Abstract
Many seasonally breeding avian species exhibit marked changes in hypothalamic content of gonadotropin-releasing vhormone 1 (GNRH1) protein that are reflective of breeding condition. We recently cloned the GNRH1 cDNA in European starlings and demonstrated that changes in GNRH1 mRNA levels occur with a time course similar to what has been observed with GNRH1 protein. However, we did not previously resolve whether these differences were attributable to changes in the number of cells expressing the gene. Herein, we investigated photoperiod-induced changes in the number and distribution of GNRH1 mRNA-expressing cells in the preoptic area of male starlings. GNRH1 mRNA-expressing cell number was significantly greater in breeding birds than in nonbreeding birds. Starlings maintained in short nonstimulatory day length (i.e., prebreeding) showed intermediate cell numbers. Detailed analysis of the rostrocaudal and mediolateral distribution revealed that breeding birds had greater numbers of cells expressing GNRH1 mRNA in the medial intermediate, mediocaudal, and lateral intermediate preoptic area compared with prebreeding and nonbreeding birds. These data demonstrate that photoperiodic changes in reproductive state in starlings are associated with region-specific alterations in the number of cells expressing the GNRH1 gene. It remains to be determined whether these changes reflect quantitative differences in gene expression among an otherwise stable population of cells or a phenotypic switch in which cells gain or lose the ability to make GNRH1 mRNA in response to environmental cues.
Keywords: gonadotropin-releasing hormone, neuroendocrinology, seasonal reproduction
Starlings exhibit marked variation in the distribution and number of cells that express gonadotropin-releasing hormone 1 (GNRH1) mRNA in the hypothalamus during differing reproductive states.
INTRODUCTION
Gonadotropin-releasing hormone 1 (GNRH1) is essential for reproduction in vertebrates [1]. GNRH1 cell populations have been reported in specific brain regions in different bird orders, including galliformes (i.e., chickens and turkey [2–6]) and passeriformes (i.e., songbirds [7–16]). Immunocytochemical studies [2, 4, 5, 9, 17–19] demonstrated that GNRH1 cells are primarily distributed bilaterally along the midline in the preoptic area (POA), extending caudodorsally to the lateral septum. GNRH1 immunoreactive fibers are present throughout the preoptic septal areas and project ventrocaudally into the median eminence [2, 4, 16, 17]. Immunoreactive fibers also project directly to two circumventricular nuclei: the organum vasculosum of the lamina terminalis (OVLT) and the subseptal organ (SSO) [6]. The secretion of GNRH1 from the median eminence stimulates the release of gonadotropins, luteinizing hormone (LH), and follicle-stimulating hormone (FSH) from the pituitary gland [20, 21], ultimately controlling gonadal function.
In seasonally breeding birds, hypothalamic GNRH1 exhibits profound changes in synthesis and secretion that regulate annual changes in reproduction [22–25]. The length of the breeding period varies across species, and in some birds the breeding season is asymmetric with changes in photoperiod [26, 27]. In these cases, annual changes in day length give rise to three physiological states: photosensitive (i.e., prebreeding), photostimulated (i.e., breeding), and photorefractory (i.e., nonbreeding) [23, 27]. Photosensitive birds have the capacity to respond to an increase in photoperiod with a dramatic growth in the gonads and an increase in gonadotropins and sex steroid hormones [23]. We refer to them as “prebreeding” birds in this article. Photostimulated birds have large gonads and marked secretion of reproductive hormones characteristic of “breeding” birds, as we refer to them herein. Finally, photorefractory birds have experienced long day length, which leads to the regression of the reproductive axis and a “nonbreeding” condition. These birds can only be restored to a photosensitive state characteristic of the prebreeding condition if they experience short day length [23].
The European starling (Sturnus vulgaris) has been a valuable species for investigating the neural mechanisms controlling seasonal breeding in birds. In short photoperiods contiguous with the winter solstice, brain GNRH1 content is low, and the gonads are in a regressed state [9, 10]. Vernal increases in photoperiod stimulate dramatic increases in GNRH1 mRNA [28] and protein expression [12, 14–16, 29–32] in the POA. The increase in GNRH1 peptide secretion results in gonadal recrudescence in breeding birds. In many species, prolonged exposure to long day length results in a decline in hypothalamic GNRH1 protein content as measured via radioimmunoassay [13, 33, 34] or immunocytochemistry [9, 10, 15, 34]. The termination of photorefractoriness occurs after exposure to short day length and is characterized by a slight increase in GNRH1 protein content [33, 34]. Indeed, there is significant variation in GNRH1 gene expression across reproductive states; however, the degree of neuroanatomical specificity that occurs in association with these overall changes in GNRH1 mRNA expression is as yet unknown. Recently, data have accumulated indicating that the POA in birds is topographically organized based on functional criteria related to the control of reproductive behavior [35]. Whether variation in the GNRH1 system induced by photoperiodic control is restricted or particularly enhanced in specialized neuroanatomical subregions has not been determined, to our knowledge.
The distribution of GNRH1 mRNA has been described in few avian species [36–38]. We recently cloned GNRH1 cDNA in two songbird species: zebra finches (Taeniopygia guttata) and starlings [28]. However, we did not determine whether the variation in GNRH1 expression is the result of changes in the number of cells expressing the mRNA, as is observed with GNRH1 protein-expressing cells [19]. Herein, we describe the topographical distribution of GNRH1 mRNA-expressing cells in the POA of male starlings from three different reproductive states. These data provide novel insight into how changes in the reproductive condition are associated with region-specific variation in GNRH1 mRNA expression in the POA. These findings thus make a significant contribution to our understanding of the neural pathways that mediate the effects of environmental stimuli on the hypothalamo-pituitary-gonadal axis.
MATERIALS AND METHODS
Animals
All birds were treated and handled in accord with institutional, state, and federal animal care guidelines and permits. European starlings were captured using a drop-down V-trap in the early spring of 2008 in Conneautville, PA (41° 45 min north latitude and 80° 22 min west longitude). After capture, starlings were transported to The Johns Hopkins University and were group housed under the natural photoperiod. Food and water were provided ad libitum for the duration of the experiment. The birds and procedures used in the present study are thoroughly described in a previous publication [28]. In brief, a total of 20 male starlings in three reproductive states were used. All birds were caught in late winter and held on short day length (11L:13D) before beginning the experiment. Prebreeding birds (n = 7) were exposed to short day length (11L:13D) for the entire duration of the experiment, breeding birds (n = 6) were exposed to long day length (16L:8D) for 4 wk, and nonbreeding birds (n = 7) were exposed to long day length for 9 wk [28]. All birds were killed by rapid decapitation, and the treatment groups were organized so as to conclude the treatment phase on the same calendar day. Brains were quickly extracted, frozen on dry ice, and placed at −80°C before sectioning for in situ hybridization.
Assessment of Reproductive State
Reproductive condition was determined via testis volume measured as V = 4/3πa2b, where a is half the width, and b is half the length. Blood samples were collected at killing via puncturing of the alar (wing) vein with a 25-gauge needle, and 300–500 μl of blood was collected into heparinized tubes. The blood samples were transferred into microfuge tubes and centrifuged at 8900 rpm for 10 min. The plasma was removed and stored in Eppendorf vials at −20° until assayed for testosterone. The serum was analyzed in duplicate (50 μl) using a commercially available 125I Coat-A-Count kit for total testosterone (Siemens Medical Solutions Diagnostics, Los Angeles, CA). The antiserum is highly specific for testosterone (i.e., 100 pg/ml) and shows negligible cross-reactivity with other steroids, including dihydrotestosterone (<3.5%), 17β-estradiol (<0.01%), and corticosterone (<0.01%). The intraassay coefficient of variation averaged 12%.
In Situ Hybridization
Brains were sectioned on a cryostat at 20-μm thickness and mounted directly onto Superfrost Plus slides (Fisher Scientific, Santa Clara, CA) and stored at −20°C until in situ hybridization. Sections were collected beginning with the tractus septomesencephalicus (TSM) split and continued every fourth section (80 μm apart) until the caudal aspect of the anterior commissure. This sampling method differs from previous immunocytochemistry procedures that consist of thicker tissue sections and prevent precise comparisons of total cell counts using the two approaches. A plasmid-containing starling GNRH1 cDNA (GenBank accession number FJ178434) corresponding to the decapeptide GNRH1-associated peptide and the 3′ untranslated region (approximately 350 base pair) was linearized with SacI (sense; Promega, Madison, WI) or NcoI (antisense; Promega) and purified using GENE-CLEAN (Qbiogene, Vista, CA). S35-labeled cRNA probes (PerkinElmer Life and Analytical Sciences, Boston, MA) were transcribed by SP6 (antisense) or T7 (sense) RNA polymerases using a MAXISCRIPT kit (Ambion, Austin, TX). Unincorporated nucleotides were separated with NucAway spin columns (Ambion). Probe quality was verified by gel electrophoresis, and probe-specific activity was determined using a scintillation counter.
GNRH1 mRNA expression in slide-mounted sections was analyzed by in situ hybridization in two separate batches, and treatment groups were counterbalanced across batches. Slides were thawed and fixed for 10 min in 4% formaldehyde (diluted from 37% formaldehyde ampules; Ted Pella, Redding, CA) in 1× PBS (Ambion). Slides were rinsed in 4× saline sodium citrate (SSC; Ambion) and then in 0.1 M triethanolamine (TEA; Sigma-Aldrich, St. Louis, MO). Tissue charge was neutralized using 0.5% acetic anhydride (Sigma-Aldrich) in 0.1 M TEA for 10 min. Slides were transferred to 2× SSC for 5 min and then dehydrated in 50%, 75%, 95%, and 100% ethanol (5 min each). The slides were rehydrated in radiolabeled riboprobe solution (2.5 × 105 cpm/ml diluted in 1× hybridization buffer [Sigma-Aldrich] with 0.01 M dithiothreitol [Sigma-Aldrich]), sealed with cover glasses, and hybridized overnight in a mineral oil bath at 60°C. After hybridization, oil was removed by two chloroform rinses and two 2× SSC rinses. Slides were then washed at 60°C to remove nonspecific probe binding as follows: 20 min in 2× SSC and 50% formamide (Sigma-Aldrich), followed by two washes for 20 min each in 0.1× SSC. Slides were then dehydrated in increasing ethanol concentrations.
Emulsion
Slides were dipped in Kodak NTB-2 Autoradiography Emulsion (Carestream Health Systems, New Haven, CT). Slides were then dried for 15–20 min before placement in light-tight slide racks. The slide racks were then wrapped loosely in black paper sealed with black electrical tape and stored at 4°C for 6 days. The slides were developed by rinsing twice in xylene for 5 min, followed by two rinses in 100% ethanol for 5 min, and then placed in Kodak D-19 Developer (Fisher Scientific) at 16°C for 5 min. Slides were rinsed in a water bath and then transferred to a Kodak Fixer (Fisher Scientific) bath for 6 min. The slides were rinsed again in water before counterstaining with 1% cresyl violet for 5 min, rinsed in 0.1 M PBS, and then serially dehydrated in ethanol and placed in xylene for 10 min. Slides were coverslipped using Permount (Fisher Scientific).
Quantification of GNRH1 Cells
The distribution and number of GNRH1 mRNA-expressing cells were determined at 10× magnification using a brightfield light microscope (Zeiss Axioskop; Carl Zeiss, Thornwood, NY). Cells were evaluated throughout the brain and manually counted in the POA beginning at the TSM split to the caudal extent of the anterior commissure. GNRH1 mRNA cells were classified using two criteria: 1) there was a the clear distribution of silver grains over cresyl-stained cell bodies, and 2) the silver grain area exceeded 50 μm2 [14]. The second criterion was selected as an initial reference to provide the basis to eliminate false-positive cells due to artifacts. We utilized a sampling procedure to provide an estimate of GNRH1 cells by using methods developed by West [39] and subsequently applied to birds in other avian investigations [40]. Briefly, the focal plane was adjusted to view the number of GNRH1-expressing cells throughout the tissue section. Because we measured cells from every fourth section, we eliminated the possibility of counting the same cell twice. This sampling interval may produce an underestimate of the total cell number; however, because the same methods were used to analyze all birds in the study, we are confident that the comparison between groups provides an accurate assessment of relative cell number and distribution. Measurements for the three-dimensional analysis for each GNRH1 cell were obtained using a charge-coupled device camera connected to a Macintosh computer (Apple Inc., Cupertino, CA). The brain images were digitized, and distance was determined using Openlab 5.0.2 (Improvision Inc., Waltham, MA). The POA can be subdivided into functional subregions based on lesion and immediate-early gene experiments conducted in relation to POA regulation of reproductive and courtship behaviors [35, 41]. For example, POA lesions in male European starlings result in decreased singing, but the degree of these effects is selective depending on the specific subregion lesioned (i.e., rostral, intermediate, or caudal POA) [41]. Guided by these findings indicating that there are functional subdivisions within the POA, we devised a method for analyzing the regional effects of photoperiod on the number of GNRH1 cells in the POA. The z-axis was calculated based on the rostrocaudal orientation determined from the plane of section beginning at the posterior aspect of the anterior commissure and continuing rostral to the TSM split. For statistical analysis, the rostrocaudal plane was divided into three subdivisions consisting of rostral, intermediate, and caudal regions, which were equally divided into 800-μm lengths. The x-axis represents the mediolateral orientation, determined from the ependymal layer of the third ventricle to the center of the cell. The mediolateral plane was parsed into two areas that represented the medial 200 μm and the lateral area greater than 200 μm from the midline. The y-axis was determined by measuring the distance from the center of each cell to the base of the brain for each section.
Statistical Analysis
We conducted one-way ANOVAs to investigate the photoperiod regulation of reproductive state, plasma testosterone concentration, and testicular volume. We conducted a 3 × 6 two-way ANOVA to analyze the effects of photoperiod on the distribution of GNRH1 cells in the POA of starlings. The two factors consisted of treatment group with three levels (prebreeding, breeding, and nonbreeding) by the six brain regions (rostromedial, rostrolateral, intermediate medial, intermediate lateral, caudomedial, and caudolateral). Tukey pairwise tests were conducted for post hoc analyses. Significance was assessed at P < 0.05.
RESULTS
Male Starling Reproductive Condition
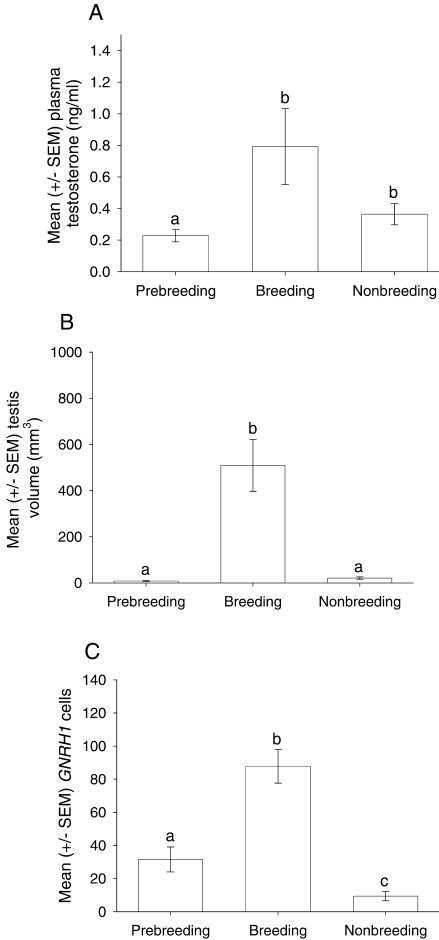
A one-way ANOVA revealed a significant difference in plasma testosterone concentration across treatment groups (F2,17 = 4.08, P < 0.05) (Fig. 1A). Post hoc analysis showed that breeding starlings had significantly greater concentration compared with prebreeding starlings (P < 0.05). However, there was no significant difference between prebreeding and nonbreeding starlings (P = 0.79) or between breeding and nonbreeding starlings (P = 0.09). There was a significant difference in testicular volume across treatment groups (F2,17 = 31.62, P < 0.001) (Fig. 1B). Tukey post hoc analysis demonstrated that breeding starlings had significantly greater volume compared with prebreeding and nonbreeding starlings (P < 0.001).
FIG. 1.
Reproductive state and comparison of GNRH1 mRNA-expressing cells in male prebreeding, breeding, and nonbreeding starlings. A) Mean ± SEM testosterone concentration. B) Mean ± SEM testicular volume. C) Mean ± SEM number of GNRH1 cells in the POA across treatment groups. Lowercase letters represent significant differences; see Results for details.
General Distribution and Photoperiodic Regulation of GNRH1 mRNA
A survey of the positively labeled cells revealed a caudodorsal pattern in the POA commencing at the TSM split toward the anterior commissure in breeding starlings. Specifically, GNRH1 mRNA was observed adjacent to the subcommissural division of the OVLT. GNRH1 mRNA was consistently observed across all treatment groups in the intermediate and caudal POA, comprising the precommissural division of the OVLT. GNRH1 mRNA was also identified symmetrically to the midline in the septal region adjacent to the supracommissural division of the OVLT (data not shown). However, these cells were sparsely distributed and did not display a clear pattern across sections or within or across treatment groups.
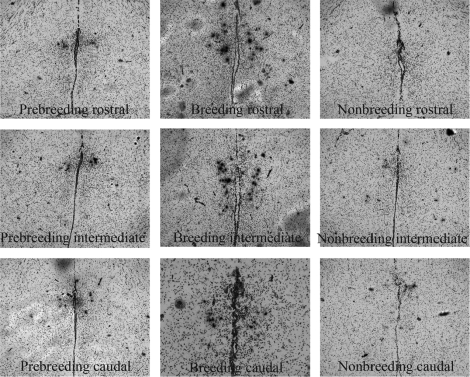
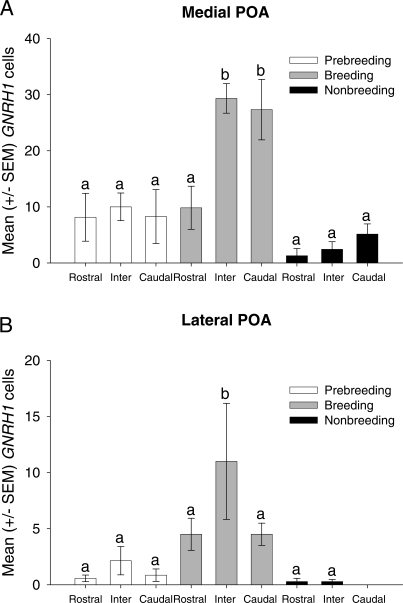
The distribution of GNRH1 mRNA varied significantly in male starlings across different reproductive states. Breeding starlings had a large number of cells (mean ± SEM, 87.83 ± 10.18) extending laterally from the ventricle throughout the entire POA (Figs. 1C and 2). In nonbreeding starlings, there were significantly fewer GNRH1 mRNA-expressing cells (mean ± SEM, 9.42 ± 2.79) (Figs. 1C and 2). Prebreeding starlings exhibited intermediate levels of GNRH1 mRNA-expressing cells (mean ± SEM, 31.00 ± 7.53) (Figs. 1C and 2). The two-way ANOVA revealed a significant main effect of treatment group (F2,102 = 39.80, P < 0.001) (Figs. 1C and 2). Tukey pairwise tests demonstrated that breeding starlings had significantly more GNRH1-expressing cells compared with prebreeding and nonbreeding starlings (P < 0.001). Moreover, prebreeding starlings had significantly more GNRH1 cells compared with nonbreeding starlings (P < 0.05). There was also a significant main effect of brain region (F5,102 = 14.73, P < 0.001). The intermediate medial POA region had significantly more GNRH1 cells compared with the other five regions (P < 0.001), and the caudomedial POA region had significantly more GNRH1 cells compared with the rostromedial, rostrolateral, intermediate lateral, and caudolateral regions (P < 0.005). There was a significant interaction between brain region and reproductive state (F10,102 = 4.37, P < 0.001) (Fig. 3, A and B). Post hoc analyses revealed that breeding starlings had more GNRH1-expressing cells in the intermediate medial POA compared with both prebreeding and nonbreeding starlings (P < 0.001) (Fig. 3A). Breeding starlings were also found to have significantly more GNRH1 cells in the caudomedial region of the POA compared with prebreeding and nonbreeding starlings (P < 0.001) (Fig. 3A). Breeding starlings also had greater numbers of GNRH1 cells in the intermediate lateral region compared with prebreeding and nonbreeding starlings (P < 0.01 and P < 0.05, respectively) (Fig. 3B). A three-dimensional reconstruction of the anatomical location of every GNRH1-expressing cell from all starlings in each treatment condition confirmed the regional changes in distribution (Fig. 4).
FIG. 2.
Representation of GNRH1 mRNA-expressing cells from coronal sections of the POA in prebreeding (left), breeding (center), and nonbreeding (right) starlings from rostral (top), intermediate (middle), and caudal (bottom) regions of the POA. Arrows indicate examples of positively labeled cells; bar = 200 μm.
FIG. 3.
Change in GNRH1 mRNA-expressing cells in different subregions in the POA. Mean ± SEM GNRH1 cell number in the medial (A) and lateral (B) POA from the rostral, intermediate (Inter), and caudal regions across treatment groups. Lowercase letters represent significant differences; see Results for details.
FIG. 4.
Three-dimensional representations demonstrating anatomical distribution of GNRH1 mRNA-expressing cells in the POA of starlings in prebreeding (n = 7), breeding (n = 6), and nonbreeding (n = 7) conditions. Graphs depict individual GNRH1 cells (indicated as black dots) from all starlings. The x-axis represents the mediolateral plane, the y-axis represents the ventrodorsal plane, and the z-axis represents the rostrocaudal plane. All measurements are in micrometers, and the z-axis corresponds to the area between the TSM split (2500 μm) and the anterior commissure (indicated as 0). Arrows along the rostrocaudal axis signify boundaries of anatomical subregions.
DISCUSSION
Herein, we show that the distribution and number of GNRH1 mRNA-expressing cells vary profoundly across reproductive states in male starlings. In breeding starlings, there is an overall increase in the number of GNRH1 mRNA-expressing cells, with the predominant increase in the intermediate and caudomedial regions of the POA compared with prebreeding and nonbreeding birds. Additionally, there was a nonsignificant trend for GNRH1 cell numbers to increase in the rostral and lateral directions in prebreeding and breeding starlings relative to nonbreeding starlings. These data suggest that the progression from prebreeding through breeding results in an increase in GNRH1 expression by cells localized primarily along the midline of the POA, with a substantial regression in these populations in association with the onset of photorefractoriness (i.e., nonbreeding condition).
The pattern of GNRH1 mRNA expression reported herein is consistent with the distribution recently reported in zebra finches [36], as well as immunocytochemical studies of GNRH1 protein-expressing cells in starlings [9] and other songbird species [12, 14, 16, 30]. In starlings, the majority of GNRH1 immunoreactive cells are localized in the subcommissural division of the OVLT, precommissural division of the OVLT, and supracommissural division of the OVLT [9]. The present analysis could not clearly differentiate between the three regions, and we therefore refer to this as the POA. Instead, we subdivided the distribution of GNRH1 mRNA in the POA equally into three distinct regions and observed that breeding starlings have increased GNRH1 cell numbers throughout the entire POA, with the vast majority observed along the midline, with a slight but nonsignificant increase in the rostral and lateral subdivisions. With the onset of a nonbreeding state, GNRH1 cell numbers are significantly down-regulated, resulting in the retention of only a small population of cells in the intermediate and caudal regions of the POA. These findings are consistent with the observed changes in GNRH1 protein measured via radioimmunoassay and immunocytochemistry in starlings [7, 9, 10, 15, 34]. Unlike many mammalian species, the control of seasonal breeding includes a gradual increase in GNRH1 due to renewed synthesis in the POA in prebreeding birds [23, 33, 34]. Subsequent photostimulation by long day length then independently increases GNRH1 synthesis and stimulates GNRH1 secretion. The findings herein suggest that hypothalamo-pituitary control of reproduction by photoperiod involves two steps: 1) an increase in GNRH1 mRNA that leads to greater GNRH1 peptide synthesis and 2) photoperiod-induced secretion of GNRH1 from the median eminence. However, further research investigating how the change in photoperiod independently regulates GNRH1 mRNA expression in the POA and GNRH1 secretion from the median eminence is required. For example, studies [42, 43] in Japanese quail have demonstrated that photoperiodically driven changes in thyroid hormone metabolism set off a sequence of cellular events that regulate GNRH1 release in the median eminence in a seasonal context. In quail, photorefractoriness related to the onset of a nonbreeding state is not associated with a marked reduction in GNRH1 [19]. It would be useful to establish whether similar photoperiodically driven effects on GNRH1 release are occurring in starlings along with the effects on GNRH1 gene expression described in this article.
It is important to note that the change in GNRH1 cell numbers detected via immunocytochemistry [9] and in situ hybridization (presented herein) is presumably not the result of a cycle of neurogenesis that is followed by apoptosis. Adult neurogenesis is widespread in the avian brain; however, it has not been observed in the POA [44, 45]. Instead, the change in GNRH1 cell numbers seems to be driven primarily by substantial alterations in GNRH1 transcription and translation within extant populations of cells, which facilitates or hinders their detection by immunocytochemistry or in situ hybridization. Whether the decline in cell numbers in nonbreeding birds represents a total inhibition of GNRH1 gene expression or simply a reduction to undetectable levels is not yet clear.
Results of several investigations have suggested that subregions of the POA are functionally specialized based on the control of reproductive behaviors and that these functional subdivisions are associated with neurochemical and hodological specializations [35]. The findings reported herein provide further evidence for a functional topography underlying the organization of the POA. We recently observed that male starlings housed with females have significantly greater numbers of GNRH1 immunoreactive cells compared with males housed alone [46]. Moreover, the increase in cell number occurred in the rostral and intermediate regions of the POA. Taken together, the findings of these studies suggest that specific subregions of the POA integrate photoperiodic and social cues. However, an alternative hypothesis from these studies it that these cells only respond to some particular cues, or it could be that the intermediate population of GNRH1 cells is significantly more plastic in general and readily changes expression patterns depending on environmental stimuli. This hypothesis is supported by findings in male ring doves (Streptopelia risoria), which demonstrate an increase in GNRH1 mRNA cell numbers in the POA for 1 h after engaging in copulatory behaviors [47]. These authors did not report where the increase in GNRH1 cells was localized.
In conclusion, the annual change in photoperiod has dramatic effects on GNRH1 mRNA expression in starlings, which underlies observed variation in reproductive physiology in this species [28]. Herein, we show that overall changes in GNRH1 mRNA reflect alterations in the number and distribution of cells expressing the GNRH1 mRNA. It appears that GNRH1 expression is up-regulated in specific subpopulations of cells within the POA to stimulate gonadal recrudescence among birds in breeding condition. Anatomical specificity of the GNRH1 expression in these cells is either restrained or actively inhibited in prebreeding or nonbreeding birds to maintain gonads in a regressed state so as to prevent reproductive efforts at inappropriate times of the year. Last, previous findings in pigs suggest that subregions of GnRH1 immunoreactive cells in the rostral POA are activated during the GnRH surge [48], and results in rats indicate that there are separate GnRH1 subgroups regulating the LH surge vs. pulsatile release [49]. It appears that functional differences in the GnRH system may be more common than previously thought.
Acknowledgments
We thank Dr. Margaret McCarthy for assistance with the emulsion protocol and Ying Wang and Adam Troyer for technical assistance.
Footnotes
1Supported by NIH/NINDS R01 NS35467 (to G.F.B.) and NIH 5R01 HD047794 (to D.J.B.). T.J.S. is an NSERC Predoctoral Fellow (PGS-D 334570). D.J.B. is a Chercheur-Boursier of the Fonds de la Recherche en Santé du Québec.
REFERENCES
- Gore AC. GnRH: The Master Molecule of Reproduction. Norwell, MA:: Kluwer Academic Publishers;; 2002. [Google Scholar]
- Mikami S, Yamada S, Hasegawa Y, Miyamoto K.Localization of avian LHRH-immunoreactive neurons in the hypothalamus of the domestic fowl (Gallus domesticus) and the Japanese quail (Coturnix coturnix). Cell Tissue Res 1988; 251: 51–58.. [DOI] [PubMed] [Google Scholar]
- Katz IA, Millar RP, King JA.Differential regional distribution and release of two forms of gonadotropin-releasing hormone in the chicken brain. Peptides 1990; 11: 443–450.. [DOI] [PubMed] [Google Scholar]
- Kuenzel WJ, Blähser S.The distribution of gonadotropin-releasing hormone (GnRH) neurons and fibers throughout the chick brain (Gallus domesticus). Cell Tissue Res 1991; 264: 481–495.. [DOI] [PubMed] [Google Scholar]
- Millam JR, Faris PL, Youngren OM, El Halawani ME, Hartman BK.Immunohistochemical localization of chicken gonadotropin-releasing hormone I and II (cGNRH1 and II) in turkey hen brain. J Comp Neurol 1993; 333: 68–82.. [DOI] [PubMed] [Google Scholar]
- Kuenzel WJ, Golden CD.Distribution and change in number of gonadotropin-releasing hormone-1 neurons following activation of the photoneuroendocrine system in the chick, Gallus gallus. Cell Tissue Res 2006; 325: 501–512.. [DOI] [PubMed] [Google Scholar]
- Dawson A, Follett BK, Goldsmith AR, Nicholls TJ.Hypothalamic gonadotropin-releasing hormone and pituitary and plasma FSH and prolactin during photostimulation and photorefractoriness in intact and thyroidectomized starlings (Sturnus vulgaris). J Endocrinol 1985; 105: 71–77.. [DOI] [PubMed] [Google Scholar]
- Blähser S, Oksche A, Farner DS.Projection of fibers immunoreactive to an antiserum against gonadoliberin (LHRH) into the pineal stalk of the white-crowned sparrow. Zonotrichia leucophrys gambelii. Cell Tissue Res 1986; 244: 193–196.. [Google Scholar]
- Foster RG, Plowman G, Goldsmith AR, Follett BK.Immunohistochemical demonstration of marked changes in the LHRH system of pre-breeding and non-breeding European starlings (Sturnus vulgaris). J Endocrinol 1987; 115: 211–220.. [DOI] [PubMed] [Google Scholar]
- Goldsmith AR, Ivings WE, Pearce-Kelly AS, Parry DM, Plowman DM, Nicholls TJ, Follett BK.Photoperiodic control of the LHRH neurosecretory system of European starlings (Sturnus vulgaris) during puberty and the onset of photorefractoriness. J Endocrinol 1989; 122: 225–268.. [DOI] [PubMed] [Google Scholar]
- Blähser S, King JA, Kuenzel WJ.Testing of arg-8-gonadotropin-releasing hormone directed antisera by immunological and immunocytochemical methods for use in comparative studies. Histochemistry 1989; 93: 39–48.. [DOI] [PubMed] [Google Scholar]
- Deviche P, Saldanha CJ, Silver R.Changes in brain gonadotropin-releasing hormone and vasoactive intestinal polypeptide-like immunoreactivity accompanying reestablishment of photosensitivity in male dark-eyed juncos (Junco hyemalis). Gen Comp Endocrinol 2000; 117: 8–19.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saldanha CJ, Deviche PJ, Silver R.Increased VIP and decreased GnRH expression in non-breeding dark-eyed juncos (Junco hyemalis). Gen Comp Endocrinol 1994; 93: 128–136.. [DOI] [PubMed] [Google Scholar]
- Hahn TP, Ball GF.Changes in brain GnRH associated with photorefractoriness in house sparrows (Passer domesticus). Gen Comp Endocrinol 1995; 99: 349–363.. [DOI] [PubMed] [Google Scholar]
- Parry DM, Goldsmith AR, Millar RP, Glennie LM.Immunocytochemical localization of GnRH precursor in the hypothalamus of European starlings during sexual maturation and photorefractoriness. J Neuroendocrinol 1997; 9: 235–243.. [DOI] [PubMed] [Google Scholar]
- Stevenson TJ, MacDougall-Shackleton SA.Season- and age-related variation in neural cGNRH1 and cGNRH1I immunoreactivity in house sparrows (Passer domesticus). Gen Comp Endocrinol 2005; 143: 33–39.. [DOI] [PubMed] [Google Scholar]
- van Gils J, Absil P, Grauwels L, Moons L, Vandesande F, Balthazart J.Distribution of luteinizing hormone-releasing hormone I and II (LHRH-I and -II) in the quail and chicken brain as demonstrated with antibodies directed against synthetic peptides. J Comp Neurol 1993; 334: 304–323.. [DOI] [PubMed] [Google Scholar]
- Jozsa R, Mess B.Immunohistochemical localization of the luteinizing hormone releasing hormone (LHRH)-containing structures in the central nervous system of the domestic fowl. Cell Tissue Res 1982; 227: 451–458.. [DOI] [PubMed] [Google Scholar]
- Foster RG, Panzica GC, Parry DM, Viglietti-Panzica C.Immunocytochemical studies on the LHRH system of the Japanese quail: influence by photoperiod and aspects of sexual differentiation. Cell Tissue Res 1988; 253: 327–335.. [DOI] [PubMed] [Google Scholar]
- Sharp PJ, Talbot RT, Main GM, Dunn IC, Fraser HM, Huskisson NS.Physiological roles of chicken LHRH-I and -II in the control of gonadotropin release in the domestic chicken. J Endocrinol 1990; 124: 291–299.. [DOI] [PubMed] [Google Scholar]
- McNaugton FJ, Dawson A, Goldsmith AR.A comparison of the responses to gonadotropin-releasing hormone of adult and juvenile, and photosensitive and photorefractory European starlings, Sturnus vulgaris. Gen Comp Endocrinol 1995; 97: 135–144.. [DOI] [PubMed] [Google Scholar]
- Sharp PJ.Photoperiodic regulation of seasonal breeding in birds. Ann N Y Acad Sci 2005; 1040: 189–199.. [DOI] [PubMed] [Google Scholar]
- Dawson A, King VM, Bentley GE, Ball GF.Photoperiodic control of seasonality in birds. J Biol Rhythms 2001; 16: 365–380.. [DOI] [PubMed] [Google Scholar]
- Ball GF.The neural integration of environmental information by seasonally breeding birds. Am Zool 1993; 33: 185–199.. [Google Scholar]
- Ball GF, Hahn TP.GnRH neuronal system in birds and their relation to the control of seasonal reproduction. Parhar IS, Sakuma Y.GnRH Neurons: Gene to Behavior Tokyo:Brain Shuppan Publishers;1997: 325–342.. [Google Scholar]
- Nicholls TJ, Goldsmith AR, Dawson A.Photorefractoriness in birds and comparison with mammals. Physiol Rev 1988; 68: 133–176.. [DOI] [PubMed] [Google Scholar]
- Dawson A, Sharp PJ.Photorefractoriness in birds: photoperiodic and non-photoperiodic control. Gen Comp Endocrinol 2007; 153: 378–384.. [DOI] [PubMed] [Google Scholar]
- Stevenson TJ, Lynch KS, Lamba P, Ball GF, Bernard DJ.Cloning of gonadotropin-releasing hormone I complementary DNAs in songbirds facilitates dissection of mechanisms mediating seasonal changes in reproduction. Endocrinology 2009; 150: 385–394.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholls TJ, Goldsmith AR, Dawson A.Photorefractoriness in European starlings: associated hypothalamic changes and the involvement of thyroid hormones and prolactin. J Exp Zool 1984; 232: 567–572.. [DOI] [PubMed] [Google Scholar]
- Cho RN, Hahn TP, MacDougall-Shackleton SA, Ball GF.Seasonal variation in brain GnRH in free-living breeding and non-breeding house finches (Carpodacus mexicanus). Gen Comp Endocrinol 1998; 109: 244–250.. [DOI] [PubMed] [Google Scholar]
- Dawson A, Talbot RT, Dunn IC, Sharp PJ.Changes in basal hypothalamic chicken gonadaotropin-releasing hormone-I and vasoactive intestinal polypeptides associated with a photo-induced cycle in gonadal maturation and prolactin secretion in intact and thyroidectomized starlings (Sturnus vulgaris). J Neuroendocrinol 2002; 14: 533–539.. [DOI] [PubMed] [Google Scholar]
- Meddle SL, Bush S, Sharp PJ, Millar RP, Wingfield JC.Hypothalamic pro-GnRH-GAP, GnRH-I and GnRH-II during the onset of photorefractoriness in the white-crowned sparrow (Zonotrichia leucophrys gambelii). J Neuroendocrinol 2006; 18: 217–226.. [DOI] [PubMed] [Google Scholar]
- Dawson A, Goldsmith AR, Nicholls TJ, Follett BK.Endocrine changes associated with the termination of photorefractoriness by short daylengths and thyroidectomy in starlings (Sturnus vulgaris). J Endocrinol 1986; 110: 73–79.. [DOI] [PubMed] [Google Scholar]
- Dawson A, Goldsmith AR.Changes in gonadotropin-releasing hormone (GnRH-I) in the preoptic area and median eminence of starlings (Sturnus vulgaris) during the recovery of photosensitivity and during photostimulation. J Reprod Fertil 1997; 111: 1–6.. [DOI] [PubMed] [Google Scholar]
- Balthazart J, Ball GF.Topography in the preoptic region: differential regulation of appetitive and consummatory male sexual behavior. Front Neuroendocrinol 2007; 28: 161–178.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ubuka T, Bentley GE.Identification, localization, and regulation of passerine GnRH-I messenger RNA. J Endocrinol 2009; 201(1):81–87.. [DOI] [PubMed] [Google Scholar]
- Dunn IC, Chen Y, Hook C, Sharp PJ, Sang HM.Characterization of the chicken preprogonadotrophin-releasing hormone-I gene. J Mol Endocrinol 1993; 11: 19–29.. [DOI] [PubMed] [Google Scholar]
- Sun YM, Dunn IC, Baines E, Talbot RT, Illing N, Millar RP, Sharp PJ.Distribution and regulation by oestrogen of fully processed and variant transcripts of gonadotropin releasing hormone I and gonadotropin releasing hormone receptor mRNAs in the male chicken. J Neuroendocrinol 2001; 13: 37–49.. [DOI] [PubMed] [Google Scholar]
- West MJ.New stereological methods for counting neurons. Neurobiol Aging 1993; 4: 275–285.. [DOI] [PubMed] [Google Scholar]
- Tramontin AD, Smith GT, Breuner CW, Brenowitz EA.Seasonal plasticity and sexual dimorphism in the avian song control system: stereological measurement of neuron density and number. J Comp Neurol 1998; 396: 186–192.. [DOI] [PubMed] [Google Scholar]
- Riters LV, Ball GF.Lesions to the medial preoptic area affect singing in the male European starlings (Sturnus vulgaris). Horm Behav 1999; 36: 276–286.. [DOI] [PubMed] [Google Scholar]
- Yamamura T, Hirunagi K, Ebihara S, Yoshimura T.Seasonal morphological changes in the neuro-glial interaction between gonadotropin-releasing hormone nerve terminals and glial endfeet in Japanese quail. Endocrinology 2004; 145: 4264–4267.. [DOI] [PubMed] [Google Scholar]
- Yamamura T, Yasuo S, Hirunagi K, Ebihara S, Yoshimura T.T-3 implantation mimics photoperiodically reduced encasement of nerve terminals by glial processes in the median eminence of Japanese quail. Cell Tissue Res 2006; 324: 175–179.. [DOI] [PubMed] [Google Scholar]
- Nottebohm F.Neuronal replacement in adulthood. Ann N Y Acad Sci 1985; 457: 143–161.. [DOI] [PubMed] [Google Scholar]
- Nottebohm F.From birdsong to neurogenesis. Sci Am 1989; 260: 74–79.. [DOI] [PubMed] [Google Scholar]
- Stevenson TJ, Bernard DJ, Ball GF. The Society for Integrative and Comparative Biology Annual Meeting 2009 Abstract Booklet-Oral Presentations. Boston:: Society for Integrative and Comparative Biology;; 2009. Photic and non-photic regulation of GNRH1 in male European starlings (Sturnus vulgaris). Abstract 29.3. [Google Scholar]
- Mantei KE, Ramakrishnan S, Sharp PJ, Buntin JD.Courtship interactions stimulate rapid changes in GnRH synthesis in male ring doves. Horm Behav 2008; 54: 669–675.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King JC, Liu E, Ronsheim PM, Slonimski M, Rubin BS.Expression of Fos within luteinizing hormone-releasing hormone neurons in relation to the steroid-induced luteinizing hormone surge in guinea pigs. Biol Reprod 1998; 58: 316–322.. [DOI] [PubMed] [Google Scholar]
- Kimura F, Funabashi T.Two subgroups of gonadotropin-releasing hormone neurons control gonadotropin secretion in rats. News Physiol Sci 1998; 13: 225–231.. [DOI] [PubMed] [Google Scholar]