Abstract
The kinetochore is a complex protein–DNA assembly that provides the mechanical linkage between microtubules and the centromere DNA of each chromosome. Centromere DNA in all eukaryotes is wrapped around a unique nucleosome that contains the histone H3 variant CENP-A (Cse4p in Saccharomyces cerevisiae). Here, we report that the inner kinetochore complex (CBF3) is required for pericentric DNA looping at the Cse4p-containing nucleosome. DNA within the pericentric loop occupies a spatially confined area that is radially displaced from the interpolar central spindle. Microtubule-binding kinetochore complexes are not involved in pericentric DNA looping but are required for the geometric organization of DNA loops around the spindle microtubules in metaphase. Thus, the mitotic segregation apparatus is a composite structure composed of kinetochore and interpolar microtubules, the kinetochore, and organized pericentric DNA loops. The linkage of microtubule-binding to centromere DNA-looping complexes positions the pericentric chromatin loops and stabilizes the dynamic properties of individual kinetochore complexes in mitosis.
INTRODUCTION
The kinetochore is a protein–DNA structure composed of >14 multiprotein complexes and CEN DNA (Bouck et al., 2008; Cheeseman and Desai, 2008). Although centromere DNA ranges in size from 0.125 kb in budding yeast, 6–9 kb in Candida albicans, 40–100 kb in Schizosaccharomyces pombe, 400 kb in Drosophila melanogaster, to 2000–4000 kb in humans (Kitagawa and Hieter, 2001; Joglekar et al., 2008), the conserved feature among all centromeres is the incorporation of the histone H3 variant CENP-A into centromeric nucleosomes. The single Cse4p nucleosome (Furuyama and Biggins, 2007) lies at the apex of the intramolecularly paired centromeric loop in budding yeast (Yeh et al., 2008). This loop reflects a cruciform configuration of pericentric chromatin that positions Cse4p on the outer surface of the chromosome. In Drosophila, C. elegans and mammalian centromeres CENP-A nucleosomes are interspersed with canonical H3-containing nucleosomes, leading to a three-dimensional structure that results in positioning CENP-A on the outside of the chromosome (Sullivan and Karpen, 2004).
The geometry provided by dispersal of CENP-A creates a platform for kinetochore protein recruitment (Moore and Roth, 2001; Sullivan and Karpen, 2004) and capture of dynamic microtubules. In budding yeast, a single microtubule is captured by the kinetochore. Sister kinetochores from the 16 chromosomes are clustered into a diffraction limited spot within the spindle after microtubule attachment and biorientation. Growing evidence suggests that the geometrical organization of the kinetochore in mitosis and meiosis is dictated by cohesion within the pericentric region (Indjeian and Murray, 2007; Sakuno et al., 2009). An alternative hypothesis is that inner centromere DNA binding complexes specify the critical geometry. The budding yeast centromere DNA binding proteins include Cse4p/CENP-A, the CBF3 complex (composed of Ndc10p, Ctf13p, Ctf14p, and Cep3p), and Mif2p/CENP-C and define the inner kinetochore. CBF3 has been shown to induce a 55° bend in centromere DNA in vitro (Pietrasanta et al., 1999). In mammalian cells, the inner centromere protein CENP-B induces a bend in alphoid satellite DNA upon binding to the CENP-B box (17-bp repeat in alphoid DNA) (Tanaka et al., 2001) and facilitates CENP-A assembly on centromeres de novo (Okada et al., 2007). Although these inner centromere DNA binding proteins are not conserved at the sequence level, their propensity to bend DNA may be indicative of a conserved function in organizing centromere DNA in a path conducive for looping.
The inner kinetochore DNA-binding complexes interact with several linker complexes, including COMA (Ctf19p, Okp1p, Mcm21p, the essential protein Ame1p), COMA-associated proteins (Mcm19p/Iml3p, Nkp1p/Nkp2p, and Chl4p), and MIND (Mtw1p, Nnf1p, Nsl1p, Dsn1p), and Spc105p. At the microtubule plus-end, the outer kinetochore complexes include NDC80 and DAM-DASH. The NDC80 complex is a microtubule-associated tetramer, containing Spc24p, Spc25p, Nuf2p/hNuf2p, and Ndc80p/Hec1p (Westermann et al., 2007; Joglekar et al., 2008). DAM-DASH is a decameric complex of essential proteins, including Ask1p, that has the ability to form rings around microtubule plus-ends in vitro (Miranda et al., 2005; Westermann et al., 2005). In vitro chromatin immunoprecipitation (ChIp) experiments and fluorescent localization dependencies have provided much insight into the hierarchical organization and epistatic relationship among these kinetochore complexes (He et al., 2001; De Wulf et al., 2003; Westermann et al., 2003, 2007). However, epistasis analysis of gene order or localization does not result in simple interpretations of complex assemblies, especially in cases where the structure may not be assembled in a linear, or single pathway (Huang and Sternberg, 2006).
We report here that the inner kinetochore complexes promote centromere DNA bending, whereas the outer microtubule binding complexes contribute to the spatial positioning of the pericentric chromatin loops. The kinetochore is assembled by linking preformed protein subcomplexes and the 16 kinetochores in haploid yeast cells cluster at microtubule plus-ends in each half spindle. On assembly into the kinetochore, the stability of individual complexes is significantly enhanced. These studies provide new insight into the mechanical structure and multiple pathways of assembly of a eukaryotic kinetochore in vivo.
MATERIALS AND METHODS
All strains (Supplemental Table 2) in this study were constructed in the YEF473A background (Bi and Pringle, 1996) unless otherwise noted, by using methods described previously (Mythreye and Bloom, 2003). All strains were maintained at 25°C, and strains containing temperature-sensitive alleles were shifted to 37°C for 3 h before imaging.
For imaging, cells were grown to logarithmic growth phase in complete medium containing glucose (YPD). Cells were harvested by centrifugation, washed with sterile water, and resuspended in 20–50 μl of sterile water. Cells were pipetted onto slabs of 25% gelatin containing minimal medium and 2% glucose as described by Yeh et al. (1995). A TE2000 microscope (Nikon, East Rutherford, NJ) with a 1.4 numerical aperture, ×100 differential interference contrast oil-immersion lens (Salmon et al., 2007) was used for imaging. Fluorescent images were acquired by taking five steps along the z-axis at 750-nm intervals. Exposure times ranged from 300 to 800 ms.
Fluorescence recovery after photobleaching (FRAP) experiments were performed by photobleaching with a 50- to 200-ms laser pulse. Five plane Z series stacks were acquired before photobleaching, immediately after photobleaching, and at 20-s or 1-min intervals thereafter. To account for photobleaching due to image acquisition, multiple images were acquired from unbleached cells as well.
For fluorescence measurements, computer generated pixel regions were drawn over kinetochore spots and analyzed as described by Hoffman et al. (2001).
To examine the conformation of centromeric chromatin in nuf2-60, ame1-4, and ndc10-1, cells were shifted to restrictive temperature (37°C) for 3 h. Chromosome conformation capture (3C) was performed as first detailed by Dekker et al. (2002). Chromatin was fixed by treating cells with formaldehyde, and all treatments thereafter were performed as described in Yeh et al. (2008).
RESULTS
Pericentric Chromatin Looping Requires the Inner Kinetochore Complex CBF3 and Not Microtubule (MT) Binding
To determine the kinetochore complexes required for pericentric chromatin looping, we used the chromosome conformation capture (3C) assay to quantitate intramolecular DNA looping. 3C experiments were performed as outlined previously (Dekker et al., 2002; Yeh et al., 2008). Chromatin structure was fixed in vivo by formaldehyde cross-linking; digested with the restriction enzyme XbaI; ligated under dilute conditions; and after reversal of the cross-links, was used as template for PCR reactions. Using DNA primers from pericentric versus arm regions (Supplemental Figure 1), the relative likelihood that chromatin will adopt a specific conformation can be determined from the amount of polymerase chain reaction (PCR) product in the linear range of the reaction (Supplemental Figure 1). In this way, we define an index of looping as the ratio of PCR products obtained from primer pairs in pericentric chromatin versus chromosome arm primers. A looping index of ∼1.0 is indicative of the random association of chromatin due to thermal motion (uncross-linked looping index; dashed line in Figure 1). In uncross-linked samples, the pericentric chromatin has a looping index of ∼1.1, indicating that thermal motion of pericentric chromatin is no more or less likely than a chromosome arm region to exhibit intramolecular looping (Yeh et al., 2008). In cross-linked samples, the pericentric chromatin from chromosomes III and XI exhibit about a 2.5-fold increase in the looping index (chrXI 2.5; Figure 1) [chrIII dashed line]; Yeh et al., 2008). The looping index reflects an increased propensity for chromatin 6- to 8-kb flanking either side of the centromere to adopt a loop conformation and not an absolute number of molecules looped. To estimate the fraction of pericentric chromatin looped in vivo, we compared the yield of PCR products from experimental samples to products from control samples with a synthetic DNA loop of known concentration (Supplemental Table 1). A minimum of 10% of the pericentric chromatin is looped in vivo.
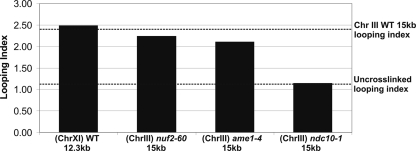
Figure 1.
Kinetochore protein requirements for centromere-loop formation. The looping index accounts for differential efficiency of PCR reactions with primers for pericentric chromatin versus chromosome arms at an equivalent ratio of input DNA. The pericentric region of chromosome III has a looping index of 2.4 (dotted line) (Yeh et al., 2008). Experimental samples (wild-type [WT], ndc10-1, ame1-4, and nuf2-60) were prepared as described previously (Yeh et al., 2008). Temperature-sensitive mutants were shifted to restrictive temperature (37°C) for 3 h before cross-linking. Random chromosomal association by using uncross-linked DNA has a looping index of 1.1 (dotted line).
Pericentric loop formation has been shown to depend upon Ndc10p, a constituent of the CBF3 complex (looping index 1.1, chrIII, ndc10-1; Figure 1) (Yeh et al., 2008). To examine whether C loop formation is dependent upon other kinetochore complexes, the looping index for 15-kb segments of pericentric DNA on chromosome III was determined in temperature-sensitive mutants of COMA (ame1-4) and the NDC80 complex (nuf2-60) grown at their restrictive temperature (37°C) for 3 h. Ame1p and Nuf2p are essential proteins of the COMA and NDC80 kinetochore complexes, respectively (DeLuca et al., 2003; Pot et al., 2005). The increase in looping index seen in wild-type (2.5; Figure 1) was unchanged in ame1-4 (2.1) and nuf2-60 (2.3). Thus, intramolecular looping of pericentric DNA is insensitive to loss of kinetochore linker complexes or MT attachment.
Density Maps Reveal the Geometry of Pericentric DNA Loops Relative to the Mitotic Spindle Axis
The organization of cohesin in a cylindrical array about the spindle predicts a specific geometric arrangement of the pericentric DNA. Pericentric DNA has been visualized through integration of a 10-kb LacO array 1.8 kb from the centromere on chromosome XV in cells containing LacI fused to green fluorescent protein (GFP) (Goshima and Yanagida, 2000). These arrays occur as diffraction-limited spots between the spindle poles in metaphase and adjacent to the poles in anaphase (Figure 2, A and C, half spindle) respectively. The pericentric LacO arrays exhibit microtubule-dependent motion toward and away from the spindle poles in metaphase (Goshima and Yanagida, 2000; He et al., 2001; Pearson et al., 2001). To determine their spatial distribution, we compiled a two-dimensional density map for LacO foci in a population of metaphase and anaphase cells. In cells where the kinetochores had bioriented, the peak intensity (pixel) of each diffraction limited focus was determined, and the coordinates of the brightest pixel were plotted relative to the spindle pole body (marked with Spc29-RFP). The distribution of x,y coordinates generates a positional density map, representing the residence frequency of the pericentric LacO array relative to the spindle pole (Figure 2, B and D). The frequency distribution in which the brightest pixel within a single diffraction limited spot was observed is indicated in the color-coded map (Figure 2, B and D, red and orange, highest density; blue and purple, least density). This technique provides the spatial distribution of a single pericentric chromatin locus within a large population.
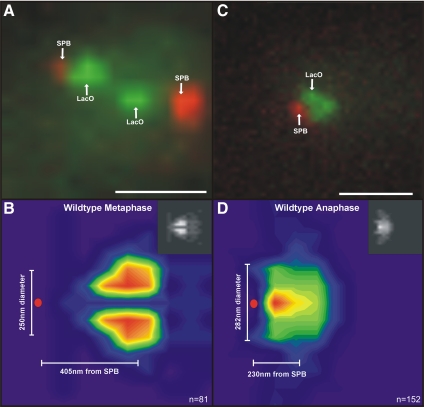
Figure 2.
Density maps of pericentric LacO to visualize the geometric arrangement of pericentric chromatin in metaphase and anaphase. The average position of 10-kb LacO DNA arrays integrated 1.8 kb from CEN15 was determined in metaphase and anaphase spindles in vivo. Metaphase (A) and anaphase (C) spindles are shown top left and right, respectively. Bar, 1 um. Spindle pole bodies are in red (Spc29-RFP), and separated pericentric LacO arrays are in green. The peak intensity (pixel) of each diffraction limited LacO foci was determined and the coordinates of the brightest pixel of each spot were plotted relative to the spindle pole body. The number and position of LacO foci were used to generate a positional heat map representing the range of motion of pericentric LacO array relative to the spindle pole (metaphase, B; anaphase D). The frequency distribution of the brightest LacO pixel is indicated in the color coded heat map below with red and orange the most likely and blue and purple least likely (metaphase, n = 81; anaphase, n = 152). B and D, insets, the insets in the top right are gray-scale density maps of the data plotted with rainbow hues. The gray-scale reduces artifacts due to the differential visual sensitivity to red, green, and blue color spectrum (Borland and Taylor, 2007).
The image of pericentric chromatin in Figure 2B represents a transverse slice through one-half of the mitotic spindle. The pericentric LacO DNA is cylindrically arrayed and radially displaced from the central spindle axis, with an average diameter of approximately 250 nm (ranging up to 480 nm). The least frequently occupied positions are the two pixels (130 nm) along the spindle axis. The LacO array occupies a position approximately 405 nm from the spindle pole, reflecting the average length of metaphase kinetochore microtubules (∼350 nm) (Pearson et al., 2001). This spatial arrangement is reminiscent of the organization of the cylinder of pericentric cohesin where cohesin is radially displaced from the central spindle (average diameter, ∼380 nm) and delimited by the length of kinetochore microtubules 350–400 nm from each pole (Yeh et al., 2008).
In anaphase, kinetochore MTs shorten to the spindle poles, and the pericentric DNA is clustered near the SPB along the spindle axis (Figure 2D). The LacO foci are no longer radially displaced from the spindle axis, and the average distance of the LacO foci is 230–280 nm in three dimensions from the spindle pole (range, 66–325 nm).
Although the outer kinetochore complex is not required for intramolecular DNA looping, microtubule attachments may contribute to the spatial organization of pericentric loops. To determine whether the integrity of the outer kinetochore is required for loop position, we examined the position of pericentric LacO in COMA (ame1-4) and NDC80 (nuf2-60) mutants. In single time point assays, the LacO arrays occur between the spindle poles, indistinguishable from wild-type cells (Supplemental Figure 2, A and D). On fluorescence averaging, LacO foci in cells containing a conditional allele of ame1-4 or nuf2-60 the pericentric chromatin is dispersed relative to wild-type (Supplemental Figure 2, B, C, E, and F). The pericentric chromatin is no longer confined to a spindle proximal location; instead, it occupies a wider area along and around the spindle. Likewise, the cylindrical array of cohesin was dispersed upon loss of the NDC80 complex (nuf2-45; Yeh et al., 2008). Thus, microtubule attachment via COMA or NDC80 is required for the geometric positioning of pericentric chromatin loops. The findings that C-loop formation (Figure 1) is separable from loop positioning (wild type, Figure 2; ame1-4, nuf2-60, Supplemental Figure 2) reveal a novel function for the outer kinetochore domain in dictating spatial organization of pericentric chromatin.
Autonomy of Functional Domains within the Kinetochore
The findings that the inner kinetochore promotes intramolecular pericentric DNA looping and that microtubule attachment dictates loop geometry reveal that the segregation apparatus is a composite structure of two dynamic polymers: DNA and microtubules. Disruption of assembled kinetochores by using temperature-sensitive alleles of the major functional subdomains (DNA bending, ndc10-1; linker domain, ame1-4; and microtubule binding, nuf2-60) reveal how the major subdomains interact with their respective polymer (DNA or microtubule) and with each other. On average, >80% of the cells exhibit loss of function for the given mutant allele at restrictive growth temperature (Figure 3, B–D). The kinetochores from each of the 16 replicated sister chromatids in yeast are clustered into two spots separated by an average of 800 nm in metaphase and segregate in unison at anaphase (He et al., 2000; Pearson et al., 2001). Several distinct patterns of kinetochore protein localization were observed at the restrictive growth temperature (Figure 3A, vi–x). The loss of fluorescence reflects the inability to observe kinetochore protein clusters in the nucleus (Figure 3Avii). Using two Cse4p molecules per kinetochore as a numerical standard, fluorescence loss represents <10 molecules clustered in a diffraction limit spot (Joglekar et al., 2006). Additional phenotypes observed include loss of kinetochore biorientation, evidenced by one diffraction limited spot per cell (Figure 3Aviii); and kinetochore declustering, defined as multiple discrete fluorescent foci around (Figure 3Aix) or along the spindle axis (Figure 3Ax). The latter classification includes linear arrays of fluorescence radiating from the cell's spindle pole bodies along the spindle axis that often extend from pole to pole.
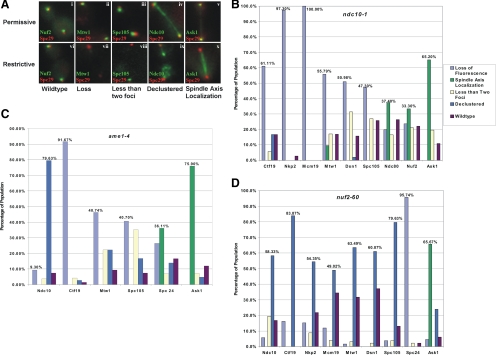
Figure 3.
Functional autonomy of kinetochore complexes. (A) Classes of kinetochore localization patterns observed using fluorescence microscopy. The protein components used to represent each kinetochore complex in this experiment are as follows: CBF3: Ndc10p; COMA: Ctf19p, Nkp2p, Mcm19p; MIND: Mtw1p, Dsn1p; SPC105: Spc105p; NDC80: Nuf2p, Spc24p, Ndc80p; and DAM-DASH: Ask1p. Each kinetochore protein-GFP fusion is shown in green, whereas spindle pole bodies (Spc29-RFP) are shown in red. Boxes i–v represent kinetochore protein localization in wild-type and temperature sensitive mutants (i, wt; ii, ndc10-1; iii, ndc10-1; iv, nuf2-60; and v, ndc10-1) grown at permissive temperature (25°C). Cells in late anaphase are shown for comparison. Boxes vi–x represents kinetochore protein localization in wild-type and temperature-sensitive mutants (vi, wt; vii, ndc10-1; viii, ndc10-1; ix, nuf2-60; and x, ndc10-1) grown at restrictive temperature (37°C) for 3 h. (B) Kinetochore protein localization in ndc10-1 mutants grown at restrictive temperature (37°C). The complete phenotype is indicated for each kinetochore protein GFP fusion (indicated on abscissa). The percentages on the graph highlight the predominant phenotype of each protein. The inner kinetochore proteins (COMA and MIND) were principally lost, whereas outer kinetochore proteins (NDC80 and DAM-DASH) redistributed to the spindle axis. (C) Kinetochore protein localization in ame1-4 mutants grown at restrictive temperature (37°C). The inner kinetochore proteins displayed either a loss of fluorescence or a declustered localization pattern. The outer kinetochore proteins Spc24p and Ask1p displayed spindle axis localization in the majority of the population. A different member of the COMA complex, Ctf19p, was undetected, indicating a complete loss of COMA in ame1-4. (D) Kinetochore protein localization in nuf2-60 mutants grown at restrictive temperature (37°C). The inner kinetochore proteins displayed a declustered phenotype. A different member of the NDC80 complex, Spc24p, was undetected, whereas the outer kinetochore protein Ask1p localized along the spindle axis in the majority of the population observed.
Microtubule Association of the Outer Kinetochore (NDC80 and DAM-DASH) in Absence of Centromeric DNA Attachment
In the absence of Ndc10p of CBF3, there is complete loss of kinetochore protein association with CEN DNA as determined by chromatin immunoprecipitation (He et al., 2001; Westermann et al., 2003; Cheeseman and Desai, 2008). However, as observed previously, inner and outer kinetochore complexes remain assembled in the absence of centromere binding (De Wulf et al., 2003) and provide a useful assay for functional autonomy of individual complexes. In ndc10-1, the outer microtubule-associated proteins (NDC80 and DAM-DASH) lose clustering and biorientation and are distributed along the length of the mitotic spindle (Supplemental Figure 3B, spindle axis ∼30% Nuf2p, Ndc80p; 66% Ask1p). Therefore, NDC80 and DAM-DASH remain bound to microtubules upon detachment of the kinetochore from CEN DNA (ndc10-1). Two proteins normally associated with the COMA complex (Nkp2p and Mcm19p) are not detectable, whereas Ctf19p of COMA and members of the MIND complex are undetectable in 50–60% of cells. Ctf19p became declustered (<20%), and the remaining MIND components (Mtw1p and Dsn1p) localized to the spindle (<10%) or retained only one focus (<20%) (Figure 4B).
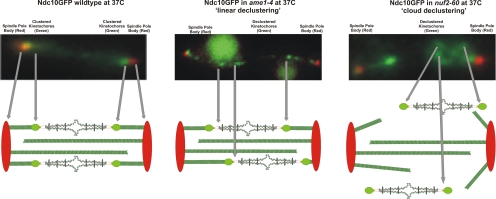
Figure 4.
Schematic representation of kinetochore declustering (Ndc10-GFP) in inner (ame1-4) versus outer (nuf2-60) kinetochore mutants. Top (left), distribution of Ndc10-GFP in anaphase. Top (center, right), types of Ndc10-GFP declustering events observed in ame1-4 (center) and nuf2-60 (right) indicate differences in the status of microtubule plus-end attachment. Linear declustering is indicative of persistent inner kinetochore protein interactions along the spindle (center). Bottom, individual kinetochore–microtubule attachment sites are organized circumferentially around the spindle microtubules (Yeh et al., 2008). A sagittal cross section of this organization in illustrated beneath each fluorescence image. Kinetochore proteins and pericentric chromatin are no longer restrained to the spindle axis in nuf2-60 (top and bottom right). Loss of kinetochore clustering in ame1-4 results in dispersion of kinetochore proteins along the spindle axis (center). Cloud declustering is indicative of loss of microtubule binding (right).
Loss of COMA Disrupts the Linkage between CBF3 DNA Binding and DAM–DASH Microtubule Binding Complexes
Inactivation of COMA (ame1-4) has been reported to disrupt the interaction between the inner and outer kinetochore, without loss of the inner kinetochore binding to centromere DNA (Pot et al., 2005). In cells containing a temperature-sensitive ame1-4 allele, kinetochore localization of another member of the COMA complex (Ctf19p) was lost in 91.7% of the population. Thus, loss of a single member of COMA results in loss of another member of the complex. In vivo, Ndc10-GFP is predominantly declustered in ame1-4 mutants (80%; Figures 3Ax and 4C). The 3C analysis (Figure 1) indicates that intramolecular looping is dependent upon Ndc10p but not COMA (ame1-4). Thus, Ndc10p remains bound and functional to CEN DNA in the absence of COMA. CBF3 looping function is therefore independent of kinetochore clustering as well. Loss of fluorescence or less than two foci was observed in other MIND proteins (Mtw1-GFP, 46% loss, 22% <2 foci) and Spc105-GFP complexes (40% loss, 37% <2 foci). The microtubule binding components (Spc24p of NDC80 and Ask1p of DAM-DASH, respectively) lost their ability to accumulate at MT plus-ends and were found predominately along the spindle axis (Figure 3C).
The declustering of kinetochore subcomplexes indicates that COMA contributes to the collection of sixteen centromeres into one diffraction-limited spot in metaphase, as proposed previously (De Wulf et al., 2003). In addition, COMA is required for the geometric displacement of pericentric DNA loops (Supplemental Figure 2, C and F). Thus COMA functions not only in stabilizing kinetochore–microtubule contacts but also is required for the arrangement of pericentric DNA loops into a cylindrical array encircling the spindle microtubules (Figure 2 and Supplemental Figure 2).
Loss of NDC80 Results in Declustering of Unit-Attachment Site Kinetochores
Nuf2p is an essential member of the NDC80 complex. In cells containing a temperature-sensitive nuf2 allele (nuf2-60), kinetochore localization of another member of the NDC80 complex (Spc24p) was lost in 95.7% of the population (Figure 3D). This result is consistent with the loss of several proteins of the NDC80 complex upon removal of a single member of the complex in mammalian cells (DeLuca et al., 2002). In the absence of the NDC80 complex, Ask1p (DAM-DASH) distributes along the spindle axis (65.7% of population; Figure 3D), indicating that NDC80 is required for the kinetochore clustering but not microtubule binding of the outer kinetochore DAM–DASH complex.
Proteins contained in the inner kinetochore (Ndc10p) were declustered in the absence of NDC80 (Fig. 3Aix and D). The patterns of the inner kinetochore components observed in nuf2-60 vary from those observed in ame1-4 mutants (Figure 4). The declustering phenotype is marked by multiple, distinct foci that surround spindle pole bodies (SPBs) or extension along the spindle axis. Multiple distinct foci that extend from pole to pole (linear declustering; Figure 4, center) are more frequently observed in ame1-4 cells (47% of the 79% declustered Ndc10p; Figure 3C). In contrast, inner kinetochores exhibiting the declustered phenotype in nuf2-60 cells usually surround one (or both) of the cell's SPBs (cloud declustering; Figure 4, right) but do not extend along the spindle axis (79% of the 58% declustered Ndc10p; Figure 3D).
The types of declustering events observed in ame1-4 versus nuf2-60 indicate differences in the status of microtubule plus-end attachment. ame1-4 cells contain chromosomes that remain attached to kinetochore microtubule plus-end (Pot et al., 2005). In ame1-4, Ndc10p remains complexed with Mtw1p (60% colocalization in cells containing Ndc10-GFP and Mtw1-Cherry; Supplemental Figure 3), but cells are unable to cluster individual attachment sites (Figure 4, center). In nuf2-60, kinetochore declustering remains localized to the area surrounding the cell's SPBs and is not restricted to the spindle axis (Figure 4, right). Previous ChIp experiments have shown that inner kinetochore proteins (Ndc10p, Ame1p, and Mtw1p) bind to CEN DNA in ndc80-1 cells (De Wulf et al., 2003). The fluorescence analysis reveals that Ndc10p largely remains complexed with Mtw1p and Spc105p in nuf2-60 mutants (66 and 64% colocalization; Supplemental Figure 3). The declustering events observed in nuf2-60 reflect retention of inner kinetochore protein complexes at the centromere that lack the ability to bind microtubule plus-ends in the absence of the outer NDC80 complex. Kinetochore proteins can therefore be recruited and retained at centromeres in microtubule-dependent or independent pathways.
Kinetochore Subdomains Are Destabilized upon Perturbation of Kinetochore Function
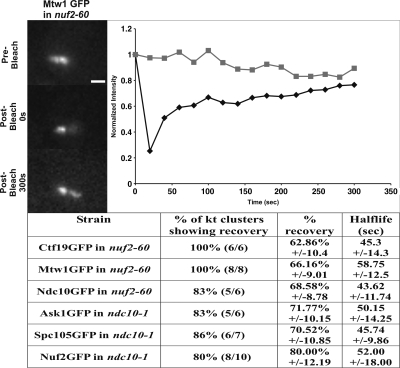
Once assembled into a fully functional kinetochore, individual kinetochore proteins are highly stable, with half-lives of recovery longer than the duration of metaphase (Pearson et al., 2004; Joglekar et al., 2006). To test whether this stability is an inherent property of the individual complexes, versus the ensemble of kinetochore proteins we used FRAP to determine protein dynamics in functional versus nonfunctional complexes. Components from each of CBF3 (Ndc10p), COMA (Ctf19p), or MIND (Mtw1p) in nuf2-60 and NDC80 (Nuf2p), Spc105p, or DAM-DASH (Ask1p) in ndc10-1 were photobleached after 3 h at restrictive temperature. Two classes of recovery were observed. One class (5/43) resembled unperturbed kinetochores in that no fluorescence recovery was observed over 5 min (Figure 5). In 38/43 cells, 67% recovery on average was detected within 5 min (Figure 5). The average halftime of recovery was 49 ± 13 s, similar to that reported for kinetochore microtubules (Maddox et al., 2000; Pearson et al., 2006). Decoupling kinetochore proteins from the centromere DNA (ndc10-1) or microtubule (nuf2-60) alters the in vivo binding affinity.
Figure 5.
In vivo dynamics of kinetochore components. Top (left), pre- and postbleach images of Mtw1-GFP in nuf2-60 cells at the restrictive temperature (37°C). Top (right), FRAP was detected within 20s after photobleaching (diamonds). Fluorescence loss of unbleached Mtw1-GFP is shown (squares). (Below) FRAP recovery values (percentage of kinetochore clusters that exhibit recovery, percentage of recovery, and recovery half-life) are shown for Ctf19p, Mtw1p, and Ndc10p in nuf2-60 and Ask1p, Spc105p, and Nuf2p in ndc10-1. Thirty-eight of 43 (88.4%) of kinetochore clusters showed an average recovery of 70 ± 9.67%, with a half-life of 49.5 ± 13.15 s.
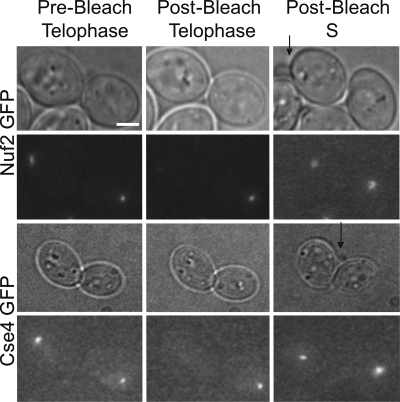
Because assembled kinetochore complexes are stable during mitosis, we were able to address the mode of kinetochore protein segregation upon centromere DNA replication. Fluorescence intensity ratios of mother to daughter kinetochore spots within one cell were compared. As shown in Table 1, in anaphase/telophase the mother/daughter kinetochore protein ratio of ∼1.00 was observed. This indicates that an equal number of fluorescent molecules are deposited to mother and daughter kinetochores after anaphase chromosome segregation. Using relative cell size to identify mother and daughter cells in an unperturbed cell cycle, the kinetochore cluster in the mother cell was photobleached in telophase (Figure 6). On average, 96 ± 7.05% of the kinetochore spot was photobleached (Table 1, Postbleach). Cells were monitored over time until DNA replication was complete, as assessed by bud emergence in the next cell cycle. The fluorescence intensity of the newly replicated 32 kinetochores in the mother relative to the unbleached 16 kinetochores in the daughter was calculated. Cse4p recovered the largest extent (1.32 ± 0.11) consistent with previous studies, indicating that new Cse4p is deposited on both replicated strands (Supplemental Figure 4) (Pearson et al., 2004). Representative inner kinetochore proteins (Dsn1p, Spc105p) recovered to 0.97 and 1.13, respectively (Table 1). The average recovery of the three inner kinetochore components (Cse4p, Spc105p, and Dsn1p) was 1.15 ± 0.12 (n = 16). The relative amounts of protein recovery after DNA duplication indicate that the MIND complex and Spc105p follow a similar mode of remodeling to Cse4p.
Table 1.
Fluorescence intensity ratios of mother/daughter kinetochore clusters obtained from FRAP experiments in which an anaphase cell with a photobleached mother kinetochore spot was observed over time until the mother cell budded and completed S phase
| Prebleach | Postbleach | Recovery | |
|---|---|---|---|
| Cse4 GFP | 0.99 ± 0.04, n = 20 | 0.03 ± 0.17, n= 15 | 1.32 ± 0.11, n = 5 |
| Dsn1 GFP | 0.99 ± 0.05, n = 20 | 0.05 ± 0.11, n = 18 | 0.97 ± 0.10, n = 4 |
| Spc105 GFP | 0.99 ± 0.05, n = 20 | 0.06 ± 0.24, n = 11 | 1.13 ± 0.14, n = 7 |
| Nuf2 GFP | 0.96 ± 0.03, n = 20 | 0.03 ± 0.32, n = 14 | 0.55 ± 0.12, n = 5 |
| Ndc80 GFP | 1.00 ± 0.04, n = 20 | 0.07 ± 0.19, n = 17 | 0.57 ± 0.06, n = 3 |
The inner kinetochore proteins (Cse4p, Dsn1p, and Spc105p) have an average ratio prebleach of 0.99 ± 0.05, n = 60; postbleach of 0.05 ± 0.14, n = 44; and recovery of 1.15 ± 0.12, n = 16. The outer kinetochore proteins Nuf2p and Ndc80p have an average ratio prebleach of 0.98± 0.04, n = 40; postbleach of 0.06 ± 0.27, n = 31; and recovery of 0.56 ± 0.18, n = 8.
Figure 6.
Pre- and postbleach images of Nuf2-GFP (top) and Cse4-GFP (bottom) after cell cycle assembly. Prebleach sister kinetochore clusters of both Nuf2-GFP and Cse4-GFP in a single late anaphase/telophase cell (left). Postbleach image in late anaphase/telophase of the mother kinetochore cluster using a 200-ms laser exposure (center). Postbleach recovery of Nuf2-GFP and Cse4-GFP fluorescence in the mother kinetochore cluster after the mother cell budded, whereas the daughter remained unbudded. Arrow marks new bud.
In contrast, the microtubule binding proteins Nuf2p and Ndc80p recovered to only half the level (∼0.50; Table 1) of the inner kinetochore components. The average recovery of Nuf2p and Ndc80p was 0.56 ± 0.18 (n = 8). Therefore, twice the amount of unbleached Cse4p, Dsn1p, and Spc105p is incorporated into new kinetochores relative to the amount of outer kinetochore proteins (Nuf2p and Ndc80p) after DNA replication. This indicates that proteins in the NDC80 complex from the previous cell cycle (bleached) are incorporated into newly assembled kinetochores. The inner centromere DNA binding complexes are replaced at each division, whereas outer microtubule binding complexes are redistributed from prior divisions onto newly duplicated kinetochores.
DISCUSSION
The 16 replicated chromosomes in budding yeast orient on the metaphase spindle in two clusters. Because individual kinetochores bind a single microtubule, the cluster of 16 kinetochores reflects a higher order level of structural organization of microtubules and pericentric DNA. The pericentric DNA is radially displaced from the central spindle and coincident with the position of pericentric cohesin (Figure 2) (Yeh et al., 2008). The inner kinetochore proteins catalyze centromere DNA looping but are not sufficient for their radial displacement. Loss of either the linker or outer domains results in disruption of the spatial restriction of kinetochores and pericentric chromatin. The linker kinetochore domains are required to cluster the 16 kinetochores into a diffraction limited spot. We propose that the segregation apparatus is a composite structure reflecting the integrated contribution of pericentric chromatin, kinetochore proteins, and microtubules. The composite of 16 clustered kinetochores in budding yeast may be analogous to multiple microtubule attachment sites within a single kinetochore in organisms with regional centromeres.
The DNA binding complex CBF3, but not COMA or NDC80, is required for the centromere DNA loop (Figure 1). In vitro, CBF3 produces an ∼55° bend in CEN DNA as assayed by atomic force microscopy (Pietrasanta et al., 1999). This bend may provide the molecular basis for intramolecular pairing of pericentric chromatin. CBF3 remains bound to the centromere in the absence of COMA (Pot et al., 2005), indicating that the looping function is intrinsic to CBF3 itself. Although there are no protein homologues of CBF3 complex in mammalian cells, recent studies regarding the role of the human kinetochore DNA-binding protein CENP-B in CEN DNA bending imply that this protein may be functionally equivalent to the CBF3 complex. Tanaka et al. (2001) showed that the binding of CENP-B to its target sequence creates a bend in the DNA of ∼59°, similar to the bend induced by CBF3 binding to the yeast centromere DNA. Therefore, the function of CBF3 in promoting intrastrand looping and extending CEN DNA on the outer surface of the chromosome may be conserved in the mammalian kinetochore via CENP-B. These proteins are structurally important for the integrity of kinetochore function in vivo and provide an essential geometry required for centromere position and accessibility to dynamic microtubules.
In metaphase, chromosome biorientation is essential for the spatial organization of the kinetochore and pericentric DNA. This requires both the linker domain consisting of the COMA, MIND, and Spc105p and the outer NDC80 and DAM–DASH complexes. MT binding is intrinsic to NDC80 but its restriction to the MT plus end requires association with centromere DNA via either CBF3 or COMA (Figure 3). Loss of NDC80 releases chromosomes from plus-end MT attachment and kinetochores are no longer restrained in a cluster near the spindle axis and disperse into multiple foci (Figure 4, right) (De Wulf et al., 2003). Likewise, the pericentric DNA loops are no longer constrained to a cylindrical array surrounding the spindle microtubules (Supplemental Figure 2). Although the inner complexes remain at the centromere, the geometrical arrangements of centromere DNA clustering and loop displacement are lost in the absence of microtubule attachment. In contrast, CBF3 is redistributed primarily to the spindle axis upon loss of COMA (Figure 4, center). NDC80 remains tethered to the centromere in MIND disrupted cells (Scharfenberger et al., 2003), indicating that microtubule attachment without COMA is not sufficient for centromere clustering and loop displacement. Thus, COMA, MIND, and Spc105p are required for the higher order geometrical arrangement of pericentric loops and efficient plus-end binding to microtubules.
Once assembled into a functional kinetochore, kinetochore proteins do not turnover and do not disassemble until the next cell cycle (Pearson et al., 2004; Joglekar et al., 2006). Loss of this structural integrity by decoupling the outer kinetochore complex (Nuf2p,Ask1p,Spc105p) from the centromere, or inner kinetochore proteins (Ndc10p, Mtw1p, and Ctf19p) from MT attachment changes the dynamics of these proteins to a behavior reminiscent of dynamic microtubule plus-ends (Figure 5). Thus, kinetochore protein stability is not intrinsic to an individual complex, rather reflects the stability of the ensemble. In addition, the difference in recovery after photobleaching in late anaphase is indicative of different modes of protein deposition into newly assembled kinetochores. The inner kinetochore proteins, including the histone H3 variant Cse4p, are completely replaced at the kinetochore upon DNA duplication (Pearson et al., 2004). In contrast, the outer microtubule binding complexes can be reincorporated from the previous cell cycle.
The kinetochore and surrounding pericentric chromatin occupy a discrete region in the mitotic spindle. The high fidelity of chromosome segregation is dependent on interactions between kinetochore and spindle components as well as the spatial organization of the major polymers, DNA, and microtubules. This finding expands upon the functionality of a eukaryotic kinetochore from a simple microtubule attachment site to a protein–DNA complex that organizes the three-dimensional structure of the centromere and pericentric chromatin. The kinetochore has classically been viewed as a protein machine designed to couple chromosome movement to depolymerizing microtubules. The work here reveals that the kinetochore and spindle microtubules dictate the geometry of pericentric DNA loops to envelop the central spindle. This structure provides a new perspective on the architecture of the centromere and how it relates to the mitotic spindle. In addition, the finding that DNA loops are an architectural component of the budding yeast spindle sheds insight into how point centromeres might function. Loops from individual chromosomes are clustered into a specific arrangement in metaphase (Figures 1 and 2). This configuration may be similar to the architecture of mammalian kinetochores with multiple microtubule attachment sites per chromosome. The major structural difference between the yeast and mammalian kinetochores is that centromere DNA loops on the outer surface of the chromosome emanate from multiple yeast chromosomes, whereas loops of mammalian DNA emanate from a single chromosome. We propose that the organization of point centromeres in budding yeast kinetochores is structurally analogous to multiple microtubule attachment site kinetochores found in higher eukaryotes.
Supplementary Material
ACKNOWLEDGMENTS
We thank members of the Bloom laboratory for critical comments and helpful suggestions. This work was supported by National Institutes of Health grant R01 GM-32238 (to K.S.B.).
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E09-05-0359) on August 5, 2009.
REFERENCES
- Bi E., Pringle J. R. ZDS1 and ZDS2, genes whose products may regulate Cdc42p in Saccharomyces cerevisiae. Mol. Cell Biol. 1996;16:5264–5275. doi: 10.1128/mcb.16.10.5264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Borland D., Taylor R. M., II Rainbow color map (still) considered harmful. IEEE Comput. Graph. Appl. 2007;March/April:14–17. doi: 10.1109/mcg.2007.323435. [DOI] [PubMed] [Google Scholar]
- Bouck D. C., Joglekar A. P., Bloom K. S. Design features of a mitotic spindle: balancing tension and compression at a single microtubule kinetochore interface in budding yeast. Annu. Rev. Genet. 2008;42:335–359. doi: 10.1146/annurev.genet.42.110807.091620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheeseman I. M., Desai A. Molecular architecture of the kinetochore-microtubule interface. Nat. Rev. Mol. Cell Biol. 2008;9:33–46. doi: 10.1038/nrm2310. [DOI] [PubMed] [Google Scholar]
- De Wulf P., McAinsh A. D., Sorger P. K. Hierarchical assembly of the budding yeast kinetochore from multiple subcomplexes. Genes Dev. 2003;17:2902–2921. doi: 10.1101/gad.1144403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dekker J., Rippe K., Dekker M., Kleckner N. Capturing chromosome conformation. Science. 2002;295:1306–1311. doi: 10.1126/science.1067799. [DOI] [PubMed] [Google Scholar]
- DeLuca J. G., Howell B. J., Canman J. C., Hickey J. M., Fang G., Salmon E. D. Nuf2 and Hec1 are required for retention of the checkpoint proteins Mad1 and Mad2 to kinetochores. Curr. Biol. 2003;13:2103–2109. doi: 10.1016/j.cub.2003.10.056. [DOI] [PubMed] [Google Scholar]
- DeLuca J. G., Moree B., Hickey J. M., Kilmartin J. V., Salmon E. D. hNuf2 inhibition blocks stable kinetochore-microtubule attachment and induces mitotic cell death in HeLa cells. J. Cell Biol. 2002;159:549–555. doi: 10.1083/jcb.200208159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuyama S., Biggins S. Centromere identity is specified by a single centromeric nucleosome in budding yeast. Proc. Natl. Acad. Sci. USA. 2007;104:14706–14711. doi: 10.1073/pnas.0706985104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goshima G., Yanagida M. Establishing biorientation occurs with precocious separation of the sister kinetochores, but not the arms, in the early spindle of budding yeast. Cell. 2000;100:619–633. doi: 10.1016/s0092-8674(00)80699-6. [DOI] [PubMed] [Google Scholar]
- He X., Asthana S., Sorger P. K. Transient sister chromatid separation and elastic deformation of chromosomes during mitosis in budding yeast. Cell. 2000;101:763–775. doi: 10.1016/s0092-8674(00)80888-0. [DOI] [PubMed] [Google Scholar]
- He X., Rines D. R., Espelin C. W., Sorger P. K. Molecular analysis of kinetochore-microtubule attachment in budding yeast. Cell. 2001;106:195–206. doi: 10.1016/s0092-8674(01)00438-x. [DOI] [PubMed] [Google Scholar]
- Hoffman D. B., Pearson C. G., Yen T. J., Howell B. J., Salmon E. D. Microtubule-dependent changes in assembly of microtubule motor proteins and mitotic spindle checkpoint proteins at PtK1 kinetochores. Mol. Biol. Cell. 2001;12:1995–2009. doi: 10.1091/mbc.12.7.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang L. S., Sternberg P. W. Genetic dissection of developmental pathways. WormBook. 2006;14:1–19. doi: 10.1895/wormbook.1.88.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Indjeian V. B., Murray A. W. Budding yeast mitotic chromosomes have an intrinsic bias to biorient on the spindle. Curr. Biol. 2007;17:1837–1846. doi: 10.1016/j.cub.2007.09.056. [DOI] [PubMed] [Google Scholar]
- Joglekar A. P., Bouck D., Finley K., Liu X., Wan Y., Berman J., He X., Salmon E. D., Bloom K. S. Molecular architecture of the kinetochore-microtubule attachment site is conserved between point and regional centromeres. J. Cell Biol. 2008;181:587–594. doi: 10.1083/jcb.200803027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joglekar A. P., Bouck D. C., Molk J. N., Bloom K. S., Salmon E. D. Molecular architecture of a kinetochore-microtubule attachment site. Nat. Cell Biol. 2006;8:581–585. doi: 10.1038/ncb1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitagawa K., Hieter P. Evolutionary conservation between budding yeast and human kinetochores. Nat. Rev. Mol. Cell Biol. 2001;2:678–687. doi: 10.1038/35089568. [DOI] [PubMed] [Google Scholar]
- Maddox P. S., Bloom K. S., Salmon E. D. The polarity and dynamics of microtubule assembly in the budding yeast Saccharomyces cerevisiae. Nat. Cell Biol. 2000;2:36–41. doi: 10.1038/71357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miranda J. J., De Wulf P., Sorger P. K., Harrison S. C. The yeast DASH complex forms closed rings on microtubules. Nat. Struct. Mol. Biol. 2005;12:138–143. doi: 10.1038/nsmb896. [DOI] [PubMed] [Google Scholar]
- Moore L. L., Roth M. B. HCP-4, a CENP-C-like protein in Caenorhabditis elegans, is required for resolution of sister centromeres. J. Cell Biol. 2001;153:1199–1208. doi: 10.1083/jcb.153.6.1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mythreye K., Bloom K. S. Differential kinetochore protein requirements for establishment versus propagation of centromere activity in Saccharomyces cerevisiae. J. Cell Biol. 2003;160:833–843. doi: 10.1083/jcb.200211116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada T., Ohzeki J., Nakano M., Yoda K., Brinkley W. R., Larionov V., Masumoto H. CENP-B controls centromere formation depending on the chromatin context. Cell. 2007;131:1287–1300. doi: 10.1016/j.cell.2007.10.045. [DOI] [PubMed] [Google Scholar]
- Pearson C. G., Gardner M. K., Paliulis L. V., Salmon E. D., Odde D. J., Bloom K. Measuring nanometer scale gradients in spindle microtubule dynamics using model convolution microscopy. Mol. Biol. Cell. 2006;17:4069–4079. doi: 10.1091/mbc.E06-04-0312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson C. G., Maddox P. S., Salmon E. D., Bloom K. Budding yeast chromosome structure and dynamics during mitosis. J. Cell Biol. 2001;152:1255–1266. doi: 10.1083/jcb.152.6.1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson C. G., Yeh E., Gardner M., Odde D., Salmon E. D., Bloom K. Stable kinetochore-microtubule attachment constrains centromere positioning in metaphase. Curr. Biol. 2004;14:1962–1967. doi: 10.1016/j.cub.2004.09.086. [DOI] [PubMed] [Google Scholar]
- Pietrasanta L. I., Thrower D., Hsieh W., Rao S., Stemmann O., Lechner J., Carbon J., Hansma H. Probing the Saccharomyces cerevisiae centromeric DNA (CEN DNA)-binding factor 3 (CBF3) kinetochore complex by using atomic force microscopy. Proc. Natl. Acad. Sci. USA. 1999;96:3757–3762. doi: 10.1073/pnas.96.7.3757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pot I., Knockleby J., Aneliunas V., Nguyen T., Ah-Kye S., Liszt G., Snyder M., Hieter P., Vogel J. Spindle checkpoint maintenance requires Ame1 and Okp1. Cell Cycle. 2005;4:1448–1456. doi: 10.4161/cc.4.10.2106. [DOI] [PubMed] [Google Scholar]
- Sakuno T., Tada K., Watanabe Y. Kinetochore geometry defined by cohesion within the centromere. Nature. 2009;458:852–858. doi: 10.1038/nature07876. [DOI] [PubMed] [Google Scholar]
- Salmon E. D., Shaw S. L., Waters J. C., Waterman-Storer C. M., Maddox P. S., Yeh E., Bloom K. A high-resolution multimode digital microscope system. Methods Cell Biol. 2007;81:187–218. doi: 10.1016/S0091-679X(06)81011-3. [DOI] [PubMed] [Google Scholar]
- Scharfenberger M., Ortiz J., Grau N., Janke C., Schiebel E., Lechner J. Nsl1p is essential for the establishment of bipolarity and the localization of the Dam-Duo complex. EMBO J. 2003;22:6584–6597. doi: 10.1093/emboj/cdg636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan B. A., Karpen G. H. Centromeric chromatin exhibits a histone modification pattern that is distinct from both euchromatin and heterochromatin. Nat. Struct. Mol. Biol. 2004;11:1076–1083. doi: 10.1038/nsmb845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka Y., Nureki O., Kurumizaka H., Fukai S., Kawaguchi S., Ikuta M., Iwahara J., Okazaki T., Yokoyama S. Crystal structure of the CENP-B protein-DNA complex: the DNA-binding domains of CENP-B induce kinks in the CENP-B box DNA. EMBO J. 2001;20:6612–6618. doi: 10.1093/emboj/20.23.6612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westermann S., Avila-Sakar A., Wang H. W., Niederstrasser H., Wong J., Drubin D. G., Nogales E., Barnes G. Formation of a dynamic kinetochore-microtubule interface through assembly of the Dam1 ring complex. Mol. Cell. 2005;17:277–290. doi: 10.1016/j.molcel.2004.12.019. [DOI] [PubMed] [Google Scholar]
- Westermann S., Cheeseman I. M., Anderson S., Yates J. R., 3rd, Drubin D. G., Barnes G. Architecture of the budding yeast kinetochore reveals a conserved molecular core. J. Cell Biol. 2003;163:215–222. doi: 10.1083/jcb.200305100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westermann S., Drubin D. G., Barnes G. Structures and functions of yeast kinetochore complexes. Annu. Rev. Biochem. 2007;76:563–591. doi: 10.1146/annurev.biochem.76.052705.160607. [DOI] [PubMed] [Google Scholar]
- Yeh E., Haase J., Paliulis L. V., Joglekar A., Bond L., Bouck D., Salmon E. D., Bloom K. S. Pericentric chromatin is organized into an intramolecular loop in mitosis. Curr. Biol. 2008;18:81–90. doi: 10.1016/j.cub.2007.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh E., Skibbens R. V., Cheng J. W., Salmon E. D., Bloom K. Spindle dynamics and cell cycle regulation of dynein in the budding yeast, Saccharomyces cerevisiae. J. Cell Biol. 1995;130:687–700. doi: 10.1083/jcb.130.3.687. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.