Abstract
The mechanistic aspects of the methylation of guanine in DNA initiated by methyl radicals that are derived from the metabolic oxidation of some chemical carcinogens remain poorly understood. In this work we investigated the kinetics and the formation of methylated guanine products by two methods: (i) the combination of •CH3 radicals and guanine neutral radicals, G(-H)•, and (ii) the direct addition of •CH3 radicals to guanine bases. The simultaneous generation of •CH3 and dG(-H)• radicals was triggered by the competitive one-electron oxidation of dimethyl sulfoxide (DMSO) and 2’-deoxyguanosine (dG) by photochemically generated sulfate radicals in deoxygenated aqueous buffer solutions (pH 7.5). The photolysis of methylcob(III)alamin to form •CH3 radicals was used to investigate the direct addition of these radicals to guanine bases. The major end-products of the radical combination reactions are the 8-methyl-dG and N2-methyl-dG products formed in a ratio of 1 : 0.7. In contrast, the methylation of dG by •CH3 radicals generates mostly the 8-methyl-dG adduct, and only minor quantities of N2-methyl-dG (1 : 0.13 ratio). The methylation of the self-complementary 5’-d(AACGCGAATTCGCGTT) duplexes was achieved by the selective oxidation of the guanines with carbonate radicals anions in the presence of DMSO as the precursor of •CH3 radicals. The methyl-G lesions formed were excised by the enzymatic digestion and identified by LC-MS/MS methods using uniformly 15N-labeled 8-methyl-dG and N2-methyl-dG adducts as internal standards. The ratio of 8-methyl-G /N2-methyl-G lesions derived from the combination of methyl radicals with G(-H)• radicals positioned in double-stranded DNA or with the free nucleoside dG(-H)• radicals were found to be similar. Utilizing the photochemical method and dipropyl or dibutyl sulfoxides as sources of alkyl radicals, the corresponding 8-alkyl-dG and N2-alkyl-dG adducts were also generated in ratios similar to those obtained with DMSO.
Introduction
The alkylation of guanine in cellular DNA can be initiated by both endogenous and exogenous electrophiles and gives rise to strongly pre-mutagenic adducts. The involvement of such adducts in malignant cell transformation and the development of cancer has been postulated.1-4 Carcinogenic nitrosamines, triazenes, alkyl methanesulfonates and related compounds induce the alkylation at different sites of guanine to form 1-, N2-, 3-, 7- and O6-alkylguanines by electrophilic addition mechanisms.5-8 The chemistry of formation and the genotoxic effects of such alkylguanine adducts have been studied extensively.1-8 In contrast, the free radical alkylation of guanine has received much less attention.9 The methylation of guanine at the C-8 position by methyl radicals derived from the reductive cleavage of tert-butylhydroperoxide10,11 and diacyl peroxides,12 or by the oxidation of methylhydrazine catalyzed by horseradish peroxidase,13 has been investigated. The methylation of 2’-deoxyguanosine (dG) initiated by a thermal or a photochemical cleavage of tert-butyl peracetate yields both C-8 (8-Me-dG) and N2 (N2-Me-dG) methylguanine adducts.14,15 However, in vivo, the metabolic activation of the carcinogen 1,2-dimethylhydrazine generates a series of methylguanine derivatives, including 7-Me-G, O6-Me-G and 8-Me-G. These entire DNA adducts have been detected in the liver and colon of rats.16-18 The mechanisms underlying the selectivities of reaction of these radicals remain poorly understood.
The methylation of guanine bases can potentially produce nine mono-methylguanine derivatives including N1, N2, N3, O6, N7, C8, and N9 adducts that are known to be stable.7,10,19 In this work we investigated the kinetics and formation of stable alkylated guanine end-products by free radical mechanisms using two methods: (i) the combination of •CH3 radicals and guanine neutral radicals, G(-H)•, and (ii) the direct addition of •CH3 radicals to guanine bases. In order to explore the combination of •CH3 and G(-H)• radicals, we devised a photochemical method for the simultaneous generation of these radicals. This method involves the one-electron oxidation of 2’-deoxyguanosine and dimethyl sulfoxide (DMSO) by photochemically generated sulfate radicals that yield, at the same time, the corresponding dG(-H)• radical20-22 and the •CH3 radical.23,24 In turn, the photolysis of methylcob(III)alamin (CH3CblIII), a known cofactor of B12-dependent methyltransferases,25 was used to generate •CH3 radicals for studies of the direct addition of these radicals to guanine bases without the simultaneous formation of G(-H)• radicals. The Co–C bond in this B12-coenzyme is very labile and undergoes efficient and selective photodissociation to form cob(II)alamin (CblII) and •CH3 radical.26-28 We found that the major end-products arising from the combination of •CH3 and G(-H)• radicals, are the 8-methyl- and N2-methyl-dG reaction products. In the case of double-stranded DNA, the guanines were oxidized to G(-H)• radicals by carbonate radical anions29 that were generated from the oxidation of bicarbonate ions by photochemically generated sulfate radicals, while the oxidation of DMSO by sulfate radicals gave rise simultaneously to methyl radicals. The ratio of 8-methyl-G /N2-methyl-G lesions derived from the combination of methyl radicals with dG(-H)• radicals or with the G(-H)• radicals positioned in double-stranded DNA were found to be similar. In contrast, the exposure of dG to •CH3 radicals generated by the photolysis of CH3CblIII results yields mostly 8-methyl-dG with a ~ 7.5 smaller proportion of the N2-methyl-dG adduct. This suggests that the direct addition of the •CH3 radical to the C8 atom of guanine can occur directly. Thus, the free radical methylation of guanine bases in DNA can occur by either the combination and/or the addition mechanisms.
Experimental Section
Materials
All chemicals (analytical grade) were used as received. Authentic standards of 1-methylguanine, 3-methylguanine, 7-methylguanine, and 8-methylguanine were purchased from Midwest Research Institute (Kansas City, MO); 1-methyl-2’-deoxyguanosine, N2-methyl-2’-deoxyguanosine, O6-methyl-2’-deoxyguanosine, and N2-ethyl-2’-deoxyguanosine were from Berry&Associates (Ann Arbor, MI); the uniformly 15N-labeled 2’-deoxyguanosine (15N > 98%) was obtained from Cambridge Isotope Laboratories (Andover, MA); methylcob(III)alamin was from Sigma – Aldrich (St. Louis, MO). Deuteration of dG at the C8 position of guanine was performed by heating dG dissolved in D2O as described by Perrier et al.;30 the integrity of C8-D-dG was confirmed by LC-MS/MS analysis.
Laser Flash Photolysis
The transient absorption spectra and kinetics of free radical reactions were monitored directly using a fully-computerized kinetic spectrometer system (~7 ns response time) described elsewhere.31 Briefly, two solutions (e.g., Na2S2O8 and dG + DMSO or CH3Cbl and dG) purged with argon to remove dissolved oxygen were forced by a small positive gas pressure (0.3 – 0.5 atm) into a mixer and then through a quartz micro flow cell (~ 100 μL) at a flow rate of 6 – 8 mL/min. The solution flow rate was controlled by two solenoid valves to provide a complete sample replacement between successive laser shots. Individual laser pulses were selected from the nanosecond pulse trains of either a 308 nm XeCl excimer laser (~60 mJ/cm2/pulse, 1 Hz) or a 355 nm Nd: Yag laser (~20 mJ/cm2/pulse, 10 Hz) by a computer-controlled electromechanical shutter. The transient absorbance was probed along a 1 cm optical path by light from a pulsed 75 W xenon arc lamp with its light beam oriented perpendicular to the laser beam. The signal was recorded by a Tektronix TDS 5052 oscilloscope operating in its high-resolution mode that typically allows for a suitable signal/noise ratio after a single laser short. All experiments were performed at room temperature (23±2 °C).
The second order rate constants of the free radical reactions were typically determined by least squares fits of the appropriate kinetic equations to the transient absorption profiles obtained in five different experiments with five different samples. Kinetic modeling was carried out using the INTKIN software developed at the Brookhaven National Laboratory by H. A. Schwarz. The Numerical Integration method used in this program is the DVODE package written by P. N. Brown and A. C. Hindmarsh, Lawrence Livermore National Laboratory, and G. D. Byrne, Exxon Research and Engineering Co.
Methylation of 2’-deoxyguanosine
The samples of either dG (0.1 μmol), DMSO (0.1 μmol) and Na2S2O8 (10 μmol) or dG (1 μmol) and CH3Cbl (0.1 μmol) in 1 mL deoxygenated phosphate buffer solutions (pH 7.5) were irradiated for fixed periods of time by a light beam from a 100 W xenon arc lamp reflected at 45° by dielectric mirrors to select the 300 – 340 nm and 350 – 400 nm spectral ranges (~ 100 mW/cm2), respectively. After irradiation, the samples were immediately subjected to reversed-phase HPLC analysis on an Agilent 1200 Series LC system (quaternary LC pump with degasser, thermostated column compartment, and diode array detector). Typical separations were performed on an analytical Zorbax SB C18 column (150 × 4.6 mm i.d.) and depending on the alkyl guanines used, gradients of 1 – 20%, 1 - 30% or 1 - 40% acetonitrile in 20 mM ammonium acetate in water (pH 7) for 60 min at a flow rate of 1 mL/min were employed. The fractions collected were evaporated under vacuum, dissolved in water and subjected to LC-MS/MS analysis.
LC-MS/MS analysis of methyl-2’-deoxyguanosines
The photoproducts were identified using an Agilent 1100 Series capillary LC/MSD Ion Trap XCT and an Agilent 1100 Series LC/VSD mono quadrupole mass spectrometer equipped with an electrospray ion source. In typical ion trap experiments 1 – 8 μl of the sample solutions were injected in a narrow bore Zorbax SB-C8 column (50 – 1 mm i. d.) and eluted by an isocratic mixture of acetonitrile and water (50 : 50) with 0.1% formic acid as the mobile phase at a flow rate of 0.2 mL/min. In mono quadrupole experiments 1 – 10 μl of the sample solutions were injected in a Zorbax SB-C8 column (150 – 4.6 mm i. d.) and eluted by an isocratic mixture of methanol and water (20 : 80) with 0.1% formic acid as the mobile phase at a flow rate of 0.75 mL/min.
Methylated guanine products in double-stranded DNA
The DNA duplexes constructed from the self-complementary 5’-d(AACGCGAATTCGCGTT) sequence (6 μM) were irradiated for 30 s by a 300 – 340 nm light beam (~100 mW/cm2) in deoxygenated buffer solutions (pH 7.5) containing 10 mM Na2S2O8, 300 mM NaHCO3 and 0.5 mM DMSO. The irradiated samples (6 nmol) were desalted by HPLC, then 15N-labeled N2-Me-dG and 8-Me-dG internal standards (1 nmol) were added and the samples were evaporated to dryness. The sample dissolved in 50 μl of sodium acetate (1 M, pH 5.2) containing 45mM ZnCl2 was incubated with 2 units of nuclease P1 and 2 units of calf intestinal phosphatase overnight at 370C. Following enzyme removal by centrifugal filtration, the Me-dG's were preliminarily separated from the nucleoside mixture by HPLC and subjected to LC-MS/MS analysis.
Results
Combination reactions of •CH3 and dG(-H)• radicals
The dG(-H)• and •CH3 radicals were produced by the competitive one-electron oxidation of dG and DMSO by photochemically generated SO4•− radicals using the reactions summarized in Table 1.
Table 1.
Reactions and rate constants relevant to the generation and combination of dG(-H)• and •CH3 radicalsa
| N | kn (M−1 s−1) | |
|---|---|---|
| 1 | S2O82- + hν → 2 SO4•– | φ308 = 0.55 (ref. 32) |
| 2 | SO4•– + SO4•– → S2O82- | (1.1±0.1)×109 (ref. 33) |
| 3 | SO4•– + dG → SO42– + dG(-H)• + H+ | (4.1±0.3)×109b |
| 4 | SO4•– + DMSO ⇆ [SO4•–... DMSO] | 1×1010b,c |
| (2.1±0.2)×107b,d | ||
| 5 | [SO4•–... DMSO] + H2O → SO42– + •CH3 + H3CSO2H + H+ | (4.2±0.4)×106b,e |
| 6 | dG + [SO4•–... DMSO] → dG(-H)• + SO42– + DMSO | (3.8±0.4)×109b |
| 7 | dG(-H)• + •CH3 → Me-dG | (4.7±0.8)×108b |
| 8 | •CH3 + •CH3 → C2H6 | (1.6±0.2)×109 (ref. 34) |
| 9 | dG(-H)• + dG(-H)• → products | (1.0±0.1)×108b |
25 mM deoxygenated phosphate buffer solution, pH 7.4.
obtained in this work.
the rate constant for complex formation, k4.
The rate constant for complex dissociation, k−4 is in units of s−1.
rate constant for •CH3 radical formation, k5 (in units of s−1).
Under our experimental conditions, the selective photodissociation of peroxodisulfate (reaction 1) induced by a single nanosecond 308 nm XeCl excimer laser pulse (60 mJ/pulse/cm2) produces ~11 μM SO4•− radicals, which can be easily identified by the prompt appearance of a broad absorption band at 445 nm (Figure 1) with an extinction coefficient of 1.6×103 M−1 cm−1.35
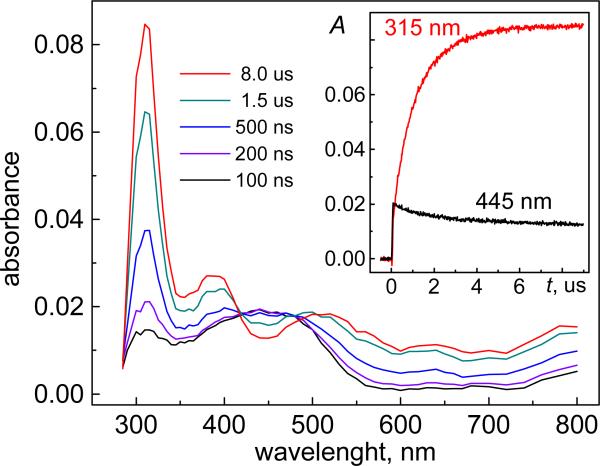
Figure 1.
Transient absorption spectra recorded at the indicated delay times after a single 308 nm laser pulse excitation of a deoxygenated buffer solution (pH 7.5) containing 0.2 mM dG and 30 mM Na2S2O8. The inset shows the time dependence of the decay of SO4•− radicals monitored at 445 nm (black), and the growth of dG(-H)• radicals recorded at 315 nm (red).
The SO4•− radicals rapidly oxidize dG to form the dG(-H)• radicals (reaction 3). This reaction was monitored by the decay of the SO4•− radicals monitored at 445 nm (black trace, inset in Figure 1), which correlates with the growth of the characteristic narrow absorption band of the dG(-H)• radicals at 315 nm (red trace) with extinction coefficient of 7.3×103 M−1 cm−1.20,36 At dG concentrations in the 0.05–0.5 mM range, the kinetic traces of the SO4•− radicals at 445 nm and dG(-H)• radicals at 315 nm can be adequately described by pseudo-first-order kinetics. The observed rate constant, k3′ = 1/τ + k3[dG]0, where τ is the lifetime of the SO4•− radicals in the absence of dG, is governed by the bimolecular recombination of sulfate radicals (reaction 2). From the slope of the k3′ vs [dG] linear plot (data not shown), the value of k3 = (4.1±0.3)×109 M−1 s−1 was obtained, which is identical to that obtained previously by the pulse radiolysis method.20 In the experiment shown in Figure 1, the yield of dG(-H)• radicals is close to quantitative, because at [dG] = 0.2 mM the value of k3[dG]0 >> 1/τ.
The one electron oxidation of DMSO by SO4•− radicals to form •CH3 radicals is very fast (reactions 4 and 5) and competes with the oxidation of dG (reaction 3). The relative fluxes of dG(-H)• and •CH3 radicals can be adjusted by varying the concentrations of dG and DMSO that allows for the exploration of the kinetics of combination of these two radicals. In our experiments, we utilized a dG concentration of 0.2 mM and two different concentrations of DMSO (0.8 mM or 4 mM) to produce dG(-H)• and •CH3 radicals in ratios of 0.45 : 0.55 and 0.25 : 0.75, respectively. These ratios were calculated under the assumption that [SO4•−]0 = [dG(-H)•]t=8μs + [•CH3]t=8μs, where the subscripts “0” and “t=8μs” refer to the earliest measured time points after the actinic laser flash and completion of the formation of dG(-H)• radicals, respectively (inset, Figure 1). Typical kinetic traces of the decay of dG(-H)• radicals recorded at 315 nm are shown in Figure 2A (red traces).
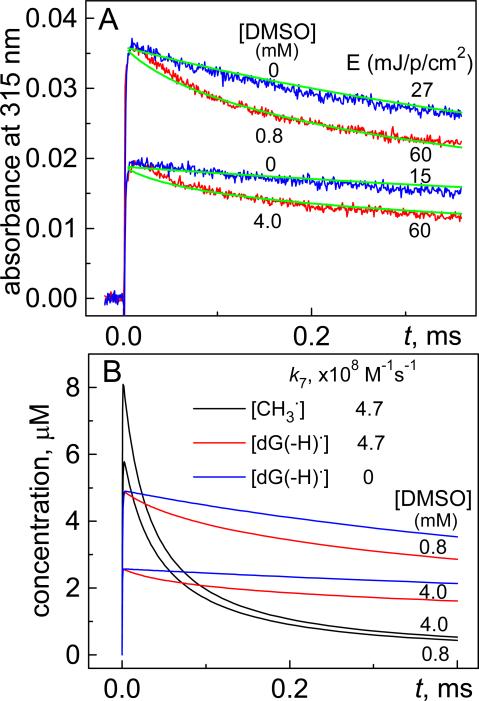
Figure 2.
(A) Transient absorption profiles of dG(-H)• radical decay at 315 nm in the presence (red traces) and absence (blue traces) of DMSO after a single 308 nm laser pulse excitation of a deoxygenated buffer solution (pH 7.5) containing 0.2 mM dG and 30 mM Na2S2O8. (B) Kinetics of dG(-H)• radical decay (red traces) and •CH3 radicals (black traces) simulated by means of the INTKIN program using the reaction rate constants 3 – 9 shown in Table 1 based on an initial, laser pulse-induced SO4•− concentration of 11 μM. The blue traces show the decay of dG(-H)• radicals with the assumption that the rate constant k7 = 0. The transient absorption profiles of dG(-H)• radicals (green traces) were calculated from the simulated kinetic decay curves shown in Panel A.
To demonstrate that the combination reaction of dG(-H)• and •CH3 radicals (reaction 7) contributes to the observed decay of dG(-H)• radicals, the kinetic traces were recorded in the absence of DMSO. In these control experiments, reduced laser pulse energies of 27 and 15 mJ/pulse/cm2 were utilized in order to provide yields of dG(-H)• radicals (blue traces, Figure 2A) similar to those in the experiments obtained in the presence of DMSO (red traces). Comparison of these kinetic profiles showed that, in the presence of •CH3 radicals, the decays of dG(-H)• radicals are markedly faster than in the control experiments without DMSO; this acceleration of reaction rates is attributed to the combination of dG(-H)• and •CH3 radicals (reaction 7) that contributes to the rates of decay of dG(-H)• that occurs by other, non-specific pathways. Overall, the adopted kinetic scheme (reactions 3 – 9), can account for the difference in the kinetics of decay of dG(-H)• radicals in the presence and absence of •CH3 radicals. The simulated kinetics describing the decay of dG(-H)• and •CH3 radicals deduced from simulations of the kinetics using the INTKIN program and the parameters summarized in Table 1, are shown in Figure 2B. The dG(-H)• decay (red traces) is correlated with the recombination of •CH3 radicals (black traces). The latter bimolecular process is characterized by the rate constant, k8 = (1.6±0.2)×109 M−1 s−1, which is ~ three times greater than the constant k7 = (4.7±0.5)×108 M−1 s−1 defining the combination reaction of dG(-H)• and •CH3 radicals. The transient absorption profiles (green curves, Figure 2A) calculated from the simulated kinetic curves of the dG(-H)• decay (Figure 2B) also agree with the experimental profiles (Figure 2A). In this kinetic scheme, the oxidation of DMSO by SO4•− radicals occurs via an intermediate [SO4•− ... DMSO] complex (reaction 4) which decomposes to form •CH3 radicals and SO42− anions (reaction 5), and oxidizes dG to form dG(-H)• radicals (reaction 6). The kinetic parameters and transient absorption spectra of this complex deduced from the laser flash photolysis experiments are discussed in greater details in the Supporting Information.
Direct reaction of •CH3 radicals with dGMP
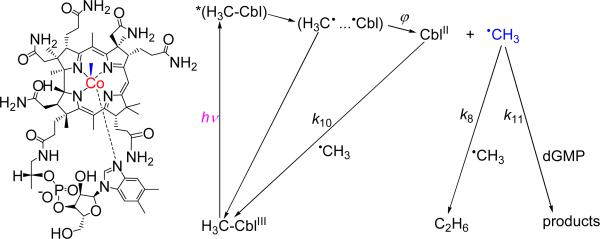
The methyl radicals were generated by the photoexcitation of methylcob(III)alamin to form cob(II)alamin and •CH3 radicals (Figure 3).
Figure 3.
Formation and fates of •CH3 radicals derived from the photolysis of methylcob(III)alamin (left). The cage escape yield, ϕ depends on the excitation wavelength and at 350 – 400 nm is about 0.3 – 0.4.26-28
The methyl radicals can decay via two competitive bimolecular reactions: (i) recombination with CblII to recover the original CH3CblIII (k10), and (ii) combination with another •CH3 radical to form ethane, C2H6 (k8). The second product of the photolysis is cob(II)alamin, which is not an oxidant and cannot abstract an electron from guanine. The •CH3 radicals can react with guanine (k11), thus creating an additional pathway for the decay of methyl radicals. Laser pulse excitation (355 nm) induces bleaching of the absorption band of CH3CblIII at 520 nm, and the rise of the absorption band of CblII at 470 nm.26-28 These properties provide unique opportunities for monitoring the kinetics of reactions of •CH3 radicals by transient absorption spectroscopy, since these radicals themselves do not have any absorption bands in the UV visible spectral range, but participate in the recovery of the CH3CblIII absorption band at 470 nm.
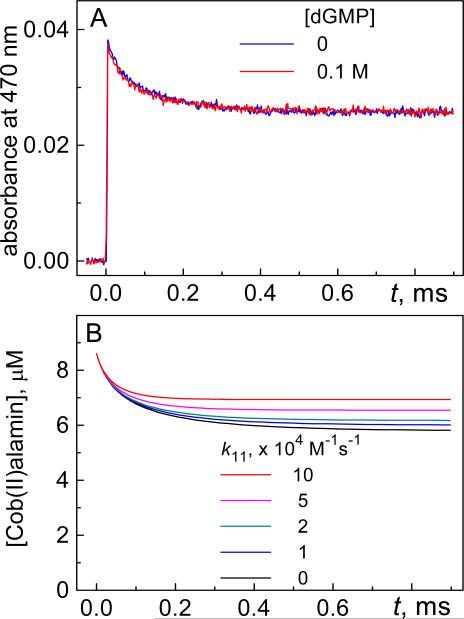
In our experiments, the deoxygenated phosphate buffer solutions (pH 7.4) containing 50 μM CH3CblIII were irradiated with 355 nm Nd: Yag laser pulses to produce a ~ 8.7 μM •CH3 radical concentration (estimated from the bleaching of the CH3CblIII absorption band at 520 nm). We found that the addition of 0.1 M 2’-deoxyguanosine 5’-monophosphate (dGMP), which has a greater solubility in aqueous solutions than dG, does not affect the transient absorption profile at 470 nm. Indeed, the kinetic profile recorded in the presence of dGMP is identical to the control kinetic trace recorded in the absence of dGMP (Figure 4).
Figure 4.
(A) Transient absorption profiles recorded at 470 nm after excitation of methylcob(III)alamin in deoxygenated buffer solutions (pH 7.4) by a single 355 nm laser pulse in the presence of 0.1 M dGMP (red trace) and in the absence (blue trace). (B) Kinetics of CblII decay dG(-H)• radicals simulated by the INTKIN program using k8 = 1.6×109 M−1 s−1, k10 = 4.7×108 M−1 s−1 and assuming an initial 8.7 μM CblII concentration generated by a single laser pulse.
The kinetics curves simulated by the INTKIN progarm show that the contribution of the reaction of •CH3 radicals with dG to the decay of CblII becomes significant if values of k11 > 2×104 M−1 s−1 are adopted. This provides a reliable lower bound for this constant (k11 ≤ 2×104 M−1 s−1).
Products of methylation of 2’-deoxyguanosine
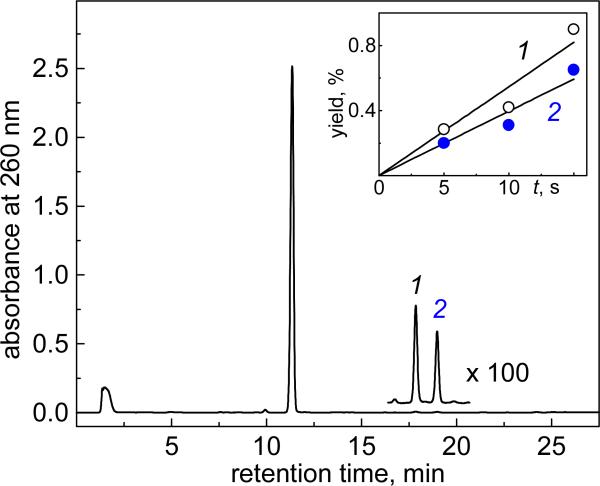
The end products generated by continuous UV irradiation of solutions of Na2S2O8, dG, and DMSO in deoxygenated buffer solutions (pH 7.5), were separated by reversed phase HPLC methods. A typical chromatogram is shown in Figure 5: two products 1 and 2 elute after the parent compound, the dG nucleoside.
Figure 5.
End-products derived from the combination of dG(-H)• and •CH3 radicals. Deoxygenated 5 mM phosphate buffer solutions (1 ml) containing 0.1 mM dG (0.1 μmol), 0.1 mM dimethyl sulfoxide (0.1 μmol) and 10 mM Na2S2O8 were illuminated with 300 – 340 nm steady-state irradiation (~100 mW/cm2) from a 100 W Xe arc lamp for 15 s. Reversed-phase HPLC elution conditions (detection of products at 260 nm): 1 – 20% gradient of acetonitrile in 20 mM ammonium acetate over 60 min. The unmodified dG elutes at 11.3 min, and 8-Me-dG and N2-Me-dG elute at 17.8 (1) and 19.0 (2) min, respectively. The S2O82− anions elute near the void volume at 1.5 min. The inset shows the dependence of the Me-dG yields on irradiation time. The product yields were estimated by integrating the areas under each elution band in the HPLC profiles.
The same distributions of final products were obtained using laser pulse excitation. In these experiments, the deoxygenated buffer solutions (pH 7.5) containing Na2S2O8, dG and DMSO were excited by a train of 308 nm XeCl excimer laser pulses (~60 mJ/cm2/pulse, 10 Hz, for 10 s) and then separated by reversed-phase HPLC methods (data not shown).
Products 1 and 2 were identified by LC-MS methods as two isomeric Me-dG products, with identical molecular ion [M+H]+ masses, and aglycone ion [BH2]+ masses. The latter were derived from the detachment of the sugar residues (- 116 Da) from the molecular ions.37-39 The masses of these ions were greater than the masses of the [M+H]+ and [BH2]+ ions derived from the parent dG by 14 Da. Increasing the irradiation time results in a gradual increase in the concentrations of methyl-dG products (inset, Figure 5), as expected for the formation of structural isomers that result from the combination of dG(-H)• and •CH3 radicals.
In contrast, the methylation of dG by •CH3 radicals derived from the photolysis of methylcob(III)alamin in deoxygenated buffer solutions in the presence of dG, generates mostly product 1, while product 2 forms in minor quantities (Figure S3, Supporting Information). We ascertained, utilizing LC-MS/MS analysis, that these products were identical to products 1 and 2 generated by the sulfate radical – DMSO method (Figure 5).
Identification of methyl-2’-deoxyguanosines
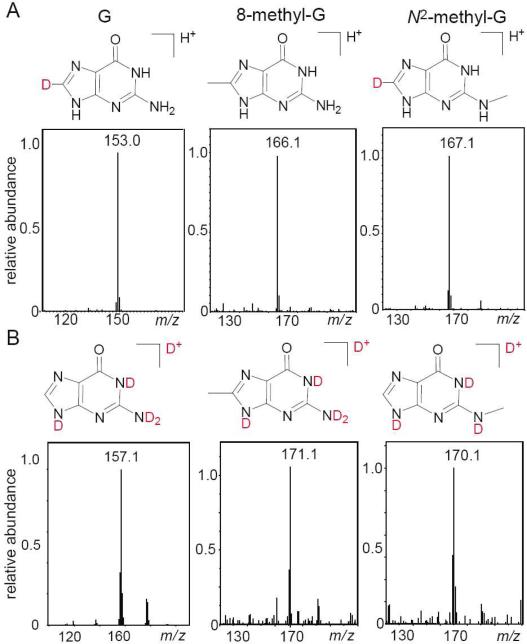
To verify the structures of the Me-dG products 1 and 2, we used C8-D-dG containing a deuterium atom at the C8 position instead of a hydrogen atom. The loss of the deuterium atom is detected in product 1, and thus provides straightforward evidence that the methyl group in this Me-dG product is linked at the C8 position (Figure 6A).
Figure 6.
Positive ion spectra of 8-methyl and N2-methyl-2’ deoxyguanosine derived from C8-D-dG (Panel A), and from normal dG with H at the C8 position, after proton exchange with D2O (Panel B). The spectra were recorded using 10 mM ammonium formate in H2O/CH3CN = 1 : 1 v/v mixtures (Panel A), and 10 mM ammonium formate in D2O/CH3CN 1 : 1 v/v (Panel B) mobile phases, respectively. The panels show the aglycone ions derived from the detachment of the sugar residues from the molecular ions.
In contrast, the mass of product 2 was greater by 1 Da unit than the mass of the control non-deuterated sample, as expected for a Me-dG product with a deuterium-substituted C8-D position.
The number of exchangeable protons in products 1 and 2 was determined from the mass spectra of the Me-dG samples after proton exchange in D2O (99.9%) by repeated lyophilization and by redissolving the products in D2O (Figure 6B). Using D2O/acetonitrile mixtures as the mobile phase in LC-MS analysis, it was found that the number of exchangeable protons in product 1 was exactly the same as in the control nucleoside dG; this result is in agreement with the methyl group positioned at C8 of dG. In the case of product 2, the number of exchangeable protons is less by one proton than in the case of the control nucleoside dG; this suggests that the methyl group is bound either at the N1 or the N2 positions. The differentiation of the N1- and N2-alkyl-dG products was achieved by co-elution with authentic standards. These experiments showed that product 2 is the N2-alkyl-dG product; in turn, co-elution with the authentic standard confirmed that product 1 is indeed the 8-alkylguanine derivative (for additional details, see Figure S4 in Supporting Information). Only monomethyl-substituted guanines were observed in our experiments because the extent of reaction did not exceed 2-3% (single hit conditions). In contrast, Constantinesco et al. found doubly-methylated N2,N2-dimethylguanosine products when guanine in a tRNA transcript was methylated by a eukaryotic tRNA methyltransferase.40 Analogous dimethylated N2-guanine adducts are not expected under our chemical reaction conditions unless the overall extent of methylation reaches much higher values.
Formation of methyl-2’-deoxyguanosines in double-stranded DNA
The selective oxidation of guanine bases in DNA by carbonate radical anions33,41,42 in the presence of DMSO, can be used to generate Me-G lesions. In these experiments, deoxygenated buffer solutions (pH 7.5) containing duplexes derived from the self-complementary sequence, 5’-d(AACGCGAATTCGCGTT), Na2S2O8, NaHCO3, and DMSO, were irradiated with continuous UV light. Under these conditions, the photochemically generated SO4•− radicals (reaction 1) induce the one-electron oxidation of DMSO (reactions 4 and 5), as well as the oxidation of HCO3− anions to yield CO3•− radicals.
| (12) |
| (13) |
| (14) |
The presence of CO3•− radicals is required for one-electron oxidation of guanine that results in the formation of G(-H)• radicals (reaction 13).33,41,42 The SO4•− radicals are unsuitable for this purpose because they can oxidize all four canonical DNA nucleobases to form the corresponding radicals.43 To avoid the direct oxidation of DNA by SO4•− radicals, we used low concentrations of oligonucleotides and relatively high concentrations of HCO3− and DMSO to provide a predominant decay of SO4•− radicals via reactions 4 and 10. Thus, the combination of the G(-H)• and •CH3 radicals to generate the methylguanine lesions (reaction 14) can be observed.
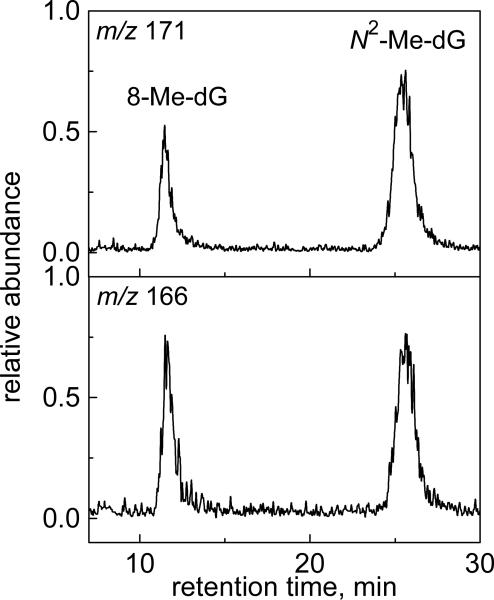
The DNA duplexes (6 μM) were irradiated for 30 s by a 300 – 340 nm light beam (~100 mW/cm2) in deoxygenated buffer solutions (pH 7.5) containing 10 mM Na2S2O8, 300 mM NaHCO3 and 0.5 mM DMSO. The methylated DNA duplexes obtained in a series of photochemical experiments were combined, desalted, mixed with the 15N-labeled 8Me-dG and N2-Me-dG authentic standards, and enzymatically digested by nuclease P1 and alkaline phosphatase. The digestion products obtained were analyzed by LC-MS/MS methods (Figure 7).
Figure 7.
LC-MS analysis of the methyl-2’-deoxyguanosines excised by enzymatic digestion of the irradiated duplexes in the presence of the 15N-labeled 8-Me-dG and N2-Me-dG standards. The extracted chromatograms were recorded in the positive mode with specific ion monitoring at m/z 166 (514N) and 171 (515N).
The extracted ion chromatograms show that 8-Me-dG and N2-Me-dG are indeed formed in double-stranded DNA and co-elute with the 15N-labeled 8-Me-dG and N2-Me-dG standards. Under these experimental conditions, the total yield of 8-Me-dG and N2-Me-dG products, calculated by integrating the areas under the relevant peaks in the ion chromatogram traces, was typically 1.2 – 1.6% based on the initial amount of the oligonucleotide sequence. These results demonstrate that the 8-Me-G and N2-Me-G lesions form in DNA duplexes with practically identical yields. We used CO3•− radicals, which selectively oxidize guanine bases33,41,42 and favor the methylation of guanine bases by a radical-radical combination mechanism. The methylation of other DNA bases by free radical mechanisms is also possible. For instance, methylation of the A, G, C, and T nucleosides has been detected in the course of tert-butyl peracetate photolysis. However, experiments along these lines were beyond the scope of this work.
Synthesis and characterization of propyl- and butyl-2’-deoxyguanosines
The photochemical method developed for the preparation of 8-Me-dG and N2-Me-dG can be successfully used for the synthesis of the higher alkylguanines, CnH2n+1-dG (n ≥ 2). In these experiments we utilized dialkyl sulfoxides with n-propyl and n-butyl groups instead of DMSO.
The reversed-phase HPLC analysis of the products obtained by continuous UV irradiation of solutions of Na2S2O8, dG and dialkyl (propyl, or butyl) sulfoxide, revealed two products eluting after the parent dG (data not shown). These products were identified by LC-MS methods as two isomeric alkyl-dG products, which have identical masses of the molecular ions, [M+H]+ and the aglycone ions, [BH2]+ derived from the detachment of the sugar residues (- 116 Da) from the molecular ions.37-39 The masses of these ions are greater by 42 Da (propyl) and 56 Da (butyl) than the masses of the [M+H]+ and [BH2]+ ions derived from the parent dG. Further analysis of the distributions of the daughter ions generated by extensive fragmentation of the [BH2]+ ions showed that the products eluting after dG, are the 8-alkyl-dG products, whereas the spectra of the products eluting after the 8-alkyl derivatives are the N2-alkyl-dG products.
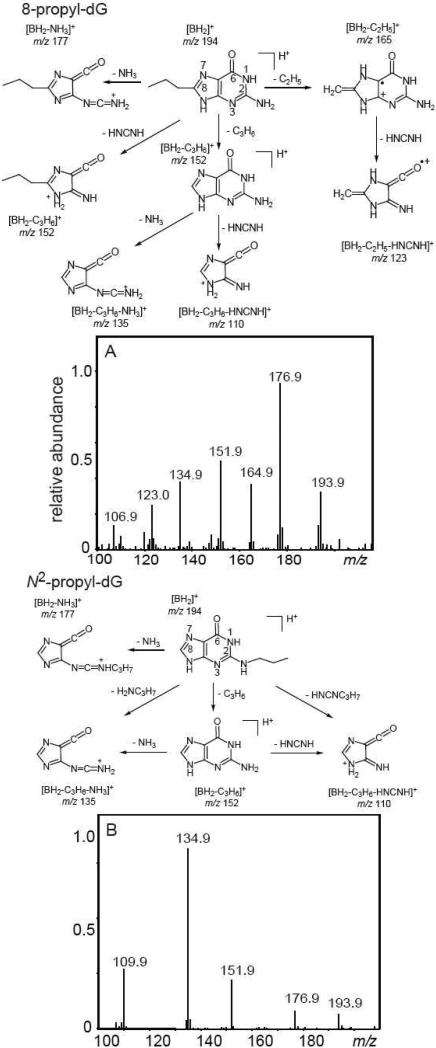
Representative positive product ion spectra of propyl-dG derivatives are shown in Figure 8.
Figure 8.
Positive product ion spectra of 8-propyl-dG (Panel A) and N2-propyl-dG (Panel B) derived from the combination of the dG(-H)• and propyl radicals that were isolated by reversed-phase HPLC methods.
The [BH2]+ ions are observed at m/z 193.9. The daughter ions at m/z 176.9 detected in both 8-propyl-dG and N2-propyl-dG arise from the expulsion of ammonia from [BH2]+ ions. The structural differentiation of propyl-dG products is complicated by the elimination of the propyl group from the [BH2]+ ions with the concomitant transfer of a hydrogen atom to the guanine base, thus resulting in a net loss of the C3H6 fragment.7,44,45 The fragmentation pathways of the protonated guanine at m/z 152 are independent of the initial site of alkylation and include expulsion of ammonia or carbodiimide to form the ions at m/z 135 and 110. In the case of the N2-propyl-G adduct, the formation of these ions can also occur via the expulsion of propylamine and N-propylcarbodiimide (Figure 8B). Indeed, the ions at m/z 135 and 110, derived from the expulsion of methylamine and N-methylcarbodiimide, were detected in the case of N2-Me-G, which does not generate the protonated guanine (Figure S5 in Supporting Information). In the case of 8-propyl-G, the ion m/z 152 can form via an alternative pathway that involves the expulsion of cyanamide (Figure 8A), because the C3H6 and HNCNH fragments have the same molecular weight of 42 Da. To demonstrate that both pathways are possible, we investigated the fragmentation of 8-butyl-dG and found two ions, [BH2-C4H8]+ (protonated guanine) at m/z 152, and [BH2-HNCNH]+ at m/z 166.0 (data not shown).
Nevertheless, the straightforward differentiation of 8-propyl-dG from other propyl-dG products is possible because of the expulsion of the C2H5 fragment from the propyl group to form a marker ion at m/z 165 detected in the case of 8-propyl-dG (Figure 8A). In contrast, this ion is not observed in N2-propyl-G (Figure 8B). Here, we propose that the ion at m/z 165 is the radical cation stabilized by the formation of the double-bond between the C8-atom of guanine and the CH2-group derived from the cleavage of the propyl group that is possible in the case of 8-alkylguanines only. Indeed, the mass of this ion at m/z 165 does not depend on the alkyl group length and this ion is observed in the mass spectra of the higher 8-alkylguanines (propyl and butyl in our experiments, or 1- and 2-hydroxyethyl in ref.46) and is not observed in other alkylguanines.7,44,45 The further fragmentation of the ions m/z 165 occurs via the expulsion of carbodiimide to form the ion m/z 123 that is a second signature of 8-alkyl-dG's.
Discussion
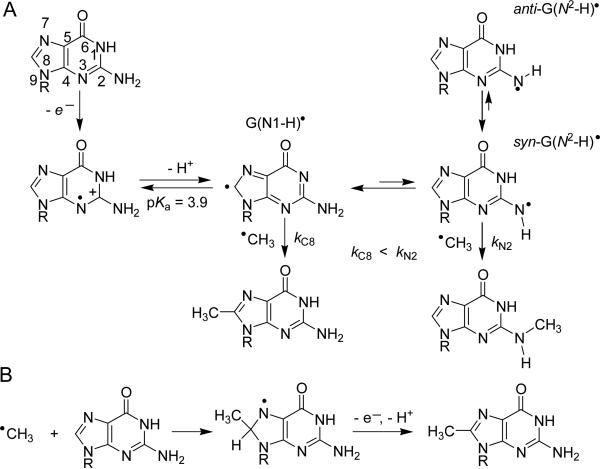
In the experiments described here, the free radical methylation of guanine was initiated by two methods: (i) combination of •CH3 and G(-H)• radicals as shown in Figure 9A, and (ii) the direct addition of •CH3 radicals to guanine bases, as summarized in Figure 9B.
Figure 9.
Free radical methylation of guanine via: combination of •CH3 and G(-H)• radicals (A), and direct addition of •CH3 radicals to guanine (B). Two conformations of G(N2–H)• radicals (syn and anti) are shown with respect to the N3 atom.
In the first method, methylation is initiated by the generation of the radical precursors (reactions 3 – 5). Pulse radiolysis studies have shown that electron abstraction from 2’-deoxyguanosine first produces the guanine radical cation (G•+), a weak acid with pKa = 3.9,20 as shown in Figure 9A. The EPR studies in aqueous solutions (pH ≤ 3) at room temperature, in combination with theoretical calculations, have shown that the G•+ radical is protonated at N1 and is deprotonated by the loss of the proton at N1 to form the G(N1–H)• neutral radical, as expected at pH > 3.47 However, in the case of the radical cation of 1-Me-dG with pKa = 4.7, deprotonation occurs via release of a proton from the exocyclic nitrogen (N2) resulting in the formation of G(N2–H)•. This suggests that, in principle, both the N1 and N2 sites can be deprotonated in guanine neutral radicals.20 The EPR studies, in combination with UV-visible spectroscopy, have shown that in frozen D2O solutions at low temperatures, guanine radicals are present mostly in the G(N1–H)• form.48 In contrast, the EPR/ENDOR studies at low temperatures have shown that G(–H)• radicals generated by ionizing radiation of the single crystals of 2’-deoxyguanosine 5’-monophosphate are present mostly in the G(N2–H)• form.49,50 The existence of two radical forms with close energies has been supported by the extensive DFT calculations for the various tautomers of G(–H)•. 48,51,52 This analysis has shown that in the gas phase the syn-G(N2–H)• tautomer with respect to the N3 atom of guanine is more stable than the anti conformer, and in turn, syn-G(N2–H)• is more stable than the G(N1–H)• tautomer by ~ 4.5 kcal/mol. However, hydration of G(–H)• by seven or more water molecules reverses the order of stabilization and G(N1–H)• becomes more stable than syn-G(N2–H)• by ~3.3 kcal/mol.48 The addition of •CH3 radicals to these tautomers that have similar energies can account for the formation of 8-Me- and N2-Me-dG products (Figure 9A). According to this mechanism, the G(N2–H)• radical with unpaired electrons on the N2 atom is expected to be more reactive than the G(N1–H)• radical (kC8 < kN2). This mechanism can explain the formation of 8-alkyl- and N2-alkyl-dG products via the combination reaction of G(–H)• with the larger alkyl radicals (propyl and butyl).
Our laser flash photolysis experiments show that the combination of •CH3 and G(-H)• radicals occurs with the rate constant k10 = 4.7×108 M−1 s−1 (Table 1). The rate constants of reaction of G(–H)• radicals with •NO2 and O2•− are very close to one another (4.5×108 M−1 s−1 and 4.7×108 M−1 s−1, respectively).53,54 However, these radicals react with the C8 and C5 positions of G(–H)• to form stable end products, which include 8-nitroguanine and 5-guanidino-4-nitroimidazole (reactions with •NO2), and 8-oxoguanine and imidazolone (reactions with O2•−). The absence of end products derived from the addition of these radicals to the N2 position of G(–H)• radicals can be accounted for by the low stabilities of the adducts formed. In turn, we did not find any 5-Me-dG products in our experiments. The formation of 5-Me-dG requires a change of hybridization of the C5 atom of guanine from sp2 to sp3, thus causing a loss of aromaticity and distortion of the shape of the guanine molecule. These factors suggest that 5-Me-dG should have a low stability. In contrast, the addition of •NO2 and O2•− to the C5 atom of G(–H)• induces fragmentation of the pyrimidine ring in guanine followed by formation of stable 5-guanidino-4-nitroimidazole and imidazolone lesions, respectively.
An alternative mechanism of free radical methylation includes the direct attack of •CH3 radicals on guanine (Figure 9B), which occurs via radical addition to the C8 position.9-15 Addition of alkyl radicals (e.g., H2NCOCH2•) to the C8 position of the purine ring of the 2’-deoxynucleosides 5’-monophosphates (dGMP and dAMP) has been clearly observed in the EPR spectra at low temperatures.55 The radical produced by the addition of •CH3 to the C8 atom of guanine should be a strong reductant, similar to 8-HO-dG(-H)• derived from the addition of •OH-radical to dG, which is readily oxidized to 8-oxoguanine by weak oxidants, such as oxygen and methylviologen.56 Our laser flash photolysis experiments show that the rate constant of the reaction between •CH3 radicals and guanine bases is less than 2×104 M−1 s−1 (Figure 4) and in the absence of efficient competitive reactions, such as reactions with O2•−,54 methyl radicals can react with guanine to form Me-dG products. Indeed, LC-MS/MS analysis showed that continuous illumination of deoxygenated solutions of methylcob(III)alamin and dG generates mostly 8-Me-dG and N2-Me-dG forms in minor quantities (Figure S2). Methylation of guanosine-5’-monophosphate (GMP) by •CH3 radicals derived from the spontaneous decomposition of [Co(NH3)5(CH3)]2+ complexes also results in the formation of 8-Me-dG.57
Our experiments showed that N2-Me-dG products form in minor proportions, because the formation of this product depends on the generation of the G(N2–H)• radical (Figure 9A). In experiments with methylcob(III)alamin, the latter radical can be generated by hydrogen atom abstraction from guanine by •CH3 radicals. The driving force of these reactions is the difference of the bond dissociation energy, BDE = 105±0.1 kcal/mol of the CH3-H bond58 and BDE = 94.3±0.5 kcal/mol of the weaker N1-H bond (also the weakest) of guanine.59 The H-atom abstraction from the 2’-deoxyribose residues by •CH3 radicals is also thermodynamically feasible.59 This reaction is supported by the observation of direct single-strand breaks in supercoiled DNA generated by •CH3 radicals produced by the photolysis of the macrocyclic methyl cobalt complex, [MeCoIII(cyclam)(H2O]2+,60 and methylcob(III)alamin.61 The rate constant of the H atom abstraction reactions estimated from the formation of single-strand breaks in plasmid DNA is 8.8×104 M−1s−1.62 The detection of methane during the photolysis of methylcob(III)alamin in the presence of nucleoside 5’-monophosphates (AMP, CMP, GMP, and TMP) and calf thymus DNA can be considered as additional support for H-atom abstraction mechanisms.61 However, investigations along these lines were beyond the scope of this work.
The mechanisms of free radical methylation of guanine bases (Figure 9) can account for the pH effects on the distributions of 8-Me-dG and N2-Me-dG products reported in earlier publications.10-12,14,15 In neutral solutions, the major products of the radical methylation reaction are 8-Me-dG and N2-Me-dG. However, in acid solutions only 8-Me-dG is formed. These observations are consistent with the free radical methylation mechanism depicted in Figure 9. In neutral solutions, guanine radicals can exist in the G(N2–H)• form, which is an essential intermediate for the formation of N2-Me-dG. The protonation of the G(–H)• radical in acid solution (the pKa of dG•+ is 3.920) suppresses the formation of the G(N2–H)• radical and thus 8-Me-dG becomes the major methylation product.
Conclusions
Free radical methylation of guanine bases occurs by two principal pathways: (i) combination of G(-H)• and •CH3 radicals, and (ii) direct addition of •CH3 radicals to guanine bases. The rate constant of radical combination, k7 = (4.7±0.5)×108 M−1 s−1, is at least 4 orders of magnitude greater than the constant for the direct addition reaction, k11 ≤ 2×104 M−1 s−1. The products of the free radical methylation reactions, the 8-methyl-G and N2-methyl-G lesions, are formed in different ratios (1 : 0.7 for the radical combination reaction and 1 : 0.13 for the direct addition reaction). Among these two lesions, 8-methyl-G is a marker of the free radical mechanism,9 whereas N2-methyl-G can be also generated by electrophilic addition mechanisms.8 In biological systems, the concentrations of free radicals are extremely low and the direct addition of •CH3 radicals to G bases is suspected to be more efficient than the bimolecular combination reaction of two radicals, the G(-H)• and the •CH3 radicals. However, the efficiency of free radical methylation is expected to be low due to the fast reaction of •CH3 radicals with oxygen.24 From this point of view, the free radical methylation reaction could be efficient in hypoxic tumor cells.63 Interestingly, 8-methyl-G lesions have been detected in the liver and the colon of rats that had been fed 1,2-dimethylhydrazine,18 a precursor of •CH3 radicals. Thus, further research is warranted on the reactivities of •CH3 radicals in cellular environments.
Supplementary Material
Acknowledgements
We thank Drs. Jean Cadet and Michael Sevilla for insightful discussion. This work was supported by the National Institute of Environmental Health and Sciences (5 R01 ES 011589-08). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute of Environmental Health and Sciences or the National Institutes of Health. This work was performed, in part using compounds provided by the National Cancer Institute's Chemical Carcinogen Reference Standards Repository operated under contract by Midwest Research Institute, NO. N02-CB-66600. Components of this work were conducted in the Shared Instrumentation Facility at NYU that was constructed with support from a Research Facilities Improvement Grant (C06 RR-16572) from the National Center for Research Resources, National Institutes of Health. The acquisition of the ion trap mass spectrometer was supported by the National Science Foundation (CHE-0234863).
Footnotes
Supporting Information Available: This information is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Loveless A. Nature. 1969;223:206–207. doi: 10.1038/223206a0. [DOI] [PubMed] [Google Scholar]
- 2.Goth R, Rajewsky MF. Proc. Natl. Acad. Sci. USA. 1974;71:639–643. doi: 10.1073/pnas.71.3.639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Choi JY, Guengerich FP. J. Biol. Chem. 2004;279:19217–19229. doi: 10.1074/jbc.M313759200. [DOI] [PubMed] [Google Scholar]
- 4.Upton DC, Wang X, Blans P, Perrino FW, Fishbein JC, Akman SA. Mutat. Res. 2006;599:1–10. doi: 10.1016/j.mrfmmm.2005.12.014. [DOI] [PubMed] [Google Scholar]
- 5.Lijinsky W. Chemistry and Biology of N-Nitroso Compounds. Cambridge University Press; Cambridge, U.K.: 1992. [Google Scholar]
- 6.Loeppky RN, Michejda CJ. Nitrosamines and Related N-Nitroso Compounds. American Chemical Society; Washington, DC.: 1994. [Google Scholar]
- 7.Lawley PD, Orr DJ, Jarman M. Biochem. J. 1975;145:73–84. doi: 10.1042/bj1450073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Blans P, Fishbein JC. Chem. Res. Toxicol. 2004;17:1531–1539. doi: 10.1021/tx0498004. [DOI] [PubMed] [Google Scholar]
- 9.Augusto O. Free Radic. Biol. Med. 1993;15:329–336. doi: 10.1016/0891-5849(93)90079-a. [DOI] [PubMed] [Google Scholar]
- 10.Maeda M, Nushi K, Kawazoe Y. Tetrahedron. 1974;30:2677–2682. [Google Scholar]
- 11.Kohda K, Tsunomoto H, Minoura Y, Tanabe K, Shibutani S. Chem. Res. Toxicol. 1996;9:1278–1284. doi: 10.1021/tx9601059. [DOI] [PubMed] [Google Scholar]
- 12.Araki M, Maeda M, Kawazoe Y. Tetrahedron. 1976;32:337–340. [Google Scholar]
- 13.Augusto O, Cavalieri EL, Rogan EG, RamaKrishna NV, Kolar C. J. Biol. Chem. 1990;265:22093–22096. [PubMed] [Google Scholar]
- 14.Zady MF, Wong JL. J. Am. Chem. Soc. 1977;99:5096–5101. doi: 10.1021/ja00457a033. [DOI] [PubMed] [Google Scholar]
- 15.Zady MF, Wong JL. J. Org. Chem. 1980;45:2373–2377. [Google Scholar]
- 16.Rogers KJ, Pegg AE. Cancer Res. 1977;37:4082–4087. [PubMed] [Google Scholar]
- 17.Herron DC, Shank RC. Cancer Res. 1981;41:3967–3972. [PubMed] [Google Scholar]
- 18.Netto LE, RamaKrishna NV, Kolar C, Cavalieri EL, Rogan EG, Lawson TA, Augusto O. J. Biol. Chem. 1992;267:21524–21527. [PubMed] [Google Scholar]
- 19.Rice JM, Dudek GO. J. Am. Chem. Soc. 1967;89:2719–2725. doi: 10.1021/ja00987a039. [DOI] [PubMed] [Google Scholar]
- 20.Candeias LP, Steenken S. J. Am. Chem. Soc. 1989;111:1094–1099. [Google Scholar]
- 21.Steenken S, Jovanovic SV. J. Am. Chem. Soc. 1997;119:617–618. [Google Scholar]
- 22.Crean C, Geacintov NE, Shafirovich V. Chem. Res. Toxicol. 2008;21:358–373. doi: 10.1021/tx700281e. [DOI] [PubMed] [Google Scholar]
- 23.Kishore K, Asmus K-D. J. Chem. Soc. Perkin Trans. 2. 1989:2079–2084. [Google Scholar]
- 24.Neta P, Grodkowski J, Ross AB. J. Phys. Chem. Ref. Data. 1996;25:709–1050. [Google Scholar]
- 25.Ludwig ML, Matthews RG. Annu. Rev. Biochem. 1997;66:269–313. doi: 10.1146/annurev.biochem.66.1.269. [DOI] [PubMed] [Google Scholar]
- 26.Endicott JF, Netzel TL. J. Am. Chem. Soc. 1979;101:4000–4002. [Google Scholar]
- 27.Chen E, Chance MR. Biochemistry. 1993;32:1480–1487. doi: 10.1021/bi00057a011. [DOI] [PubMed] [Google Scholar]
- 28.Shiang JJ, Walker LA, Anderson NA, Cole AG, Sension RJ. J. Phys. Chem. B. 1999;103:10532–10539. [Google Scholar]
- 29.Pacher P, Beckman JS, Liaudet L. Physiol. Rev. 2007;87:315–424. doi: 10.1152/physrev.00029.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Perrier S, Hau J, Gasparutto D, Cadet J, Favier A, Ravanat JL. J. Am. Chem. Soc. 2006;128:5703–5710. doi: 10.1021/ja057656i. [DOI] [PubMed] [Google Scholar]
- 31.Shafirovich V, Dourandin A, Huang W, Luneva NP, Geacintov NE. J. Phys. Chem. B. 1999;103:10924–10933. [Google Scholar]
- 32.Ivanov KL, Glebov EM, Plyusnin VF, Ivanov YV, Grivin VP, Bazhin NM. J. Photochem. Photobiol. A. 2000;133:99–104. [Google Scholar]
- 33.Shafirovich V, Dourandin A, Huang W, Geacintov NE. J. Biol. Chem. 2001;276:24621–24626. doi: 10.1074/jbc.M101131200. [DOI] [PubMed] [Google Scholar]
- 34.Hickel B. J. Phys. Chem. 1975;79:1054–1059. [Google Scholar]
- 35.McElroy WJ. J. Phys. Chem. 1990;94:2435–2441. [Google Scholar]
- 36.Steenken S, Jovanovic SV, Bietti M, Bernhard K. J. Am. Chem. Soc. 2000;122:2373–2374. [Google Scholar]
- 37.Sannes-Lowery KA, Mack DP, Hu P, Mei H-Y, Loo JA. J. Am. Soc. Mass Spectrom. 1997;8:90–95. [Google Scholar]
- 38.Ni J, Mathews MAA, McCloskey JA. Rapid. Commun. Mass Spectrom. 1997;11:535–540. doi: 10.1002/(SICI)1097-0231(199704)11:6<535::AID-RCM898>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- 39.Wang PP, Bartlett MG, Martin LB. Rapid. Commun. Mass Spectrom. 1997;11:846–856. [Google Scholar]
- 40.Constantinesco F, Benachenhou N, Motorin Y, Grosjean H. Nucleic Acids Res. 1998;26:3753–3761. doi: 10.1093/nar/26.16.3753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Joffe A, Geacintov NE, Shafirovich V. Chem. Res. Toxicol. 2003;16:1528–1538. doi: 10.1021/tx034142t. [DOI] [PubMed] [Google Scholar]
- 42.Crean C, Geacintov NE, Shafirovich V. Angew. Chem., Int. Ed. Engl. 2005;44:5057–5060. doi: 10.1002/anie.200500991. [DOI] [PubMed] [Google Scholar]
- 43.Candeias LP, Steenken S. J. Am. Chem. Soc. 1993;115:2437–2440. [Google Scholar]
- 44.Singh R, Kaur B, Farmer PB. Chem. Res. Toxicol. 2005;18:249–256. doi: 10.1021/tx049793j. [DOI] [PubMed] [Google Scholar]
- 45.Singh R, Farmer PB. Carcinogenesis. 2006;27:178–196. doi: 10.1093/carcin/bgi260. [DOI] [PubMed] [Google Scholar]
- 46.Nakao LS, Augusto O. Chem. Res. Toxicol. 1998;11:888–894. doi: 10.1021/tx9800351. [DOI] [PubMed] [Google Scholar]
- 47.Bachler V, Hildenbrand K. Radiat. Phys. Chem. 1992;40:59–68. [Google Scholar]
- 48.Adhikary A, Kumar A, Becker D, Sevilla MD. J. Phys. Chem. B. 2006;110:24171–24180. doi: 10.1021/jp064361y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hole EO, Sagstuen E. Radiat. Res. 1987;109:190–205. [PubMed] [Google Scholar]
- 50.Hole EO, Nelson WH, Sagstuen E, Close DM. Radiat. Res. 1992:119–138. [PubMed] [Google Scholar]
- 51.Wetmore SD, Boyd RJ, Eriksson LA. J. Phys. Chem. B. 1998;102:9332–9343. [Google Scholar]
- 52.Mundy J, Colvin ME, A. QA. J. Phys. Chem. A. 2002;106:10063–10071. [Google Scholar]
- 53.Misiaszek R, Crean C, Geacintov NE, Shafirovich V. J. Am. Chem. Soc. 2005;127:2191–2200. doi: 10.1021/ja044390r. [DOI] [PubMed] [Google Scholar]
- 54.Misiaszek R, Crean C, Joffe A, Geacintov NE, Shafirovich V. J. Biol. Chem. 2004;279:32106–32115. doi: 10.1074/jbc.M313904200. [DOI] [PubMed] [Google Scholar]
- 55.Wang W, Razskazovskii Y, Sevilla MD. Int. J. Radiat. Biol. 1997;71:387–399. doi: 10.1080/095530097144003. [DOI] [PubMed] [Google Scholar]
- 56.Candeias LP, Steenken S. Chem. Eur. J. 2000;6:475–484. doi: 10.1002/(sici)1521-3765(20000204)6:3<475::aid-chem475>3.0.co;2-e. [DOI] [PubMed] [Google Scholar]
- 57.Kofod P. J. Inorg. Biochem. 2004;98:1978–1980. doi: 10.1016/j.jinorgbio.2004.08.014. [DOI] [PubMed] [Google Scholar]
- 58.Luo YR. Handbook of Bond Dissociation Energies in Organic Compounds. CRC Press; Boca Raton, Florida.: 2003. [Google Scholar]
- 59.Steenken S, Jovanovic SV, Candeias LP, Reynisson J. Chem. Eur. J. 2001;7:2829–2833. doi: 10.1002/1521-3765(20010702)7:13<2829::aid-chem2829>3.0.co;2-n. [DOI] [PubMed] [Google Scholar]
- 60.Riordan CG, Wei P. J. Am. Chem. Soc. 1994;116:2189–2190. [Google Scholar]
- 61.Tanaka M, Ohkubo K, Fukuzumi S. J. Photochem. Photobiol. A. 2008;197:94–100. [Google Scholar]
- 62.Milligan JR, Ward JF. Radiation Res. 1994;137:295–299. [PubMed] [Google Scholar]
- 63.Brown JM, Wilson WR. Nature Rev. 2004;4:437–447. doi: 10.1038/nrc1367. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.