Abstract
In mammalian spermatocytes, cell division cycle protein 2 (CDC2)/cyclin B1 and the chaperone heat shock protein A2 (HSPA2) are required for the G2→M transition in prophase I. Here, we demonstrate that in primary spermatocytes, linker histone chaperone testis/embryo form of nuclear autoantigenic sperm protein (tNASP) binds the heat shock protein HSPA2, which localizes on the synaptonemal complex of spermatocytes. Significantly, the tNASP-HSPA2 complex binds linker histones and CDC2, forming a larger complex. We demonstrate that increasing amounts of tNASP favor tNASP-HSPA2-CDC2 complex formation. Binding of linker histones to tNASP significantly increases HSPA2 ATPase activity and the capacity of tNASP to bind HSPA2 and CDC2, precluding CDC2/cyclin B1 complex formation and, consequently, decreasing CDC2/cyclin B1 kinase activity. Linker histone binding to NASP controls the ability of HSPA2 to activate CDC2 for CDC2/cyclin B1 complex formation; therefore, tNASP's role is to provide the functional link between linker histones and cell cycle progression during meiosis.
Keywords: CDC2/cyclin B1, cell cycle, HSPA2, linker histones, meiosis, NASP, spermatogenesis, synamptonemal complex, testis
Linker histone binding to nuclear autoantigenic sperm protein (NASP) controls the ability of HSPA2 to activate CDC2 for CDC2/cyclin B1 complex formation; therefore, tNASP's role in the synaptonemal complex is to provide a link between DNA repair and cell cycle progression during meiosis.
INTRODUCTION
During male meiosis, the synapsed homologous chromosomes undergo extensive nucleosome remodeling when a testis-specific “histone code” is generated, combining both non-tissue-specific histone variants and testis-specific members [1]. In pachytene spermatocytes, histone variant H3.3 is incorporated into sex chromosome nucleosomes, and the testis-specific linker histone variant HIST1H1T (also known as H1T) is incorporated throughout the chromosomes [2]. As the synapsed chromosomes reorganize, they require the chaperones associated with nucleosome remodeling and DNA repair events [3], eventually signaling the cell cycle program to indicate the completion of these tasks. Signals to the cell cycle program are generally known as meiotic recombination or pachytene checkpoints (CHKs) that link the repair of double-strand DNA breaks through a series of kinase activation pathways to cyclin-dependent kinases (CDKs) [4, 5]. In yeast, Wee1 and Mik1 kinases inhibit cell division cycle protein 2 (CDC2; also known as CDK1) activation by phosphorylation of tyrosine 15, and Mek1 (CHK2) kinase promotes inhibition by phosphorylation of CDC25 [5]. In mammals, CDC2/cyclin B1 kinase activity in pachytene spermatocytes is required for the G2→M transition in prophase I [6, 7]. The G2→M transition also requires cyclin A, which may partner with CDK1 or CDK2 during meiosis [8–10]. Furthermore, the formation of DNA double-strand breaks (DSBs) that occur at the beginning of meiotic recombination induces cyclin A1/CDK2 activation, which has downstream effects on XRCC6 (also known as Ku70) [11], a DNA repair protein that can regulate the choice of repair pathways [12]. Concomitantly with DSBs, pachytene spermatocytes require CDK2 for the proper formation of the synaptonemal complex [13], which has as one of the components of its lateral elements a testis chaperone, heat shock protein A2 (HSPA2) [14]. HSPA2 is necessary for the activation of CDC2 (CDK1) to form the active CDC2/cyclin B1 complex [14–17], and as such it may provide a link between the synapsed chromosomes and the cell cycle component CDC2.
Previous studies on the linker histone chaperone, nuclear autoantigenic sperm protein (NASP), found that NASP associates with heat shock protein HS90AA1 (also known as HSP90) in the cytoplasm of mouse germ cells, which facilitates NASP's loading of H1 histones for transport to the nucleus [18]. Inside the nucleus, NASP has been reported in the chromatin assembly factor 1, subunit A (CHAF1A; also known as CAF-1) multichaperone complex during nucleosome remodeling [19], and it has been shown to be required for progression from G1 to S phase [20, 21]. Cells normally regulate the amount of NASP present in the cell because either the loss of NASP [21] or the overexpression of NASP [20] delays progression through the G1/S border, and targeted mutation of the Nasp gene causes early embryonic lethality [21]. In addition to being essential for progression through the G1/S border, NASP is phosphorylated after DNA damage by irradiation of U2OS cells [22], and it binds to XRCC6/XRCC5 (Ku70/Ku80)/DNA-PK in HeLa cells [23], implying that NASP is present during DSB repair [24]. Recent in vitro chromatin reconstitution experiments have demonstrated that NASP facilitates the incorporation of linker histones onto nucleosome arrays [25].
In this report, we have extended our study of the physiological role of the testis/embryo form of nuclear autoantigenic sperm protein (tNASP) and report that during the first meiotic division in mouse germ cells, tNASP is associated with testis HSPA2 and localized on the synaptonemal complex. Moreover, linker histone binding to tNASP increases HSPA2 ATPase activity, subsequently increasing binding of CDC2 to the H1-tNASP-HSPA2 complex and concomitantly decreasing the amount of active CDC2/cyclin B1. Because active CDC2/cyclin B1 complexes are required for the G2→M transition during meiosis and H1-tNASP controls the ATPase activity of HSPA2 and the ability to bind CDC2, tNASP's role in the synaptonemal complex may be to provide the functional link between linker histones and cell cycle progression during meiosis.
MATERIALS AND METHODS
Materials
All of the chemicals and reagents used in this study were of molecular biology grade. The restriction enzymes were purchased from Roche Applied Science (Indianapolis, IN). Purification of plasmid DNA and PCR products was carried out using QIAprep Miniprep and QIAquick PCR purification kits (Qiagen, Valencia, CA), and sequencing was performed at the University of North Carolina at Chapel Hill automated sequencing facility. Mouse monoclonal anti-CDC2 antibody (sc-54), rabbit polyclonal anti-CDC2 antibody (sc-954), and rabbit polyclonal anti-cyclin B1 antibody (H-433) were purchased from Santa Cruz Biotechnology (Santa Cruz, CA). Rabbit polyclonal anti-HSPA2 antibody was a gift from Dr. E.M. Eddy (Laboratory of Reproductive and Developmental Toxicology, National Institute of Environmental Health Sciences, National Institutes of Health, Research Triangle Park, NC). Rabbit antiserum to full-length recombinant mouse tNASP (AAB87567) and goat antiserum to full-length human tNASP (AAH10105) were custom made by Bethyl Laboratories (Montgomery, TX). Mouse monoclonal Penta-His Antibody was purchased from Qiagen.
Construction of Expression Vectors
The entire coding sequence of mouse Nasp (nucleotides 92-2406; GenBank accession number AF034610) was amplified from mouse testis Quick-clone cDNA (Clontech, Palo Alto, CA) as described previously [20]. The entire coding sequences of mouse Hspa2 (nucleotides 1-1902; GenBank accession number M20567), mouse histone Hist1h1t (also known as H1t; nucleotides 56-682; GenBank accession number NM_010377), mouse histone Hist1h1a (also known as H1a; nucleotides 50-688; GenBank accession number NM_030609), and mouse Cdc2a (nucleotides 25-918; GenBank accession number M38724) were amplified from mouse testis Quick-clone cDNA (Clontech), cloned into pEXP5-CT/TOPO vector (Invitrogen, Carlsbad, CA), expressed in BL21-AI cells (Invitrogen), and purified using Ni-NTA Agarose (Qiagen).
Affinity Chromatography
Affinity chromatography was carried out with the recombinant CDC2 or tNASP proteins coupled to Aminolink Plus Coupling Gel (Pierce, Rockford, IL). After the remaining binding sites were blocked by primary amine-containing buffer, proteins of interest, lysates, or their combinations were mixed with tNASP-coupled beads or CDC2-coupled beads and incubated. After extensive washing, the gel with coupled proteins was boiled in nonreducing SDS containing sample buffer, separated by SDS-PAGE, blotted, and probed with the appropriate antibody. Amount of bound proteins was calculated from background-subtracted images using TL100 software (Nonlinear Dynamics, Newcastle Upon Tyne, U.K.).
Immunoprecipitation
Lysates or protein mixtures were cleared by incubation with 15 μl of UltraLink Immobilized Protein A/G (Pierce). Precipitations were carried out with mouse monoclonal anti-CDC2 antibody, mouse monoclonal anti-histone H1 antibody, rabbit polyclonal anti-cyclin B1 antibody, affinity-purified goat polyclonal anti-tNASP antibody, or mouse monoclonal anti-5His antibody. Aliquots of precleared lysates were incubated with antibody for 1 h at 4°C with constant shaking. Antibody-antigen complexes were collected by 15 μl of Ultra Link Immobilized Protein A/G (Pierce) and eluted by boiling in reducing (β-mercaptoethanol) SDS-PAGE sample buffer. Samples were separated in 10%–20% SDS-polyacrylamide gels (Bio-Rad Laboratories, Richmond, CA) by electrophoresis.
Spermatogenic Cells, Testis Lysate, and Testicular Tissue
Germ cells from male mice (60–80 days after birth) were isolated, separated, and briefly cultured as described previously [26]. Decapsulated mouse testes were subjected to sequential action of collagenase and trypsin. Intercellular connections were disrupted by intense pipetting. After the cell mixture was filtered through the 80-μm mesh, the cells were grown in plastic flasks for 16–18 h to allow Sertoli cells to adhere to the dish and be selectively discarded as the germ cells were harvested. This procedure resulted in a mixed germ cell population. All experiments on mice conformed to the relevant regulatory standards approved by the University of North Carolina at Chapel Hill Institutional Animal Care and Use Committee. Testis lysate was obtained from decapsulated mouse testes using an electric Tissue-Tearor (BioSpec Products, Bartlesville, OK) with extraction in T-PER Tissue Protein Extraction Reagent (Pierce). For histological study, mouse testes were fixed in Bouin solution, paraffin embedded, and stained with anti-tNASP antibody or control antibody absorbed with antigen as described previously [27].
Surface-Spread Meiotic Nuclei Preparation
A mixed mouse germ cell population, obtained as described above, was washed twice with minimum essential medium, and long-tipped glass pipettes were used to draw small drops of cell suspension and place them on the surface of a 0.5% solution of NaCl in distilled water [28]. The spread cells were collected by touching the surface with poly-l-lysine-coated slides and fixed in 0.4% paraformaldehyde (0.5 mM borate buffer, pH 8.2, containing 0.03% SDS) for 4 min, followed by a 5-min incubation in a 0.5 mg/ml solution of sodium borohydride in distilled water. After washing twice in 50 mM Tris buffer (pH 7.6), slides were washed with 0.05% Triton X-100 (90 sec), washed in PBS twice, and incubated in primary antibody for 1 h and secondary antibody for 1 h. To detect tNASP and HSPA2 simultaneously, chromosome spreads were first stained with goat anti-NASP antibody and secondary donkey anti-goat IgG-Alexa Fluor 568 (Molecular Probes, Eugene, OR), fixed a second time in paraformaldehyde 6 min [29], and after thorough washing in PBS, samples were stained with primary anti-HSPA2 antibody and with secondary goat anti-rabbit immunoglobulin G (IgG)-AlexaFluor 488 (Molecular Probes, Eugene, OR) antibody. Staining with 4′,6′-diamidino-2-phenylindole (1:40 000) was employed for DNA detection. Slides were covered with Prolong Gold Antifade reagent (Invitrogen, Eugene, OR) and examined with a Zeiss fluorescence microscope. Secondary antibody controls, included in all experiments, were negative (data not shown).
Preparation of Nuclear Fraction of Mouse Spermatogenic Cells
Harvested mouse spermatogenic cells were washed in PBS, followed by washes in Earle Balanced Salt Solution, and were resuspended in 10 volumes of RBS buffer (10 mM Tris-HCl, pH 7.4; 10 mM NaCl; and 3 mM MgCl2) as described previously [18]. Nuclei were prepared essentially as described previously [30]. Microscopic examination of the nuclei and Western blotting (staining for histones) of nuclear and cytoplasmic fractions confirmed their separation.
Mass Spectrometry Identification of tNASP-Binding Partners
The identification of tNASP-binding partners has been described previously [18, 23]. Mass spectrometry identification was done in the University of North Carolina at Chapel Hill Proteomics Core Laboratory.
Colorimetric Determination of ATPase Activity (Malachite Green Assay)
The amount of inorganic phosphate in solution was determined by a malachite green assay based on the formation of a phosphomolybdate complex as described previously [31], with some modifications. Typical protein concentrations were 0.5 μM for HSPA2, CDC2, tNASP, and HIST1H1T. The reaction was carried out in assay buffer (40 mM HEPES, pH 7.7; and 5 mM MgCl2) containing 125 μM adenosine-5′-triphosphate disodium salt (Calbiochem, San Diego, CA), which resulted in an intermediate-level reading to detect the effect of added proteins on HSPA2 ATPase activity. Proteins to be tested were mixed and incubated at 37°C. Stock solutions of malachite green (Sigma, St. Louis, MO), polyvinyl alcohol USP (Sigma), ammonium molybdate (Mallinckrodt, St. Louis, MO), and water were mixed in the ratio 2:1:1:2, and 80 μl was added to 50-μl reaction mixture in the wells of a 96-well clear-bottom plate. Immediately after adding malachite green reagent, 10 μl of 34% sodium citrate was added to the reaction to prevent nonenzymatic hydrolysis of ATP in the presence of acidic malachite green reagent, causing an increase in color. The optical density at 620 nm was measured on the multimode microplate reader Synergy-2 (BioTech, Winooski, VT). The intrinsic hydrolysis of ATP (the reading from ATP in the reaction buffer) was subtracted from the total reading. To compare the results from different experiments, the opitical density at 620 nm readings were translated into inorganic phosphate molar concentrations using a phosphate standard curve of KH2PO4, which was prepared for each experiment. Significance was determined by the Student t-test.
In Vitro CDC2 Kinase Assay
The CDC2 complexes were immunoprecipitated from the nuclear fraction of mouse spermatogenic cell lysate or from testis lysate with antiserum to NASP or antiserum to cyclin B1. UltraLink Immobilized Protein A/G (Pierce) was added and incubated for 1 h at 4°C with rocking. The immunoprecipitate was collected by centrifugation and washed six times in cold immunoprecipitation buffer (50 mM Tris, pH 7.5; 150 mM NaCl; 0.1% Nonidet P-40; 1 mM ethylenediaminetetraacetic acid, pH 8.0; 1 mM PMSF; 10 mg/ml soybean trypsin inhibitor; 10 μg/ml leupeptin; and 10 μg/ml aprotinin) and three times in kinase assay buffer (50 mM Hepes, pH 7.5; 10 mM MgCl2; 80 mM α-glycerophosphate, pH 7.3; 20 mM ethylene glycol tetraacetic acid; 1 mM dithiothreitol; and 10 mM ATP [17]). Histone H1 kinase activity of CDC2 was assayed in the kinase assay buffer (50 μl) with the addition of 10 μl of inhibitor cocktail (20 mM protein kinase C inhibitor peptide, 2 mM protein kinase A inhibitor peptide, and 2 mM Compound R24571, which inhibits kinases except CDC2/cyclin B1; Upstate, Lake Placid, NY), calf thymus histone H1 (50 μg per reaction; Calbiochem) and ATP γ-32P (0.1 mCi/ml; MP Biomedicals, Irvine, CA). The reactions were incubated at 30°C for 30 min. Samples were separated by SDS-PAGE and stained with Coomassie blue. Radioactive bands were detected in a PhosphorImager Storm-860 (Molecular Dynamics, Piscataway, NJ). To determine the effect of NASP and histone HIST1H1T/HIST1H1A on the kinase activity of the CDC2-cyclin B1 complex, mouse testis lysate was incubated 30 min at room temperature with added recombinant mouse tNASP (2 μM) or mouse histone HIST1H1T (6 μM) or a combination of both proteins. A separate identical experiment was conducted for recombinant mouse HIST1H1A. The cyclin B1 complexes were immunoprecipitated with antiserum to cyclin B1 and assayed as described above.
RESULTS
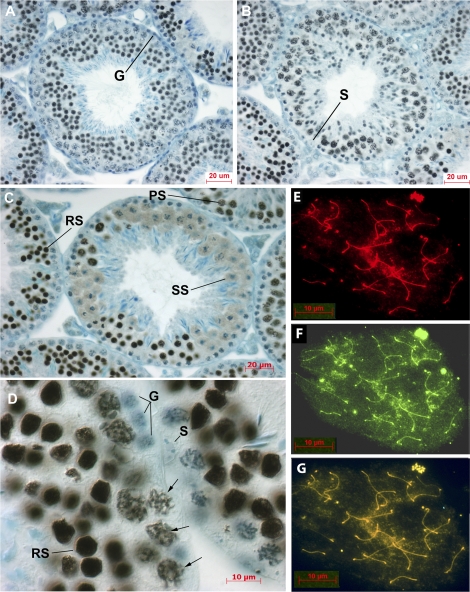
tNASP first appears at the leptotene/zygotene stage of primary spermatocytes, as described during spermatogenesis in the rabbit almost 20 yr ago by Welch and O'Rand (see figure 1 in Welch and O'Rand [32]). Similarly, in the mouse, tNASP is first detected by antibody staining in the nuclei of leptotene/zygotene spermatocytes, and tNASP staining continues to increase in intensity in pachytene spermatocytes. Figure 1, A and B, shows seminiferous tubules in stages VI and IX, respectively, demonstrating tNASP in leptotene, pachytene, and spermatid stages. Spermatogonia (G) and Sertoli cells (S) do not stain for tNASP. Figure 1C shows a stage XII tubule with diplotene and M-phase spermatocytes and the apparent exclusion of tNASP staining from the condensed chromatin of cells proceeding through meiosis to become secondary spermatocytes (SS). Round spermatids (RS) and pachytene spermatocytes (PS) can be seen staining for tNASP in adjacent tubules. At higher magnification in Figure 1D, spermatogonia (G) and Sertoli cells (S) clearly do not stain for tNASP, whereas round spermatids (RS) are intensely stained. In pachytene spermatocytes, tNASP can be seen associated with the chromatin in the nuclei (Fig. 1D, arrows). Examination of mouse chromosome spreads confirms the observation that tNASP is present on the chromatin of primary spermatocytes, along the lengths of all the synaptonemal complexes (Fig. 1E).
FIG. 1.
Localization of tNASP in mouse seminiferous tubules and on meiotic chromosomes. A and B) Sections through tubules at stage VI (A) and stage IX (B) show tNASP in leptotene, pachytene, and spermatid stages. Spermatogonia (G) and Sertoli cells (S) do not stain for tNASP. C) A stage XII tubule with diplotene and M-phase spermatocytes proceeding through meiosis to become secondary spermatocytes (SS). Round spermatids (RS) and pachytene spermatocytes (PS) can be seen staining for tNASP in adjacent tubules. D) Higher-magnification image of a seminiferous tubule shows tNASP localized in round spermatid nuclei (RS) and on chromatin in pachytene spermatocytes (arrows). Sertoli cell (S) and spermatogonia (G) nuclei are negative. Control staining with anti-NASP antibody absorbed with antigen was negative (data not shown). E–G) Surface-spread meiotic chromosomes demonstrate the localization of tNASP and HSPA2 in mouse primary spermatocytes. E) Staining for tNASP. F) Staining for HSPA2. G) Double staining for tNASP and HSPA2 on chromosomal spreads of mouse spermatogenic cells. Yellow/orange staining demonstrates the colocalization of tNASP and HSPA2. Bars = 20 μm (A–C) and 10 μm (D–G).
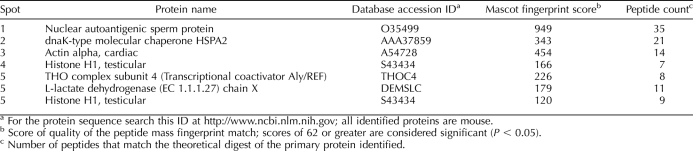
NASP's binding partners, HSP90 and H1 linker histones, including the testis-specific linker histone HIST1H1T, have been previously identified in affinity chromatography and 3,3′-dithiobis sulfosuccinimidylpropionate cross-linking studies by mass spectrometry of protein spots after 2-dimensional SDS-PAGE [18, 23]. In this study, we have identified testis-specific heat shock protein 70 (HSPA2) as an additional tNASP-binding partner in the mouse testis by immunoprecipitation with anti-NASP antibody and mass spectrometry of protein spots after SDS-PAGE (Supplemental Fig. S1, available online at www.biolreprod.org, and Table 1).
TABLE 1.
Identification of protein spots.
Figure 1F demonstrates that HSPA2 is localized on the lateral elements of the synaptonemal complex during prophase of the first meiotic division in mouse chromosome spreads, as reported previously [14, 33]. Double staining for tNASP and HSPA2 on the chromosomal spreads of mouse spermatogenic cells demonstrates that tNASP and HSPA2 colocalize (Fig. 1G). Therefore, we conclude from these initial observations that HSPA2 and tNASP may be bound together on the synaptonemal complex in primary spermatocytes. The crucial role of HSPA2 in the regulation of the meiotic cell cycle in mouse spermatogenesis [17] led us to undertake further studies on the tNASP-HSPA2 complex.
Association of tNASP with HSPA2 and CDC2
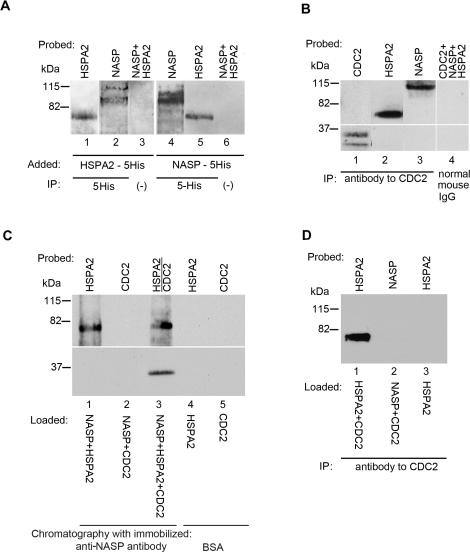
Using histidine-tagged (5His) recombinant proteins to facilitate purification, we added recombinant histidine-tagged HSPA2 (HSPA2-5His; 1 μM) to the nuclear fraction of mouse germ cells and found that endogenous tNASP (nonrecombinant) is immunoprecipitated by anti-histidine tag antibodies (anti-5His; Fig. 2A, lanes 1 and 2). Similarly, endogenous HSPA2 present in the nuclear fraction of mouse germ cells is immunoprecipitated by anti-5His antibodies when recombinant tNASP-5His is added (1 μM) to the nuclear fraction (Fig. 2A, lanes 4 and 5). Control immunoprecipitations without added 5His antibody demonstrated that NASP and HSPA2 do not bind nonspecifically to protein A/G (Fig. 2A, lanes 3 and 6).
FIG. 2.
tNASP-HSPA2 binding in the nuclear fraction of mouse spermatogenic cells. A) Lanes 1 and 2: HSPA2-5His was added to a mouse germ cell nuclear fraction and immunoprecipitated (IP) with anti-5His antibody. Both recombinant HSPA2 (lane 1) and endogenous (nonrecombinant) tNASP (lane 2) were precipitated. In control (lane 3) without anti-5His antibody, neither HSPA2 nor tNASP was immunoprecipitated. Lanes 4 and 5: tNASP-5His was added to a mouse germ cell nuclear fraction and immunoprecipitated with anti-5His antibody. Both recombinant tNASP (lane 4) and endogenous (nonrecombinant) HSPA2 (lane 5) were precipitated. In control (lane 6) without anti-5His antibody, neither HSPA2 nor tNASP was immunoprecipitated. B) Immunoprecipitation of CDC2 (lane 1), HSPA2 (lane 2), and tNASP (lane 3) from the nuclear fraction of mouse spermatogenic cells by anti-CDC2 antibody. Control immunoprecipitation with normal mouse IgG demonstrated that CDC2, tNASP, and HSPA2 (lane 4) did not bind nonspecifically. C) Affinity chromatography with Aminolink-coupled recombinant tNASP bound recombinant HSPA2 (lane 1) but not recombinant CDC2 (lane 2). When HSPA2 and CDC2 were preincubated before addition to the tNASP affinity column, the complex was bound and eluted (lane 3). Control affinity chromatography with Aminolink-coupled bovine serum albumin demonstrated that recombinant HSPA2 (lane 4) and recombinant CDC2 (lane 5) did not bind nonspecifically to Aminolink. D) Anti-CDC2 antibody immunoprecipitates recombinant HSPA2 premixed with recombinant CDC2 (lane 1); however, it does not immunoprecipitate recombinant tNASP premixed with recombinant CDC2 (lane 2). The control (lane 3) shows that anti-CDC2 does not immunoprecipitate recombinant HSPA2.
HSPA2 binds CDC2 in mouse spermatogenic cells, and the binding interaction has been characterized as necessary for CDC2 kinase activation in mouse testes [14–16]. Therefore, we asked whether or not CDC2 is associated with tNASP and HSPA2 in mouse germ cells. Immunoprecipitation from the germ cell nuclear fraction with anti-CDC2 antibodies precipitated CDC2, HSPA2, and tNASP (Fig. 2B, lanes 1–3). A control immunoprecipitation with normal mouse IgG (Santa Cruz Biotechnology) demonstrated that CDC2, HSPA2, and tNASP did not bind nonspecifically (Fig. 2B, lane 4). From these results, we conclude that tNASP and HSPA2 are binding partners in the nuclear fraction of mouse germ cells, and that CDC2 is present in the complex.
To confirm the interactions found in nuclear lysates, in vitro experiments were carried out. Affinity chromatography with Aminolink-coupled recombinant tNASP demonstrated that recombinant HSPA2 binds to recombinant tNASP (Fig. 2C, lane 1); however, recombinant CDC2 does not (Fig. 2C, lane 2). When HSPA2 and CDC2 recombinant proteins were preincubated together and then applied to the tNASP affinity column, both proteins were eluted (Fig. 2C, lane 3). A control affinity chromatography with Aminolink-coupled bovine serum albumin demonstrated that recombinant HSPA2 (Fig. 2C, lane 4) and recombinant CDC2 (Fig. 2C, lane 5) did not bind nonspecifically. CDC2, HSPA2, and tNASP interaction was further confirmed by immunoprecipitation with anti-CDC2 antibodies. In the presence of recombinant CDC2, anti-CDC2 antibodies can immunoprecipitate recombinant HSPA2 but not recombinant tNASP (Fig. 2D, lanes 1 and 2), unless CDC2-HSPA2 and tNASP are preincubated with each other (data not shown). Control immunoprecipitation (Fig. 2D, lane 3) demonstrates that anti-CDC2 antibody does not recognize HSPA2. Consequently, tNASP and HSPA2 are binding partners in the nuclear fraction of mouse germ cells, and CDC2 is bound to HSPA2 in the tNASP-HSPA2 complex; however, tNASP does not directly bind CDC2 and only interacts with CDC2 through its binding to HSPA2.
tNASP Competes with Cyclin B1 for CDC2 in Mouse Spermatocytes
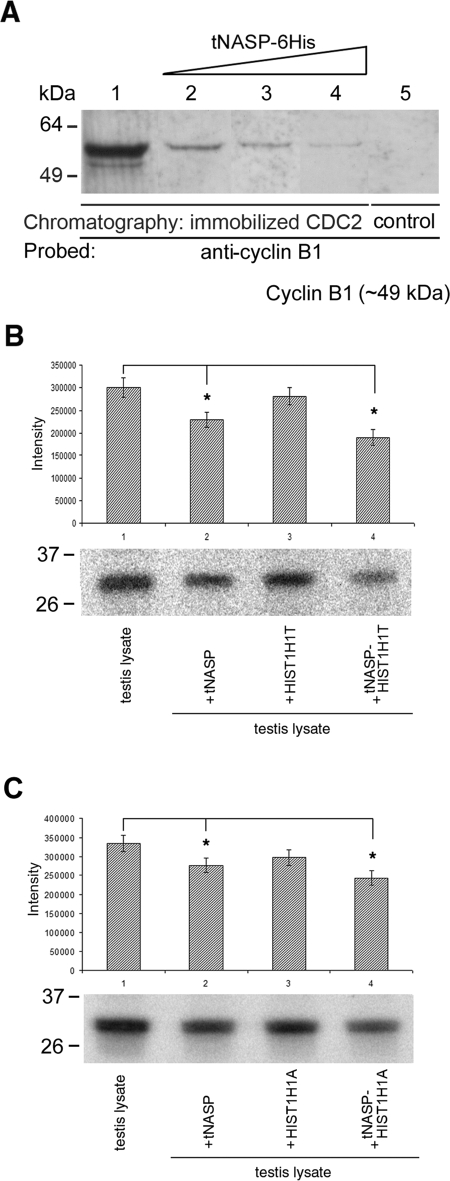
Cyclin B1 is the regulatory subunit of CDC2, and together they act as an M-phase-specific kinase [6]. To determine the effect of tNASP on the formation of the CDC2/cyclin B1 complex, we added increasing amounts of recombinant tNASP to the nuclear fraction of mouse spermatogenic cells, incubated the fraction with Aminolink-coupled CDC2, and eluted the column. In controls in which no tNASP was added to the nuclear lysate, abundant cyclin B1 bound to the column (Fig. 3A, lane 1). Increasing amounts of recombinant tNASP significantly decreased the amount of cyclin B1 bound to CDC2 (Fig. 3A, lanes 2–4). Immunoprecipitation with control Aminolink Plus Coupling Gel (no CDC2 bound) did not show any nonspecific cyclin B1 binding to the column (Fig. 3A, lane 5). Additionally, when recombinant tNASP was added to mouse testis lysates, followed by immunoprecipitation with anti-cyclin B1, the CDC2/cyclin B1 kinase activity was significantly lower in tNASP-treated samples compared with untreated controls (Fig. 3, B and C, lanes 1 and 2). When a mixture of tNASP and histone HIST1H1T (Fig. 3B, lane 4) or tNASP and histone HIST1H1A (Fig. 3C, lane 4) was added to the lysate, a further significant decrease in kinase activity was observed. There was no difference between testis-specific histone HIST1H1T and histone HIST1H1A modulation of CDC2/cyclin B1 activity. Adding only histone HIST1H1T (Fig. 3B, lane 3) or histone HIST1H1A (Fig. 3C, lane 3) did not affect CDC2/cyclin B1 kinase activity in the testis lysate. We conclude from these experiments that increasing amounts of tNASP successfully compete with cyclin B1 for CDC2 and that tNASP and the tNASP-H1 complex significantly lower CDC2/cyclin B1 kinase activity in the lysate.
FIG. 3.
A) The presence of increasing amounts of tNASP decreases binding between CDC2 and cyclin B1 in nuclear fractions from mouse spermatogenic cells. Eluates from a CDC2 affinity column loaded with a nuclear fraction from mouse spermatogenic cells only (lane 1) or a nuclear fraction preincubated with increasing amounts of recombinant tNASP at 5 μM (lane 2), 10 μM (lane 3), and 25 μM (lane 4) show a decrease in cyclin B1 binding. The control using affinity beads without CDC2 cross-linked shows no cyclin B1 binding (lane 5). B and C) PhosphorImager detection of 32P-labeled histone indicating that CDC2/cyclin B1 kinase activity decreases in the presence of tNASP and H1 histones. B) tNASP (lane 2), HIST1H1T (lane 3), or tNASP + HIST1H1T (lane 4) was added to mouse testis lysates, and CDC2/cyclin B1 complexes were immunoprecipitated using anti-cyclin B1 antibody. After immunoprecipitation, the kinase activity was determined as a measure of the presence of 32P-labeled histone. The control (lane 1) consisted of a testis lysate alone. *Significant differences in the kinase activity: control vs. tNASP (P < 0.01); control vs. tNASP-HIST1H1T (P < 0.003); and tNASP vs. tNASP-HIST1H1T (P < 0.03). Data are represented as mean ± SD. C) Experiments identical to B were performed using histone HIST1H1A instead of HIST1H1T. Lane 1: control (testis lysate alone). tNASP (lane 2), HIST1H1A (lane 3), or tNASP + HIST1H1A (lane 4) was added to mouse testis lysates, and CDC2/cyclin B1 complexes were immunoprecipitated using anti-cyclin B1 antibody. After immunoprecipitation, the kinase activity was determined as a measure of the presence of 32P-labeled histone. *Significant differences in the kinase activity: control vs. tNASP (P < 0.02); control vs. tNASP-HIST1H1A (P < 0.006); and tNASP vs. tNASP-HIST1H1T (P < 0.05). Data are represented as mean ± SD.
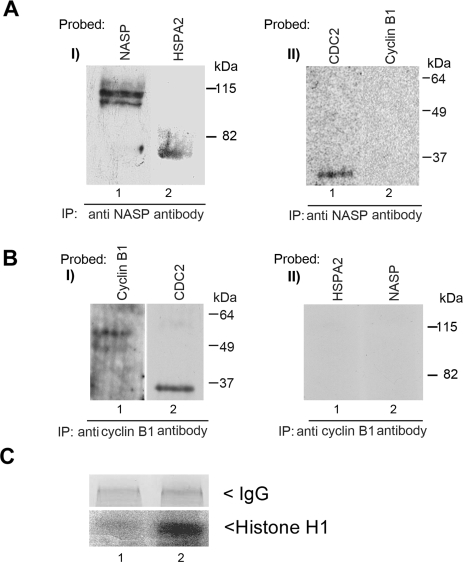
CDC2 Exists in Two Populations: Associated with tNASP-HSPA2 and with Cyclin B1
If CDC2 cannot simultaneously bind tNASP and cyclin B1, there must be two populations of CDC2. Accordingly, two different immunoprecipitations were carried out with anti-NASP and anti-cyclin B1 antibodies. HSPA2, tNASP, and CDC2 are all coprecipitated with anti-NASP antibodies (Fig. 4A), whereas only cyclin B1 and CDC2 were precipitated with anti-cyclin B1 antibodies (Fig. 4B). Assays of the immunoprecipitated fractions obtained above demonstrated that little or no kinase activity was detected in CDC2 obtained from the anti-NASP immunoprecipitates (Fig. 4C, lane 1). Significant histone H1 kinase activity was only detected in CDC2 obtained from the anti-cyclin B1 immunoprecipitate (Fig. 4C, lane 1).
FIG. 4.
A) Immunoprecipitation (IP) from the nuclear fraction of mouse spermatogenic cells with anti-NASP antibody precipitated tNASP (I, lane 1), and coimmunoprecipitated HSPA2 (I, lane 2) and CDC2 (II, lane 1) but not cyclin B1 (II, lane 2). B) Immunoprecipitation from the nuclear fraction of mouse spermatogenic cells with anti-cyclin B1 antibody precipitated cyclin B1 (I, lane 1) and CDC2 (I, lane 2) but not HSPA2 (II, lane 1) or tNASP (II, lane 2). C) PhosphorImager detection of 32P-labeled histone indicating specific kinase activity of CDC2 containing immunoprecipitates. Immunoprecipitation with anti-NASP antibody does not have any kinase activity in the precipitate (lane 1); however, when the immunoprecipitation is done with anti-cyclin B1 antibody (lane 2), a strongly radioactive H1 band demonstrates specific kinase activity in the precipitate. Control panel demonstrates equal amounts of antibodies (IgG) used.
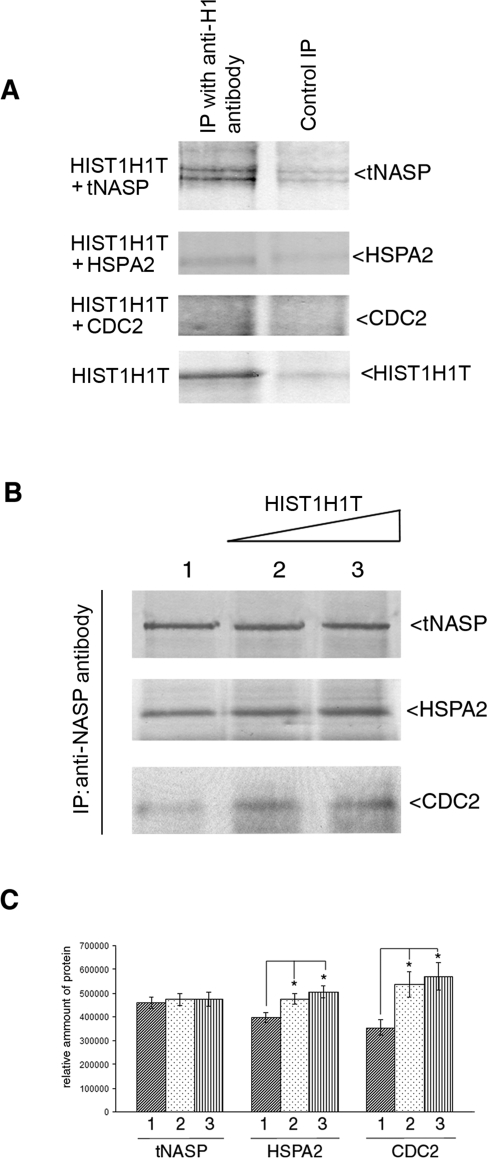
Histone HIST1H1T Increases Binding Between tNASP and HSPA2 and CDC2
One of the NASP's multiple cellular functions is thought to be that of an H1 linker histone chaperone, and we asked whether the testis-specific linker histone, HIST1H1T, could influence the binding between tNASP and HSPA2 in mouse spermatogenic cells. In vitro binding analysis demonstrates that histone HIST1H1T coimmunoprecipitates with tNASP but not with HSPA2 or CDC2 (Fig. 5A). Immunoprecipitation from the mouse testis lysate with anti-NASP antibody resulted in immunoprecipitation of tNASP, HSPA2, and CDC2 (Fig. 5B, lane 1), and as shown above (Figs. 2 and 4). When increasing amounts of recombinant HIST1H1T were added (3 and 6 μM), a significant increase in the amounts of coimmunoprecipitated HSPA2 and CDC2 occurred, whereas the amount of immunoprecipitated tNASP did not change significantly (Fig. 5, B, lanes 2 and 3, and C). We conclude from these experiments that there is an HIST1H1T-dependent change in stoichiometry of the tNASP-HSPA2-CDC2 complex, and it leads to increased HSPA2 and, consequently, CDC2 binding to tNASP.
FIG. 5.
HIST1H1T affects the equilibrium of tNASP-HSPA2-CDC2 complex formation. A) Histone HIST1H1T binds to tNASP but not HSPA2 or CDC2 in vitro. Recombinant HIST1H1T was mixed with recombinant tNASP, HSPA2, CDC2, or HIST1H1T. All lanes were immunoprecipitated (IP) with anti-H1 antibody and stained with their respective antisera (IP with anti-H1). In negative controls (Control IP), HIST1H1T was not added except in the bottom panel (HIST1H1T), where HIST1H1T and a nonrelevant control antibody (anti-CYR61) were added. B) Increasing amounts of HIST1H1T were added to mouse testis lysates and tNASP immunoprecipitated. As HIST1H1T increased, HSPA2 and CDC2 increased: Lane 1: no HIST1H1T was added; lanes 2 and 3: 3 and 6 μM of recombinant HIST1H1T were added, respectively. C) Analysis of bands from B demonstrating significant changes (*) in the quantity of immunoprecipitated HSPA2 and CDC2 in the presence of increasing HIST1H1T. HSPA2: lane 1 vs. lane 2 (P < 0.02), lane 1 vs. lane 3 (P < 0.006). CDC2: lane 1 vs. lane 2 (P < 0.02), lane 1 vs. lane 3 (P < 0.02). The y-axis represents arbitrary units. Data are represented as mean ± SD.
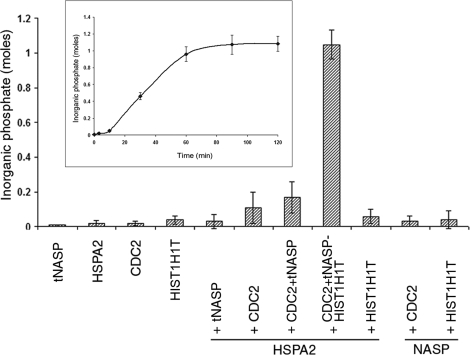
ATPase Activity of HSPA2 Is Increased by tNASP-HIST1H1T
Previous studies on tNASP-HSP90 interaction found that tNASP increased HSP90 ATPase activity [18]. Therefore, we tested the effect of tNASP on the ATPase activity of HSPA2. Recombinant mouse HSPA2 displayed negligible ATPase activity (Fig. 6), and in contrast to tNASP-HSP90 interaction, recombinant tNASP added to HSPA2 in a 1:1 molar ratio had no effect on the ATPase activity of HSPA2 (Fig. 6). Addition of CDC2 or tNASP + CDC2 to HSPA2 resulted in only slight increases in HSPA2 ATPase activity (Fig. 6). However, when tNASP-HIST1H1T + CDC2 was added to HSPA2 (1:1:1:1 molar ratio), a significant increase in HSPA2 ATPase activity was observed (Fig. 6). Control experiments with different combinations of proteins (HSPA2 + HIST1H1T, tNASP + CDC2, tNASP-HIST1H1T) showed only negligible ATPase activity (Fig. 6). Kinetic analysis of HSPA2-mediated ATP hydrolysis in the presence of tNASP, CDC2, and HIST1H1T in excess ATP shows that the amount of liberated inorganic phosphate increases during the first 60 min and reaches a plateau after that. The maximum amount of inorganic phosphate liberated constituted approximately the same molar amount as HSPA2 and tNASP present in the reaction (Fig. 6, inset).
FIG. 6.
The HIST1H1T-tNASP-HSPA2-CDC2 complex shows ATPase activity. ATPase activity of HSPA2 is presented as moles of inorganic phosphate liberated by 1 mol of HSPA2 after 60 min of incubation at 37°C in the presence of the indicated proteins. Data are represented as mean ± SD. Inset shows kinetics of HSPA2-mediated ATP hydrolysis in the presence of tNASP, CDC2, and HIST1H1T. Graph presents the moles of inorganic phosphate liberated by 1 mol of HSPA2 during 120 min of incubation at 37°C. Reaction plateau occurs after 60 min of incubation at 37°C.
DISCUSSION
This study has demonstrated that tNASP in the nuclei of mouse spermatogenic cells binds the testis heat shock protein HSPA2, and they colocalize on the synaptonemal complexes of spermatocytes during meiotic prophase I (Fig. 1). HSPA2 was reported previously to be a component of the lateral elements of the synaptonemal complex of pachytene spermatocytes [14, 33] and necessary for the activation of CDC2 by cyclin B1 [16, 17]. We have confirmed that HSPA2 and CDC2 coprecipitate from mouse germ cell nuclei [17] and shown that immunoprecipitation of either tNASP or HSPA2 resulted in the coprecipitation of the other component. Although CDC2 does not bind tNASP, CDC2 coprecipitated with HSPA2 and tNASP (Fig. 2), indicating that HSPA2 binds tNASP and CDC2 simultaneously in a complex that is part of the lateral elements of the synaptonemal complex. The interaction of linker histones with tNASP changed the stoichiometry of the tNASP-HSPA2 complex formation in favor of an increased capacity of tNASP to bind HSPA2 and, consequently, CDC2.
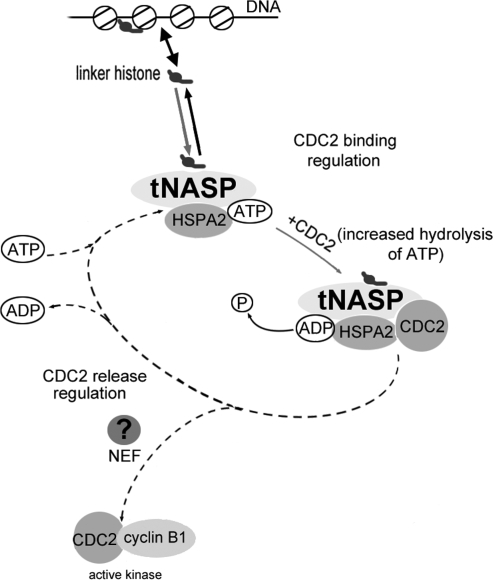
A number of positive and negative regulators of HSP70 ATPase activity have been identified in mammalian cells [34–36]. Figure 7 presents a hypothetical schematic of HSPA2 chaperone cycle dynamics through tNASP-histone HIST1H1T binding during meiosis. NASP has at least three tetratricopeptide repeats, which are known to bind the EEVD domain of heat shock proteins, but NASP lacks a typical J domain, which is essential for J proteins reported as stimulators of HSP70 ATPase activity [38]. However, the absence of a J domain was reported previously in RESA proteins [39], which are known stimulators of HSP70 ATPase activity. The chaperone activity of HSPA2 as well as other HSP70 heat shock proteins is controlled by their nucleotide-binding domain, which binds and hydrolyses ATP, inducing conformational changes in the substrate-binding domain [37, 40]. We found that the ATPase activity of unstimulated HSPA2 was very low and that activity increased after binding of CDC2 to HSPA2 (Fig. 6), which was most likely specific substrate stimulation, as reported previously for other substrates [41, 42]. Increased binding of CDC2, the reported substrate of HSPA2, was an immediate result of the binding of ATP by HSPA2. Binding between ATP and HSPA2 has been reported to open the substrate-binding domain located on the C terminus [43]. The HSPA2 hydrolysis of ATP that was demonstrated in our study was triggered by CDC2 binding (Fig. 6). The binding of the linker histone-tNASP complex to HSPA2 in the presence of the substrate CDC2 significantly increased the ATP hydrolysis rate by HSPA2. The mechanisms by which linker histone-tNASP stimulates the ATPase activity of HSPA2 remain to be elucidated.
FIG. 7.
Regulation of HSPA2 chaperone cycle dynamics by linker histone-tNASP (modified from Bukau et al. [37] with permission from Elsevier). The cycle regulation starts with the association of linker histones with tNASP and the formation of a linker histone-tNASP-HSPA2 complex. The binding of CDC2 and increased ATP hydrolysis by HSPA2 causes the “locking in” of the substrate (CDC2) and formation of a linker histone-tNASP-HSPA2-CDC2 complex. The predicted release of CDC2 and ADP from the complex would mediate formation of active CDC2/cyclin B1 kinase (dotted line) and depend on nucleotide exchange factors, which are still to be identified.
Upon release from HSPA2, CDC2 gains the ability to complex with cyclin B1 during meiosis I of mouse spermatogenesis. It has been reported previously that ADP/ATP exchange determines the substrate's association and dissociation rates from HSP70 [41]. Successful release of substrate from HSP70 proteins is linked to the discharge of ADP, which is typically regulated by nucleotide exchange factors (Fig. 7). In addition to the known nucleotide exchange factors for HSP70 proteins, GRPE [44] and BAG1 [36, 45], other proteins have been found to act as nucleotide exchange factors for HSP70 proteins: yeast proteins Lhs1, Sls1/Sil1, and Fes1, and mammalian proteins SIL1 (BAP) and HSPBP1 [43]. Different nucleotide exchange factors are often specific to a single protein, and a specific one for HSPA2 remains to be discovered. The coordinated effect of ATPase activity coupled to substrate binding and stimulation of ADP/ATP exchange is balanced in vivo and achieved by coregulation of gene expression [41]. Formation of the complete complex of linker histone-tNASP-HSPA2-CDC2 significantly activated ATP hydrolysis by HSPA2 and resulted in increased binding of CDC2 to HSPA2 in primary spermatocytes. The lack of dissociation of CDC2 from HSPA2 in our in vitro experiments indicates that other signals (nucleotide exchange factor?) are needed to release CDC2 from HSPA2 in its active form [46]. In the absence of this signal, the presence of only NASP-HIST1H1T causes activation of the initial part of the HSPA2 chaperone cycle (binding CDC2 to HSPA2) but a subsequent decline in the number of functionally active CDC2-cyclin B1 complexes.
NASP has been shown to exchange linker histones with DNA [20], and therefore we speculate that shifting the binding equilibrium of H1-tNASP toward DNA and away from HSPA2 may release the CDC2 substrate. In HSPA2 knockout mice [16], CDC2 was not converted to its active form, and no CDC2/cyclin B1 kinase activity was found. Similarly, increasing amounts of tNASP inhibit CDC2/cyclin B1 kinase activity. Therefore, we would expect that the overexpression of tNASP in the testis would inhibit the progression of meiosis through the G2/M border.
In the mammalian testis, tNASP is a chaperone for linker histones (somatic and HIST1H1T), transporting them from the cytoplasm to the nucleus [18], where they are bound or exchanged for an existing linker histone on the DNA [20]. In pachytene spermatocytes, HIST1H1T, the predominant linker histone, is exchanged for the somatic H1 histones present in spermatogonia [47]. HIST1H1T differs from somatic forms in its secondary DNA-binding site, which lowers its affinity for DNA and results in a less condensed chromatin structure [48]. Phosphorylation of linker histones by CDKs [49] is thought to play a critical role in the decondensation of chromatin, and linker histones may influence promoter access for transcription factors [50] and nonspecific repression of transcription [51]. Consequently, tNASP's role in the synaptonemal complex may be to efficiently link H1 signaling from the chromatin to the cell cycle signaling complex CDC2/cyclin B1. CDC2 (CDK1) has been shown to regulate alternative DSB repair pathways in both G1 and G2 [52], and CDC2/cyclin B1 has been shown to be required for homologous recombination [53]. In providing such a signaling link, tNASP may facilitate displacement of linker histones during homologous recombination and DNA repair.
In this study, we found that histone HIST1H1T and HIST1H1A significantly increase tNASP's binding to HSPA2-CDC2, and therefore increase the inhibition of formation of the CDC2/cyclin B1 complexes. In pachytene spermatocytes, this linker histone-mediated cell cycle inhibition effect could be required for the cell to complete repair of DSBs during homologous recombination. Human NASP is known to be phosphorylated on T464 and S421 as a result of DNA damage [22], and therefore DSBs during meiotic recombination may influence tNASP's ability to bind HSPA2. We speculate that the signaling pathway from DNA status to the cell cycle stage may very well include an H1-tNASP component for fine tuning of the active nucleosome-remodeling processes [25] by CDC2 activity regulation. Linker histone modulation of DNA replication [54] could be through a significantly reduced expression of the CDC2 gene [55]. Active CDC2/cyclin B1 complexes (maturation-promoting factor) are required for the G2→M transition during the meiotic and mitotic cell cycles [6, 56, 57].
Supplementary Material
Acknowledgments
The authors thank Gail Grossman for preparation of histological samples and the University of North Carolina-Duke Proteomics Center for mass spectrometry identification of tNASP-binding partners.
Footnotes
1Supported by the Eunice Kennedy Shriver National Institute of Child Health and Human Development/National Institutes of Health through cooperative agreement U54 (HD35041), as part of the Specialized Cooperative Centers Program in Reproduction and Infertility Research.
REFERENCES
- Govin J, Caron C, Lestrat C, Rousseaux S, Khochbin S.The role of histones in chromatin remodelling during mammalian spermiogenesis. Eur J Biochem 2004; 271: 3459–3469.. [DOI] [PubMed] [Google Scholar]
- van der Heijden GW, Derijck AA, Pósfai E, Giele M, Pelczar P, Ramos L, Wansink DG, van der Vlag J, Peters AH, de Boer P.Chromosome-wide nucleosome replacement and H3.3 incorporation during mammalian meiotic sex chromosome inactivation. Nat Genet 2007; 39: 251–258.. [DOI] [PubMed] [Google Scholar]
- Groth A, Rocha W, Verreault A, Almouzni G.Chromatin challenges during DNA replication and repair. Cell 2007; 128: 721–733.. [DOI] [PubMed] [Google Scholar]
- Hochwagen A, Amon A.Checking your breaks: surveillance mechanisms of meiotic recombination. Curr Biol 2006; 16: R217–R228.. [DOI] [PubMed] [Google Scholar]
- Pérez-Hidalgo L, Moreno S, San-Segundo PA.Regulation of meiotic progression by the meiosis-specific checkpoint kinase Mek1 in fission yeast. J Cell Sci 2003; 116: 259–271.. [DOI] [PubMed] [Google Scholar]
- Chapman DL, Wolgemuth DJ.Regulation of M-phase promoting factor activity during development of mouse male germ cells. Dev Biol 1994; 165: 500–506.. [DOI] [PubMed] [Google Scholar]
- Draetta G, Beach D.Activation of cdc2 protein kinase during mitosis in human cells: cell cycle-dependent phosphorylation and subunit rearrangement. Cell 1988; 54: 17–26.. [DOI] [PubMed] [Google Scholar]
- Liu D, Liao C, Wolgemuth DJ.A role for cyclin A1 in the activation of MPF and G2-M transition during meiosis of male germ cells in mice. Dev Biol 2000; 224: 388–400.. [DOI] [PubMed] [Google Scholar]
- Liu D, Matzuk MM, Sung WK, Guo Q, Wang P, Wolgemuth DJ.Cyclin A1 is required for meiosis in the male mouse. Nat Genet 1998; 20: 377–380.. [DOI] [PubMed] [Google Scholar]
- Nickerson HD, Joshi A, Wolgemuth DJ.Cyclin A1-deficient mice lack histone H3 serine 10 phosphorylation and exhibit altered aurora B dynamics in late prophase of male meiosis. Dev Biol 2007; 306: 725–735.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller-Tidow C, Ji P, Diederichs S, Potratz J, Baumer N, Kohler G, Cauvet T, Choudary C, van der Meer T, Chan WY, Nieduszynski C, Colledge WH, et al. The cyclin A1-CDK2 complex regulates DNA double-strand break repair. Mol Cell Biol 2004; 24: 8917–8928.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierce AJ, Hu P, Han M, Ellis N, Jasin M.Ku DNA end-binding protein modulates homologous repair of double-strand breaks in mammalian cells. Genes Dev 2001; 15: 3237–3242.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortega S, Prieto I, Odajima J, Martín A, Dubus P, Sotill R, Barbero JL, Malumbres M, Barbacid M.Cyclin-dependent kinase 2 is essential for meiosis but not for mitotic cell division in mice. Nat Genet 2003; 35: 25–31.. [DOI] [PubMed] [Google Scholar]
- Eddy E.Role of heat shock protein HSP70–2 in spermatogenesis. Rev Reprod 1999; 4: 23–30.. [DOI] [PubMed] [Google Scholar]
- Dix D, Allen J, Collins B, Poorman-Allen P, Mori C, Blizard D, Brown P, Goulding E, Strong B, Eddy E.HSP70–2 is required for desynapsis of synaptonemal complexes during meiotic prophase in juvenile and adult mouse spermatocytes. Development 1997; 124: 4595–4603.. [DOI] [PubMed] [Google Scholar]
- Dix DJ, Allen JW, Collins BW, Mori C, Nakamura N, Poorman-Allen P, Goulding EH, Eddy EM.Targeted gene disruption of Hsp70–2 results in failed meiosis, germ cell apoptosis, and male infertility. Proc Natl Acad Sci U S A 1996; 93: 3264–3268.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu D, Dix D, Eddy E.HSP70–2 is required for CDC2 kinase activity in meiosis I of mouse spermatocytes. Development 1997; 124: 3007–3014.. [DOI] [PubMed] [Google Scholar]
- Alekseev OM, Widgren EE, Richardson RT, O'Rand MG.Association of NASP with HSP90 in mouse spermatogenic cells: stimulation of ATPase activity and transport of linker histones into nuclei. J Biol Chem 2005; 280: 2904–2911.. [DOI] [PubMed] [Google Scholar]
- Groth A, Ray-Gallet D, Quivy JP, Lukas J, Bartek J, Almouzni G.Human Asf1 regulates the flow of S phase histones during replicational stress. Mol Cell 2005; 17: 301–311.. [DOI] [PubMed] [Google Scholar]
- Alekseev OM, Bencic DC, Richardson RT, Widgren EE, O'Rand MG.Overexpression of the linker histone-binding protein tNASP affects progression through the cell cycle. J Biol Chem 2003; 278: 8846–8852.. [DOI] [PubMed] [Google Scholar]
- Richardson RT, Alekseev OM, Grossman G, Widgren EE, Thresher R, Wagner EJ, Sullivan KD, Marzluff WF, O'Rand MG.Nuclear autoantigenic sperm protein (NASP), a linker histone chaperone that is required for cell proliferation. J Biol Chem 2006; 281: 21526–21534.. [DOI] [PubMed] [Google Scholar]
- Matsuoka S, Ballif BA, Smogorzewska A, McDonald ER, III, Hurov KE, Luo J, Bakalarski CE, Zhao Z, Solimini N, Lerenthal Y, Shiloh Y, Gygi SP, et al. ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage. Science 2007; 316: 1160–1166.. [DOI] [PubMed] [Google Scholar]
- Alekseev OM, Richardson RT, Pope MR, O'Rand MG.Mass spectrometry identification of NASP binding partners in HeLa cells. Proteins 2005; 61: 1–5.. [DOI] [PubMed] [Google Scholar]
- Sancar A, Lindsey-Boltz LA, Unsal-Kacmaz K, Linn S.Molecular mechanisms of mammalian DNA repair and the DNA damage checkpoints. Annu Rev Biochem 2004; 73: 39–85.. [DOI] [PubMed] [Google Scholar]
- Finn RM, Browne K, Hodgson KC, Ausio J.sNASP, a histone H1-specific eukaryotic chaperone dimer that facilitates chromatin assembly. Biophys J 2008; 95: 1314–1325.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Brien D.Isolation, separation, and short-term culture of spermatogenic cells. Methods Toxicol 1993; 3: 246–264.. [Google Scholar]
- Sivashanmugam P, Richardson R, Hall S, Hamil K, French F, O'Rand M.Cloning and characterization of an androgen-dependent acidic epididymal glycoprotein/CRISP1-like protein from the monkey. J Androl 1999; 20: 384–393.. [PubMed] [Google Scholar]
- Dresser M, Pisetsky D, Warren R, McCarty G, Moses M.A new method for the cytological analysis of autoantibody specificities using whole-mount, surface-spread meiotic nuclei. J Immunol Methods 1987; 104: 111–121.. [DOI] [PubMed] [Google Scholar]
- Hoek M, Stillman B.Chromatin assembly factor 1 is essential and couples chromatin assembly to DNA replication in vivo. Proc Natl Acad Sci U S A 2003; 100: 12183–12188.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spector D, Goldman R, Leinwand L.Cells: A Laboratory Manual, vol. 1 New York:Cold Spring Harbor Laboratory Press;1998: 43.7–43.11.. [Google Scholar]
- Aherne W, Maloney A, Prodromou C, Rowlands MG, Hardcastle A, Boxall K, Clarke P, Walton MI, Pearl L, Workman P.Assays for HSP90 and inhibitors. Methods Mol Med 2003; 85: 149–161.. [DOI] [PubMed] [Google Scholar]
- Welch J, O'Rand M.Characterization of a sperm-specific nuclear autoantigenic protein. II. Expression and localization in the testis. Biol Reprod 1990; 43: 569–578.. [DOI] [PubMed] [Google Scholar]
- Allen JW, Dix DJ, Collins BW, Merrick BA, He C, Selkirk JK, Poorman-Allen P, Dresser ME, Eddy EM.HSP70–2 is part of the synaptonemal complex in mouse and hamster spermatocytes. Chromosoma 1996; 104: 414–421.. [DOI] [PubMed] [Google Scholar]
- Brehmer D, Rüdiger S, Gässler CS, Klostermeier D, Packschies L, Reinstein J, Mayer MP, Bukau B.Tuning of chaperone activity of Hsp70 proteins by modulation of nucleotide exchange. Nat Struct Biol 2001; 8: 427–432.. [DOI] [PubMed] [Google Scholar]
- Hafizur RM, Yano M, Gotoh T, Mori M, Terada K.Modulation of chaperone activities of Hsp70 and Hsp70–2 by a mammalian DnaJ/Hsp40 homolog, DjA4. J Biochem 2004; 135: 193–200.. [DOI] [PubMed] [Google Scholar]
- Sondermann H, Scheufler C, Schneider C, Hohfeld J, Hartl F-U, Moarefi I.Structure of a Bag/Hsc70 complex: convergent functional evolution of Hsp70 nucleotide exchange factors. Science 2001; 291: 1553–1557.. [DOI] [PubMed] [Google Scholar]
- Bukau B, Weissman J, Horwich A.Molecular chaperones and protein quality control. Cell 2006; 125: 443–451.. [DOI] [PubMed] [Google Scholar]
- Zhao X, Braun A, Braun J.Biological roles of neural J proteins. Cell Mol Life Sci 2008; 65: 2385–2396.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bork P, Sander C, Valencia A, Bukau B.A module of the DnaJ heat shock proteins found in malaria parasites. Trends Biochem Sci 1992; 17: 129 [DOI] [PubMed] [Google Scholar]
- Minami Y, Hohfeld J, Ohtsuka K, Hartl FU.Regulation of the heat-shock protein 70 reaction cycle by the mammalian DnaJ homolog, Hsp40. J Biol Chem 1996; 271: 19617–19624.. [DOI] [PubMed] [Google Scholar]
- Bukau B, Horwich AL.The Hsp70 and Hsp60 chaperone machines. Cell 1998; 92: 351–366.. [DOI] [PubMed] [Google Scholar]
- Theyssen H, Schuster HP, Packschies L, Bukau B, Reinstein J.The second step of ATP binding to DnaK induces peptide release. J Mol Biol 1996; 263: 657–670.. [DOI] [PubMed] [Google Scholar]
- Mayer M, Bukau B.Hsp70 chaperones: cellular functions and molecular mechanism. Cell Mol Life Sci 2005; 62: 670–684.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao B, Davis JE, Craig EA.Mge1 functions as a nucleotide release factor for Ssc1, a mitochondrial Hsp70 of Saccharomyces cerevisiae. J Mol Biol 1997; 265: 541–552.. [DOI] [PubMed] [Google Scholar]
- Briknarova K, Takayama S, Brive L, Havert ML, Knee DA, Velasco J, Homma S, Cabezas E, Stuart J, Hoyt DW, Satterthwait AC, Llinas M, et al. Structural analysis of BAG1 cochaperone and its interactions with Hsc70 heat shock protein. Nat Struct Mol Biol 2001; 8: 349–352.. [DOI] [PubMed] [Google Scholar]
- Hermand D, Westerling T, Pihlak A, Thuret JY, Vallenius T, Tiainen M, Vandenhaute J, Cottarel G, Mann C, Mäkelä T.Specificity of Cdk activation in vivo by the two Caks Mcs6 and Csk1 in fission yeast. EMBO J 2001; 20: 82–90.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meistrich ML, Bucci LR, Trostle-Weige PK, Brock WA.Histone variants in rat spermatogonia and primary spermatocytes. Dev Biol 1985; 112: 230–240.. [DOI] [PubMed] [Google Scholar]
- Ramesh S, Bharath MMS, Chandra NR, Rao M.A K52Q substitution in the globular domain of histone H1t modulates its nucleosome binding properties. FEBS Lett 2006; 580: 5999–6006.. [DOI] [PubMed] [Google Scholar]
- Hale TK, Contreras A, Morrison AJ, Herrera RE.Phosphorylation of the linker histone H1 by CDK regulates its binding to HP1a. Mol Cell 2006; 22: 693–699.. [DOI] [PubMed] [Google Scholar]
- Song X, Gorovsky MA.Unphosphorylated H1 is enriched in a specific region of the promoter when CDC2 is down-regulated during starvation. Mol Cell Biol 2007; 27: 1925–1933.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weintraub H.Assembly and propagation of repressed and derepressed chromosomal states. Cell 1985; 42: 705–711.. [DOI] [PubMed] [Google Scholar]
- Ira G, Pellicioli A, Balijja A, Wang X, Fiorani S, Carotenuto W, Liberi G, Bressan D, Wan L, Hollingsworth NM, Haber JE, Foiani M.DNA end resection, homologous recombination and DNA damage checkpoint activation require CDK1. Nature 2004; 431: 1011–1017.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caspari T, Murray JM, Carr A.Cdc2-cyclin B kinase activity links Crb2 and Rqh1-topoisomerase III. Genes Dev 2002; 16: 1195–1208.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Z, Sittman D, Brown D, Munshi R, Leno G.Histone H1 modulates DNA replication through multiple pathways in Xenopus egg extract. J Cell Sci 1997; 110: 2745–2758.. [DOI] [PubMed] [Google Scholar]
- Brown D, Alexander B, Sittman D.Differential effect of H1 variant overexpression on cell cycle progression and gene expression. Nucl Acids Res 1996; 24: 486–493.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandeis M, Rosewell I, Carrington M, Crompton T, Jacobs MA, Kirk J, Gannon J, Hunt T.Cyclin B2-null mice develop normally and are fertile whereas cyclin B1-null mice die in utero. Proc Natl Acad Sci U S A 1998; 95: 4344–4349.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunphy WG, Brizuela L, Beach D, Newport J.The Xenopus cdc2 protein is a component of MPF, a cytoplasmic regulator of mitosis. Cell 1988; 54: 423–431.. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.