expression (Fig. 2g). Together, these data indicate that the PRDM16-C/EBP-β transcriptional complex specifically plays a critical role in the initiation of myoblast-brown fat switch. This strongly suggests that PRDM16 acts in myf5-positive myoblastic precursors, at least in part, by coactivation of C/EBP-β to induce the expression of PPARγ and PGC-1α. Subsequently, PRDM16 coactivates PPARγ and PGC-1α through direct binding events, which drives a complete brown fat differentiation program (Supplementary Fig. 10).
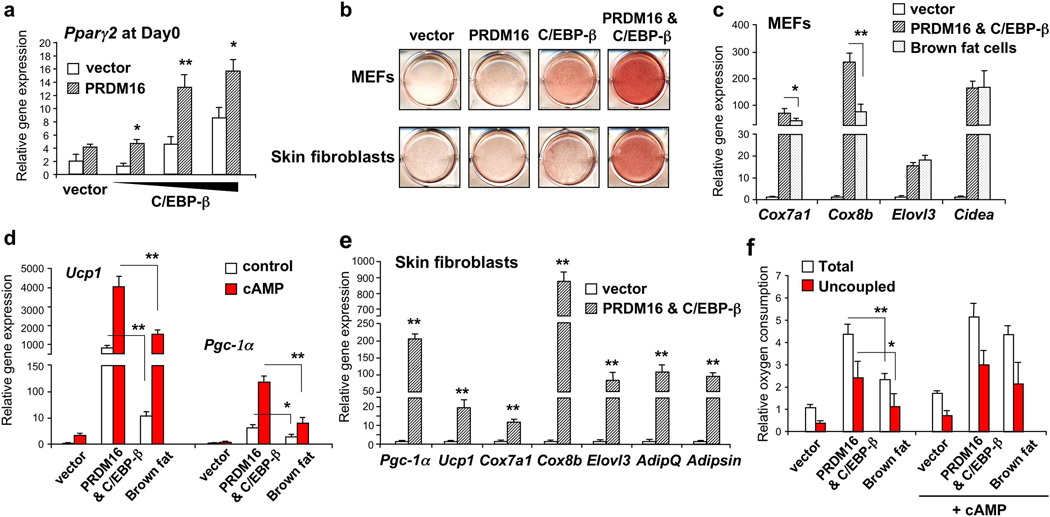
This mechanistic model suggests a critical question: Are the two factors sufficient to reconstitute a brown fat program in naïve cells? To this end, PRDM16 and C/EBP-β were ectopically expressed in mouse embryonic fibroblasts (MEFs) or primary skin fibroblasts with no inherent adipose or brown fat character. As shown in Fig. 3a, Pparγ2 mRNA expression was synergistically induced by PRDM16 and C/EBP-β in a dose-dependent manner in undifferentiated fibroblasts. After 6–8 days under adipogenic conditions, both MEFs and skin fibroblasts expressing these two factors uniformly differentiated into lipid-filled adipocytes, as shown by Oil-Red-O staining (Fig. 3b). The single factors alone were not sufficient to robustly stimulate the differentiated state. Gene expression studies showed that PRDM16 and C/EBP-β powerfully induced mRNA levels of brown fat genes including Cox7a1 (70-fold), Cox8b (260-fold), Elovl3 (16-fold) and Cidea (170-fold) to levels comparable with or even higher than those seen in bona-fide immortalized brown fat cells (Fig. 3c). Importantly, as in authentic brown fat cells, mRNA level of thermogenic genes such as Pgc-1α and Ucp1 were further enhanced by cAMP treatment (Fig. 3d). The mechanism underlying the augmentation of cAMP effects in the engineered brown fat cells remains unknown. To our surprise, mRNA level of those genes at the basal state were activated to levels seen in cAMP-stimulated brown fat cells. Lastly, the two factors were able to induce the brown fat gene program from primary mouse skin fibroblasts (Fig. 3e) and human skin fibroblasts isolated from newborn foreskin (Supplementary Fig. 11).
Figure 3. Reconstitution of the brown fat gene program in fibroblasts through PRDM16 and C/EBP-β.
a, Pparγ2 expression in undifferentiated MEFs expressing indicated viral vectors. n=3. b, Immortalized MEFs or skin fibroblasts expressing indicated viral vectors were stained with Oil-Red-O 6–8 days after inducing adipocyte differentiation. c, BAT-selective gene expression. d, Thermogenic gene expression. The cells were treated with cAMP for 4 hours. n=4. e, BAT-selective gene expression in primary skin fibroblasts expressing vector or PRDM16 and C/EBP-β. n=3. f, Total and uncoupled cellular respiration in differentiated brown fat cells and the MEFs expressing vector or PRDM16 and C/EBP-β. The cells were treated with dbcAMP for 12 hours. n=3; error bars are s.e.m.; *P<0.05. **P<0.01.
An important characteristic of brown fat cells is their extraordinarily high rates of respiration, particularly uncoupled respiration in response to cAMP. As shown in Fig. 3f, engineered brown fat cells induced by these two factors have significantly higher levels of total and uncoupled respiration than control cells, by 4.4 and 6.5 fold, respectively, at the basal state. It is notable that the engineered cells have greater basal respiration, both total and uncoupled, than bona-fide brown fat cells. However, while the bona-fide brown fat cells can further increase both total and uncoupled respiration (by 85% and 90%, respectively) in response to cAMP, engineered brown fat cells already seem to be at their maximal respiration. That these cells are responsive to cAMP is shown by the fact that expression of thermogenic genes such as such as Pgc-1α and Ucp1, are induced by cAMP treatment (Fig. 3d). Hence, some other aspect of the respiratory apparatus, unknown at this point, seems to be limiting in the engineered brown fat cells.
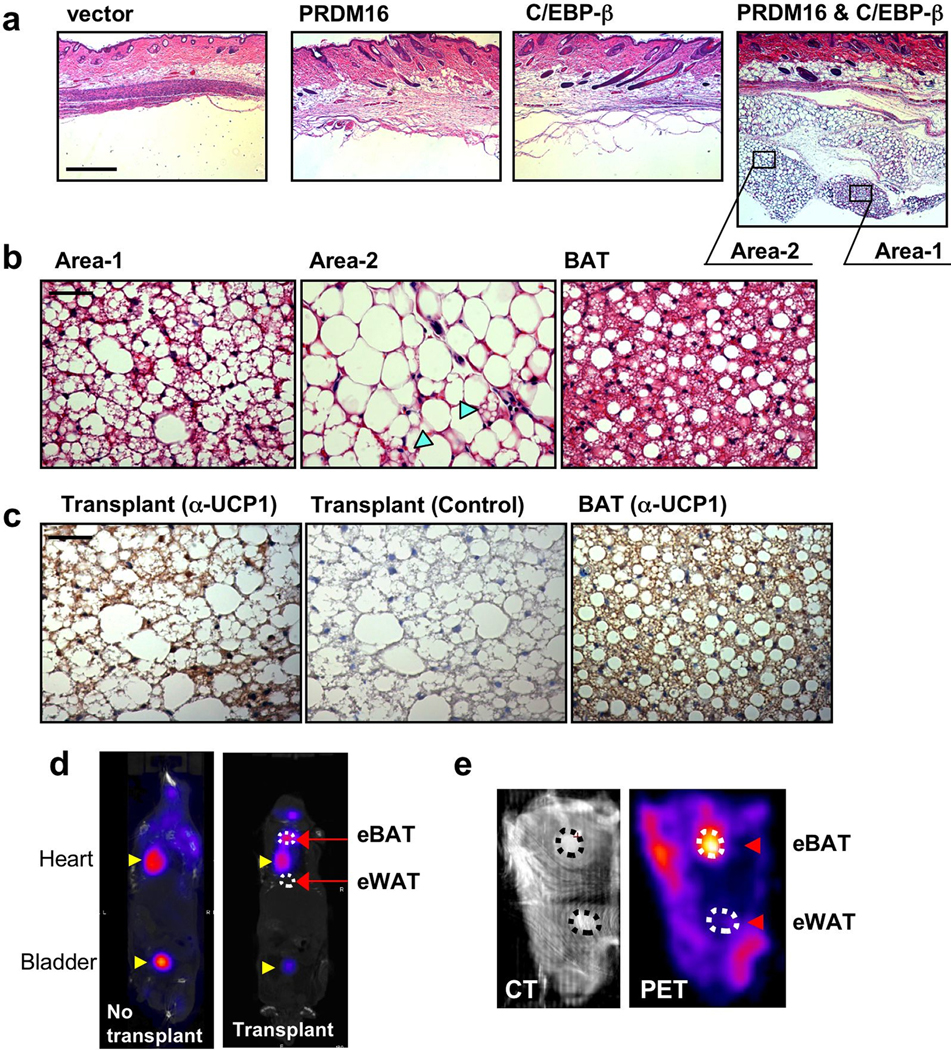
The finding that the combination of PRDM16 and C/EBP-β is sufficient to reconstitute a near complete brown fat program offers an opportunity for controlling brown fat levels and function in vivo. We conducted transplantation studies, as originally developed by Green and Kehinde26, using undifferentiated MEFs expressing either: vector, PRDM16, C/EBP-β or combination of the two factors. As shown in hematoxylin and eosin staining (Fig. 4a), the cells expressing vector nor either PRDM16 or C/EBP-β alone did not form visible fat tissues. In contrast, the cells expressing both PRDM16 and C/EBP-β formed very distinct fat pads in vivo. Notably, at high magnification, the engineered fat tissue induced by the two factors contained “multilocular” fat cells, a morphological characteristic of brown fat in vivo (Fig. 4b). The population of mulilocular fat cells (area 1) is mixed with regions of “unilocular” fat cells (area 2). Importantly, immunohistochemical analyses showed that the engineered adipose tissue was UCP1 positive both in the multilucolar and unilocular fat cells (Fig. 4c).
Figure 4. Generation of functional brown adipose tissue in vivo through expression of PRDM16 and C/EBP-β.
a, Fat pads from transplanted MEFs expressing indicated viral vectors were stained by H&E. Scale bar, 500 µm. b, High magnification images of H&E staining in the transplants expressing PRDM16 and C/EBP-β, and endogenous BAT. Arrow-heads show multilocular fat cells. Scale bar, 50 µm. c, Immunohistochemistry to detect UCP1 expression in the transplant (left, anti-UCP1; middle, negative control) and BAT (right). Scale bar, 50 µm. d, PET/CT image of mice with engineered BAT (eBAT) and engineered WAT (eWAT). e, CT image (left) and PET image (right) of mouse skin with the eBAT and eWAT.
To further characterize the activity of engineered brown fat tissue in vivo, we utilized positron emission tomography (PET) with fluorodeoxyglucose (18FDG), which has recently been used to detect active BAT in adult humans4–7. This technique measures glucose uptake, with brown fat functioning in vivo as an active “sink” for glucose. To this end, we engineered two adipose tissues with similar sizes in the same nude mice: a “brown” fat tissue induced by PRDM16 and C/EBP-β and a “white” fat tissue induced by PPARγ alone as a control (Supplementary Fig. 12a). Induction of BAT-selective genes by PRDM16 and C/EBP-β were confirmed in the cultured cells by RT-PCR (Supplementary Fig. 12b). As shown in Fig. 4d, PET scanning detected a signal in mice from the engineered BAT. To enhance the sensitivity and specificity of the PET signal from the engineered fat tissues, the skin with these fat tissues attached were removed and scanned. The combination of computed tomography (CT) image (Fig. 4e, left) and PET image (Fig. 4e, right) clearly showed that the PET signal was detected from the engineered BAT, but not from the engineered WAT. These results indicate that the engineered brown fat cells function as a sink for active glucose disposal. Given the incredible capacity of BAT to dissipate stored chemical energy and thus counteract obesity, we are optimistic that the PRDM16 pathway can be used to drive brown fat development in vivo in a therapeutic setting. Certainly natural or synthetic compounds that can induce PRDM16 in white fat precursors or in myoblastic cells could have great value in human metabolic disease. Alternatively, as shown here, autologous transplantation of engineered brown fat induced by PRDM16 and C/EBP-β in amounts that are both clinically acceptable and therapeutically useful may well be possible. Future experiments must define the optimal conditions to achieve maximal angiogenesis, innervation and resulting energy expenditure from autologous transplants.
METHODS SUMMARY
Cell Culture
Immortalized brown fat cells have been described previously27. Mouse embryonic fibroblasts were isolated from E13.5 C57/Bl6 embryos, and immortalized according to the established methods28. R2F primary skin fibroblasts isolated from human newborn foreskin were a kind gift from Dr. J.G. Rheinwald (Harvard Medical School) and cultured following the methods described elsewhere29. Adipocyte differentiation in fibroblasts was induced with medium containing 5 µM dexamethosone, 850 nM insulin, 1 nM T3 and 1 µM rosiglitazone.
Identification of the PRDM16 transcriptional complex
Immortalized brown fat cells stably expressing flag-tagged PRDM16 were homogenized to prepare nuclear extracts. The nuclear extracts were incubated with flag M2 agarose (Sigma), washed in a binding buffer, and eluted by incubating with flag peptide12. The immunoprecipitated proteins were digested by trypsin and subjected to reverse-phase liquid chromatography with tandem mass spectrometry (LC-MS/MS), using a high resolution hybrid mass spectrometer (LTQ-Orbitrap, Thermo Scientific) with TOP10 method18.
Animal experiments
All animal experiments were performed according to procedures approved by Beth Israel Deaconess Medical Center Institutional Animal Care and Use Committee. C/EBP-β null mice (Cebpbtm1Vpo/J) were obtained from the Jackson Laboratory. For transplantation studies, immortalized MEFs, transduced with retroviral PRDM16 and/or C/EBP-β, were implanted subcutaneously into 7–9 week old male nude mice (NCr-Foxn1nu)13,26. For PET scanning studies, MEFs expressing retroviral PPARγ were implanted as control. After 4–6 weeks, fat pads were dissected for histological analysis. MicroPET/CT scanning was performed using the Mosaic HP microPET in conjunction with the microCT of the NanoSPECT/CT (Philips), 60 min after the injection of 18FDG.
Full Materials and Methods
Cell Culture
Immortalized brown fat cells have been described previously27. Mouse embryonic fibroblasts were isolated from E13.5 C57/Bl6 embryos (Jackson Laboratory), and immortalized according to the methods established by Todaro and Green28. Mouse dermal fibroblasts were obtained from Millipore. R2F primary skin fibroblasts isolated from human newborn foreskin were kind gift from Dr. J.G. Rheinwald (Harvard Medical School) and cultured as per methods described elsewhere29. HEK293 cells and C2C12 cells were obtained from ATCC. Adipocyte differentiation in C2C12 cells was induced by treating confluent cells in DMEM containing 10% FBS, 0.5 mM isobutylmethylxanthine, 125 nM indomethacin, 5 µM dexamethasone, 850 nM insulin, 1 nM T3, and 1 µM rosiglitazone. Two days after induction, cells were switched to the maintenance medium containing 10% FBS, 850 nM insulin, 1 nM T3, 1 µM rosiglitazone. Adipocyte differentiation in fibroblasts was induced with medium containing 5 µM dexamethosone, 850 nM insulin, 1 nM T3 and 1 µM rosiglitazone. For cAMP treatment, cells were incubated with 10 µM forskolin or 0.5 mM dibutyryl cyclic AMP. All chemicals for cell culture were obtained from Sigma unless otherwise indicated.
DNA constructs and viruses production
Deletion mutants of flag-tagged PRDM16 were amplified by PCR using full length PRDM16 as a template, and subcloned into pMSCV-puro retroviral vector (Stratagene). Various fragments of GST-fused PRDM16 fragments (1–223, 224–454, 455–680, 680–880, 881–1038, and 1039–1176) were described previously12. Myc-tagged C/EBP-β constructs31 were kind gifts from Dr. S.R. Farmer (Boston University). The sequences used for retroviral shRNA expression vectors targeting C/EBP-β were 5’-GCC CTG AGT AAT CAC TTA AAG-3’ (shβ -1) and 5’-CCG GGC CCT GAG TAA TCA C-3’ (shβ -2). The corresponding double-stranded DNA sequences were ligated into pSUPER-Retro (Oligoengine) for retroviral expression. For retrovirus production, Phoenix packaging cells32 were transfected at 70% confluence by calcium phosphate method with 10 µg retroviral vectors. After 48 hours, the viral supernatant was harvested and filtered. Cells were incubated overnight with the viral supernatant, supplemented with 8 µg/ml polybrene. Subsequently, puromycin (PRDM16), hygromycin (C/EBP-β) or G418 (shRNAs) were used for selection. Fibroblasts expressing both PRDM16 and C/EBP-β were selected by puromycin and hygromycin to ensure expression of both constructs.
Affinity purification of PRDM16 transcriptional complex
Immortalized brown fat cells stably expressing flag-tagged wild type, PR-domain deletion mutant, and ZF-1 deletion mutant of PRDM16 or an empty vector were grown to confluence. The cells were homogenized to prepare nuclear extracts12. The nuclear extracts were incubated overnight with flag M2 agarose (Sigma), washed in a binding buffer (180 mM KCl), and then eluted by incubating with 1×flag peptide (0.2 mg/ml). The eluted materials were TCA precipitated, separated in a 4%–20% acrylamide gradient gel and visualized by silver stain, as described previously12.
Mass spectrometry
The immunoprecipitated proteins were precipitated with methanol/chloroform, and precipitates were dissolved in 50 mM Tris-Cl, pH 7.5, containing 8 M urea, 50 mM EDTA and 0.005% n-dodecylβ-D-maltoside (DM). Proteins were reduced with dithiothreitol (DTT) and alkylated with iodoacetamide. After diluting urea concentration to 1 M with 50 mM Tris-Cl, pH 7.5, containing 0.005% DM, trypsin was added and proteins were digested in solution at 37°C for 12 hours. Reaction was stopped with formic acid (FA), and the resultant peptides were desalted with StageTips33. Desalted peptides were subjected to reverse-phase liquid chromatography with tandem mass spectrometry (LC-MS/MS) using a high resolution hybrid mass spectrometer (LTQ-Orbitrap, Thermo Scientific) with TOP10 method, as described previously18. The obtained data were searched against IPI mouse database30. Proteins were identified with at least two unique valid peptides, and the false discovery rate was estimated to be 0% using target-decoy approach34.
Protein interaction analysis
HEK293 cells expressing PRDM16 or C/EBPs were harvested 24 hours after transfection. Total cell lysates were incubated overnight at 4 °C with flag M2 agarose, washed and eluted with flag peptide. The eluted materials were analyzed by Western blot using antibodies for C/EBP-α, C/EBP-β, and C/EBP-δ (Santa Cruz). For in vitro binding assays, various fragments of the GST–fusion PRDM16 fragments were purified as described previously12. [35S]-labeled proteins were made with a TNT reticulocyte lysate kit (Promega). Equal amounts of GST fusion proteins (2 µg) were incubated overnight at 4°C with in vitro translated proteins in a binding buffer containing 20mM HEPES pH 7.7, 300 mM KCl, 2.5 mM MgCl2, 0.05% NP40, 1 mM DTT, and 10% glycerol. The sepharose beads were then washed five times with the binding buffer. Bound proteins were separated by SDS-PAGE and analyzed by autoradiography.
Gene expression analysis
Total RNA was isolated from cells or tissues using Trizol (Invitrogen). Reverse transcriptase reactions were performed using cDNA reverse transcription kit (Applied Biosystems). The sequences of primers used in this study are found in supplementary table 2. Quantitative real-time PCR was performed with SYBR green fluorescent dye using an ABI9300 PCR machine. TATA-binding protein (TBP) served as an internal control.
Microarray analysis
Total RNA was isolated from undifferentiated C2C12 cells transduced with scr or sh-CEBP-β, together with PRDM16 or vector control. Array hybridization and scanning were performed by the Dana-Farber Cancer Institute Core Facility using Affymetrix GeneChip Mouse Genome 430 2.0 arrays according to established methods35. The array data were analyzed using the DNA-Chip Analyzer (dChip) software36. The statistical significance of differences in gene expression was assessed by unpaired t-test (p< 0.05).
Reporter gene assay
PGC-1α (-2kb) promoter linked to a luciferase reporter was transiently cotransfected with PRDM16 and/or C/EBP-β expression plasmids in brown preadipocytes using Lipofectamine 2000 (Invitrogen). Forty-eight hours after the transfection, cells were harvested and reporter gene assays were carried out using the Dual Luciferase Kit (Promega). Transfection efficiency was normalized by measuring expression of Renilla Luciferase.
Cellular respiration assay
Immortalized brown fat cells or MEFs transduced with retroviral PRDM16 and C/EBP-β or an empty vector were grown to confluence and induced to differentiate. At day 6 or 7 of differentiation, oxygen consumption was measured as described previously12,13. For cAMP-induced respiration assays, fully differentiated fat cells were incubated with 0.5 mM dibutyryl cyclic AMP for 12 hours prior to measuring oxygen consumption.
Animals
All animal experiments were performed according to procedures approved by by Beth Israel Deaconess Medical Center Institutional Animal Care and Use Committee. C/EBP-β null mice (Cebpbtm1Vpo/J) were obtained from the Jackson Laboratory. For transplantation studies, male NCR-nude mice (NCr-Foxn1nu) were obtained from Taconic.
Cell transplantations
Immortalized MEFs (3× 107) were transduced with retroviral PRDM16, C/EBP-β, vector control, or combination of PRDM16 and C/EBP-β, and implanted subcutaneously into 7–9 weeks old male nude mice (n=6 mice/group), according to the methods described previously13,26. For PET scanning studies, MEFs expressing retroviral PPAR-γ alone were implanted as a control. After 4–6 weeks, fat pads were carefully dissected and fixed in 4% paraformaldehyde for histological analysis. For Immonohistochemistry, paraffin-embedded sections were incubated with anti-UCP1 antibody (Chemicon), followed by detection using the ABC Vectastain-Elite kit (Vector Labs) according to manufacturer’s instructions.
PET/CT imaging
18FDG (100 µCi) was injected intravenously to animals acclimated for at least 48 hours to a room temperature. Animals were imaged or sacrificed at 1 h post-injection in the Longwood small animal imaging facility of Harvard medical school. PET/CT imaging was performed using a Minerve anesthesia bed moved between a Philips Mosaic HP small animal scanner and a Bioscan CT scanner, and co-registered using custom fiducial markers. The acquired data was reconstructed by InVivoScope software (Bioscan).
Acknowledgements
We are grateful to Dr. S.R. Farmer (Boston University), Dr. J. Rheinwald (Harvard Medical School), and Dr. P.F. Johnson (National Cancer Institute) for providing cells and other reagents, Dr. R. Gupta (Dana-Farber Cancer Institute) for his critical comments on the manuscript, and Dr. J.Y Choi and E. Naseri and for their assistance. S.K is supported by AHA scientist development grant (0930125N). P.S is supported by an NIH grant (DK081605). This work was supported by grants from the Picower Foundation and NIH (DK31405) to BMS, NIH HG3456 and GM67945 to S.P.G, and NIH/NCRR shared instrumentation grant S10-RR-023010.
Footnotes
Supplementary Information contains Supplementary Figures 1–12 and Tables 1–2. Microarray data has been deposited in Gene Expression Omnibus (GEO): GSE15895.
References
- 1.Cannon B, Nedergaard J. Brown adipose tissue: function and physiological significance. Physiol Rev. 2004;84:277–359. doi: 10.1152/physrev.00015.2003. [DOI] [PubMed] [Google Scholar]
- 2.Seale P, Kajimura S, Spiegelman BM. Transcriptional control of brown adipocyte development and physiological function--of mice and men. Genes Dev. 2009;23:799–797. doi: 10.1101/gad.1779209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Seale, et al. PRDM16 Controls a Brown Fat/Skeletal Muscle Switch. Nature. 2008;454:961–967. doi: 10.1038/nature07182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nedergaard J, Bengtsson T, Cannon B. Unexpected evidence for active brown adipose tissue in adult humans. Am J Physiol Endocrinol Metab. 2007;293:E444–E452. doi: 10.1152/ajpendo.00691.2006. [DOI] [PubMed] [Google Scholar]
- 5.Cypess AM, et al. Identification and importance of brown adipose tissue in adult humans. N Engl J Med. 2009;360:1509–1517. doi: 10.1056/NEJMoa0810780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.van Marken Lichtenbelt WD, et al. Cold-activated brown adipose tissue in healthy men. N Engl J Med. 2009;360:1500–1508. doi: 10.1056/NEJMoa0808718. [DOI] [PubMed] [Google Scholar]
- 7.Virtanen KA, et al. Functional brown adipose tissue in healthy adults. N Engl J Med. 360:1518–1525. doi: 10.1056/NEJMoa0808949. [DOI] [PubMed] [Google Scholar]
- 8.Hansen JB, et al. Retinoblastoma protein functions as a molecular switch determining white versus brown adipocyte differentiation. Proc Natl Acad Sci USA. 2004;101:4112–4117. doi: 10.1073/pnas.0301964101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Scime A, et al. Rb and p107 regulate preadipocyte differentiation into white versus brown fat through repression of PGC-1alpha. Cell Metab. 2005;2:283–295. doi: 10.1016/j.cmet.2005.10.002. [DOI] [PubMed] [Google Scholar]
- 10.Leonardsson G, et al. Nuclear receptor corepressor RIP140 regulates fat accumulation. Proc Natl Acad Sci USA. 2004;101:8437–8442. doi: 10.1073/pnas.0401013101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cederberg A, et al. FOXC2 is a winged helix gene that counteracts obesity, hypertriglyceridemia, and diet-induced insulin resistance. Cell. 2001;106:563–573. doi: 10.1016/s0092-8674(01)00474-3. [DOI] [PubMed] [Google Scholar]
- 12.Kajimura S, et al. Regulation of the brown and white fat gene programs through a PRDM16/CtBP transcriptional complex. Genes Dev. 2008;22:1397–1409. doi: 10.1101/gad.1666108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Seale P, et al. Transcriptional control of brown fat determination by PRDM16. Cell Metab. 2007;6:38–54. doi: 10.1016/j.cmet.2007.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tontonoz P, Hu E, Spiegelman BM. Stimulation of adipogenesis in fibroblasts by PPAR gamma 2, a lipid- activated transcription factor. Cell. 1994;79:1147–1156. doi: 10.1016/0092-8674(94)90006-x. [DOI] [PubMed] [Google Scholar]
- 15.Tontonoz P, Spiegelman BM. Fat and beyond: the diverse biology of PPARgamma. Annu Rev Biochem. 2008;77:289–312. doi: 10.1146/annurev.biochem.77.061307.091829. [DOI] [PubMed] [Google Scholar]
- 16.Mochizuki N, et al. A novel gene, MEL1, mapped to 1p36.3 is highly homologous to the MDS1/EVI1 gene and is transcriptionally activated in t(1;3)(p36;q21)-positive leukemia cells. Blood. 2000;96:3209–3214. [PubMed] [Google Scholar]
- 17.Shing DC, et al. Overexpression of sPRDM16 coupled with loss of p53 induces myeloid leukemias in mice. J Clin Invest. 2007;117:3696–3707. doi: 10.1172/JCI32390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Haas W, et al. Optimization and use of peptide mass measurement accuracy in shotgun proteomics. Mol Cell Proteomics. 2006;5:1326–1337. doi: 10.1074/mcp.M500339-MCP200. [DOI] [PubMed] [Google Scholar]
- 19.Wu Z, Xie Y, Bucher NL, Farmer SR. Conditional ectopic expression of C/EBP beta in NIH-3T3 cells induces PPAR gamma and stimulates adipogenesis. Genes Dev. 1995;9:2350–2363. doi: 10.1101/gad.9.19.2350. [DOI] [PubMed] [Google Scholar]
- 20.Farmer SR. Transcriptional control of adipocyte formation. Cell Metab. 2006;4:263–273. doi: 10.1016/j.cmet.2006.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Descombes P, Schibler U. A liver-enriched transcriptional activator protein, LAP, and a transcriptional inhibitory protein, LIP, are translated from the same mRNA. Cell. 1991;67:569–579. doi: 10.1016/0092-8674(91)90531-3. [DOI] [PubMed] [Google Scholar]
- 22.Wang H, et al. CCAAT/enhancer binding protein-beta is a transcriptional regulator of peroxisome-proliferator-activated receptor-gamma coactivator-1alpha in the regenerating liver. Mol Endocrinol. 2008;22:1596–1605. doi: 10.1210/me.2007-0388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tanaka T, Yoshida N, Kishimoto T, Akira S. Defective adipocyte differentiation in mice lacking the C/EBPbeta and/or C/EBPdelta gene. EMBO J. 1997;16:7432–7443. doi: 10.1093/emboj/16.24.7432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Carmona MC, et al. Defective thermoregulation, impaired lipid metabolism, but preserved adrenergic induction of gene expression in brown fat of mice lacking C/EBPbeta. Biochem J. 2005;389:47–56. doi: 10.1042/BJ20050009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Screpanti I, et al. Lymphoproliferative disorder and imbalanced T-helper response in C/EBP beta-deficient mice. EMBO J. 1995;14:1932–1941. doi: 10.1002/j.1460-2075.1995.tb07185.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Green H, Kehinde O. Formation of normally differentiated subcutaneous fat pads by an established preadipose cell line. J Cell Physiol. 1979;101:169–171. doi: 10.1002/jcp.1041010119. [DOI] [PubMed] [Google Scholar]
- 27.Uldry M, et al. Complementary action of the PGC-1 coactivators in mitochondrial biogenesis and brown fat differentiation. Cell Metab. 2006;3:333–341. doi: 10.1016/j.cmet.2006.04.002. [DOI] [PubMed] [Google Scholar]
- 28.Todaro GJ, Green H. Quantitative studies of the growth of mouse embryo cells in culture and their development into established lines. J Cell Biol. 1963;17:299–313. doi: 10.1083/jcb.17.2.299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rheinwald JG, et al. A two-stage, p16(INK4A)- and p53-dependent keratinocyte senescence mechanism that limits replicative potential independent of telomere status. Mol Cell Biol. 2002;22:5157–5172. doi: 10.1128/MCB.22.14.5157-5172.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kersey PJ, et al. The International Protein Index: an integrated database for proteomics experiments. Proteomics. 2004;4:1985–1988. doi: 10.1002/pmic.200300721. [DOI] [PubMed] [Google Scholar]
- 31.Bezy O, Vernochet C, Gesta S, Farmer SR, Kahn CR. TRB3 blocks adipocyte differentiation through the inhibition of C/EBPbeta transcriptional activity. Mol Cell Biol. 2007;27:6818–6831. doi: 10.1128/MCB.00375-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kinsella TM, Nolan GP. Episomal vectors rapidly and stably produce high-titer recombinant retrovirus. Hum Gene Ther. 1996;7:1405–1413. doi: 10.1089/hum.1996.7.12-1405. [DOI] [PubMed] [Google Scholar]
- 33.Rappsilber J, Mann M, Ishihama Y. Protocol for micro-purification, enrichment, pre-fractionation and storage of peptides for proteomics using StageTips. Nat Protoc. 2007;2:1896–1906. doi: 10.1038/nprot.2007.261. [DOI] [PubMed] [Google Scholar]
- 34.Elias JE, Gygi SP. Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry. Nat. Methods. 2007;4:207–214. doi: 10.1038/nmeth1019. [DOI] [PubMed] [Google Scholar]
- 35.Lockhart DJ, et al. Expression monitoring by hybridization to high-density oligonucleotide arrays. Nat Biotechnol. 1996;14:1675–1680. doi: 10.1038/nbt1296-1675. [DOI] [PubMed] [Google Scholar]
- 36.Li C, Wong WH. Model-based analysis of oligonucleotide arrays: expression index computation and outlier detection. Proc Natl Acad Sci U S A. 2001;98:31–36. doi: 10.1073/pnas.011404098. [DOI] [PMC free article] [PubMed] [Google Scholar]