Abstract
Onset of proteotoxicity is linked to change in the subcellular location of proteins that cause misfolding diseases. Yet, factors that drive changes in disease protein localization and the impact of residence in new surroundings on proteotoxicity are not entirely clear. To address these issues, we examined aspects of proteotoxicity caused by Rnq1-green fluorescent protein (GFP) and a huntingtin's protein exon-1 fragment with an expanded polyglutamine tract (Htt-103Q), which is dependent upon the intracellular presence of [RNQ+] prions. Increasing heat-shock protein 40 chaperone activity before Rnq1-GFP expression, shifted Rnq1-GFP aggregation from the cytosol to the nucleus. Assembly of Rnq1-GFP into benign amyloid-like aggregates was more efficient in the nucleus than cytosol and nuclear accumulation of Rnq1-GFP correlated with reduced toxicity. [RNQ+] prions were found to form stable complexes with Htt-103Q, and nuclear Rnq1-GFP aggregates were capable of sequestering Htt-103Q in the nucleus. On accumulation in the nucleus, conversion of Htt-103Q into SDS-resistant aggregates was dramatically reduced and Htt-103Q toxicity was exacerbated. Alterations in activity of molecular chaperones, the localization of intracellular interaction partners, or both can impact the cellular location of disease proteins. This, in turn, impacts proteotoxicity because the assembly of proteins to a benign state occurs with different efficiencies in the cytosol and nucleus.
INTRODUCTION
Protein misfolding and accumulation of amyloid-like aggregates are the hallmarks of a broad class of protein conformational diseases, which include Alzheimer's disease, frontotemporal dementia, spongiform encephalopathies, and polyglutamine expansion diseases (Carrell and Lomas, 1997). Factors that initiate protein aggregation and disease onset include inheritance of mutant proteins, aberrant protein processing, expansion of glutamine- or alanine-rich tracts of amino acids, and environmental stress (Chiti and Dobson, 2006). The amino acid sequence of individual, disease-causing proteins defines the neurodegenerative disorder as well as the affected brain regions. Although it is unclear why some neuronal populations are more vulnerable to toxicity and others are more apt to efficiently manage protein misfolding events (Taylor et al., 2002). The differential susceptibility of various neuronal subtypes to proteotoxicity indicates that the cellular environment plays a major role in dictating whether a misfolded disease protein is toxic (Balch et al., 2008; Morimoto, 2008).
The exact nature of the neurotoxic conformer formed by individual, disease proteins is not clear (Caughey and Lansbury, 2003). Aggregates formed by disease proteins can sequester essential cellular proteins (Burke et al., 1996; Steffan et al., 2000), inhibit the activity of the proteasome (Bence et al., 2001), or both and thereby cause cell death. Yet, the extent of disease protein aggregation does not always correlate with the observed pathology (Caughey and Lansbury, 2003; Haass and Selkoe, 2007) and small oligomers formed by disease proteins have been implicated as the toxic species (Kayed et al., 2003; Haass and Selkoe, 2007; Treusch et al., 2009).
A primary mechanism to prevent the accumulation of toxic protein species is chaperone-dependent suppression of disease protein aggregation and subsequent degradation (Muchowski and Wacker, 2005; Cohen et al., 2006). Emerging evidence demonstrates that the conversion of disease proteins into ordered, benign aggregates also serves to prevent the accumulation of toxic protein oligomers (Behrends et al., 2006; Cheng et al., 2007). Paradoxically, the ability of disease proteins to assemble into ordered benign aggregates is critically dependent upon molecular chaperone action (Behrends et al., 2006; Douglas et al., 2008). Thus, molecular chaperones can protect against proteotoxicity by either suppressing or promoting protein aggregation (Douglas et al., 2009).
Cell stress or subtle age-dependent changes in the activity of chaperone networks seem to enable pools of disease proteins to escape surveillance and accumulate as toxic conformers (Morimoto, 2008). Epigenetic factors control molecular chaperone expression, and the capacity of chaperone networks in different cell types and subcellular compartments is variable (Morimoto, 2008). Therefore, selective vulnerability to conformational disease could result from differences in the neuron's ability to promote efficient flux of disease proteins through degradation and aggregation pathways.
Components in the cytosol and nucleus have different capacities to buffer the presence of mutant disease proteins, but the mechanism behind this occurrence is unknown (Ross, 1997). For example, normal Huntingtin protein (Htt) contains a 25-residue polyglutamine (polyQ) sequence within exon I and predominately resides within the cytosol (De Rooij et al., 1996). Huntington's disease is caused by the expansion of the polyQ region in Htt beyond 39 residues and is associated with the nuclear accumulation of mutant Htt in striatal neurons (DiFiglia et al., 1997; Saudou et al., 1998). Conversion of mutant Htt to a toxic species in the nucleus may occur through its interactions with new nuclear neighbors (Duennwald et al., 2006). In addition, the cytoplasmic chaperonin TriC, which functions to assemble mutant Htt into benign aggregates (Behrends et al., 2006), is not present in the nucleus (Kim et al., 1994). Therefore, when mutant Htt enters the nucleus, it seems to face a combination of negative environmental influences that enable it to assume a toxic conformation. However, whether interactions with environmental factors drive a change in the cellular location of mutant Htt is not clear.
To define how the cellular environment modulates the accumulation of proteotoxic species, we study features of Rnq1 and Htt toxicity in yeast. The yeast protein Rnq1 belongs to a class of glutamine/asparagine-rich (Q/N) proteins that exist in native and alternate, self-propagating amyloid-like forms (Sondheimer and Lindquist, 2000). Although benign at low levels, elevated expression of exogenous Rnq1 or Rnq1-green fluorescent protein (GFP) induces cell death (Douglas et al., 2008). Formation of toxic Rnq1 species requires the presence of pre-existing [RNQ+]/[PIN+] prions (Derkatch et al., 1997; Douglas et al., 2008). Rnq1 toxicity is suppressed by increasing the levels of the type II heat-shock protein (Hsp)40 chaperone, Sis1, which enhances Rnq1 assembly into SDS-resistant aggregates and reduces the accumulation of a detergent-soluble Rnq1 species (Douglas et al., 2008). These data demonstrate that chaperone-dependent conversion of a toxic protein into amyloid-like aggregates can serve as a protective mechanism. Thus, efficient flux through amyloid assembly pathways seems to strongly influence whether cells tolerate disease protein expression.
Yeast prions act as environmental factors that promote the conversion of proteins into alternate conformational states (Meriin et al., 2002; Gokhale et al., 2005). In particular, [RNQ+] prions expose surfaces that template the conversion of endogenous yeast prions (Derkatch et al., 1997) and foreign prions (Taneja et al., 2007) into amyloid-like states. Furthermore, [RNQ+] prions act via an ill-defined mechanism to facilitate the conformational switching of the polyQ expanded exon-1 fragment from huntingtin's protein (Htt-103Q) from a benign to toxic state (Meriin et al., 2002). Thus, the study of interactions between Rnq1 and Htt-103Q provides an excellent model to understand how members of a disease protein's neighborhood influence whether it becomes benign or toxic.
To investigate how a disease protein's neighborhood influences its toxicity, we examined the impact of relocating Rnq1-GFP aggregates from the cytosol to the nucleus on cell death caused by Rnq1 and Htt-103Q. Shifting localization of Rnq1-GFP from the cytosol to the nucleus dramatically enhanced the accumulation of SDS-resistant Rnq1 aggregates and suppressed Rnq1 toxicity. Surprisingly, nuclear Rnq1 aggregates sequestered Htt-103Q and interfered with the formation of SDS-resistant Htt-103Q aggregates. The resultant accumulation of detergent-soluble forms of Htt-103Q correlated with a dramatic increase in Htt toxicity. Changes in Rnq1-GFP and Htt-103Q localization reciprocally correlate with the ability of the cytosol and nucleus to package Rnq1 and Htt into benign SDS-resistant aggregates.
MATERIALS AND METHODS
Strains, Plasmids, and Antibodies
Yeast strains W303, YEF473A, YEF473B, BY4741, and 10B-H49 were used to take advantage of different genetic markers, gene integrations, gene deletions, and karyogamy defects. W303, MAT a and α, can1-100 ade2-1, his3-11,15 leu2-3112 ura3-1 trp1-1; YEF473A, MAT a, trp1Δ63, leu2Δ, ura3-52, his3Δ200, lys2-8Δ1, SSF1-GFP∷KANR; YEF473B, Mat a, trp1Δ63, leu2Δ, ura3-52, his3Δ200, lys2-8Δ1, NUP49-GFP∷HygB; 10B-H49 MATα, ρ° ade2-1, lys1-1, his3-11,15, leu2-3112, kar1-1, ura3∷KANR; BY4741 MAT a, his3Δ, leu2Δ, met15Δ, ura3Δ, SIS1-GFP∷HIS3; BY4741 MAT a, his3Δ, leu2Δ, met15Δ, ura3Δ, NUP188-GFP∷HIS3 (Huh et al., 2003). All strains harbored Rnq1 in its [RNQ+] prion form and the generation of isogenic [rnq-] strains was accomplished via sequential passage of cells on plates containing 3 mM guanidinium-HCl (Douglas et al., 2008). Strains were transformed with plasmids and cultured in synthetic media as described previously (Douglas et al., 2008). CuSO4 was present in all media at a final concentration of 0.25 μM, which drove constitutive low level expression of all proteins under control of the copper-inducible promoter element (CUP1). Commercial antibodies including α-Hsp104, α-Nop1, α-GFP, α-FLAG, and α-PGK1 were obtained from Assay Designs (Ann Arbor, MI), EnCor Biotechnology (Gainesville, FL), Roche Diagnostics (Indianapolis, IN), Sigma-Aldrich (St. Louis, MO), and Invitrogen (Carlsbad, CA), respectively. Polyclonal antibodies including α-Sis1 and α-Ssa1 were generated by our laboratory. The polyclonal Rnq1 antibody was a kind gift from the Lindquist laboratory. Supplemental Table 1 contains a detailed list of yeast plasmids and their construction.
Sytox Dye Exclusion
W303α [RNQ+] cells were sequentially transformed with pRS414-SIS1 or pRS414 and then pRS416-RNQ1 or pRS416. Transformants were grown overnight to an OD600 of 0.2–0.4 in synthetic media containing raffinose as the carbon source. Galactose (2%) was added to the media to induce Rnq1 expression from the galactose-inducible promoter element (GAL1) in pRS416-RNQ1. After 5 h of induced Rnq1 overexpression, 1 OD600 unit of cells was collected, and Sytox permeability was measured with a FLUOstar fluorometer (BMG Labtech, Offenburg, Germany) as described previously (Zakrzewska et al., 2007).
Fluorescence Microscopy
Rnq1-GFP or Rnq1-monomeric red fluorescent protein (mRFP) as well as Htt-25Q-GFP and Htt-103Q-GFP were expressed under control of the indicated inducible promoters. Images were captured from live or fixed cells in synthetic liquid media with an E600 fluorescence microscope (Nikon, Tokyo, Japan), and images were processed with MetaMorph (Molecular Devices, Sunnyvale, CA) and Photoshop (Adobe Systems, Mountain View, CA). The nuclear envelope was delineated in YEF473B or BY4741 [RNQ+] cells via visualization of the nuclear pore proteins Nup49-GFP or Nup188-GFP, respectively. The localization of the nucleolus was monitored in YEF473A [RNQ+] cells by visualization of Ssf1-GFP.
Sis1-mediated localization of Rnq1-GFP to the nucleus was observed when W303α [RNQ+] cells were sequentially transformed with pRS414-GPD-SIS1 and then pRS316-CUP1-RNQ1-GFP. Cells harboring both plasmids were cultured in synthetic media overnight at 30°C to an OD600 of 0.5–2. Additional CuSO4 was added to overnight cultures at final concentration of 1 μM, which allowed for Rnq1-GFP expression and visualization. In the order of expression experiments, W303α [RNQ+] cells harbored two distinct plasmids that expressed two proteins from either the GAL1 or CUP1 promoters. Cells were grown overnight at 30°C in synthetic media containing raffinose (2%) to OD600 of 0.5–1.5. CuSO4 was added to overnight cultures at final concentration of 1 and 10 μM to visualize the respective Rnq1-GFP and Rnq1-mRFP proteins under control of the CUP1 promoter. Media were then supplemented with galactose (2%) to induce expression of the indicated proteins under control of the GAL1 promoter. After 1-h incubations, cells were collected and photographed live or fixed.
Cells were stained with 4,6-diamidino-2-phenylindole (DAPI) to visualize nuclear DNA. Cells were first fixed with 4% formaldehyde for 30 min in culture, washed twice with phosphate-buffered saline (PBS), and permeabilized by incubation in 0.1% PBS Triton X-100 for 10 min. Cells were then washed twice with PBS, incubated for 10 min with DAPI (1 μg/ml), and again washed twice with PBS before visualization.
Cytoduction
W303a [RNQ+] cells that harbored pRS416-GAL1-Rnq1-GFP-NLS were grown overnight in synthetic media containing raffinose (2%). Once cells reached an OD600 of 0.5–1.5, media was supplemented with galactose (2%) and incubated for 2 h. Cells were then mated with the 10B-H49α [rnq-] strain that is defective in karyogamy (Sondheimer and Lindquist, 2000). Before mating, 10B-H49 cells harboring pRS315-CUP-RNQ1-mRFP, were cultured overnight at 30°C in synthetic media. 10B-H49 cells at an OD600 of 0.5–1.5 were supplemented with 50 μM CuSO4 and cultured for an additional 2 h before they were mated. Mating was conducted with 2 OD600 units of cells from either mating type that were washed with sterile double distilled H2O, mixed and incubated on YPD plates. Incubations were performed at 30°C for either 30 min or 2 h, after which cells were scraped and washed with sterile PBS. Mated cells were fixed in formaldehyde and stained with DAPI before visualization.
Analysis of Rnq1 and Htt-103Q Toxicity
Cell growth on agarose plates was monitored in [RNQ+] and [rnq-] W303α strains that harbored pRS416-RNQ1-GFP, pRS416-RNQ1-GFP-NLS, pYES2-Flag-Htt-103Q-GFP or pYES2-Flag-Htt-103Q-GFP-NLS under control of the GAL1 promoter. In the order of expression experiments, W303α [RNQ+] cells were transformed with two plasmids that expressed the indicated proteins under control of the GAL1 or CUP1 promoter. Cells were grown overnight in synthetic media containing 2% raffinose and 1 μM CuSO4 to an OD600 of 1–2 before fivefold dilutions were spotted onto plates containing 2% galactose. Cu++ was present at a final concentration of 0.25 μM in agarose plates to allow for modest, nontoxic expression of the indicated Rnq1 proteins off the CUP1 promoter. Plates were incubated for 3–5 d at 30°C before being photographed.
Analysis of SDS-resistant Htt-103Q and Rnq1 Aggregates
Monitoring the assembly of Rnq1 and Htt-103Q into SDS-resistant aggregates was performed as described previously by semidenaturing detergent agarose gel electrophoresis (SDD-AGE) (Kryndushkin et al., 2003; Douglas et al., 2008) and filter trap analysis (Scherzinger et al., 1997). Total protein levels from cell extracts were standardized by a protein determination assay (DC protein assay kit; Bio-Rad Laboratories, Hercules, CA). Polyvinylidene difluoride or cellulose acetate membranes were probed with α-GFP or α-FLAG to visualize Rnq1-GFP or FLAG-Htt-103Q-GFP. Monitoring protein levels by Western blot was performed as described previously (Douglas et al., 2008). Quantitation of band intensity was determined using QuantityOne 4.6.1 software (Bio-Rad Laboratories).
Rnq1 Coimmunoprecipitation with Htt
Expression of Htt-103Q-GFP in W303α [RNQ+] cells harboring pYES3-Htt-103Q-GFP was induced by supplementation with 2% galactose. Nondenaturing cell extracts were generated 2 h later in buffer containing 50 mM HEPES, pH 7.4, 150 mM NaCl, 1 mM EDTA, 0.1% Triton X-100, 1 mM phenylmethysulfonyl fluoride (PMSF), and a protease inhibitor cocktail (Roche Diagnostics). Coimmunoprecipitations were performed as described previously (Fan et al., 2004). In brief, α-Rnq1 was added to lysates for 1 h before supplementation with preblocked protein G-beads. Htt-103Q-GFP was immunoprecipitated with endogenous Rnq1 and detected with α-GFP by Western blot. Strains that were used in coimmunoprecipitation experiments with exogenous GFP-tagged Rnq1 and Htt-103Q-GFP, were constructed as follows. W303α [RNQ+] cells were transformed with pYES3-Htt-103Q-GFP and pRS316-RNQ1-GFP or pRS316-RNQ1-GFP-NLS. The expression of Rnq1-GFP and Rnq1-GFP-NLS was control of the CUP1 promoter. Three hours after the addition of 50 μM CuSO4 and 2 h after addition of galactose (2%) to cultures, cell extracts were prepared under the nondenaturing conditions described above. Rnq1-GFP and Rnq1-GFP-NLS were immunoprecipitated with α-Rnq1 and probed by Western blot for the presence of Htt-25Q-GFP or Htt-103Q-GFP. All coimmunoprecipitation experiments were preformed in the presence of Triton X-100, which prevents nonspecific Rnq1 aggregation. Under these conditions, neither Rnq1 nor Htt-103Q was pelletable at low speed. Rnq1 precipitation was only observed in presence of the indicated antibodies.
Size Exclusion Chromatography
Nondenaturing extracts from W303α [RNQ+] cells were prepared for analysis by size-exclusion chromatography as described previously (Behrends et al., 2006). In brief, 250 ODs of mid-log phase cells were lysed by glass bead disruption in 1.2 ml of buffer containing 25 mM Tris-HCl, pH 7.5, 50 mM KCl, 10 mM Mg2Cl, 1 mM EDTA, 5% glycerol, 1% Triton X-100, 1 mM PMSF, and a protease inhibitor cocktail (Roche Diagnostics). Proteins in 500 ml of extract were resolved on a Sephacryl S-200 Hi-Prep column (GE Healthcare, Pittsburgh, PA). The ability of Triton detergent in nondenaturing lysis buffers to lyse the nucleus was assayed by monitoring the liberation of the resident nucleolar protein Nop1 from pellet to supernatant fractions after a 13,000 × g spin (data not shown).
Sis1-dependent suppression of Rnq1-GFP toxicity is associated with enhanced formation of a high-molecular-weight SDS-resistant species and depletion of low-molecular-weight pools (Douglas et al., 2008). Small oligomeric intermediates of amyloid assembly pathways have been implicated as a toxic protein species (Kayed et al., 2003; Haass and Selkoe, 2007). However, Rnq1-GFP migrates at the same low-molecular-weight peak in extracts from [rnq-] cells (Douglas et al., 2008) which do not assemble Rnq1 into amyloid-like aggregates. In addition, Rnq1 seems to be a natively unfolded (data not shown). So the low-molecular pools of Rnq1-GFP detected by gel filtration are most likely unassembled monomers but could also represent small oligomers such as a dimer or trimer.
Isolation of Nuclei by Differential Centrifugation
[RNQ+] W303α strains that harbored either pRS414-GPD or pRS414-GPD-SIS1 were grown overnight in synthetic media at 30°C. Cultures (25 ml) were diluted to an OD600 of 0.4 in 300 ml of synthetic media. After a 4- to 6-h incubation period, 250 OD units of cells were collected and frozen in liquid nitrogen. Frozen samples were thawed on ice and spheroplasts were created via 45-min incubations at 30°C in the presence of 2 mg/ml Zymolyase-100T (MP Biomedicals, Solon, OH). Cell extracts were generated by homogenization of spheroplasts in buffer containing 18% Ficoll-400, 20 mM Tris-HCl, pH 7.5, 20 mM KCl, 5 mM MgCl2, 3 mM dithiothreitol, 1 mM EDTA, 1 mM PMSF, and protease inhibitor cocktail (Roche Diagnostics). Cell extracts were spun at 3000 × g for 5 min to clear unbroken cells, and the resulting supernatant was spun at 13,000 × g for 20 min to pellet the nuclei. The supernatant and pellet fractions from the 13,000 × g spin were boiled in sample buffer. Proteins were resolved by SDS-polyacrylamide gel electrophoresis (PAGE) and detected by Western blot analysis.
RESULTS
Sis1-mediated Suppression of Rnq1 Toxicity Is Accompanied by Movement of Rnq1-GFP Aggregates to the Nucleus
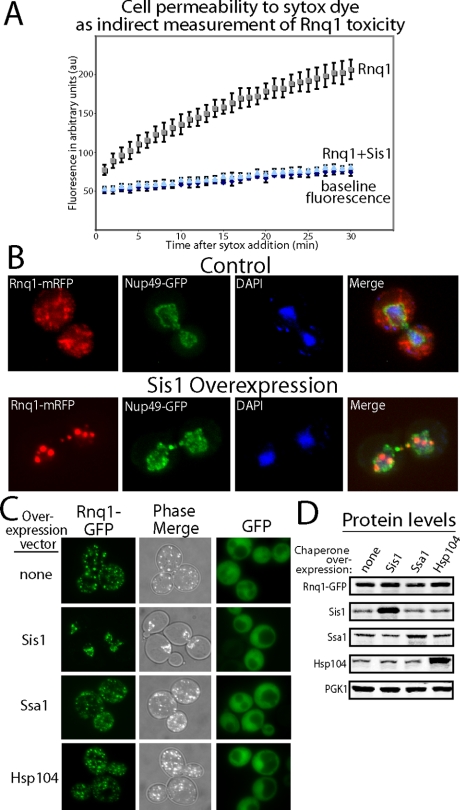
To explore how changes in chaperone activity within the cellular environment impact toxicity of disease proteins, we examined the ability of the Hsp40 Sis1 to subdue the rapid onset of Rnq1 toxicity in relation to the subcellular distribution of the Rnq1-GFP aggregates. The yeast plasma membrane becomes permeable to Sytox dye (Zakrzewska et al., 2007) within 5 h of inducing Rnq1 overexpression (Figure 1A). Elevating Sis1 expression within the physiological levels observed during heat stress prevents Rnq1 from permeablizing the yeast plasma membrane to Sytox, which correlates with suppression of Rnq1-induced cell death (Douglas et al., 2008). It is interesting that, in ∼50% of cells, Sis1-dependent suppression of Rnq1 toxicity was associated with a dramatic change in the localization of fluorescent foci containing Rnq1-mRFP from the cytosol to the nucleus (Figure 1B). Rnq1-mRFP that accumulated within the nucleus does not seem to be nucleolar because nuclear Rnq1-mRFP foci do not colocalize with the nucleolar marker Ssf1-GFP (Supplemental Figure 1).
Figure 1.
Suppression of Rnq1 toxicity by Sis1 correlates with the nuclear accumulation of Rnq1 aggregates. (A) Sytox dye uptake into W303 [RNQ+] cells expressing Rnq1 with and without excess Sis1. Rnq1 overexpression was induced for 5 h before the addition of Sytox dye, at which point fluorescent readings were taken every min for 30 min. (B) Rnq1-mRFP localization in the nucleus of YEF473B [RNQ+] cells resultant from Sis1 overexpression. Fluorescent images show the cellular distribution of Rnq1-mRFP, which was overexpressed for 15 h from the CUP1 promoter due to the presence of 10 μM CuSO4 in media. Nup49-GFP is a marker of the nuclear envelope boundary. DAPI denotes the localization of nuclear DNA. (C) Effect of chaperone overexpression on the cellular distribution of Rnq1-GFP in W303 [RNQ+] cells. Fluorescent images show the cellular distribution of Rnq1-GFP, which was overexpressed for 15 h from the CUP1 promoter due to 1 μM CuSO4 present in media. The GFP panel on the right shows the localization of GFP under conditions describes on the left. (D) Western blot analysis of the indicated proteins from cell extracts in C.
The link between suppression of Rnq1-GFP toxicity by Sis1 and the relocation of exogenous Rnq1 aggregates to the nucleus is intriguing because, until now, changes in the nucleocytoplasmic distribution of disease-causing proteins has been associated with the onset of cell death (DiFiglia et al., 1997; Saudou et al., 1998; Neumann et al., 2006). Type II Hsp40s such as Sis1 have been implicated in mediating the accumulation of a range of proteins in the nucleus (Cheng et al., 2008; Zhang et al., 2008). Thus, we sought to establish the concept that molecular chaperones can impact proteotoxicity by influencing the cellular location of the model disease protein Rnq1-GFP.
In control studies we demonstrated that the effect of Sis1 on Rnq1 aggregate localization was a specific result of Sis1 action on Rnq1-GFP. The Hsp70 Ssa1 as well as Hsp104 play important roles in [RNQ+] prion propagation (Sondheimer and Lindquist, 2000; Schwimmer and Masison, 2002) but do not suppress Rnq1 toxicity (Douglas et al., 2008). Accordingly, overexpression of either of these chaperones did not detectably alter the subcellular distribution or expression level of Rnq1-GFP (Figure 1, C and D). In addition, elevating Sis1 levels did not have a pleiotropic effect on nucleocytoplasmic traffic. Sis1 overexpression did not alter the cellular location of GFP (Figure 1C, right column), Sup35-GFP (Supplemental Figure 2, A–C) (Summers et al., 2008), or a GFP reporter of nucleocytoplasmic transport that contains both nuclear localization signal (NLS) and nuclear export signal (NES) moieties (Supplemental Figure 2D) (Stade et al., 1997). Furthermore, elevating Sis1 levels did not alter the architecture of the nuclear envelope (Figure 1B and Supplemental Figure 3).
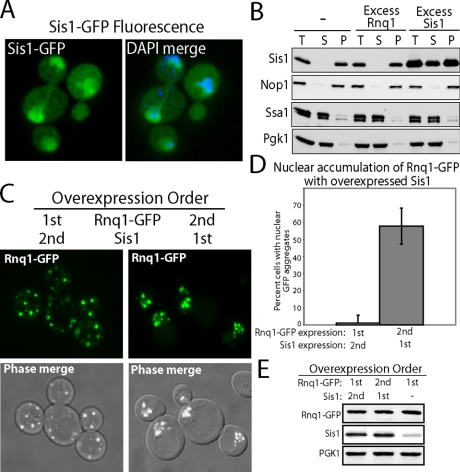
To understand how elevation of Sis1 impacts the localization of Rnq1-GFP, the cellular compartmentalization of Sis1 was examined. Consistent with previous reports (Luke et al., 1991), Sis1-GFP can support cell viability (data not shown) and almost all of it localized to the nucleus (Figure 2A). Endogenous Sis1 cofractionated with the nuclear marker Nop1 and not the cytosolic marker PGK1 or Hsp70 Ssa1 (Figure 2B). On overexpression, large pools of Sis1 were now detected in both the cytosol and nucleus (Figure 2B). Furthermore, Sis1-GFP and Rnq1-mRFP colocalized with each other in the nucleus when coexpressed together (Supplemental Figure 4).
Figure 2.
The impact of the order of Sis1 and Rnq1-GFP overexpression on Rnq1-GFP localization (A) The localization of Sis1-GFP in BY4741 [RNQ+] cells was monitored by fluorescence microscopy. Fixed cells were permeabilized and stained with DAPI in the merge. (B) Cofractionation of Sis1 with nuclei in cell extracts. Differential centrifugation was performed on native cell extracts from W303 [RNQ+] cells to determine the amount of Sis1 that could pellet with the nuclear marker Nop1 after centrifugation at 13,000 × g. Western blot analysis shows the levels of the indicated protein in the total (T), supernatant (S), or pellet (P) fractions. Sis1 was overexpressed from the constitutively active GPD promoter. Untagged Rnq1 was overexpressed from the GAL1 promoter via addition of 2% galactose to cell cultures. (C) Fluorescent images show the cellular distribution of Rnq1-GFP that was expressed from the CUP1 promoter at low, nontoxic levels due to the presence of 1 μM CuSO4 in media. Sis1 was overexpressed from the constitutively active GPD promoter. Rnq1-GFP and Sis1 overexpression constructs were individually introduced into W303 [RNQ+] cells via sequential transformations, and the order of transformation of each protein was designated as 1st or 2nd. Rnq1-GFP fluorescence was overlaid onto phase images. (D) Quantitation of Rnq1-GFP nuclear compartmentalized populations of 200 cells from three separate transformations. Values are expressed in percentage of total cells counted and the error bars reflect the SD. The p value was <0.0001. (E) Expression levels of the indicated proteins by Western blot analysis.
At this point, we also attempted to analyze the effect of modulating Sis1 levels on the subcellular localization of endogenous Rnq1. However, this was technically difficult because endogenous Rnq1 is expressed at low levels, and the antibodies that were generated against Rnq1 are insufficient for use in localization studies carried out by indirect immunofluorescence. Endogenous Rnq1 present in [RNQ+] and [rnq-] cells formed large aggregates when cell extracts were prepared in the absence of detergent for fractionation by differential centrifugation (data not shown). Thus, we were unable to determine the localization of endogenous Rnq1 or the effect of Sis1 expression on the localization of endogenous Rnq1. Nevertheless, Rnq1-GFP can propagate as a prion similar to endogenous Rnq1 (Derkatch et al., 2001). Thus, experiments conducted with Rnq1-GFP provide useful information on basic aspects of molecular chaperone action in cellular pathways for assembly of amyloid-like aggregates.
Data obtained thus far suggest that Sis1 overexpression can suppress Rnq1 toxicity via mechanisms that involve its action in both the cytosol and nucleus. Cytosolic Sis1 levels are low and seem insufficient to support efficient assembly of Rnq1-GFP into benign amyloid-like aggregates. Sis1 overexpression dramatically increases its cytosolic pool, which, in turn, may account for its ability to increase the efficient conversion of soluble Rnq1 into benign SDS-resistant aggregates and suppress Rnq1 toxicity (Douglas et al., 2008). Yet, the majority of Sis1 is localized to the nucleus. Therefore, increasing the ratio of Sis1 to Rnq1-GFP also may be protective because it depletes the cytosol of toxic Rnq1-GFP species by enhancing Rnq1-GFP aggregation in the nucleus.
The localization data shown in Figure 1, B and C, was obtained in a yeast strain that was sequentially transformed with a plasmid that expressed Sis1 from a glyceraldehyde-3-phosphate dehydrogenase promoter element (GPD) and then either Rnq1-mRFP or Rnq1-GFP expression plasmid. Because cytosolic levels of Sis1 were normally very low, we wondered whether the timing of Sis1 overexpression relative to elevation of Rnq1 levels would have an impact on the site of Rnq1-GFP aggregation. To address this question, the order of Sis1 and Rnq1-GFP expression was varied by sequentially introducing expression plasmids for these proteins in yeast in different orders (Figure 2, C and D). When Rnq1-GFP was expressed first, large Rnq1-GFP aggregates accumulated in the cytosol and subsequent Sis1 overexpression did not redistribute aggregates to the nucleus. In contrast, when Sis1 levels were elevated before Rnq1-GFP expression, the nuclear accumulation of Rnq1-GFP aggregates was clearly evident (Figure 2, C and D).
These data support the notion that Sis1 acts on both cytosolic and nuclear Rnq1-GFP aggregates. Yet, the level of Sis1 at the time when Rnq1-GFP is induced has a profound impact on the cellular location of Rnq1-GFP aggregation. When pools of Rnq1-GFP are present in the cytosol before elevation of Sis1, Sis1 enhances the assembly of native Rnq1 into pre-existing aggregates that seem too large to enter the nucleus. Yet, when Sis1 levels are elevated at the time of induced Rnq1-GFP expression, formation of nuclear Rnq1-GFP aggregates predominates in ∼50% of cells examined. Thus, changes in the ratio of Sis1 to Rnq1-GFP can have dramatic effects on whether amyloid-like aggregate assembly occurs within the cytosol or the nucleus. Regarding Rnq1-GFP, such chaperone-dependent changes in localization seem to be part of the cellular mechanism for suppression of its proteotoxicity.
The Nuclear Environment Favors [RNQ+]-dependent Rnq1-GFP Aggregation
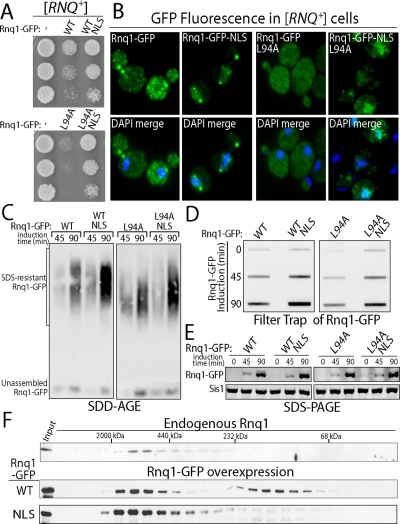
We suggest that Sis1-dependent relocalization of Rnq1-GFP aggregates to the nucleus is protective against Rnq1-GFP toxicity. However, Sis1 overexpression dramatically increases cytosolic pools of Sis1 and Sis1 can suppress Rnq1-GFP toxicity under conditions where Rnq1-GFP aggregation predominates in the cytosol. Thus, we sought to directly test whether localization of Rnq1-GFP to the nucleus protects against Rnq1-toxicity in absence of Sis1 overexpression. We examined the toxicity of Rnq1-GFP and Rnq1-GFP L94A that were tagged with an NLS from simian virus-40 (Kalderon et al., 1984). The Rnq1 L94A allele contains a point mutation in the Sis1-binding site in the nonprion domain of Rnq1 and is highly toxic because its conversion into amyloid-like Rnq1 aggregates is inefficient (Douglas et al., 2008). Rnq1-GFP-NLS and Rnq1-GFP-NLS L94A were dramatically less toxic than forms without the NLS tag (Figure 3A). Decreased toxicity of NLS-tagged Rnq1-GFP and Rnq1-GFP L94A correlated with their accumulation in the nucleus (Figure 3B). It is interesting that filter trap and SDD-AGE analysis demonstrated that SDS-resistant pools of NLS-tagged forms of Rnq1-GFP and Rnq1-GFP L94A accumulated to 1.5-fold higher levels than the non–NLS-tagged forms, but their expression levels were similar at all time points tested (Figure 3, C–E).
Figure 3.
Rnq1-GFP-NLS is less toxic than Rnq1-GFP in W303 [RNQ+] cells. (A) Analysis of toxicity in fivefold serial dilutions of W303 [RNQ+] cells grown on plates supplemented with galactose to induce Rnq1 expression from the GAL1 promoter. (B) Localization of Rnq1-GFP-NLS and Rnq1-GFP via fluorescence microscopy. Fixed cells were permeabilized and stained with DAPI in the merge. (C) SDD-AGE analysis of the time course for assembly of SDS-resistant Rnq1-GFP aggregates after induction of Rnq1-GFP expression with 2% galactose. (D) Filter trap analysis of Rnq1-GFP conversion to an SDS-resistant species at the indicated time after induced expression. Rnq1-GFP retained on the cellulose acetate filter was detected with α-GFP. In quantitations, Rnq1-GFP-NLS band intensity was compared with the Rnq1-GFP signal at the 90-min time point. (E) Time course of Rnq1-GFP expression determined by Western blot. (F) Populations of exogenous Rnq1-GFP and endogenous Rnq1 species were determined by size exclusion chromatography 4 h after the induced expression of either Rnq1-GFP or Rnq1-GFP-NLS via supplementation of media with 2% galactose. Input corresponds to 10% of the total lysate protein loaded. Quantitation of signal intensity in each peak was compared with the total signal from the sum of all fractions.
Toxicity caused by Rnq1-GFP overexpression correlates with the accumulation of a low-molecular-weight pool of Rnq1-GFP that migrates on gel filtration columns with an apparent mobility of approximately of 175–200 kDa (Douglas et al., 2008). The exact nature of this unassembled pool of Rnq1 is not clear; yet, its accumulation is a marker for Rnq1 toxicity. We took advantage of this occurrence as a tool to monitor the impact of the nuclear environment on Rnq1-GFP assembly and toxicity. Under toxic conditions, ∼55% of total Rnq1-GFP migrated as a high-molecular-weight species and the other 45% behaved as a lower molecular weight species. In contrast, almost all Rnq1-GFP-NLS behaved as a high-molecular-weight species and unassembled pools of Rnq1-GFP-NLS were difficult to detect (Figure 3F).
The dramatic decrease in unassembled Rnq1 pools correlates well with the reduced toxicity of Rnq1-GFP-NLS. This effect does not seem related to degradation of Rnq1 because levels of total Rnq1-GFP and Rnq1-GFP-NLS are similar (Figure 3E). When Rnq1-GFP accumulates in the nucleus the decrease in unassembled pools of Rnq1 seems to occur in response to the observed 1.5-fold increase in the accumulation of SDS-resistant Rnq1-GFP aggregates (Figure 3, C–E). Rnq1-GFP aggregates were shown in previous studies to bind thioflavin-T and thus seem amyloid like (Douglas et al., 2008). These data suggest that conversion of Rnq1-GFP into SDS-resistant amyloid-like aggregates is more efficient in the nucleus than cytosol. Thus, accumulation of Rnq1-GFP in the nuclear environment, via a mechanism that is independent of Sis1 overexpression, enables yeast to tolerate Rnq1-GFP expression.
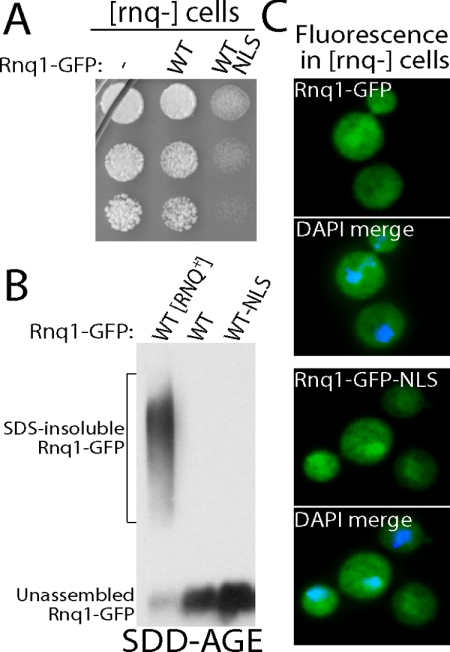
Rnq1-GFP-NLS Is Toxic in the [rnq-] Cells
We interpret our data to suggest that the nuclear localized Rnq1 is less toxic to cells because the nuclear environment is more efficient than the cytosol at assembling Rnq1-GFP into benign aggregates. Yet, the depletion of soluble cytosolic Rnq1-GFP via its nuclear accumulation could be protective in the absence of Rnq1-GFP aggregation. Hence, we examined the effect of Rnq1-GFP-NLS overexpression on growth of [rnq-] cells where its aggregation does not occur. Rnq1-GFP-NLS, but not Rnq1-GFP, was toxic to [rnq-] cells (Figure 4A). Rnq1-GFP overexpression is not toxic in [rnq-] cells because [RNQ+] prions are required to template the formation of toxic Rnq1 species (Douglas et al., 2008). Thus, we were surprised to observe Rnq1-GFP-NLS exhibit a toxic gain of function in [rnq-] cells. Rnq1-GFP-NLS toxicity in [rnq-] cells could not be rescued by Sis1 overexpression (data not shown), and Rnq1-GFP-NLS was not observed to form SDS-resistant aggregates (Figure 4B). Identical results were obtained with Rnq1-GFP-NLS L94A (data not shown). Rnq1-GFP-NLS was found in both the nucleus and cytosol of [rnq-] cells (Figure 4C), so its sequestration to the nucleus seems to be coupled to its [RNQ+] prion-dependent aggregation. At present, it is not clear why Rnq1-GFP-NLS, but not Rnq1-GFP, is toxic to [rnq-] cells. The observed toxicity may result from an increase in the pool size of unassembled Rnq1-GFP in the nucleus. Nevertheless, efficient [RNQ+] prion-dependent assembly of Rnq1-GFP into an aggregated species seems coupled to depletion of Rnq1-GFP from the cytosol. We suggest that these coupled events represent a pathway for Rnq1-GFP detoxification.
Figure 4.
Targeting Rnq1-GFP to the nucleus is toxic in W303 [rnq-] cells. (A) Fivefold serial dilutions of W303 [rnq-] cells harboring the indicated Rnq1-GFP proteins were grown on agarose plates containing galactose. (B) The ability of different Rnq1-GFP proteins to assemble into SDS-resistant aggregates was monitored in [rnq-] cells by SDD-AGE analysis. The left lane shows Rnq1-GFP from W303 [RNQ+] cell extracts. (C) Fluorescent distribution of different Rnq1-GFP proteins whose expression was driven by 2% galactose for 4 h in [rnq-] cells. Fixed cells were permeabilized and stained with DAPI in the merge image.
Nuclear Rnq1-GFP Aggregates Act In Trans to Detoxify Rnq1 L94A
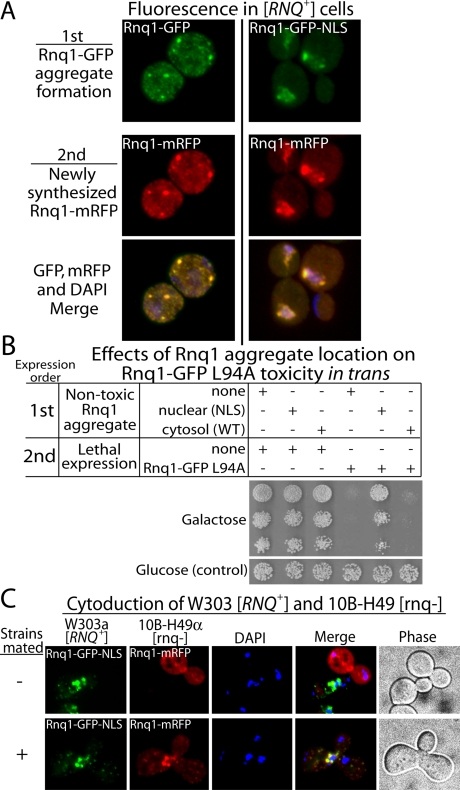
Rnq1 aggregation seems to prevent the accumulation of toxic Rnq1 conformers in the cytosol by trapping soluble Rnq1-GFP in the nucleus. If this is true, then the location of pre-existing Rnq1 aggregate pools should dictate the site where soluble Rnq1 is converted into [RNQ+] prion-dependent aggregates. To test this prediction, we asked whether an NLS-tagged pool of Rnq1-GFP aggregates could act in trans to concentrate nascent Rnq1 in the nucleus and thereby suppress Rnq1 toxicity (Figure 5). Rnq1-GFP-NLS or Rnq1-GFP under control of the CUP1 promoter was first expressed at low nontoxic levels, which allowed for the assembly of GFP-tagged Rnq1 into either nuclear or cytosolic aggregates, respectively (Figure 5A). Different forms of Rnq-GFP were expressed for 15 h to allow for distinct compartmentalization within the cytosol or nucleus, then expression of Rnq1-mRFP under control of the GAL1 promoter was induced for 1 h and fluorescence of the different tagged proteins was visualized. The subcellular location of Rnq1-mRFP was dependent upon the localization of preformed GFP-tagged Rnq1 aggregates (Figure 5A). Nearly the entire pool of newly synthesized Rnq1-mRFP became colocalized with pools of nuclear Rnq1 aggregates that accumulated during the prior expression of Rnq1-GFP-NLS (Figure 5A). It is important that the severe growth defects caused by GAL1-dependent Rnq1-GFP L94A overexpression were suppressed through prior low-level expression of Rnq1-GFP-NLS (Figure 5B). Therefore, [RNQ+] prion-dependent Rnq1-GFP aggregates can act in trans to trap Rnq1-mRFP in the nucleus, and this event is coupled to suppression of Rnq1 toxicity.
Figure 5.
Expression of Rnq1-GFP-NLS promotes the nuclear accumulation of Rnq1-mRFP and suppresses Rnq1 L94A toxicity in W303 [RNQ+] cells. (A) Prior expression of Rnq1-GFP-NLS leads to the accumulation of Rnq1-mRFP in the nucleus. Rnq1-GFP and Rnq1-GFP-NLS were expressed for 15 h at low, nontoxic levels from the CUP1 promoter due to the presence of 1 μM CuSO4 in media. Subsequent expression of Rnq1-mRFP from the GAL1 promoter was induced by the addition of 2% galactose. After 1 h, the location of the GFP and mRFP signals was determined in fixed, DAPI-stained cells by fluorescence microscopy. (B) Analysis of cell growth in fivefold serial dilutions of W303 [RNQ+] cells that harbored the indicated constructs. Cells were grown overnight in synthetic media containing 1 μM CuSO4 to allow for Rnq1-GFP or Rnq1-GFP-NLS expression from the CUP1 promoter. Cells were then plated on agar that contained glucose (the control) or galactose to induce Rnq1-GFP L94A expression to toxic levels. (C) Rnq1-GFP-NLS in the nucleus of cytoduced W303a [RNQ+] cells sequestered Rnq1-mRFP from the cytosol of 10B-H49α [rnq-] cells. Rnq1-GFP-NLS was expressed from the GAL1 promoter and Rnq1-mRFP expression was under control of the CUP1 promoter. Two hours before mixture of different mating types, W303 and 10B-H49 cell cultures harboring the indicated Rnq1 constructs, were supplemented with 2% galactose and 50 μM CuSO4, respectively. Cell fusion was analyzed 30 min (top row, control) and 2 h (bottom row) after cell mixture. Merge images represent the combination of all fluorescent channels.
To further demonstrate the concept that pre-existing pools of amyloid-like assemblies can trap native amyloidogenic proteins in a specific location, we conducted cytoduction experiments (Figure 5C). Cytoduction refers to an experimental situation where haploid yeast initiate the mating process, fuse their plasma membranes, and mix cytosolic contents, but fail to undergo karyogamy because one of the tester stains is defective in nuclear fusion (Conde and Fink, 1976). This scenario allowed us to monitor the fate of soluble Rnq1-mRFP in the cytosol of [rnq-] cells that was introduced into [RNQ+] cells, which harbored pre-existing nuclear Rnq1 aggregates formed by Rnq1-GFP-NLS (Figure 5C). GFP-tagged Rnq1 aggregates present in the nucleus of [RNQ+] cells depleted the cytosol of [rnq-] cells of soluble Rnq1-mRFP and drove the assembly of a hybrid aggregate that contained both GFP and mRFP. Thus, nuclear Rnq1 aggregates do indeed trap soluble Rnq1 in the nucleus by driving its fusion into large assemblies whose size constraints prevent export back to the cytosol. In this situation, the nuclear envelope acts as a sieve that helps suppress Rnq1 toxicity by sequestering nascent Rnq1 in the nucleus.
Nuclear Rnq1 Aggregates Concentrate Htt-103Q in the Nucleus and Exacerbate Htt Toxicity
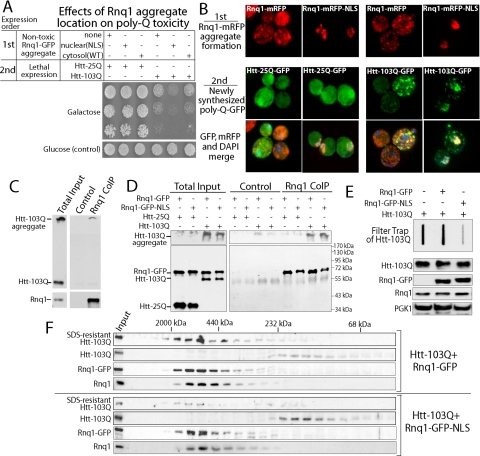
[RNQ+] prions promote Htt-103Q toxicity in yeast (Meriin et al., 2002) and movement of polyQ-expanded Htt protein from the cytosol to the nucleus of neurons and yeast correlates with exacerbated toxicity (DiFiglia et al., 1997; Schaffar et al., 2004). There is also evidence to suggest that intermolecular interaction between Htt-103Q and environmental factors can impact its subcellular localization (Wang et al., 2009). Thus, we examined whether interactions with nuclear Rnq1 aggregates could impact Htt-103Q localization and toxicity (Figure 6). To accomplish this, the effect that expression of low, nontoxic levels of Rnq1-GFP or Rnq1-GFP-NLS had on Htt-103Q toxicity was determined. These different forms of Rnq1 were expressed first for 15 h from the CUP1 promoter at low nontoxic levels. Then expression of Htt-25Q and Htt-103Q was induced from the GAL1 promoter and the influence of Rnq1 aggregate location on Htt toxicity was evaluated (Figure 6A). Growth of yeast was not impacted by expression of Htt-25Q regardless of whether Rnq1-GFP aggregates were predominantly localized in the cytosol or nucleus. Yet, in dramatic contrast to what was observed with Rnq1 toxicity, the presence of Rnq1-GFP aggregates in the nucleus exacerbated Htt-103Q toxicity. The effect of Rnq1 localization on Htt toxicity was dependent upon the presence of the [RNQ+] prion conformer because Htt-103Q expression was not toxic in [rnq-] cells regardless of whether low nontoxic levels of Rnq1-GFP or Rnq1-GFP-NLS was present (Supplemental Figure 5).
Figure 6.
The presence of Rnq1-GFP aggregates in the nucleus of W303 [RNQ+] cells exacerbates Htt-103Q toxicity. (A) Analysis of cell growth in fivefold serial dilutions of [RNQ+] cells, which harbored the indicated constructs. Rnq1-GFP and Rnq1-GFP-NLS were first expressed for 15 h at nontoxic levels from a CUP1 promoter due to the presence of 1 μM CuSO4 in the culture. Then, Htt-25Q-GFP and Htt-103Q-GFP were expressed at toxic levels from the GAL1 promoter on agarose plates containing 2% galactose. (B) The impact of Rnq1-mRFP-NLS expression on the cellular localization of Htt-25Q-GFP and Htt-103Q-GFP. Rnq1-mRFP or Rnq1-mRFP-NLS was expressed for 15 h at low nontoxic levels from the CUP1 promoter due to the presence of 10 μM CuSO4 in media. Subsequent expression of Htt-25Q-GFP and Htt-103Q-GFP from the GAL1 promoter was induced by the addition of 2% galactose. After 1 h, the location of the GFP and mRFP signals was determined in fixed, DAPI-stained cells by fluorescence microscopy. (C) Coprecipitation of endogenous Rnq1 and Htt-103Q-GFP in complex with each other. Extracts were generated from W303 [RNQ+] cells expressing Htt-103Q-GFP and endogenous Rnq1 was precipitated under native conditions with α-Rnq1. Precipitates were resolved by SDS-PAGE and Western blots were probed with α-GFP or α-Rnq1 to detect Htt-103Q-GFP or endogenous Rnq1, respectively. (D) Coprecipitation of Htt-103Q-GFP in complex with Rnq1-GFP and Rnq1-GFP-NLS. Nondenaturing cell extracts were generated from W303 [RNQ+] cells. Rnq1-GFP and Rnq1-GFP-NLS were precipitated by the addition α-Rnq1 and Western blots were probed with α-GFP (bottom) or α-FLAG (top) to detect the different forms of Rnq1 and Htt. (E) Filter trap analysis of SDS-resistant Htt-103Q aggregates. Htt-103Q retained on the cellulose acetate filter was detected with α-FLAG. Bottom panels are western blots of cell extracts used in the filter trap assay. In quantitations, Htt-103Q band intensity from cells expressing Rnq1-GFP and Rnq1-GFP-NLS was compared with the Htt-103Q signal in the absence of exogenous Rnq1. (F) Analysis of Htt-103Q assembly status in W303 [RNQ+] cells. Rnq1-GFP or Rnq1-GFP-NLS under control of the CUP1 promoter was expressed for 15 h at low nontoxic levels by the presence of 1 μM CuSO4 in cell cultures. Then Htt-103Q was expressed for 2 h by the addition of 2% galactose. Proteins in cell extracts were resolved by size exclusion chromatography. Top gels in each set of panels show the quantity of SDS-resistant Htt-103Q that was incapable of gel entry. The second set of panels from the top shows Htt-103Q-GFP which was able to migrate into gels. The third set of panels from the top shows the mobility of either Rnq1-GFP or Rnq1-GFP-NLS. The bottom set of panels shows the mobility of endogenous Rnq1. Input represents 10% of the total amount of protein detected in extracts. Quantitation of Htt-103Q signal intensity in each peak was compared with the total signal from the sum of all fractions.
To explain the increase in Htt toxicity observed in the presence of nuclear Rnq1 aggregates, we asked whether the cellular location of the Rnq1 aggregates controlled the location of Htt-103Q (Figure 6B). Rnq1-mRFP or Rnq1-mRFP-NLS were expressed for 15 h at low nontoxic levels from the CUP1 promoter to permit the formation of respective cytosolic and nuclear pools of Rnq1-mRFP aggregates. Then, expression of Htt-25Q-GFP or Htt-103Q-GFP from the GAL1 promoter was induced for 1 h and cells were visualized. The site of Rnq1-mRFP localization had no effect on the solubility or distribution of Htt-25Q-GFP (Figure 6B, left columns). Conversely, nuclear Rnq1 aggregates colocalized with Htt-103Q-GFP and seemed to sequester Htt-103Q-GFP in the nucleus (Figure 6B, right columns). Thus, [RNQ+] prion dependent Rnq1-GFP aggregates seem to control the cellular localization Htt-103Q.
To evaluate the specificity of Rnq1's ability to relocate Htt-103Q to the nucleus, we examined the ability of Rnq1-mRFP aggregates to alter the localization of a different interaction partner Het-s. Het-s is a Podospora anserina prion that lacks a polyQ stretch and forms amyloid-like aggregates in a [RNQ+] prion-dependent manner when expressed in yeast. (Taneja et al., 2007). Het-s-GFP was localized throughout the cell in the presence or absence of Rnq1-GFP-NLS, and we did not observe colocalization between Rnq1-mRFP-NLS and Het-s-GFP (Supplemental Figure 6).
To determine whether a physical interaction between Rnq1 and Htt-103Q occurs, we analyzed whether they could form an immunoprecipitable complex. First, endogenous Rnq1 and Htt-103Q-GFP present in [RNQ+] cell extracts was demonstrated to coprecipitate (Figure 6C). SDS-soluble forms of Htt-103Q-GFP protein that are capable of migrating into the SDS-PAGE gels were not coprecipitated with Rnq1. Yet, the SDS-insoluble Htt-103Q aggregates, which were incapable of entry into the SDS-PAGE gel, precipitated with Rnq1 and could be observed as immunoreactive bands at the top of Western blots. Thus, endogenous Rnq1 and Htt103Q are present in a complex with each other.
We were also able to detect exogenous Rnq1-GFP and Rnq1-GFP-NLS in coimmunoprecipitates with Htt-103Q aggregates (Figure 6D). The form of Htt-103Q present in complex with Rnq1-GFP and Rnq1-GFP-NLS was an aggregated SDS-resistant forms that could not enter SDS-PAGE gels. Thus, it seems the Rnq1-GFP and Htt-103Q form a hetero-oligomeric complex, and the subcellular location of Rnq1 helps control where such a complex accumulates.
To explore the mechanism for exacerbation of Htt-103Q toxicity, we investigated how nuclear Rnq1 aggregates affected the assembly status of Htt-103Q (Figure 6E). Accumulation of exogenous Rnq1-GFP aggregates in the cytosol had no significant effect on the assembly of Htt-103Q into an SDS-resistant species. Yet, when nuclear Rnq1-GFP aggregates were present, the conversion of Htt-103Q into tight SDS-resistant aggregates was reduced by 80% (Figure 6E). It should be noted that under these same conditions, Htt-103Q expression had no effect on the assembly of endogenous Rnq1 into high-molecular-weight aggregates (Figure 6F, bottom).
The accumulation of a small oligomeric form of polyQ-expanded Htt in the size range of a dimer or trimer is linked to Htt toxicity in yeast (Behrends et al., 2006). Thus, we asked whether decreased formation of SDS-resistant Htt-103Q assemblies observed in the presence of nuclear Rnq1 aggregates was associated with the accumulation of similar detergent-soluble Htt-103Q oligomers (Figure 6F). Under control conditions, 70% of Htt-103Q in cell extracts migrated on a size exclusion column as a high-molecular-weight species that was SDS-resistant and could not enter into SDS-PAGE gels. The other 30% of Htt-103Q detected behaved as an SDS-sensitive form that could enter into SDS-PAGE gels and showed up as a small oligomer. Yet, the presence of nuclear Rnq1 aggregates dramatically reduced the formation of high molecular weight Htt-103Q assemblies. This was accompanied by a corresponding increase in the accumulation of SDS-sensitive oligomeric Htt-103Q. Therefore, increased potency of Htt toxicity observed in the presence of exogenous nuclear Rnq1 aggregates is associated with a dramatic decrease in the conversion of Htt-103Q into an SDS-resistant species and the accumulation of small Htt-103Q oligomers.
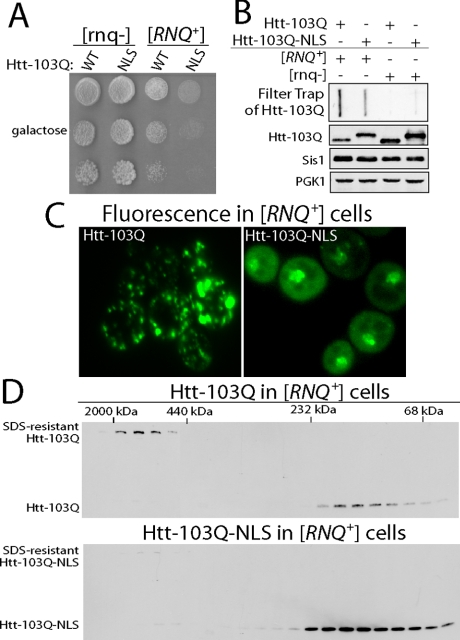
At this point, we sought to evaluate whether interaction with [RNQ+] prion-dependent Rnq1 aggregates or the relocation of Htt-103Q to the nucleus was the major factor that decreased the cells capacity to detoxify Htt-103Q. To approach this question, an NLS was fused to the C terminus of the GFP moiety on Htt-103Q-GFP and the ability of Htt-103Q-GFP-NLS to kill yeast and form SDS-resistant aggregates was determined (Figure 7, A and B). Similar to Htt-103Q, Htt-103Q-GFP-NLS was not toxic to [rnq-] cells but was dramatically more toxic to [RNQ+] cells than Htt-103Q-GFP. Increased toxicity of Htt-103Q-GFP-NLS correlated with a 60% decrease in the formation of SDS-resistant aggregates. This is interesting because Htt-103Q-GFP does not form SDS-resistant aggregates in [rnq-] cells and it is not toxic (Figure 7B).
Figure 7.
Htt-103Q-GFP-NLS forms SDS-resistant aggregates with reduced efficiency and is dramatically more toxic than Htt-103Q-GFP in W303 [RNQ+] cells. (A) Analysis of cell growth in fivefold serial dilutions of W303 [RNQ+] and [rnq-] cells harboring the indicated forms of Htt-103Q that were expressed from the GAL1 promoter. (B) Filter trap analysis of SDS-resistant Htt-103Q and Htt-103Q-GFP-NLS aggregates. Htt-103Q chimeras were overexpressed for 4 h in W303 [RNQ+] and [rnq-] cells. Bottom panels are Western blots of cell extracts used in the filter trap assay. In quantitations, Htt-103Q-NLS band intensity was compared with the Htt-103Q signal from [RNQ+] cell extracts. (C) Cellular localization of Htt-103Q-GFP and Htt-103Q-GFP-NLS in W303 [RNQ+] cells as determined by fluorescence microscopy. The expression of Htt fusion proteins was driven by the addition of 2% galactose for 4 h before fluorescent images were obtained. (D) Comparison of Htt-103Q-GFP and Htt-103Q-GFP-NLS assembly status in W303 [RNQ+] cells by size exclusion chromatography. Htt-103Q chimeras were expressed for 2 h in cells harboring only endogenous Rnq1.
Altering the extent to which Htt-103Q-GFP-NLS forms SDS-resistant aggregates correlated well with its accumulation in the nucleus (Figure 7C). Yet, the aggregation pattern of Htt-103Q-GFP and Htt-103Q-GFP-NLS was different. Htt-103Q-GFP formed large foci that were localized throughout [RNQ+] cells. In contrast, Htt-103Q-GFP-NLS was predominantly nuclear and its cytosolic forms did not form distinct foci.
The accumulation of Htt-103Q-GFP-NLS in the nucleus and its inefficient conversion to an SDS-resistant aggregate was accompanied by a significant increase in the amount of detergent-soluble low-molecular-weight Htt-103Q that was detected by size-exclusion chromatography (Figure 7D). These results are very similar to those observed when Htt-103Q-GFP is attracted the nucleus via interaction with Rnq1-GFP-NLS. Thus, conversion of Htt-103Q-GFP into an SDS-resistant species is less efficient in the nuclear environment, and this effect correlates with enhanced toxicity. Furthermore, nuclear Rnq1-GFP can act to exacerbate Htt-103Q-GFP toxicity by attracting it to an environment that has reduced capacity for it detoxification.
DISCUSSION
Alteration in the cellular localization of disease proteins is associated with the onset of cell death in protein aggregation diseases (Davies et al., 1997; Ross, 1997; Neumann et al., 2006). Yet, mechanisms that mediate such changes in cellular localization and the reasons for the compartment specific proteotoxicity are not clear. A large pool of the yeast Hsp40 Sis1 is localized to the nucleus, and we found that subtle increases in Sis1 activity shifted the localization of the aggregation pathway for the model disease protein Rnq1-GFP from the cytosol to the nucleus. This relocation correlated with reduced toxicity and occurred when elevation of chaperone expression preceded elevation of disease protein levels. Increased Sis1 expression did not cause a general perturbation of nucleocytoplasmic transport. Instead, increased Hsp40 activity seemed to promote assembly of nascent Rnq1-GFP into nuclear Rnq1 aggregates, which depleted the cytosol of soluble species. Thus, it is possible that well documented age-dependent changes in the activity of chaperone networks (Morimoto, 2008) influence proteotoxicty by altering the cellular location where disease protein aggregation occurs.
Mutant disease proteins that contain expanded polyQ tracts are the cause of at least nine protein-misfolding diseases (Zoghbi and Orr, 2000; Ross, 2002). Yet, numerous functional proteins also contain short polyQ tracts. PolyQ-expanded Htt is proposed to cause cell death via sequestration of essential proteins that contain short polyQ tracts into aggregates and inactivation of such proteins through altering their conformation (Burke et al., 1996; Steffan et al., 2000; Dunah et al., 2002). Yet, polyQ aggregation is also proposed to be a protective mechanism. Aggregation, toxicity, or both of polyQ-expanded Htt is often triggered through interaction with neighboring proteins (Perez et al., 1998; Duennwald et al., 2006). The presence of amyloid-like [RNQ+] prions is required for Htt toxicity in yeast (Meriin et al., 2002), and we found that the cellular location of Rnq1-GFP aggregates strongly influenced Htt-103Q-GFP localization and toxicity. Rnq1-GFP-NLS aggregates concentrated Htt-103Q-GFP to the nucleus, hindered conversion of Htt-103Q-GFP into SDS-resistant aggregates, and increased Htt toxicity. Thus, changes in the localization of an intracellular interaction partner can influence the cellular compartment where Htt-103Q accumulates. When Htt-103Q accumulates in the nucleus, it seems more toxic because its conversion into a tightly aggregated SDS-resistant species is less efficient. One reason why changes in localization can impact toxicity is that components in the cytosol and nucleus differentially package distinct disease proteins into benign states.
Several factors may explain the apparent changes in the extent of Rnq1 and Htt-103Q aggregation when this process occurs in the nucleus instead of the cytosol. Sis1 is required for efficient assembly of [RNQ+] prions (Douglas et al., 2008). It also acts in conjunction with Hsp104 to fragment [RNQ+] prions into smaller seeds required for propagation (Aron et al., 2007; Tipton et al., 2008). The presence of [RNQ+] prion seeds is required for Rnq1 toxicity (Douglas et al., 2008). We found that the majority of endogenous Sis1 is localized to the nucleus, whereas Hsp104 is present in both the nucleus and cytosol (Tkach and Glover, 2008). Thus, movement of Rnq1-GFP to the nucleus may favor the accumulation of benign Rnq1-GFP aggregates because the level of Sis1 and Hsp104 are optimal for Rnq1-GFP aggregate formation in this location.
Compartmental-specific differences in chaperone levels also may provide an explanation for why formation of SDS-resistant Htt aggregates is disfavored in the nucleus. TriC is a ringed chaperonin required for polyQ detoxification (Behrends et al., 2006; Kitamura et al., 2006; Tam et al., 2006) that is localized primarily in the cytosol (Kim et al., 1994). Accumulation of Htt-103Q in the nucleus may therefore partition it away from TriC and limit its conversion into a benign aggregate. It is also possible that encounters between Htt-103Q and new nuclear neighbors interferes with their function or hinders Htt-103Q detoxification (Duennwald et al., 2006).
Sis1 can suppress Rnq1 toxicity when its levels are elevated before Rnq1-GFP expression or when Rnq1-GFP and Sis1 are coexpressed simultaneously. In all cases, suppression of Rnq1 toxicity is associated with reductions in soluble pools of Rnq1-GFP and accumulation of SDS-resistant Rnq1-GFP aggregates. Yet, change in the localization of Rnq1-GFP aggregates is observed when Sis1 levels were elevated before expression of Rnq1-GFP. Rnq1-GFP-NLS was dramatically less toxic and nuclear Rnq1-GFP-NLS aggregates formed in [RNQ+] cells could act in trans to sequester Rnq1-GFP in the nucleus. Thus, elevation of Sis1 is likely to shift the site of Rnq1-GFP aggregation to the nucleus because it promotes fusion of nascent Rnq1 to nuclear Rnq1 aggregates. This event seems to occur in ∼50% of cells examined. However, for technical reasons, we were unable to monitor the effect of Sis1 overexpression on the cellular location of endogenous Rnq1. Nevertheless, it is clear that changes in Sis1 levels alter Rnq1-GFP localization and studies with Rnq1-GFP-NLS demonstrate that accumulation of Rnq1-GFP in the nucleus is protective.
Rnq1-GFP is detected in both the cytosol and nucleus of [rnq-] and [RNQ+] cells. In addition, Rnq1-GFP-NLS is not exclusively localized to the nucleus of [rnq-] cells. Thus, we surmise that Rnq1-GFP normally traffics in and out of the nucleus. Increasing nuclear Sis1 may therefore trap Rnq1-GFP in the nucleus by facilitating fusion of Rnq1-GFP monomers into nuclear Rnq1 aggregates. Yet, type II Hsp40s are known to help retain proteins in the nucleus (Cheng et al., 2008; Zhang et al., 2008), so it also possible that Sis1 has a direct impact on the nucleocytoplasmic trafficking of Rnq1. Indeed, the nonprion domain of Rnq1 contains a hydrophobic Sis1 binding site (Douglas et al., 2008) that closely resembles a canonical nuclear export signal. Sis1 binding to Rnq1 may therefore negatively regulate Rnq1 nuclear export and thereby increase nuclear Rnq1 levels. However, in [rnq-] cells, where Rnq1-GFP aggregation is not observed, Sis1 overexpression does not enhance Rnq1-GFP accumulation in the nucleus. We attempted to test the hypothesis that Sis1 may regulate nuclear export of Rnq1 but had difficulty because mutation of the putative NES within Rnq1 hindered Sis1 binding and drove Rnq1-GFP to aggregate in the cytosol (data not shown). Regardless of the mechanism, it is clear that Hsp40s modulate the cellular location of where disease proteins are assembled into aggregates and this can impact their toxicity.
Native Rnq1 is normally benign in the cytosol of [rnq-] cells and interactions with [RNQ+] prions are required to promote toxicity (Douglas et al., 2008). Yet, Rnq1-GFP-NLS is toxic to [rnq-] cells. In [rnq-] cells, Rnq1-GFP-NLS is not observed to self-associate when in the cytosol or the nucleus. Therefore, an increased propensity to aggregate does not seem related to the observed toxicity. The mechanism for Rnq1-GFP-NLS toxicity in [rnq-] cell is not clear. Yet, it is possible that the nuclear environment contains interaction partners that are either inactivated by Rnq1-GFP-NLS or are capable of templating Rnq1-GFP-NLS into an alternate toxic conformation (Duennwald et al., 2006). An important aspect of these data are that they further demonstrate that [RNQ+]-dependent conversion of Rnq1-GFP-NLS into an amyloid-like aggregate is essential for detoxification. However, it remains obscure as to why Rnq1-GFP is benign when in the cytosol but toxic when accumulation in the nucleus of [rnq-] cells is favored. Future studies will focus on identification of differences in the networks of chaperones and interacting factors that reciprocally buffer Rnq1 and Htt toxicity in the cytosol and nucleus.
Supplementary Material
ACKNOWLEDGMENTS
We thank Susan Lindquist and Sebastian Treusch for helpful discussions during early stages of this study. We also thank Karsten Weis, Kerry Bloom, Susan Lindquist, Takashi Toda, Susan Liebman, and Michael Sherman for gifts of reagents. We thank Michael and Sarah Douglas for critical reading of the manuscript. This work was supported, in whole or in part, by a predoctoral fellowship from the American Heart Association (to P.M.D.), National Institutes of Health predoctoral training grant 5T32GM008581-09 (to D.W.S.), and National Institutes of Health grant 5R01GM067785 (to D.M.C.).
Abbreviations used:
- CUP1
copper-inducible promoter element
- GAL1
galactose-inducible promoter element
- GPD
glyceraldehyde-3-phosphate dehydrogenase promoter element
- Htt
Huntingtin
- mRFP
monomeric red fluorescent protein
- NES
nuclear export signal
- NLS
nuclear localization signal
- SDD-AGE
semidenaturing detergent agarose gel electrophoresis.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E09-02-0170) on August 5, 2009.
REFERENCES
- Aron R., Higurashi T., Sahi C., Craig E. A. J-protein co-chaperone Sis1 required for generation of [RNQ+] seeds necessary for prion propagation. EMBO J. 2007;26:3794–3803. doi: 10.1038/sj.emboj.7601811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balch W. E., Morimoto R. I., Dillin A., Kelly J. W. Adapting proteostasis for disease intervention. Science. 2008;319:916–919. doi: 10.1126/science.1141448. [DOI] [PubMed] [Google Scholar]
- Behrends C., et al. Chaperonin TRiC promotes the assembly of polyQ expansion proteins into nontoxic oligomers. Mol. Cell. 2006;23:887–897. doi: 10.1016/j.molcel.2006.08.017. [DOI] [PubMed] [Google Scholar]
- Bence N. F., Sampat R. M., Kopito R. R. Impairment of the ubiquitin-proteasome system by protein aggregation. Science. 2001;292:1552–1555. doi: 10.1126/science.292.5521.1552. [DOI] [PubMed] [Google Scholar]
- Burke J. R., Enghild J. J., Martin M. E., Jou Y. S., Myers R. M., Roses A. D., Vance J. M., Strittmatter W. J. Huntingtin and DRPLA proteins selectively interact with the enzyme GAPDH. Nat. Med. 1996;2:347–350. doi: 10.1038/nm0396-347. [DOI] [PubMed] [Google Scholar]
- Carrell R. W., Lomas D. A. Conformational disease. Lancet. 1997;350:134–138. doi: 10.1016/S0140-6736(97)02073-4. [DOI] [PubMed] [Google Scholar]
- Caughey B., Lansbury P. T. Protofibrils, pores, fibrils, and neurodegeneration: separating the responsible protein aggregates from the innocent bystanders. Annu. Rev. Neurosci. 2003;26:267–298. doi: 10.1146/annurev.neuro.26.010302.081142. [DOI] [PubMed] [Google Scholar]
- Cheng I. H., et al. Accelerating amyloid-beta fibrillization reduces oligomer levels and functional deficits in Alzheimer disease mouse models. J. Biol. Chem. 2007;282:23818–23828. doi: 10.1074/jbc.M701078200. [DOI] [PubMed] [Google Scholar]
- Cheng X., Belshan M., Ratner L. Hsp40 facilitates nuclear import of the human immunodeficiency virus type 2 Vpx-mediated preintegration complex. J. Virol. 2008;82:1229–1237. doi: 10.1128/JVI.00540-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiti F., Dobson C. M. Protein misfolding, functional amyloid, and human disease. Annu. Rev. Biochem. 2006;75:333–366. doi: 10.1146/annurev.biochem.75.101304.123901. [DOI] [PubMed] [Google Scholar]
- Cohen E., Bieschke J., Perciavalle R. M., Kelly J. W., Dillin A. Opposing activities protect against age-onset proteotoxicity. Science. 2006;313:1604–1610. doi: 10.1126/science.1124646. [DOI] [PubMed] [Google Scholar]
- Conde J., Fink G. R. A mutant of Saccharomyces cerevisiae defective for nuclear fusion. Proc. Natl. Acad. Sci. USA. 1976;73:3651–3655. doi: 10.1073/pnas.73.10.3651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies S. W., Turmaine M., Cozens B. A., DiFiglia M., Sharp A. H., Ross C. A., Scherzinger E., Wanker E. E., Mangiarini L., Bates G. P. Formation of neuronal intranuclear inclusions underlies the neurological dysfunction in mice transgenic for the HD mutation. Cell. 1997;90:537–548. doi: 10.1016/s0092-8674(00)80513-9. [DOI] [PubMed] [Google Scholar]
- De Rooij K. E., Dorsman J. C., Smoor M. A., Den Dunnen J. T., Van Ommen G. J. Subcellular localization of the Huntington's disease gene product in cell lines by immunofluorescence and biochemical subcellular fractionation. Hum. Mol. Genet. 1996;5:1093–1099. doi: 10.1093/hmg/5.8.1093. [DOI] [PubMed] [Google Scholar]
- Derkatch I. L., Bradley M. E., Hong J. Y., Liebman S. W. Prions affect the appearance of other prions: the story of [PIN(+)]. Cell. 2001;106:171–182. doi: 10.1016/s0092-8674(01)00427-5. [DOI] [PubMed] [Google Scholar]
- Derkatch I. L., Bradley M. E., Zhou P., Chernoff Y. O., Liebman S. W. Genetic and environmental factors affecting the de novo appearance of the [PSI+] prion in Saccharomyces cerevisiae. Genetics. 1997;147:507–519. doi: 10.1093/genetics/147.2.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiFiglia M., Sapp E., Chase K. O., Davies S. W., Bates G. P., Vonsattel J. P., Aronin N. Aggregation of huntingtin in neuronal intranuclear inclusions and dystrophic neurites in brain. Science. 1997;277:1990–1993. doi: 10.1126/science.277.5334.1990. [DOI] [PubMed] [Google Scholar]
- Douglas P. M., Summers D. W., Cyr D. M. Molecular chaperones antagonize proteotoxicity by differentially modulating protein aggregation pathways. Prion. 2009;3:51–58. doi: 10.4161/pri.3.2.8587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douglas P. M., Treusch S., Ren H. Y., Halfmann R., Duennwald M. L., Lindquist S., Cyr D. M. Chaperone-dependent amyloid assembly protects cells from prion toxicity. Proc. Natl. Acad. Sci. USA. 2008;105:7206–7211. doi: 10.1073/pnas.0802593105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duennwald M. L., Jagadish S., Giorgini F., Muchowski P. J., Lindquist S. A network of protein interactions determines polyglutamine toxicity. Proc. Natl. Acad. Sci. USA. 2006;103:11051–11056. doi: 10.1073/pnas.0604548103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunah A. W., Jeong H., Griffin A., Kim Y. M., Standaert D. G., Hersch S. M., Mouradian M. M., Young A. B., Tanese N., Krainc D. Sp1 and TAFII130 transcriptional activity disrupted in early Huntington's disease. Science. 2002;296:2238–2243. doi: 10.1126/science.1072613. [DOI] [PubMed] [Google Scholar]
- Fan C. Y., Lee S., Ren H. Y., Cyr D. M. Exchangeable chaperone modules contribute to specification of type I and type II Hsp40 cellular function. Mol. Biol. Cell. 2004;15:761–773. doi: 10.1091/mbc.E03-03-0146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gokhale K. C., Newnam G. P., Sherman M. Y., Chernoff Y. O. Modulation of prion-dependent polyglutamine aggregation and toxicity by chaperone proteins in the yeast model. J. Biol. Chem. 2005;280:22809–22818. doi: 10.1074/jbc.M500390200. [DOI] [PubMed] [Google Scholar]
- Haass C., Selkoe D. J. Soluble protein oligomers in neurodegeneration: lessons from the Alzheimer's amyloid beta-peptide. Nat. Rev. Mol. Cell Biol. 2007;8:101–112. doi: 10.1038/nrm2101. [DOI] [PubMed] [Google Scholar]
- Huh W. K., Falvo J. V., Gerke L. C., Carroll A. S., Howson R. W., Weissman J. S., O'Shea E. K. Global analysis of protein localization in budding yeast. Nature. 2003;425:686–691. doi: 10.1038/nature02026. [DOI] [PubMed] [Google Scholar]
- Kalderon D., Roberts B. L., Richardson W. D., Smith A. E. A short amino acid sequence able to specify nuclear location. Cell. 1984;39:499–509. doi: 10.1016/0092-8674(84)90457-4. [DOI] [PubMed] [Google Scholar]
- Kayed R., Head E., Thompson J. L., McIntire T. M., Milton S. C., Cotman C. W., Glabe C. G. Common structure of soluble amyloid oligomers implies common mechanism of pathogenesis. Science. 2003;300:486–489. doi: 10.1126/science.1079469. [DOI] [PubMed] [Google Scholar]
- Kim S., Willison K. R., Horwich A. L. Cystosolic chaperonin subunits have a conserved ATPase domain but diverged polypeptide-binding domains. Trends Biochem. Sci. 1994;19:543–548. doi: 10.1016/0968-0004(94)90058-2. [DOI] [PubMed] [Google Scholar]
- Kitamura A., Kubota H., Pack C. G., Matsumoto G., Hirayama S., Takahashi Y., Kimura H., Kinjo M., Morimoto R. I., Nagata K. Cytosolic chaperonin prevents polyglutamine toxicity with altering the aggregation state. Nat. Cell Biol. 2006;8:1163–1170. doi: 10.1038/ncb1478. [DOI] [PubMed] [Google Scholar]
- Kryndushkin D. S., Alexandrov I. M., Ter-Avanesyan M. D., Kushnirov V. V. Yeast [PSI+] prion aggregates are formed by small Sup35 polymers fragmented by Hsp104. J. Biol. Chem. 2003;278:49636–49643. doi: 10.1074/jbc.M307996200. [DOI] [PubMed] [Google Scholar]
- Luke M. M., Sutton A., Arndt K. T. Characterization of SIS1, a Saccharomyces cerevisiae homologue of bacterial dnaJ proteins. J. Cell Biol. 1991;114:623–638. doi: 10.1083/jcb.114.4.623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meriin A. B., Zhang X., He X., Newnam G. P., Chernoff Y. O., Sherman M. Y. Huntington toxicity in yeast model depends on polyglutamine aggregation mediated by a prion-like protein Rnq1. J. Cell Biol. 2002;157:997–1004. doi: 10.1083/jcb.200112104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morimoto R. I. Proteotoxic stress and inducible chaperone networks in neurodegenerative disease and aging. Genes Dev. 2008;22:1427–1438. doi: 10.1101/gad.1657108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muchowski P. J., Wacker J. L. Modulation of neurodegeneration by molecular chaperones. Nat. Rev. Neurosci. 2005;6:11–22. doi: 10.1038/nrn1587. [DOI] [PubMed] [Google Scholar]
- Neumann M., et al. Ubiquitinated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Science. 2006;314:130–133. doi: 10.1126/science.1134108. [DOI] [PubMed] [Google Scholar]
- Perez M. K., Paulson H. L., Pendse S. J., Saionz S. J., Bonini N. M., Pittman R. N. Recruitment and the role of nuclear localization in polyglutamine-mediated aggregation. J. Cell Biol. 1998;143:1457–1470. doi: 10.1083/jcb.143.6.1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross C. A. Intranuclear neuronal inclusions: a common pathogenic mechanism for glutamine-repeat neurodegenerative diseases? Neuron. 1997;19:1147–1150. doi: 10.1016/s0896-6273(00)80405-5. [DOI] [PubMed] [Google Scholar]
- Ross C. A. Polyglutamine pathogenesis: emergence of unifying mechanisms for Huntington's disease and related disorders. Neuron. 2002;35:819–822. doi: 10.1016/s0896-6273(02)00872-3. [DOI] [PubMed] [Google Scholar]
- Saudou F., Finkbeiner S., Devys D., Greenberg M. E. Huntingtin acts in the nucleus to induce apoptosis but death does not correlate with the formation of intranuclear inclusions. Cell. 1998;95:55–66. doi: 10.1016/s0092-8674(00)81782-1. [DOI] [PubMed] [Google Scholar]
- Schaffar G., Breuer P., Boteva R., Behrends C., Tzvetkov N., Strippel N., Sakahira H., Siegers K., Hayer-Hartl M., Hartl F. U. Cellular toxicity of polyglutamine expansion proteins: mechanism of transcription factor deactivation. Mol. Cell. 2004;15:95–105. doi: 10.1016/j.molcel.2004.06.029. [DOI] [PubMed] [Google Scholar]
- Scherzinger E., Lurz R., Turmaine M., Mangiarini L., Hollenbach B., Hasenbank R., Bates G. P., Davies S. W., Lehrach H., Wanker E. E. Huntingtin-encoded polyglutamine expansions form amyloid-like protein aggregates in vitro and in vivo. Cell. 1997;90:549–558. doi: 10.1016/s0092-8674(00)80514-0. [DOI] [PubMed] [Google Scholar]
- Schwimmer C., Masison D. C. Antagonistic interactions between yeast [PSI(+)] and [URE3] prions and curing of [URE3] by Hsp70 protein chaperone Ssa1p but not by Ssa2p. Mol. Cell. Biol. 2002;22:3590–3598. doi: 10.1128/MCB.22.11.3590-3598.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sondheimer N., Lindquist S. Rnq 1, an epigenetic modifier of protein function in yeast. Mol. Cell. 2000;5:163–172. doi: 10.1016/s1097-2765(00)80412-8. [DOI] [PubMed] [Google Scholar]
- Stade K., Ford C. S., Guthrie C., Weis K. Exportin 1 (Crm1p) is an essential nuclear export factor. Cell. 1997;90:1041–1050. doi: 10.1016/s0092-8674(00)80370-0. [DOI] [PubMed] [Google Scholar]
- Steffan J. S., Kazantsev A., Spasic-Boskovic O., Greenwald M., Zhu Y. Z., Gohler H., Wanker E. E., Bates G. P., Housman D. E., Thompson L. M. The Huntington's disease protein interacts with p53 and CREB-binding protein and represses transcription. Proc. Natl. Acad. Sci. USA. 2000;97:6763–6768. doi: 10.1073/pnas.100110097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Summers D. W., Douglas P. M., Ren H. Y., Cyr D. M. The type I HSP40 YDJ1 utilizes a farnesyl moiety and zinc finger like-region to suppress prion toxicity. J. Biol. Chem. 2009;284:3628–3639. doi: 10.1074/jbc.M807369200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tam S., Geller R., Spiess C., Frydman J. The chaperonin TRiC controls polyglutamine aggregation and toxicity through subunit-specific interactions. Nat. Cell Biol. 2006;8:1155–1162. doi: 10.1038/ncb1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taneja V., Maddelein M. L., Talarek N., Saupe S. J., Liebman S. W. A non-Q/N-rich prion domain of a foreign prion, [Het-s], can propagate as a prion in yeast. Mol. Cell. 2007;27:67–77. doi: 10.1016/j.molcel.2007.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor J. P., Hardy J., Fischbeck K. H. Toxic proteins in neurodegenerative disease. Science. 2002;296:1991–1995. doi: 10.1126/science.1067122. [DOI] [PubMed] [Google Scholar]
- Tipton K. A., Verges K. J., Weissman J. S. In vivo monitoring of the prion replication cycle reveals a critical role for Sis1 in delivering substrates to Hsp104. Mol. Cell. 2008;32:584–591. doi: 10.1016/j.molcel.2008.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tkach J. M., Glover J. R. Nucleocytoplasmic trafficking of the molecular chaperone Hsp104 in unstressed and heat-shocked cells. Traffic. 2008;9:39–56. doi: 10.1111/j.1600-0854.2007.00666.x. [DOI] [PubMed] [Google Scholar]
- Treusch S., Cyr D. M., Lindquist S. Amyloid deposits: protection against toxic protein species? Cell Cycle. 2009;8:1668–1674. doi: 10.4161/cc.8.11.8503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Meriin A. B., Zaarur N., Romanova N. V., Chernoff Y. O., Costello C. E., Sherman M. Y. Abnormal proteins can form aggresome in yeast: aggresome-targeting signals and components of the machinery. FASEB J. 2009;23:451–463. doi: 10.1096/fj.08-117614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zakrzewska A., Boorsma A., Delneri D., Brul S., Oliver S. G., Klis F. M. Cellular processes and pathways that protect Saccharomyces cerevisiae cells against the plasma membrane-perturbing compound chitosan. Eukaryot. Cell. 2007;6:600–608. doi: 10.1128/EC.00355-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Yang Z., Cao Y., Zhang S., Li H., Huang Y., Ding Y. Q., Liu X. The Hsp40 family chaperone protein DnaJB6 enhances Schlafen1 nuclear localization which is critical for promotion of cell-cycle arrest in T-cells. Biochem. J. 2008;413:239–250. doi: 10.1042/BJ20071510. [DOI] [PubMed] [Google Scholar]
- Zoghbi H. Y., Orr H. T. Glutamine repeats and neurodegeneration. Annu. Rev. Neurosci. 2000;23:217–247. doi: 10.1146/annurev.neuro.23.1.217. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.