Abstract
Purpose
Loss of heterozygosity (LOH) at chromosome 18q frequently occurs late during colon cancer development and is inversely associated with microsatellite instability (MSI). 18q LOH has been reported to predict shorter survival in patients with colorectal cancer, whereas MSI-high status has been associated with superior prognosis. However, it is unclear whether 18q LOH in colorectal cancer has any prognostic implication independent of MSI status and other potential predictors of clinical outcome.
Patients and Methods
Among 555 non–MSI-high colorectal cancers (stage I to IV) in two independent prospective cohort studies, we examined 18q LOH in relation to other molecular events and patient survival. Cox proportional hazard models computed hazard ratio of death, adjusted for clinical and tumoral characteristics, including KRAS, BRAF, PIK3CA, β-catenin, p53, CpG island methylator phenotype, LINE-1 methylation, and John Cunningham (JC) virus T antigen.
Results
In multivariate logistic regression, 18q LOH was independently associated with JC virus T antigen (odds ratio [OR] = 1.93; P = .0077), body mass index ≥ 30 kg/m2 (obesity; OR = 2.01; P = .014), high tumor grade (OR = 0.40; P = .018), KRAS mutation (OR = 0.66; P = .40), and LINE-1 hypomethylation (for a 30% decrease; OR = 1.92; P = .045). Five-year colorectal cancer–specific survival was 75% among patients with 18q LOH-positive tumors and 74% among those with 18q LOH-negative tumors (log-rank P = .80). Five-year overall survival was 70% among patients with 18q LOH-positive tumors and 68% among those with 18q LOH-negative tumors (log-rank P = .54). Multivariate analysis did not show prognostic significance of 18q LOH.
Conclusion
In our large prospective study of patients with non–MSI-high colorectal cancer, 18q LOH or allelic imbalance was not associated with patient survival.
INTRODUCTION
Colorectal cancers develop through a multistep carcinogenic process in which genetic and epigenetic events sequentially accumulate.1 Allelic loss at chromosome 18q, which can be assessed by loss of heterozygosity (LOH) analysis, frequently occurs late in the carcinogenic process. There are many candidate tumor suppressor genes in 18q, including DCC, SMAD4 (DPC4), SMAD2, and CABLES1.2-4 18q LOH has been inversely associated with microsatellite instability (MSI),5 which is an important molecular classifier in colorectal cancer.6,7
The presence of tumoral 18q LOH has been associated with an inferior survival in colorectal cancer in some studies,5,8-11 though not all.12-15 Many of the previous studies assessing the prognostic significance of 18q LOH8,9,12,16 did not consider the potential confounding effect of MSI, which is typically associated with improved patient survival.17 Moreover, a meta-analysis of research articles on 18q LOH and clinical outcome in colorectal cancers showed strong evidence for publication bias,18 in that small studies with null results had a higher likelihood of being unpublished, as compared with small studies with “significant” results. Although three large-scale studies (N > 300) have failed to show independent prognostic significance of 18q LOH in colorectal cancer,13-15 one study with 279 informative cases of colorectal cancer did find that 18q LOH was independently associated with a significant reduction in patient survival.5 Additional well-conducted large-scale studies are necessary to draw definitive conclusions.
In this study using more than 500 stage I to IV colorectal cancers identified in two independent, prospective cohort studies, we examined the prognostic significance and molecular associations of 18q LOH in non–MSI-high tumors. Because we concurrently assessed related molecular variables such as the CpG island methylator phenotype (CIMP), LINE-1 hypomethylation, p53, and BRAF and KRAS mutations, we could evaluate the effect of 18q LOH independent of these potential confounders.
PATIENTS AND METHODS
Study Population
We used the databases of two independent prospective cohort studies: the Nurses' Health Study (N = 121,701 women observed since 1976)19,20 and the Health Professionals Follow-Up Study (N = 51,529 men observed since 1986).20 Every 2 years, participants have been sent follow-up questionnaires to identify newly diagnosed cancer. We collected paraffin-embedded tissue blocks from hospitals where patients underwent tumor resections.20 We excluded cases preoperatively treated with radiation and/or chemotherapy. Tissue sections from all colorectal cancer cases were reviewed by the pathologist (S.O.). Tumor grade was categorized as high (poor or undifferentiation) versus low (moderate or well differentiation), defined as ≤ 50% versus more than 50% glandular area, respectively. We excluded 62 non–MSI-high tumors because of lack of informative 18q LOH results. On the basis of availability of tissue specimens and informative results of 18q LOH, we included a total of 555 non–MSI-high colorectal cancers diagnosed up to 2002. Written informed consent was obtained from all study subjects. This study was approved by the human subjects committees at Brigham and Women's Hospital and the Harvard School of Public Health.
Measurement of Mortality
Patients were observed until death or June 30, 2006, whichever came first. Ascertainment of deaths included reporting by the family or postal authorities. In addition, the names of persistent nonresponders were searched in the National Death Index. The cause of death was assigned by physicians blinded to information on lifestyle exposures and tumoral molecular changes.
DNA Extraction and Pyrosequencing of KRAS, BRAF, and PIK3CA
DNA from paraffin-embedded tissue was extracted, and pyrosequencing targeted for KRAS codons 12 and 13,21 BRAF codon 600,22 and PIK3CA exons 9 and 20 were performed.23
MSI and 18q LOH Analysis
MSI status was determined using 10 microsatellite markers (D2S123, D5S346, D17S250, BAT25, BAT26, BAT40, D18S55, D18S56, D18S67, and D18S487).24 MSI high was defined as the presence of instability in ≥ 30% of the markers, MSI low was defined as the presence of instability in 10% to 29% of the markers, and microsatellite stability was defined as no unstable marker. In this study, we examined non–MSI-high tumors to assess the clinical and molecular correlates with 18q LOH, because MSI high, if present, often made interpretation of 18q microsatellite markers difficult or impossible, and because MSI-high tumors rarely exhibited 18q LOH.
For LOH analysis, we duplicated polymerase chain reaction (PCR) reaction and electrophoresis for 18q markers (D18S55 at 18q22.1, D18S56 at 18q12.2-3, D18S67 at 18q12.2-3, and D18S487 at 18q21.1) to exclude allele dropouts of one of two alleles.24 LOH at each locus (D18S55, D18S56, D18S67, or D18S487) was defined as ≥ 40% reduction of one of two allele peak heights in tumor DNA relative to normal DNA. 18q LOH positivity was defined as the presence of LOH in any of the 18q markers. 18q LOH negativity was strictly defined as the presence of at least two informative markers and the absence of LOH.
Real-Time PCR for CpG Island Methylation and Pyrosequencing to Measure LINE-1 Methylation
Sodium bisulfite treatment on tumor DNA and subsequent real-time PCR (MethyLight) assays were validated and performed.25 We quantified promoter methylation in eight CIMP-specific genes (CACNA1G, CDKN2A, CRABP1, IGF2, MLH1, NEUROG1, RUNX3, and SOCS1).26-28 CIMP high was defined as six or more of eight methylated promoters using the eight-marker CIMP panel, and CIMP low/0 was defined as 0 to five methylated promoters, according to the previously established criteria.27 To accurately quantify relatively high LINE-1 methylation levels, we used pyrosequencing as previously described.29
Immunohistochemistry for p53, β-catenin, and John Cunningham Virus T Antigen
Tissue microarrays were constructed,20,30 and immunohistochemistry was performed as previously described for John Cunningham virus T antigen (JCVT),31 p53,32 and β-catenin.33 All immunohistochemically stained slides were interpreted by a pathologist (p53 by S.O., β-catenin and JCVT by K.N.) unaware of other data. A random sample of 118 to 402 tumors were re-examined for each marker by a second observer (p53 by K.N., β-catenin by S.O., and JCVT by Y.B.) unaware of other data, and the concordance between the two observers was 87% for p53 (κ = 0.75, n = 118), 83% for β-catenin (κ = 0.65, n = 402), and 87% for JCVT (κ = 0.74, n = 147), indicating substantial agreement.
Statistical Analysis
We used SAS program (version 9.1, SAS Institute, Cary, NC). All P values were two-sided, and statistical significance was set at P < .05. Nonetheless, P values were conservatively interpreted when conducting multiple hypothesis testing. For categoric data, the χ2 test (or Fisher's exact test if appropriate) was performed. The κ coefficient was calculated to assess an agreement between the two observers in immunohistochemical analyses. We also examined the possibility of a nonlinear relation of body mass index (BMI) or LINE-1 methylation with 18q LOH, nonparametrically with restricted cubic spines.34
To assess independent relations of multiple variables with 18q LOH, a multivariate logistic regression analysis was performed. Odds ratio (OR) was adjusted for sex, age (continuous), BMI (< 30 v ≥ 30 kg/m2), family history of colorectal cancer (present v absent), tumor location (proximal v distal), tumor stage (I to II v III to IV), tumor grade (low v high), CIMP status (high v low/0), MSI status (low v microsatellite stability), LINE-1 methylation (continuous), BRAF, KRAS, PIK3CA, p53, β-catenin, and JCVT. For BMI, tumor grade, and JCVT, we assigned separate indicator (“missing”) variables to those cases with missing data. For cases missing LINE-1 data, we assigned the mean LINE-1 methylation level. For cases with missing information in any of the other variables (tumor location, tumor stage, CIMP, BRAF, KRAS, PIK3CA, and p53), we included those cases in a majority category to not use more than 1 df for each variable in a multivariate model. We confirmed that excluding cases with missing information in any of the variables did not substantially alter results (data not shown).
For survival analysis, the Kaplan-Meier method and log-rank test were used. For analyses of colorectal cancer–specific mortality, deaths not resulting from colorectal cancer were censored. To assess independent effect of 18q LOH on mortality, we constructed a multivariate, stage-matched (stratified) Cox proportional hazard model to compute a hazard ratio (HR). Tumor stage (I, IIA, IIB, IIIA, IIIB, IIIC, IV, unknown) was used as a matching (stratifying) variable, and other adjustment was performed as in the multivariate logistic regression. The proportionality of hazards assumption was satisfied by evaluating time-dependent variables, which were the cross-product of the 18q LOH variable and survival time (P = .31 for colon cancer–specific mortality; P = .077 for overall mortality). An interaction was assessed by including the cross-product of the 18q LOH variable and another variable of interest in a multivariate Cox model, and the Wald test was performed.
RESULTS
18q LOH in Non–MSI-High Colorectal Cancer
Using 555 non–MSI-high colorectal cancers with informative 18q LOH data, we detected LOH in at least one 18q marker in 362 tumors (65%; Table 1). Tumors with 18q LOH were associated with obesity (BMI ≥ 30 kg/m2; P = .018), distal colon (P = .025), low tumor grade (P = .0060), low-level LINE-1 methylation (P = .040), wild-type KRAS (P = .015), and JCVT (P = .0004).
Table 1.
Clinical and Molecular Features of Non–MSI-High Colorectal Cancer According to 18q LOH Status
| Clinical or Molecular Feature | Total |
18q LOH Negative |
18q LOH Positive |
P | |||
|---|---|---|---|---|---|---|---|
| No. | % | No. | % | No. | % | ||
| Total No. of patients | 555 | 193 | 362 | ||||
| Sex | .36 | ||||||
| Male, HPFS | 244 | 44 | 90 | 47 | 154 | 43 | |
| Female, NHS | 311 | 56 | 103 | 53 | 208 | 57 | |
| Age, years | .27 | ||||||
| Mean | 66.8 | 67.3 | 66.5 | ||||
| SD | 8.3 | 8.4 | 8.2 | ||||
| Body mass index, kg/m2 | .018 | ||||||
| < 30 | 424 | 82 | 154 | 88 | 270 | 80 | |
| ≥ 30 | 90 | 18 | 21 | 12 | 69 | 20 | |
| Family history of colorectal cancer in first-degree relative(s) | .55 | ||||||
| No | 428 | 77 | 146 | 76 | 282 | 78 | |
| Yes | 127 | 23 | 47 | 24 | 80 | 22 | |
| Year of diagnosis | .42 | ||||||
| Before 1995 | 223 | 40 | 82 | 42 | 141 | 39 | |
| 1995 to 2002 | 332 | 60 | 111 | 58 | 221 | 61 | |
| Tumor location | .025 | ||||||
| Proximal, cecum to transverse | 217 | 40 | 89 | 48 | 128 | 36 | |
| Distal colon, splenic flexure to sigmoid | 178 | 33 | 50 | 27 | 128 | 36 | |
| Rectum | 144 | 27 | 48 | 26 | 96 | 27 | |
| AJCC tumor stage | .26 | ||||||
| I | 131 | 24 | 43 | 22 | 88 | 24 | |
| IIA | 122 | 22 | 42 | 22 | 80 | 22 | |
| IIB | 13 | 2.3 | 7 | 3.6 | 6 | 1.7 | |
| IIIA | 24 | 4.3 | 12 | 6.2 | 12 | 3.3 | |
| IIIB | 75 | 14 | 19 | 9.8 | 56 | 15 | |
| IIIC | 64 | 12 | 23 | 12 | 41 | 11 | |
| IV | 84 | 15 | 29 | 15 | 55 | 15 | |
| Unknown | 42 | 7.6 | 18 | 9.3 | 24 | 6.6 | |
| Tumor grade | .0060 | ||||||
| Low | 506 | 93 | 167 | 89 | 339 | 95 | |
| High | 36 | 6.6 | 20 | 11 | 16 | 4.5 | |
| MSI status | .80 | ||||||
| MSS | 495 | 89 | 173 | 90 | 322 | 89 | |
| MSI low | 60 | 11 | 20 | 10 | 40 | 11 | |
| CIMP, No. of methylated CIMP markers | .095 | ||||||
| CIMP 0 | 272 | 50 | 82 | 44 | 190 | 54 | |
| CIMP low (1–5) | 233 | 43 | 90 | 48 | 143 | 41 | |
| CIMP high (6–8) | 34 | 6.3 | 14 | 7.5 | 20 | 5.7 | |
| LINE-1 methylation, % | .040 | ||||||
| Mean | 60.7 | 61.9 | 60.1 | ||||
| SD | 9.3 | 9.5 | 9.2 | ||||
| BRAFmutation | .45 | ||||||
| Negative | 497 | 91 | 170 | 90 | 327 | 92 | |
| Positive | 48 | 8.8 | 19 | 10 | 29 | 8.2 | |
| KRASmutation | .015 | ||||||
| Negative | 330 | 60 | 101 | 53 | 229 | 63 | |
| Positive | 224 | 40 | 91 | 47 | 133 | 37 | |
| PIK3CA mutation | .11 | ||||||
| Negative | 438 | 85 | 145 | 82 | 293 | 87 | |
| Positive | 75 | 15 | 32 | 18 | 43 | 13 | |
| p53 expression | .071 | ||||||
| Negative | 292 | 53 | 110 | 59 | 182 | 51 | |
| Positive | 254 | 47 | 77 | 41 | 177 | 49 | |
| β-catenin activation* | .30 | ||||||
| Inactive (score 0-1) | 207 | 43 | 75 | 46 | 132 | 41 | |
| Active (score 2-5) | 277 | 57 | 88 | 54 | 189 | 59 | |
| JC virus T antigen | .0004 | ||||||
| Negative | 285 | 60 | 120 | 71 | 165 | 54 | |
| Positive | 190 | 40 | 50 | 29 | 140 | 46 | |
NOTE. Table indicates the proportion of tumors with a specific clinical, pathologic, or molecular feature in all patients or patients with 18q LOH-negative tumors or 18q LOH-positive tumors.
Abbreviations: MSI, microsatellite instability; LOH, loss of heterozygosity; HPFS, Health Professionals Follow-Up Study; NHS, Nurses' Health Study; SD, standard deviation; AJCC, American Joint Committee on Cancer; MSS, microsatellite stable; CIMP, CpG island methylator phenotype; JC, John Cunningham.
β-catenin was assessed by immunohistochemistry and activation score was calculated as previously described.33
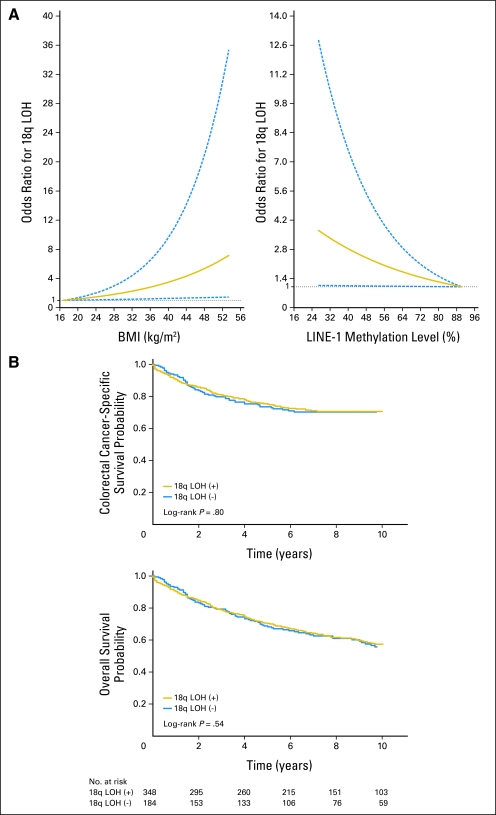
We examined the possibility of nonlinear relations of BMI and LINE-1 methylation with 18q LOH, nonparametrically with restricted cubic spines34 (Fig 1A). This flexible method allowed us to examine the relation to 18q LOH without any categorization of BMI or LINE-1 methylation level. It was evident that odds for 18q LOH increased as BMI increased or as LINE-1 methylation level decreased.
Fig 1.
18q loss of heterozygosity (LOH) in colorectal cancer. (A) Smoothing spline plot of odds ratio (OR) for 18q LOH in non–microsatellite instability (MSI) –high colorectal cancer according to body mass index (BMI) (left panel) and LINE-1 methylation level (right panel). Patients with low BMI or with high-level LINE-1 methylation were used as a reference group; 95% CIs are indicated by blue dashed lines. (B) Kaplan-Meier curves for colorectal cancer–specific survival (top panel) and overall survival (bottom panel) according to 18q LOH status in non–MSI-high colorectal cancer.
Multivariate Analysis for Relations With 18q LOH
In multivariate logistic regression analysis (Table 2), 18q LOH was independently associated with JCVT (OR = 1.93; 95% CI, 1.19 to 3.13; P = .0077), BMI ≥ 30 kg/m2 (OR = 2.01; 95% CI, 1.15 to 3.52; P = .014), and LINE-1 hypomethylation (for a 30% decrease; OR = 1.92; 95% CI, 1.02 to 3.62; P = .045), and inversely with high tumor grade (OR = 0.40; 95% CI, 0.19 to 0.86; P = .018) and KRAS mutation (OR = 0.66; 95% CI, 0.44 to 0.98; P = .040). Nonetheless, considering multiple hypothesis testing, any of these associations could be a chance event.
Table 2.
Multivariate Analysis of the Relations With 18q LOH in 555 Non–MSI-High Colorectal Cancers
| Variable Independently Associated With 18q LOH | Multivariate OR | 95% CI | P |
|---|---|---|---|
| JC virus T antigen | 1.93 | 1.19 to 3.13 | .0077 |
| BMI ≥ 30 kg/m2v < 30 kg/m2 | 2.01 | 1.15 to 3.52 | .014 |
| High tumor grade v low grade | 0.40 | 0.19 to 0.86 | .018 |
| KRAS mutation | 0.66 | 0.44 to 0.98 | .040 |
| LINE-1 hypomethylation, 30% decrease as a unit | 1.92 | 1.02 to 3.62 | .045 |
NOTE. The multivariate logistic regression model included age, sex, family history of colorectal cancer, tumor location, stage, MSI (low v microsatellite stability), CIMP, BRAF, PIK3CA, p53, β-catenin, and the variables listed in the table.
Abbreviations: LOH, loss of heterozygosity; MSI, microsatellite instability; OR, odds ratio; JC, John Cunningham; BMI, body mass index.
18q LOH and Patient Survival in Non–MSI-High Colorectal Cancer
Using our cohort database, we previously demonstrated that molecular features in colon cancer, such as BRAF mutation, fatty acid synthase expression, PIK3CA mutation, and LINE-1 hypomethylation, were significantly associated with prognosis.35-38 We assessed the influence of 18q LOH status on patient survival. During follow-up of 532 patients who were eligible for survival analysis, there were a total of 239 deaths, including 155 colorectal cancer–specific deaths. Five-year colorectal cancer–specific survival was 75% among patients with 18q LOH-positive tumors and 74% among those with 18q LOH-negative tumors (log-rank P = .80; Fig 1B). Five-year overall survival was 70% among patients with 18q LOH-positive tumors and 68% among those with 18q LOH-negative tumors (log-rank P = .54).
In univariate Cox regression analysis, compared with patients with 18q LOH-negative tumors, patients with 18q LOH-positive tumors experienced similar colorectal cancer–specific mortality (HR = 0.96; 95% CI, 0.69 to 1.33; Table 3). We examined the prognostic effect of 18q LOH after adjusting for potential predictors of patient survival, including age, sex, BMI, family history of colorectal cancer, year of diagnosis, tumor location, stage, grade, CIMP, LINE-1 hypomethylation, p53, β-catenin, KRAS, BRAF, and PIK3CA. Compared with patients with 18q LOH-negative tumors, patients with 18q LOH-positive tumors experienced similar colorectal cancer–specific mortality (multivariate HR = 1.18; 95% CI, 0.81 to 1.71). When overall mortality was used as an end point, results were similar (Table 3).
Table 3.
18q LOH Status and Patient Mortality in 532 Stage I to IV Non–MSI-High Colorectal Cancer
| 18q LOH Status | Total No. | Colorectal Cancer-Specific Mortality |
Overall Mortality |
||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Events | Person-Years | Univariate HR | 95% CI | Stage-Matched HR | 95% CI | Multivariate HR | 95% CI | Events | Person-Years | Univariate HR | 95% CI | Stage-Matched HR | 95% CI | Multivariate HR | 95% CI | ||
| 18q LOH negative | 184 | 55 | 1,404 | 1 | Referent | 1 | Referent | 1 | Referent | 87 | 1,404 | 1 | Referent | 1 | Referent | 1 | Referent |
| 18q LOH positive | 348 | 100 | 2,659 | 0.96 | 0.69 to 1.33 | 1.00 | 0.71 to 1.41 | 1.18 | 0.81 to 1.71 | 152 | 2,659 | 0.92 | 0.71 to 1.20 | 0.94 | 0.72 to 1.24 | 1.03 | 0.77 to 1.38 |
NOTE. The multivariate, stage-matched (stratified) Cox regression model included age, year of diagnosis, sex, family history of colorectal cancer, tumor location, tumor grade, KRAS, BRAF, PIK3CA,p53, β-catenin, JC virus T antigen, LINE-1 methylation, MSI (low v microsatellite stability), and CpG island methylator phenotype.
Abbreviations: LOH, loss of heterozygosity; MSI, microsatellite instability; HR, hazard ratio.
In addition, we examined whether LOH status at each of the four microsatellite loci could predict patient survival. LOH at any of the 18q loci was not significantly associated with survival (data not shown).
Prognostic Effect of 18q LOH in Strata of Tumor Stage
We examined the prognostic effect of 18q LOH in strata of tumor stage (Table 4). Patient mortality did not significantly differ according to 18q LOH status in any stage strata. The effect of 18q LOH on mortality did not significantly differ according to tumor stage (P interaction = .50 for colorectal cancer–specific mortality and P interaction = .76 for overall mortality).
Table 4.
18q LOH and Patient Mortality in Non–MSI-High Colorectal Cancer in Strata of Tumor Stage
| AJCC Tumor Stage | Total No. | HR for Colorectal Cancer–Specific Mortality in 18q LOH-Positive Patients* |
HR for Overall Mortality in 18q LOH-Positive Patients* | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Univariate HR | 95% CI | Multivariate HR | 95% CI | Univariate HR | 95% CI | Multivariate HR | 95% CI | ||
| I to II | 262 | 1.31 | 0.58 to 2.98 | 1.24 | 0.54 to 2.85 | 0.92 | 0.58 to 1.47 | 0.89 | 0.55 to 1.43 |
| III | 160 | 0.78 | 0.43 to 1.40 | 0.76 | 0.42 to 1.38 | 0.91 | 0.56 to 1.47 | 0.93 | 0.56 to 1.52 |
| IV | 82 | 0.93 | 0.58 to 1.48 | 1.35 | 0.80 to 2.28 | 0.89 | 0.55 to 1.42 | 1.34 | 0.80 to 2.25 |
| Pfor interaction (18q LOH and stage)† | .32 | .50 | .86 | .76 | |||||
NOTE. The multivariate Cox regression model included the 18q LOH variable stratified by tumor stage, age, year of diagnosis, sex, family history of colorectal cancer, tumor location, tumor grade, KRAS, BRAF, PIK3CA,p53, β-catenin, JC virus T antigen, LINE-1 methylation, MSI (low v microsatellite stability), and CpG island methylator phenotype.
Abbreviations: LOH, loss of heterozygosity; MSI, microsatellite instability; AJCC, American Joint Committee on Cancer; HR, hazard ratio.
Versus 18q LOH-negative patients as a referent.
For assessing a potential interaction, tumor stage was dichotomized as I to II v III to IV.
Prognostic Effect of 18q LOH in Strata of Tumor Location
We examined the prognostic effect of 18q LOH in strata of tumor location (Table 5). The effect of 18q LOH on mortality did not significantly differ according to tumor location (P interaction = .17 for colorectal cancer–specific mortality and P interaction = .91 for overall mortality).
Table 5.
18q LOH and Patient Mortality in Non–MSI-High Colorectal Cancer in Strata of Tumor Location
| Tumor Location | Total No. | HR for Colorectal Cancer–Specific Mortality in 18q LOH-Positive Patients* | HR for Overall Mortality in 18q LOH-Positive Patients* |
||||||
|---|---|---|---|---|---|---|---|---|---|
| Stage-Matched HR | 95% CI | Multivariate HR | 95% CI | Stage-Matched HR | 95% CI | Multivariate HR | 95% CI | ||
| Rectum | 138 | 1.34 | 0.66 to 2.75 | 1.46 | 0.70 to 3.07 | 1.13 | 0.64 to 1.98 | 1.10 | 0.62 to 1.96 |
| Distal colon, splenic flexure to sigmoid | 174 | 1.30 | 0.59 to 2.88 | 1.78 | 0.78 to 4.05 | 0.88 | 0.53 to 1.47 | 1.01 | 0.60 to 1.71 |
| Proximal colon, cecum to transverse colon | 217 | 0.87 | 0.54 to 1.39 | 0.95 | 0.57 to 1.58 | 0.92 | 0.62 to 1.38 | 1.01 | 0.66 to 1.56 |
| P for interaction (18q LOH and location)† | .26 | .17 | .81 | .91 | |||||
NOTE. The multivariate, stage-matched (stratified) Cox regression model included the 18q LOH variable stratified by location, age, year of diagnosis, sex, family history of colorectal cancer, tumor grade, KRAS, BRAF, PIK3CA,p53, β-catenin, JC virus T antigen, LINE-1 methylation, MSI (low v microsatellite stability), and CpG island methylator phenotype.
Abbreviations: LOH, loss of heterozygosity; MSI, microsatellite instability; HR, hazard ratio.
Versus 18q LOH-negative patients as a referent.
For assessing a potential interaction, tumor location was dichotomized as proximal v distal (including rectum).
Stratified Analysis of 18q LOH and Patient Mortality
We further examined whether the influence of 18q LOH on colorectal cancer–specific and overall mortality was modified by any of the other potential predictors of patient survival (including age, sex, family history of colorectal cancer, BMI, year of diagnosis, tumor grade, and status of CIMP, LINE-1 hypomethylation, KRAS, BRAF, PIK3CA, p53, and β-catenin). None of the strata showed a significant differential effect of 18q LOH on mortality (all P interaction > .10). Notably, the effect of 18q LOH did not significantly differ between the two independent cohort studies (Pinteraction = .89).
DISCUSSION
We performed this study to examine the prognostic significance of 18q LOH in non–MSI-high colorectal cancer. Because 18q LOH and MSI high are almost mutually exclusive,5 and MSI high is associated with better outcome,17 the association between 18q LOH and poor prognosis might be simply due to the enrichment of MSI-high tumors within 18q LOH-negative cases. Thus it is necessary to examine 18q LOH status in non–MSI-high colorectal cancers to assess the independent prognostic significance of 18q LOH. We found that 18q LOH was not associated with survival among 532 patients with non–MSI-high colorectal cancer. Moreover, the presence of 18q LOH did not significantly influence patient survival within any individual stratum of pathologic stage or any stratum of clinical and molecular predictors of patient outcome.
Examining prognostic factors is important in colon cancer research.39-43 The presence of 18q LOH was associated with recurrence or mortality in colorectal cancer in earlier studies.5,8,9,16,44,45 However, many of the previous studies on prognostic significance of 18q LOH8,9,12,16 did not consider a confounding effect of MSI. Moreover, a meta-analysis on 18q LOH and outcome in colorectal cancers demonstrated evidence for publication bias.18 Large studies (total N > 300) could be published even with null results,13-15 whereas smaller studies were selected for publication based on “significant” results. Smaller studies with nonsignificant results experienced a higher likelihood of being unpublished, leading to publication bias toward a deviation from the null hypothesis.
In the largest study to date, Roth et al15 failed to show independent prognostic significance of 18q LOH in more than 1,200 patients with stage II and III colon cancer. Our current study (N = 532 non–MSI-high tumors), the second largest to date, is in agreement with Roth et al,15 as well as the two other large studies conducted by Halling et al13 (N = 432 non–MSI-high tumors) and Barratt et al14 (N = 279 non–MSI-high tumors). Although, in univariate analyses,Barratt et al14 found an improved survival with 18q LOH and Roth et al15 reported an inferior survival in the subset of patients with stage II cancer, both studies found that the prognostic influence of 18q LOH was no longer apparent in multivariate analyses that adjusted for MSI and other relevant parameters. In notable exception, Watanabe et al5 obtained informative 18q LOH data in 279 colon cancers and showed that 18q LOH was independently associated with poor outcome. Moreover, Watanabe et al5 found a significant relation between 18q LOH and poor survival in stage III non–MSI-high tumors (N = 148), although no such relation was found in stage II tumors. We should be aware that all of these large studies5,13-15 (including our study) used different markers and criteria, which could confound the results and cause the somewhat discrepant findings. It would be necessary to develop a consensus panel to assess 18q LOH in colorectal cancer in future trials. Nonetheless, the frequencies of 18q LOH seem relatively comparable between these investigations. Moreover, we demonstrated the significant associations of 18q LOH with JCVT, KRAS, and LINE-1 hypomethylation, supporting the validity of our 18q LOH assessment. LINE-1 hypomethylation and JCVT have previously been related with chromosomal instability.29,31,46-49
We recognize potential limitations in our analysis. Data on cancer treatment in our cohorts were limited. Nonetheless, it is unlikely that use of chemotherapy considerably differed according to 18q LOH status, because such data were not typically available for treatment decision making. In addition, beyond cause of mortality, data on cancer recurrences were not available in these cohorts. Nonetheless, given that the median survival for metastatic colon cancer was approximately 10 to 12 months during much of the time period of this study,37 colorectal cancer–specific survival should be a reasonable surrogate for cancer-specific outcomes.
There are advantages in using the database of the two independent prospective cohort studies, the Nurses' Health Study and Health Professionals Follow-Up Study, to examine prognostic significance of 18q LOH and its relationship with tumoral and host factors. Anthropometric measurements, family history of cancer, other clinical information, pathologic and tumor staging data, and tumoral molecular features were prospectively collected and entered into the database, blinded to patient outcome. Cohort participants who developed colorectal cancer were treated at hospitals throughout the United States and thus were more representative of colorectal cancers in the general population than studies based on a single to few hospitals. Tumor specimen procurement rate has been approximately 60%, and there were no demographic differences between cases with tumor tissue analyzed and those without tumor tissue analyzed.20 In addition, our extensive database of clinical, pathologic, and molecular features enabled us to simultaneously adjust for other predictors of patient outcome.
Beyond the influence of 18q LOH on patient prognosis, there remain limited data on the ability of 18q LOH to predict the benefit from treatment. Among the aforementioned large, published studies that assessed the prognostic significance of 18q LOH,5,13-15 only the study by Barratt et al14 examined whether 18q LOH predicted benefit from fluorouracil-based adjuvant therapy. For patients with LOH at one 18q locus (D18S61), adjuvant fluorouracil did not improve survival, whereas among subjects with no LOH at D18S61, adjuvant fluorouracil did confer a significant survival benefit (Pinteraction = .02).14 An ongoing, large clinical trial (Eastern Cooperative Oncology Group Trial 5202) is prospectively examining whether MSI and 18q LOH can define the optimal use of postoperative adjuvant chemotherapy in patients with curatively resected stage II colon cancer. In that study, patients with either MSI-high tumors or non–MSI-high tumors with retention of 18q will not receive postoperative therapy. In contrast, patients with non–MSI-high tumors that demonstrate 18q LOH will receive postoperative chemotherapy (fluorouracil, leucovorin, and oxaliplatin) with or without bevacizumab.
In summary, our large cohort study did not find a significant prognostic impact for 18q LOH in patients with colorectal cancer or any subset of this study population. Future studies are needed to confirm these results.
Acknowledgment
The Acknowledgment is included in the full-text version of this article, available online at www.jco.org. It is not included in the PDF version (via Adobe® Reader®).
We thank the Nurses' Health Study and Health Professionals Follow-Up Study cohort participants who have generously agreed to provide us with biologic specimens and information through responses to questionnaires, hospitals, and pathology departments throughout the United States for providing us with tumor tissue materials.
Footnotes
Supported by the National Institutes of Health (Grants No. P01 CA87969 to S. Hankinson, P01 CA55075 to W. Willett, P50 CA127003 to C.S.F., K07 CA122826 to S.O., and K07 CA97992 to J.A.M.), the Bennett Family Fund, and the Entertainment Industry Foundation National Colorectal Cancer Research Alliance. K.N. was supported by a fellowship grant from the Japan Society for Promotion of Science. Funding agencies did not have any role in the design of the study; the collection, analysis, or interpretation of the data; the decision to submit the manuscript for publication; or the writing of the manuscript.
The content of this article is solely the responsibility of the authors and does not necessarily represent the official views of the National Cancer Institute or National Institutes of Health.
Authors' disclosures of potential conflicts of interest and author contributions are found at the end of this article.
AUTHORS' DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
The author(s) indicated no potential conflicts of interest.
AUTHOR CONTRIBUTIONS
Conception and design: Shuji Ogino, Charles S. Fuchs
Financial support: Shuji Ogino, Charles S. Fuchs
Administrative support: Gregory J. Kirkner, Charles S. Fuchs
Provision of study materials or patients: Shuji Ogino, Katsuhiko Nosho, Natsumi Irahara, Kaori Shima, Yoshifumi Baba, Gregory J. Kirkner, Jeffrey A. Meyerhardt, Charles S. Fuchs
Collection and assembly of data: Shuji Ogino, Katsuhiko Nosho, Natsumi Irahara, Kaori Shima, Yoshifumi Baba, Gregory J. Kirkner, Jeffrey A. Meyerhardt, Charles S. Fuchs
Data analysis and interpretation: Shuji Ogino, Katsuhiko Nosho, Natsumi Irahara, Kaori Shima, Yoshifumi Baba, Gregory J. Kirkner, Jeffrey A. Meyerhardt, Charles S. Fuchs
Manuscript writing: Shuji Ogino, Katsuhiko Nosho, Jeffrey A. Meyerhardt, Charles S. Fuchs
Final approval of manuscript: Shuji Ogino, Katsuhiko Nosho, Natsumi Irahara, Kaori Shima, Yoshifumi Baba, Gregory J. Kirkner, Jeffrey A. Meyerhardt, Charles S. Fuchs
REFERENCES
- 1.Grady WM, Carethers JM. Genomic and epigenetic instability in colorectal cancer pathogenesis. Gastroenterology. 2008;135:1079–1099. doi: 10.1053/j.gastro.2008.07.076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fearon ER, Vogelstein B. A genetic model for colorectal tumorigenesis. Cell. 1990;61:759–767. doi: 10.1016/0092-8674(90)90186-i. [DOI] [PubMed] [Google Scholar]
- 3.Thiagalingam S, Lengauer C, Leach FS, et al. Evaluation of candidate tumour suppressor genes on chromosome 18 in colorectal cancers. Nat Genet. 1996;13:343–346. doi: 10.1038/ng0796-343. [DOI] [PubMed] [Google Scholar]
- 4.Park do Y, Sakamoto H, Kirley SD, et al. The Cables gene on chromosome 18q is silenced by promoter hypermethylation and allelic loss in human colorectal cancer. Am J Pathol. 2007;171:1509–1519. doi: 10.2353/ajpath.2007.070331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Watanabe T, Wu TT, Catalano PJ, et al. Molecular predictors of survival after adjuvant chemotherapy for colon cancer. N Engl J Med. 2001;344:1196–1206. doi: 10.1056/NEJM200104193441603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jass JR. Classification of colorectal cancer based on correlation of clinical, morphological and molecular features. Histopathology. 2007;50:113–130. doi: 10.1111/j.1365-2559.2006.02549.x. [DOI] [PubMed] [Google Scholar]
- 7.Ogino S, Goel A. Molecular classification and correlates in colorectal cancer. J Mol Diagn. 2008;10:13–27. doi: 10.2353/jmoldx.2008.070082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jen J, Kim H, Piantadosi S, et al. Allelic loss of chromosome 18q and prognosis in colorectal cancer. N Engl J Med. 1994;331:213–221. doi: 10.1056/NEJM199407283310401. [DOI] [PubMed] [Google Scholar]
- 9.Ogunbiyi OA, Goodfellow PJ, Herfarth K, et al. Confirmation that chromosome 18q allelic loss in colon cancer is a prognostic indicator. J Clin Oncol. 1998;16:427–433. doi: 10.1200/JCO.1998.16.2.427. [DOI] [PubMed] [Google Scholar]
- 10.Zhou W, Goodman SN, Galizia G, et al. Counting alleles to predict recurrence of early-stage colorectal cancers. Lancet. 2002;359:219–225. doi: 10.1016/S0140-6736(02)07448-2. [DOI] [PubMed] [Google Scholar]
- 11.Diep CB, Thorstensen L, Meling GI, et al. Genetic tumor markers with prognostic impact in Dukes' stages B and C colorectal cancer patients. J Clin Oncol. 2003;21:820–829. doi: 10.1200/JCO.2003.05.190. [DOI] [PubMed] [Google Scholar]
- 12.Carethers JM, Hawn MT, Greenson JK, et al. Prognostic significance of allelic lost at chromosome 18q21 for stage II colorectal cancer. Gastroenterology. 1998;114:1188–1195. doi: 10.1016/s0016-5085(98)70424-x. [DOI] [PubMed] [Google Scholar]
- 13.Halling KC, French AJ, McDonnell SK, et al. Microsatellite instability and 8p allelic imbalance in stage B2 and C colorectal cancers. J Natl Cancer Inst. 1999;91:1295–1303. doi: 10.1093/jnci/91.15.1295. [DOI] [PubMed] [Google Scholar]
- 14.Barratt PL, Seymour MT, Stenning SP, et al. DNA markers predicting benefit from adjuvant fluorouracil in patients with colon cancer: A molecular study. Lancet. 2002;360:1381–1391. doi: 10.1016/s0140-6736(02)11402-4. [DOI] [PubMed] [Google Scholar]
- 15.Roth A, Tejpar S, Yan P, et al. State-specific prognostic value of molecular markers in colon cancer: Results of the translational study on the PETACC 3 - EORTC 40993-SAKK 60-00 trial. J Clin Oncol. 2009;27(suppl; abstr 4002):15s. doi: 10.1200/JCO.2009.23.3452. [DOI] [PubMed] [Google Scholar]
- 16.Jernvall P, Makinen MJ, Karttunen TJ, et al. Loss of heterozygosity at 18q21 is indicative of recurrence and therefore poor prognosis in a subset of colorectal cancers. Br J Cancer. 1999;79:903–908. doi: 10.1038/sj.bjc.6690144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Popat S, Hubner R, Houlston RS. Systematic review of microsatellite instability and colorectal cancer prognosis. J Clin Oncol. 2005;23:609–618. doi: 10.1200/JCO.2005.01.086. [DOI] [PubMed] [Google Scholar]
- 18.Popat S, Houlston RS. A systematic review and meta-analysis of the relationship between chromosome 18q genotype, DCC status and colorectal cancer prognosis. Eur J Cancer. 2005;41:2060–2070. doi: 10.1016/j.ejca.2005.04.039. [DOI] [PubMed] [Google Scholar]
- 19.Colditz GA, Hankinson SE. The Nurses' Health Study: Lifestyle and health among women. Nat Rev Cancer. 2005;5:388–396. doi: 10.1038/nrc1608. [DOI] [PubMed] [Google Scholar]
- 20.Chan AT, Ogino S, Fuchs CS. Aspirin and the risk of colorectal cancer in relation to the expression of COX-2. N Engl J Med. 2007;356:2131–2142. doi: 10.1056/NEJMoa067208. [DOI] [PubMed] [Google Scholar]
- 21.Ogino S, Kawasaki T, Brahmandam M, et al. Sensitive sequencing method for KRAS mutation detection by Pyrosequencing. J Mol Diagn. 2005;7:413–421. doi: 10.1016/S1525-1578(10)60571-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ogino S, Kawasaki T, Kirkner GJ, et al. CpG island methylator phenotype-low (CIMP-low) in colorectal cancer: Possible associations with male sex and KRAS mutations. J Mol Diagn. 2006;8:582–588. doi: 10.2353/jmoldx.2006.060082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nosho K, Kawasaki T, Ohnishi M, et al. PIK3CA mutation in colorectal cancer: Relationship with genetic and epigenetic alterations. Neoplasia. 2008;10:534–541. doi: 10.1593/neo.08336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ogino S, Brahmandam M, Cantor M, et al. Distinct molecular features of colorectal carcinoma with signet ring cell component and colorectal carcinoma with mucinous component. Mod Pathol. 2006;19:59–68. doi: 10.1038/modpathol.3800482. [DOI] [PubMed] [Google Scholar]
- 25.Ogino S, kawasaki T, Brahmandam M, et al. Precision and performance characteristics of bisulfite conversion and real-time PCR (MethyLight) for quantitative DNA methylation analysis. J Mol Diagn. 2006;8:209–217. doi: 10.2353/jmoldx.2006.050135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ogino S, Cantor M, Kawasaki T, et al. CpG island methylator phenotype (CIMP) of colorectal cancer is best characterised by quantitative DNA methylation analysis and prospective cohort studies. Gut. 2006;55:1000–1006. doi: 10.1136/gut.2005.082933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ogino S, Kawasaki T, Kirkner GJ, et al. Evaluation of markers for CpG island methylator phenotype (CIMP) in colorectal cancer by a large population-based sample. J Mol Diagn. 2007;9:305–314. doi: 10.2353/jmoldx.2007.060170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Weisenberger DJ, Siegmund KD, Campan M, et al. CpG island methylator phenotype underlies sporadic microsatellite instability and is tightly associated with BRAF mutation in colorectal cancer. Nat Genet. 2006;38:787–793. doi: 10.1038/ng1834. [DOI] [PubMed] [Google Scholar]
- 29.Ogino S, Kawasaki T, Nosho K, et al. LINE-1 hypomethylation is inversely associated with microsatellite instability and CpG methylator phenotype in colorectal cancer. Int J Cancer. 2008;122:2767–2773. doi: 10.1002/ijc.23470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ogino S, Brahmandam M, kawasaki T, et al. Combined analysis of COX-2 and p53 expressions reveals synergistic inverse correlations with microsatellite instability and CpG island methylator phenotype in colorectal cancer. Neoplasia. 2006;8:458–464. doi: 10.1593/neo.06247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nosho K, Shima K, Kure S, et al. JC virus T-antigen in colorectal cancer is associated with p53 expression and chromosomal instability, independent of CpG island methylator phenotype. Neoplasia. 2009;11:87–95. doi: 10.1593/neo.81188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ogino S, Kawasaki T, Kirkner GJ, et al. Loss of nuclear p27 (CDKN1B/KIP1) in colorectal cancer is correlated with microsatellite instability and CIMP. Mod Pathol. 2007;20:15–22. doi: 10.1038/modpathol.3800709. [DOI] [PubMed] [Google Scholar]
- 33.Kawasaki T, Nosho K, Ohnishi M, et al. Correlation of beta-catenin localization with cyclooxygenase-2 expression and CpG island methylator phenotype (CIMP) in colorectal cancer. Neoplasia. 2007;9:569–577. doi: 10.1593/neo.07334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Durrleman S, Simon R. Flexible regression models with cubic splines. Stat Med. 1989;8:551–561. doi: 10.1002/sim.4780080504. [DOI] [PubMed] [Google Scholar]
- 35.Ogino S, Nosho K, Meyerhardt JA, et al. Cohort study of fatty acid synthase expression and patient survival in colon cancer. J Clin Oncol. 2008;26:5713–5720. doi: 10.1200/JCO.2008.18.2675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ogino S, Nosho K, Kirkner GJ, et al. CpG island methylator phenotype, microsatellite instability, BRAF mutation and clinical outcome in colon cancer. Gut. 2009;58:90–96. doi: 10.1136/gut.2008.155473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ogino S, Nosho K, Kirkner GJ, et al. A cohort study of tumoral LINE-1 hypomethylation and prognosis in colon cancer. J Natl Cancer Inst. 2008;100:1734–1738. doi: 10.1093/jnci/djn359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ogino S, Nosho K, Kirkner GJ, et al. PIK3CA mutation is associated with poor prognosis among patients with curatively resected colon cancer. J Clin Oncol. 2009;27:1477–1484. doi: 10.1200/JCO.2008.18.6544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Braun MS, Richman SD, Quirke P, et al. Predictive biomarkers of chemotherapy efficacy in colorectal cancer: Results from the UK MRC FOCUS trial. J Clin Oncol. 2008;26:2690–2698. doi: 10.1200/JCO.2007.15.5580. [DOI] [PubMed] [Google Scholar]
- 40.Cohen SJ, Punt CJ, Iannotti N, et al. Relationship of circulating tumor cells to tumor response, progression-free survival, and overall survival in patients with metastatic colorectal cancer. J Clin Oncol. 2008;26:3213–3221. doi: 10.1200/JCO.2007.15.8923. [DOI] [PubMed] [Google Scholar]
- 41.Di Nicolantonio F, Martini M, Molinari F, et al. Wild-type BRAF is required for response to panitumumab or cetuximab in metastatic colorectal cancer. J Clin Oncol. 2008;26:5705–5712. doi: 10.1200/JCO.2008.18.0786. [DOI] [PubMed] [Google Scholar]
- 42.Mitry E, Fields AL, Bleiberg H, et al. Adjuvant chemotherapy after potentially curative resection of metastases from colorectal cancer: A pooled analysis of two randomized trials. J Clin Oncol. 2008;26:4906–4911. doi: 10.1200/JCO.2008.17.3781. [DOI] [PubMed] [Google Scholar]
- 43.Weiser MR, Landmann RG, Kattan MW, et al. Individualized prediction of colon cancer recurrence using a nomogram. J Clin Oncol. 2008;26:380–385. doi: 10.1200/JCO.2007.14.1291. [DOI] [PubMed] [Google Scholar]
- 44.Martínez-López E, Abad A, Font A, et al. Allelic loss on chromosome 18q as a prognostic marker in stage II colorectal cancer. Gastroenterology. 1998;114:1180–1187. doi: 10.1016/s0016-5085(98)70423-8. [DOI] [PubMed] [Google Scholar]
- 45.Sarli L, Bottarelli L, Bader G, et al. Association between recurrence of sporadic colorectal cancer, high level of microsatellite instability, and loss of heterozygosity at chromosome 18q. Dis Colon Rectum. 2004;47:1467–1482. doi: 10.1007/s10350-004-0628-6. [DOI] [PubMed] [Google Scholar]
- 46.Matsuzaki K, Deng G, Tanaka H, et al. The relationship between global methylation level, loss of heterozygosity, and microsatellite instability in sporadic colorectal cancer. Clin Cancer Res. 2005;11:8564–8569. doi: 10.1158/1078-0432.CCR-05-0859. [DOI] [PubMed] [Google Scholar]
- 47.Suzuki K, Suzuki I, Leodolter A, et al. Global DNA demethylation in gastrointestinal cancer is age dependent and precedes genomic damage. Cancer Cell. 2006;9:199–207. doi: 10.1016/j.ccr.2006.02.016. [DOI] [PubMed] [Google Scholar]
- 48.Rodriguez J, Frigola J, Vendrell E, et al. Chromosomal instability correlates with genome-wide DNA demethylation in human primary colorectal cancers. Cancer Res. 2006;66:8462–9468. doi: 10.1158/0008-5472.CAN-06-0293. [DOI] [PubMed] [Google Scholar]
- 49.Ricciardiello L, Baglioni M, Giovannini C, et al. Induction of chromosomal instability in colonic cells by the human polyomavirus JC virus. Cancer Res. 2003;63:7256–7262. [PubMed] [Google Scholar]