Abstract
Purpose
Chromosomal aberrations are a hallmark of multiple myeloma but their global prognostic impact is largely unknown.
Patients and Methods
We performed a genome-wide analysis of malignant plasma cells from 192 newly diagnosed patients with myeloma using high-density, single-nucleotide polymorphism (SNP) arrays to identify genetic lesions associated with prognosis.
Results
Our analyses revealed deletions and amplifications in 98% of patients. Amplifications in 1q and deletions in 1p, 12p, 14q, 16q, and 22q were the most frequent lesions associated with adverse prognosis, whereas recurrent amplifications of chromosomes 5, 9, 11, 15, and 19 conferred a favorable prognosis. Multivariate analysis retained three independent lesions: amp(1q23.3), amp(5q31.3), and del(12p13.31). When adjusted to the established prognostic variables (ie, t(4;14), del(17p), and serum β2-microglobulin [Sβ2M]), del(12p13.31) remained the most powerful independent adverse marker (P < .0001; hazard ratio [HR], 3.17) followed by Sβ2M (P < .0001; HR, 2.78) and the favorable marker amp(5q31.3) (P = .0005; HR, 0.37). Patients with amp(5q31.3) alone and low Sβ2M had an excellent prognosis (5-year overall survival, 87%); conversely, patients with del(12p13.31) alone or amp(5q31.3) and del(12p13.31) and high Sβ2M had a very poor outcome (5-year overall survival, 20%). This prognostic model was validated in an independent validation cohort of 273 patients with myeloma.
Conclusion
These findings demonstrate the power and accessibility of molecular karyotyping to predict outcome in myeloma. In addition, integration of expression of genes residing in the lesions of interest revealed putative features of the disease driving short survival.
INTRODUCTION
Almost all multiple myeloma (MM) patients harbor genetic lesions. Evaluation of these chromosomal aberrations by conventional karyotype, by fluorescent in situ hybridization (FISH), by comparative genomic hybridization, or by single nucleotide polymorphism (SNP) –based mapping arrays have revealed their prognostic value. Both numerical and structural abnormalities impact prognosis. Hyperdiploidy, characterized by multiple trisomies of chromosomes 3, 5, 7, 9, 11, 15, 19, and 21, is identified in 50% to 60% of myeloma patients and imparts longer survival.1–3 Conversely, structural abnormalities such as del(13) detected in approximately 50% of patients,4–6 del(16q) reported in approximately 20% of patients,7 del(17p) detectable in approximately 10% of patients,8–10 gain of 1q21 observed in approximately 30% to 43% of patients,11–13 or translocation involving immunoglobulin heavy chain locus t(4;14)(p16.3;q32) found in 14% to 20% of patients,14–15 and t(14;16)(q32;q23) seen in 2% to 10% of patients confer a higher risk of death.16–17
Importantly, while these targeted genetic abnormalities have prognostic impact individually or in combination, the high-resolution MM genome profiles provided some indications of the prognostic values of DNA copy-number abnormalities (CNAs).18
However, a global multivariate survival model that takes account of all recurrent CNAs harbored by a large cohort of newly diagnosed uniformly treated MM is still missing. This study sought to identify recurrent genetic lesions associated with prognosis using SNP-based mapping array technology that allows high-resolution detection of DNA copy number changes in order to develop a powerful survival model based on chromosomal abnormalities in MM.
PATIENTS AND METHODS
Patients
Both initial and validation cohorts examined in this study comprised patients with newly diagnosed myeloma younger than 66 years. All patients were treated according to Intergroupe Francophone du Myélome (IFM) 99 clinical trials in the IFM centers. Comparison of clinical parameters, cytogenetic abnormalities and outcome showed the absence of bias between SNP and nonSNP subsets across the separate trials.
Genomic and Interphase Cytogenetic Analysis
DNA extracted from CD138+ purified plasma cells was genotyped with Affymetrix GeneChip Human Mapping 500K Array Set (Affymetrix Inc, Santa Clara, CA) according to the manufacturer's instructions. The median call rates were 96.77% and 97.35%, respectively, for Affymetrix 250k Nsp and Sty arrays. FISH arrays have already been reported in a previous study.10 Concordance between 500K SNPs array and FISH copy number estimates was verified.
SNP Microarray Analysis Procedures
We used the dChip software (www.dchip.org) to normalize all the arrays of 192 MM samples and 10 normal blood samples and compute model-based signal values.19,20 A molecular karyotype was established with the average value of the SNPs intensities in each cytoband and used to identify hyperdiploid patients (47 chromosomes or more). Segmentation analyses were performed by applying the circular binary segmentation algorithm to the above log2 ratios data to identify CNAs for each sample.21
RESULTS
Recurrent Copy Number Alterations in MM
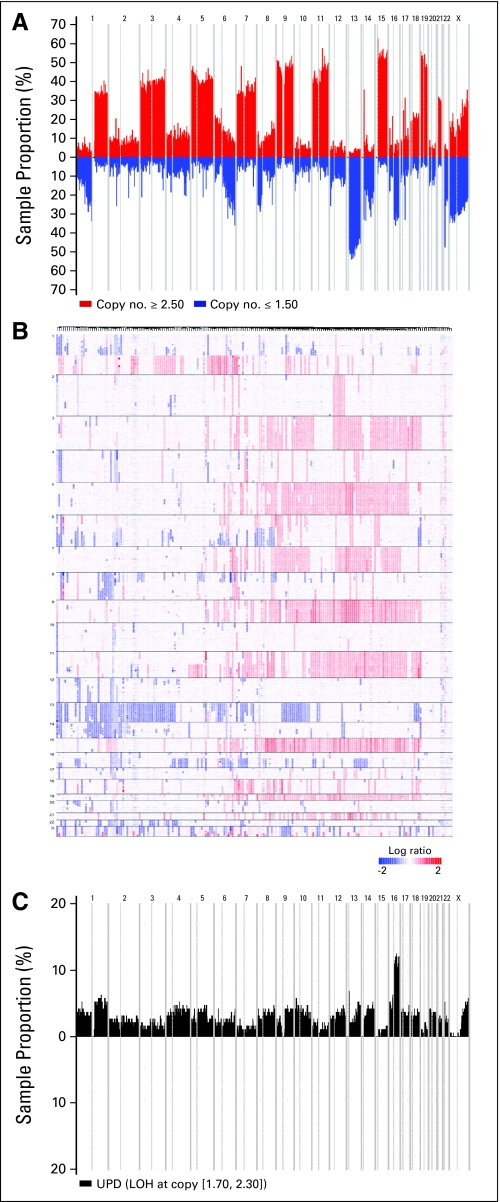
We used high-resolution 500K SNP mapping arrays to examine CNAs of malignant plasma cells from the initial cohort of 192 MM (Appendix Fig A1A). DNA copy number alterations of all chromosomes using 500K SNP mapping arrays in 192 MM revealed distinct patterns: loss or gain of the chromosome, loss or gain of a whole arm, interstitial losses or gains (Appendix Fig A1B). We first investigated these different patterns at the cytoband-level of each chromosome for each patient. Analysis of the most frequent lesions (> 10%) revealed two main groups. The first group encompasses almost exclusively (with the exception of chromosome 11) either gain or loss of entire chromosome or interstitial gain or loss of the flanking centromeric region (Table 1). This group includes gain of chromosomes 3, 5, 7, 9, 11, 15, 18, 19, 21, and loss of chromosomes 13, 22, and X (in female patients). While the second group is composed of genetic lesions that affect gain or loss of subchromosomal material, including amplification of 1q and 6p and deletion of 1p, 6q, 8p, 12p, 14q, 16p, 16q, and 20p. Hierarchical clustering of MM according to group 1 and group 2 abnormalities indicated that one major branch (approximately 50% of the samples) of the dendrogram captures the majority of hyperdiploid MM. This pattern is mainly related to gains of chromosomes 5, 9, 15, 19, and groups a lower fraction of deaths. In contrast, other CNAs patterns (mainly loss of submaterial chromosomes) appeared inhomogeneous. However, clustering analysis identified three separated branches of approximately 20 patients having either a loss of 16p/16q, or a loss of 13/14q or a loss 12p. These data mirror well the skyline recurrence plot for CNAs using microarray-based comparative genomic hybridization (array_CGH)18 as well as CNAs identified either by metaphase CGH22–24 or by high-density SNP arrays.25 However, metaphase CGH analysis underestimated CNAs frequencies compared to high-density data sets (for example, 1p loss, 8p loss, 7 gain, 9 gain, 15 gain, 19 gain) this is probably due to the hybridization method used (slides with human metaphase chromosome spreads versus microarrays) and the dramatic difference of resolution (5 to 10 megabases v 5 to 10 kb). Importantly, recurrent 12p and 20p losses were only identified in our study, probably because of the larger size of our cohort. We also investigated uniparental disomy (UPD) status (acquired or constitutive) by identifying loss of heterozygosity (LOH) from 500K SNP data set of MM in region with normal diploid copy number (2 ± 0.30). We identified only one frequent UPD (> 10%) among the 192 patients (Appendix Fig A1C). This region of UPD encompassed the whole arm of 16q and was previously reported with a lower frequency (7%).7
Table 1.
Frequency of Genetic Lesions That Were Present in More Than 10% of Samples
| Genetic Lesions | |||||
|---|---|---|---|---|---|
| Group 1 |
Group 2 |
||||
| Chromosome | Status | Frequency (%) | Chromosome Arm | Status | Frequency (%) |
| 3 | Gain | 36.1 | 1p | Loss | 24 |
| 5 | Gain | 38.5 | 1q | Gain | 31.3 |
| 7 | Gain | 29.5 | 6p | Gain | 15.1 |
| 9 | Gain | 41.7 | 6q | Loss | 20.8 |
| 11 | Gain | 34.7 | 8p | Loss | 22 |
| 13 | Loss | 44.8 | 11q (only) | Gain | 10.5 |
| 15 | Gain | 49 | 12p | Loss | 13 |
| 18 | Gain | 15.6 | 14q | Loss | 23.4 |
| 19 | Gain | 42.9 | 16p | Loss | 14.7 |
| 21 | Gain | 21.2 | 16q | Loss | 28.3 |
| 22 | Loss | 17.1 | 20p | Loss | 10.4 |
| X (women) | Loss | 49.5 | |||
Survival Impact of Genomic Lesions
Univariate analysis performed on the initial cohort revealed that in addition to clinical parameters: albumin (< 35 g/L), creatinine (> 180 μmol/L), serum β2-microglobulin (Sβ2M; ≥ 5.5 mg/L), genetic abnormalities detected by FISH: t(4;14), del(13) and del(17p) (> 60%) and hyperdiploidy detected by SNP array (≥ 47 chromosome), a large number of the genetic lesions identified were correlated with survival.
Given that our goal was to identify for each chromosome the minimum common genetic lesions (MCGL) with the most significant prognostic impact, we conducted univariate Cox analyses and retained MCGL highly significantly associated with overall survival (P < .001). We defined 612 amplifications or deletions located in 12 chromosomes. Two regions of amplification at 1q21.3 and 1q23.3 containing 22 MCGL, three regions of amplification at 5q31.3, 5q33.1, and 5q33.2 containing five MCGL, and one region of deletion at 12p31.31 containing two MCGL were below permutation and resampling tests thresholds (P < .05) with an individual false discovery rate lower than 1% and a frequency in agreement with the previous dChip recurrence plot and SNP-based karyotype.
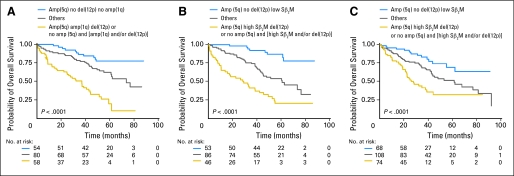
A further analysis to eliminate redundant MCGL identified only three MCGL located at 1q23.3, 5q31.3, and 12p13.31 retaining their significance in affecting overall survival (Table 2). Kaplan-Meier curves according to the presence or absence of these variables clearly identified three large subsets of patients that had different outcomes (Fig 1A). Patients with amp(5q31.3) only had the longest overall survival (5-year overall survival, 77.5%), while patients with any two risk factors amp(1q23.3) or del(12p13.31) or absence of amp(5q31.3) had the shortest overall survival (median, 33.7 months). The intermediate group with a median overall survival of 73.6 months had only one of these three features. Overlap between these three lesions was limited to 11%. del(12p13.31) was independent of both amp(1q23.3) and amp(5q31.3) whereas the last two were significantly associated. Furthermore, 5q and 1q abnormalities were strongly associated with hyperdiploidy and gain or loss of subchromosomal material, respectively. In order to evaluate the prognostic value of these DNA segments regarding to all parameters significantly associated with survival, we performed multivariate analysis. The results showed that amp(1q23.3) lost its significance while only amp(5q31.3) and del(12p13.31) along with Sβ2M retained strong independent prognostic influence (Table 3). An alternative multivariate analysis including the established prognostic variables; Sβ2M (≥ 5.5 mg/L) and combination of t(4;14) and del(17p) (≥ 60%) into the model first and the three CNAs variables second confirmed the added value of amp(5q31.3) and del(12p13.31) in terms of routine clinical use (Table 4). del(17p) (≥ 60%) did not retain its significance in multivariate analyses probably because of its low frequency in patients with myeloma (< 8%). Importantly, these analyses provide an original simple but powerful prognostic model based on one favorable genetic parameter, amp(5q31.3) (hazard ratio [HR], 0.37; P = .0005), one adverse genetic marker, del(12p13.31) (HR, 3.17; P < .0001), and one clinical parameter, high Sβ2M (≥ 5.5 mg/L; HR, 2.78; P < .0001). Patients with presence of amp(5q31.3), without del(12p13.31) and low Sβ2M levels (representing 29% of the cohort) had a particularly good prognosis with a 5-year overall survival of 87% (95% CI, 73.4 to 94.0) while the presence of amp(5q31.3) and del(12p13.31) and high Sβ2M levels or the absence of amp(5q31.3) and either the presence of del(12p13.31) or high Sβ2M levels or both (representing 25% of the cohort) conferred a poor prognosis with a median overall survival of 28.7 months (Fig 1B). High-risk patients have a nine-fold hazard ratio (95% CI, 4.21 to 19.96; P < .0001) of death compared with low-risk patients. The model performed similarly across the clinical trials and remained a highly significant variable when adjusted for the protocols (HR, 2.71; P < .0001). The prognostic value of this model was validated by applying 12p13.31 and 5q31.3 FISH probes in an independent validation cohort of 273 newly diagnosed MM (Fig 1C). High-risk patients predicted by the FISH-based model (30% of patients) have a median overall survival of 27.7 months and their relative risk of death is increased in by a factor of 4 (95% CI, 2.18 to 7.19; P < .0001) compared with low-risk patients. When competed with the 15-gene signature,26 both predictors remain highly significant independent variables. The combination of copy number-based and expression-based models is able to identify patients at low risk (59% of patients) with a 4-year survival rate of 76% and patients at high risk (10% of patients) with a 4-year survival rate of only 19%.
Table 2.
Univariate and Multivariate Analyses of amp(1q23.3), amp(5q31.3), and del(12p13.31) in the Initial Cohort
| CNA | Start Position (kb) | Size (kb) | No. of Cases | Univariate Analysis |
Adjusted Cox |
||||
|---|---|---|---|---|---|---|---|---|---|
| HR | 95% CI | P | HR | 95% CI | P | ||||
| amp(1q23.3) | 162 551 530 | 142 | 58 | 2.24 | 1.46 to 3.44 | .0002 | 1.90 | 1.23 to 2.94 | .0039 |
| amp(5q31.3) | 140 901 595 | 702 | 70 | 0.33 | 0.19 to 0.56 | < .0001 | 0.37 | 0.22 to 0.63 | .0002 |
| del(12p13.31) | 6 398 330 | 208 | 22 | 3.03 | 1.74 to 5.27 | < .0001 | 2.32 | 1.33 to 4.06 | .0032 |
Abbreviations: CNA, copy-number abnormality; HR, hazard ratio.
Fig 1.
Prognostic impact of copy-number abnormality–based model in initial and validation cohorts. (A) Initial cohort stratified according to amp(1q23.3), amp(5q31.3), del(12p13.31) by single nucleotide polymorphism (SNP) array. (B) Initial cohort stratified according to amp(5q31.3), del(12p13.31) by SNP array and serum β2-microglobulin (Sβ2M) ≥ 5.5 mg/L. (C) Validation cohort stratified according to amp(5q31.3), del(12p13.31) by fluorescent in situ hybridization and Sβ2M ≥ 5.5 mg/L.
Table 3.
Contribution of Sβ2M to the Copy-Number Abnormalities Model
| Prognostic Variable | Multivariate Analysis (stepwise Cox model) |
||
|---|---|---|---|
| Hazard Ratio | 95% CI | P | |
| Sβ2M, ≥ 5.5 v < 5.5 mg/L | 2.78 | 1.78 to 4.35 | < .0001 |
| amp(5q31.3), yes v no | 0.37 | 0.21 to 0.64 | .0005 |
| del(12p13.31), yes v no | 3.17 | 1.81 to 5.56 | < .0001 |
Abbreviation: Sβ2M, serum β2-microglobulin.
Table 4.
Added Value of Copy-Number Abnormalities to the Established Prognostic Model
| Prognostic Variable | Multivariate Analysis (Cox model) | ||
|---|---|---|---|
| Hazard Ratio | 95% CI | P | |
| Sβ2M, ≥ 5.5 mg/L | 2.99 | 1.88 to 4.74 | < .0001 |
| t(4;14)_del(17p) | 1.66 | 0.97 to 2.86 | .066 |
| amp(5q31.3) | 0.45* | 0.25 to 0.80 | .0070 |
| del(12p13.31) | 2.61* | 1.44 to 4.72 | .0015 |
Abbreviation: Sβ2M, serum β2-microglobulin.
Stepwise analysis.
Candidate Genes Residing Within the Three DNA Lesions
We firstly investigated the association between pattern of expression of genes residing within the three lesions and presence or absence of CNAs. Then, resident genes with significant correlations with CNAs were tested for association with overall survival. Finally, genes satisfying both criteria were considered as potential gene targets within the genetic lesion, and Kaplan-Meier analysis of the initial cohort according to each gene risk (divided into quartiles) was performed.
We found five highly significant prognostic genes (P < .0001); four upregulated, ILF2, ADAR, ALDH9A1, and UBAP2L resided on 1q23.3 that identified high-risk patients in the upper quartile and one downregulated gene, CD27, mapped to12p13.31 that identified patients in the lower quartile with very poor outcome. Thus integration of DNA copy number analysis, expression profile, and clinical outcome provided potential cancer-relevant targets. These included one gene postulated to be important in MM pathogenesis, CD27,27 whose diminution of expression is associated with disease progression28–31 as well as genes not known to be associated with myelomagenesis. ILF2 that encodes NF45, the regulatory subunit of NF90/NF110 complexes32 that have been demonstrated to stimulate gene expression at multiple levels, including transcription, translation, post-transcriptional stabilization, and nuclear exportation33 and ADAR that encodes ADAR1, a RNA-editing enzyme involved in the conversion of adenosines to inosines.34,35 Especially noteworthy, ADAR1 interacts with NF90 complexes through double-stranded RNA and enhances NF90-mediated gene expression.33
Major Contribution of Chromosome 5 Gain to Hyperdiploidy Prognostic Impact
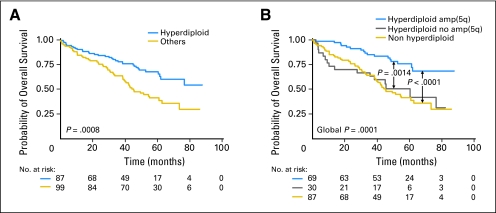
Hyperdiploidy (≥ 47 chromosomes) observed in 99 patients (53.2%) conferred a good prognosis (Fig 2A). Given that recent studies have clearly shown a biologic heterogeneity among hyperdiploid patients associated with a different outcome,3,18 we examined whether gain at 5q31, the most favorable prognosis marker compared to other gains at 9p21, 15q12-q13, and 19q13, identified a subgroup with different outcome. Hyperdiploid cases with 5q31 gain had a better outcome than hyperdiploid MM lacking this feature (P = .0014; Fig 2B). Furthermore, outcome of hyperdiploid patients without 5q gain and non hyperdiploid patients was not different (P > .8). In our series, all tients with 5q31 gain were hyperdiploid except one. Thus, our results suggest that hyperdiploidy with 5q31 gain is a distinct entity that drives a more favorable prognosis.
Fig 2.
Prognostic impact of (A) hyperdiploidy and (B) amp(5q31.3) within hyperdiploid patients.
DISCUSSION
We have shown that DNA copy-number analysis using high-density SNP arrays provides a molecular karyotype for every patient and leads to the identification of novel chromosomal abnormalities that impact prognosis. Our SNP array data mirror well the skyline recurrence plot using microarray-based comparative genomic hybridization for chromosomal aberrations derived from 67 newly diagnosed MM.18 High-resolution karyotyping is particularly relevant in MM since almost all patients (98% in this study) harbored chromosomal lesions and required only a small amount of purified myeloma cells (approximately 100,000 cells). In myeloma with lack of proliferative clone makes conventional cytogenetics informative in only limited number of patients (approximately 30% of patients), while use of interphase FISH circumvented the analysis to only very select loci in the genome. Use of SNP array as described here makes the technique applicable in most of the patients with evaluation of myeloma cells irrespective of their cell cycle status as well as investigation of the whole genome. Standardization of the technique and requirement of relatively smaller number of myeloma cells now provide general applicability of this technique in future.
We present here the first prognostic model based on frequent genome-wide DNA copy-number lesions in newly diagnosed MM. Univariate analyses revealed genetic lesions located on chromosomes 1, 5, 8q, 9, 11, 12p, 14q, 15q, 16q, 19q, 20p, 22q, significantly associated with survival (P < .001). After testing the robustness of these selected DNA segments by permutation and resampling only MCGL located at 1q, 5q, and 12p were retained in a multivariate predictive model.
The main interest of this model resides in its ability to identify patients who greatly benefit from high-dose therapy (patients with amp(5q31.3) alone and low Sβ2M; 5-year overall survival, 87%). Of note, the prognostic impact of amp(5q31.3) over-rides that of hyperdiploidy and 5q lesion is easier to assess than hyperdiploid status. In addition, our model identifies high-risk patients (25% of patients) with a survival equivalent to that of gene expression models.26,36 Comparison of both CNA- and expression-based models suggest that these molecular predictors should be combined to form the most accurate prognostic model for MM. The CNA-based model is more discriminating than FISH-based models that are by definition targeted on combination of specific abnormalities such as t(4;14), del(17p), or del(16q) in the context of high or low Sβ2M.7,10,37 For example, high-risk patients defined either by our CNA-based model or by del(17p) and/or a t(4;14) (approximately 20% of patients) had median overall survival of 29 months and 37 months, respectively. del(12p) is the strongest prognostic parameter in our cohort, this lesion has been previously reported in both newly diagnosed1,25 andrelapsed MM patients.38 del(12p) frequency calculated from Smadja et al1 (11.6%) is comparable with our results (13%). The strong prognostic value of del(12p) emerged solely in our study probably for several reasons including: the size of the cohort, the resolution of the genetic method used, and the median follow-up period available in the cohort.
Our results from LOH and copy-number analyses of 16q region in the 192 patients are in agreement with a recent study of 55 newly diagnosed patients with using the same Affymetrix platform.7 We identified 16q LOH in 38.3% of patients. The entire or interstitial deletions represent 27.8% (53 of 191) of patients and UPD represent 10.5% (20 of 191). Univariate analysis identified two MCGL located at 16q12 and 16q23 significantly associated with adverse overall survival (P < .001), but their stability was not strong enough (permutation and resampling P = 0.3; false discovery rate > 30%) to enter the multivariate model. In addition, Cox analysis revealed that UDP16q has no prognostic impact in our cohort (P = .015).
In addition to the strong prognostic impact of specific DNA copy-number changes revealed by high resolution SNP array, combination with expression profiles provides novel features of disease pathogenesis. NF45 stabilizes double-stranded RNA-binding protein NF90 complexes and reduction of NF45 or NF90 induces cell growth retardation.32 NF90 binds to vascular endothelial growth factor 3′ untranslated mRNA and contributes to hypoxia-induced vascular endothelial growth factor expression and tumorigenesis in vivo in a breast carcinoma.39 Of particular interest, recent results demonstrated an essential role of ADAR1 in hematopoiesis, where it acts to suppress interferon signaling and to block premature apoptosis.40 The authors suggest that the ADAR1-NF90 interaction may be critical in this process. These recent data support the hypothesis that concerted overexpression of ADAR1 and NF45 could contribute to MM pathophysiology.
CD27 is a member of the tumor necrosis factor receptor superfamily and like many other members of this family activates c-jun kinase and both the classical and alternative nuclear factor-κB pathways.41 We can hypothesize that diminution of CD27 in high-risk patients reduces NF-kB pathway activity in these patients. This hypothesis is supported by recent studies showing that the proliferation and MMSET-spike subgroups of poor prognosis relative to the other MM subgroups had low expression NF-kB signature.42,43 In the same way, relative underexpression of CD27 in CD-1 compared to CD-2 myeloma subgroups is associated with a lower NF-kB signature.42 These results suggest that therapeutic strategies targeting the NF-kB pathway may not be tailored for high-risk patients defined by molecular profiling.
In conclusion, high-resolution karyotyping using SNP or CGH arrays is a powerful investigative tool to detect novel lesions that impact prognosis in MM (this study7,27), thereby improving accuracy of the established predictive models. Molecular karyotype will occupy rapidly a place of choice in the armory of biologic tools used for prognostication and patient adapted-risk therapeutic strategy.
Supplementary Material
Acknowledgment
We thank Elise Béguet, Magali Martin, Emilie Ollivier, and Nathalie Roi for excellent technical expertise.
Appendix
Affymetrix CEL files for the 192 CD138+ myeloma plasma cells, and the 10 blood samples used as diploid reference samples for copy number analysis have been deposited in NCBI Gene Expression Omnibus (GEO, http://www.ncbi.nlm.nih.gov/geo/) and are accessible through GEO Series accession number GSE12896.
Fig A1.
(A) Recurrence of copy-number abnormalities (CNAs) across 192 multiple myeloma (MM) in chromosomal order. Red or blue bars denote gain or loss of chromosome material. (B) dChip median-smoothed log ratio copy number heatmap showing all chromosomes. Each column represents a case and SNPs are arranged from 1p(tel) to Xq(tel) from top to the bottom. Blue represents deletion and red amplification. (C) Recurrence of uniparental disomy across the samples.
Footnotes
Supported in part by grants from the Ligue contre le Cancer (Equipe Labellisée); Grant No. SPORE P50 CA100707-04 (H.A.L., K.C.A., N.C.M.) from the National Cancer Institute; by the Department of Veterans Affairs merit review award; and Grants No. RO1-129295 (N.C.M.), PO1-78378 (K.C.A., N.C.M.), and RO1-50947 (K.C.A.) from the National Institutes of Health.
Written on behalf of the Intergroupe Francophone du Myélome.
Authors' disclosures of potential conflicts of interest and author contributions are found at the end of this article.
AUTHORS' DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
The author(s) indicated no potential conflicts of interest.
AUTHOR CONTRIBUTIONS
Conception and design: Hervé Avet-Loiseau, Florence Magrangeas, Kenneth C. Anderson, Nikhil C. Munshi, Stéphane Minvielle
Provision of study materials or patients: Jean-Luc Harousseau, Michel Attal, Gerald Marit, Claire Mathiot, Thierry Facon, Philippe Moreau
Collection and assembly of data: Florence Magrangeas, Wilfried Gouraud, Stéphane Minvielle
Data analysis and interpretation: Hervé Avet-Loiseau, Cheng Li, Catherine Charbonnel, Loïc Campion, Nikhil C. Munshi, Stéphane Minvielle
Manuscript writing: Stéphane Minvielle
Final approval of manuscript: Hervé Avet-Loiseau, Cheng Li, Florence Magrangeas, Philippe Moreau, Loïc Campion, Nikhil C. Munshi, Stéphane Minvielle
REFERENCES
- 1.Smadja NV, Bastard C, Brigaudeau C, et al. Hypodiploidy is a major prognostic factorin multiple myeloma. Blood. 2001;98:2229–2238. doi: 10.1182/blood.v98.7.2229. [DOI] [PubMed] [Google Scholar]
- 2.Fonseca R, Debes-Marun CS, Picken EB, et al. The recurrent IgH translocations are highly associated with nonhyperdiploid variant multiple myeloma. Blood. 2003;102:2562–2567. doi: 10.1182/blood-2003-02-0493. [DOI] [PubMed] [Google Scholar]
- 3.Chng WJ, Kumar S, Vanwier S, et al. Molecular dissection of hyperdiploid multiple myeloma by gene expression profiling. Cancer Res. 2007;67:2982–2989. doi: 10.1158/0008-5472.CAN-06-4046. [DOI] [PubMed] [Google Scholar]
- 4.Zojer N, Königsberg R, Ackermann J, et al. Deletion of 13q14 remains an independent adverse prognostic variable in multiple myeloma despite its frequent detection by interphase fluorescence in situ hybridization. Blood. 2000;95:1925–1930. [PubMed] [Google Scholar]
- 5.Facon T, Avet-Loiseau H, Guillerm G, et al. Chromosome 13 abnormalities identified by FISH analysis and serum β2-microglobulin produce a very powerful myeloma staging system for patients receiving high dose therapy. Blood. 2001;97:1566–1571. doi: 10.1182/blood.v97.6.1566. [DOI] [PubMed] [Google Scholar]
- 6.Fonseca R, Harrington D, Oken MM, et al. Biological and prognostic significance of interphase fluorescence in situ hybridization detection of chromosome 13 abnormalities (Δ13) in multiple myeloma: An Eastern Cooperative Oncology Group study. Cancer Res. 2002;62:715–720. [PubMed] [Google Scholar]
- 7.Jenner MW, Leone PE, Walker BA, et al. Gene mapping and expression analysis of 16q loss of heterozygosity identifies WWOX and CYLD as being important in clinical outcome in multiple myeloma. Blood. 2007;110:3291–3300. doi: 10.1182/blood-2007-02-075069. [DOI] [PubMed] [Google Scholar]
- 8.Drach J, Ackermann J, Fritz E, et al. Presence of a p53 gene deletion in patients with multiple myeloma predicts for short survival after conventional-dose chemotherapy. Blood. 1998;92:802–809. [PubMed] [Google Scholar]
- 9.Chang H, Qi C, Yi QL, et al. p53 gene deletion detected by fluorescence in situ hybridization is an adverse prognostic factor for patients with multiple myeloma following autologous stem cell transplantation. Blood. 2005;105:358–360. doi: 10.1182/blood-2004-04-1363. [DOI] [PubMed] [Google Scholar]
- 10.Avet-Loiseau H, Attal M, Moreau P, et al. Genetic abnormalities and survival in multiple myeloma: The experience of the Intergroupe Francophone du Myelome. Blood. 2007;109:3489–3495. doi: 10.1182/blood-2006-08-040410. [DOI] [PubMed] [Google Scholar]
- 11.Fonseca R, Van Wier SA, Chng WJ, et al. Prognostic value of chromosome 1q21 gain by fluorescent in situ hybridization and increase CKS1B expression in myeloma. Leukemia. 2006;20:2034–2040. doi: 10.1038/sj.leu.2404403. [DOI] [PubMed] [Google Scholar]
- 12.Chang H, Ning Y, Qi X, et al. Chromosome 1p21 deletion is a novel prognostic marker in patients with multiple myeloma. Br J Haematol. 2007;139:51–54. doi: 10.1111/j.1365-2141.2007.06750.x. [DOI] [PubMed] [Google Scholar]
- 13.Hanamura I, Stewart JP, Huang Y, et al. Frequent gain of chromosome band 1q21 in plasma-cell dyscrasias detected by fluorescence in situ hybridization: Incidence increases from MGUS to relapsed myeloma. Blood. 2006;108:1724–1732. doi: 10.1182/blood-2006-03-009910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Moreau P, Facon T, Leleu X, et al. Recurrent 14q32 translocations determine the prognosis of multiple myeloma, especially in patients receiving intensive chemotherapy. Blood. 2002;100:1579–1583. doi: 10.1182/blood-2002-03-0749. [DOI] [PubMed] [Google Scholar]
- 15.Keats JJ, Reiman T, Maxwell CA, et al. In multiple myeloma, t(4;14)(p16;q32) is an adverse prognostic factor irrespective of FGFR3 expression. Blood. 2003;101:1520–1529. doi: 10.1182/blood-2002-06-1675. [DOI] [PubMed] [Google Scholar]
- 16.Fonseca R, Blood E, Rue M, et al. Clinical and biologic implications of recurrent genomic aberrations in myeloma. Blood. 2003;101:4569–4575. doi: 10.1182/blood-2002-10-3017. [DOI] [PubMed] [Google Scholar]
- 17.Avet-Loiseau H, Facon T, Grosbois B, et al. Oncogenesis of multiple myeloma: 14q32 and 13q chromosomal abnormalities are not randomly distributed, but correlate with natural history, immunological features, and clinical presentation. Blood. 2002;99:2185–2191. doi: 10.1182/blood.v99.6.2185. [DOI] [PubMed] [Google Scholar]
- 18.Carrasco DR, Sukhdeo K, Protopopova M, et al. The differentiation and stress response factor XBP-1 drives multiple myeloma pathogenesis. Cancer Cell. 2006;11:349–360. doi: 10.1016/j.ccr.2007.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lin M, Wei LJ, Sellers WR, et al. DChipSNP: Significance curve and clustering of SNP-array-based loss-of-heterozygosity data. Bioinformatics. 2004;20:1233–1240. doi: 10.1093/bioinformatics/bth069. [DOI] [PubMed] [Google Scholar]
- 20.Beroukhim R, Lin M, Park Y, et al. Inferring loss-of-heterozygosity from unpaired tumors using high-density oligonucleotide SNP arrays. PLoS Comput Biol. 2006;5:e41. doi: 10.1371/journal.pcbi.0020041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Olshen AB, Venkatraman ES, Lucito R, et al. Circular binary segmentation for the analysis of array-based DNA copy number data. Biostatistics. 2004;5:557–572. doi: 10.1093/biostatistics/kxh008. [DOI] [PubMed] [Google Scholar]
- 22.Cigudosa JC, Rao PH, Calasanz MJ, et al. Characterization of nonrandom chromosomal gains and losses in multiple myeloma by comparative genomic hybridization. Blood. 1998;91:3007–3010. [PubMed] [Google Scholar]
- 23.Liebisch P, Viardot A, Bassermann N, et al. Value of comparative genomic hybridization and fluorescence in situ hybridization for molecular diagnostics in multiple myeloma. Br J Haematol. 2003;122:193–201. doi: 10.1046/j.1365-2141.2003.04417.x. [DOI] [PubMed] [Google Scholar]
- 24.Gutierrez NC, Garcia JL, Hernandez JM, et al. Prognostic and biologic significance of chromosomal imbalances assessed by comparative genomic hybridization in multiple myeloma. Blood. 2004;104:2661–2666. doi: 10.1182/blood-2004-04-1319. [DOI] [PubMed] [Google Scholar]
- 25.Walker BA, Leone PE, Jenner MW, et al. Integration of global SNP-based mapping and expression arrays reveals key regions, mechanisms, and genes important in the pathogenesis of multiple myeloma. Blood. 2006;108:1733–1743. doi: 10.1182/blood-2006-02-005496. [DOI] [PubMed] [Google Scholar]
- 26.Decaux O, Lodé L, Magrangeas F, et al. Prediction of survival in multiple myeloma based on gene expression profiles reveals cell cycle and chromosomal instability signatures in high-risk patients and hyperdiploid signatures in low-risk patients: A study of the Intergroupe Francophone du Myelome. J Clin Oncol. 2008;26:4798–4805. doi: 10.1200/JCO.2007.13.8545. [DOI] [PubMed] [Google Scholar]
- 27.Katayama Y, Sakai A, Oue N, et al. A possible role for the loss of CD27-CD70 interaction in myelomagenesis. Br J Haematol. 2003;120:223–234. doi: 10.1046/j.1365-2141.2003.04069.x. [DOI] [PubMed] [Google Scholar]
- 28.Zhan F, Hardin J, Kordsmeier B, et al. Global gene expression profiling of multiple myeloma, monoclonal gammopathy of undetermined significance, and normal bone marrow plasma cells. Blood. 2002;99:1745–1757. doi: 10.1182/blood.v99.5.1745. [DOI] [PubMed] [Google Scholar]
- 29.Guikema JE, Hovenga S, Vellenga E, et al. CD27 is heterogeneously expressed in multiple myeloma: Low CD27 expression in patients with high-risk disease. Br J Haematol. 2003;121:36–43. doi: 10.1046/j.1365-2141.2003.04260.x. [DOI] [PubMed] [Google Scholar]
- 30.Moreau P, Robillard N, Jégo G, et al. Lack of CD27 in myeloma delineates different presentation and outcome. Br J Haematol. 2006;132:168–170. doi: 10.1111/j.1365-2141.2005.05849.x. [DOI] [PubMed] [Google Scholar]
- 31.Morgan TK, Zhao S, Chang KL, et al. Low CD27 expression in plasma cell dyscrasias correlates with high-risk disease: An immunohistochemical analysis. Am J Clin Pathol. 2006;126:545–551. doi: 10.1309/ELGMGX81C2UTP55R. [DOI] [PubMed] [Google Scholar]
- 32.Guan D, Altan-Bonnet N, Parrott AM, et al. Nuclear factor 45 (NF45) is a regulatory subunit of complexes with NF90/110 involved in mitotic control. Mol Cell Biol. 2008;28:4629–4641. doi: 10.1128/MCB.00120-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nie YL, Ding PN, Kao R, et al. ADAR1interacts with NF90 through double-stranded RNA and regulates NF90-mediated gene expression independently of RNA editing. Mol Cell Biol. 2005;25:6956–6963. doi: 10.1128/MCB.25.16.6956-6963.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Patterson JB, Samuel CE. Expression and regulation by interferon of a double-stranded-RNA-specific adenosine deaminase from human cells: Evidence for two forms of the deaminase. Mol Cell Biol. 1995;15:5376–5388. doi: 10.1128/mcb.15.10.5376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Herbert A, Wagner S, Nickerson JA, et al. Induction of protein translation by ADAR1 within living cell nuclei is not dependent on RNA editing. Mol Cell. 2002;10:1235–1246. doi: 10.1016/s1097-2765(02)00737-2. [DOI] [PubMed] [Google Scholar]
- 36.Shaughnessy JD, Jr, Zhan F, Burington B, et al. A validated gene expression model of high-risk multiple myeloma is defined by deregulated expression of genes mapping to chromosome 1. Blood. 2007;109:2276–2284. doi: 10.1182/blood-2006-07-038430. [DOI] [PubMed] [Google Scholar]
- 37.Moreau P, Attal M, Garban F, et al. Heterogeneity of t(4;14) in multiple myeloma. Long-term follow-up of 100 cases treated with tandem transplantation in IFM99 trials. Leukemia. 2007;21:2020–2024. doi: 10.1038/sj.leu.2404832. [DOI] [PubMed] [Google Scholar]
- 38.Bryant B, Danaee H, Lichter D, et al. High-resolution assessment of chromosomal gains and losses in multiple myeloma tumors from bortezomib clinical trial. J Clin Oncol. 2008;26(suppl):471s. abstr 8570. [Google Scholar]
- 39.Vumbaca F, Phoenix KN, Rodriguez-Pinto D, et al. Double-stranded RNA-binding protein regulates vascular endothelial growth factor mRNA stability, translation, and breast cancer angiogenesis. Mol Cell Biol. 2008;28:772–783. doi: 10.1128/MCB.02078-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hartner JC, Walkley CR, Lu J, et al. ADAR1 is essential for the maintenance of hematopoiesis and suppression of interferon signaling. Nat Immunol. 2009;10:109–115. doi: 10.1038/ni.1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xie P, Kraus ZJ, Stunz LL, et al. Roles of TRAF molecules in B lymphocyte function. Cytokine Growth Factor Rev. 2008;19:199–207. doi: 10.1016/j.cytogfr.2008.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhan F, Huang Y, Colla S, et al. The molecular classification of multiple myeloma. Blood. 2006;108:2020–2028. doi: 10.1182/blood-2005-11-013458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Annunziata CM, Davis RE, Demchenko Y, et al. Frequent engagement of the classical and alternative NF-kappaB pathways by diverse genetic abnormalities in multiple myeloma. Cancer Cell. 2007;12:115–130. doi: 10.1016/j.ccr.2007.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.