Abstract
We previously used a human artificial chromosome (HAC) with a synthetic kinetochore that could be targeted with chromatin modifiers fused to tetracycline repressor to show that targeting of the transcriptional repressor tTS within kinetochore chromatin disrupts kinetochore structure and function. Here we show that the transcriptional corepressor KAP1, a downstream effector of the tTS, can also inactivate the kinetochore. The disruption of kinetochore structure by KAP1 subdomains does not simply result from loss of centromeric CENP-A nucleosomes. Instead it reflects a hierarchical disruption of the outer kinetochore, with CENP-C levels falling before CENP-A levels and, in certain instances, CENP-H being lost more readily than CENP-C. These results suggest that this novel approach to kinetochore dissection may reveal new patterns of protein interactions within the kinetochore.
INTRODUCTION
The centromere/kinetochore is one of the most complex cellular substructures, with more than 80 protein components described to date (reviewed in Carroll and Straight, 2006; Cheeseman and Desai, 2008; Fukagawa, 2008; Vagnarelli et al., 2008). These components perform the complex job of attaching chromosomes to the mitotic spindle; ensuring that those attachments are correct; signaling to delay mitotic progression if they are not, and regulating the movements of the chromosomes toward the spindle poles in anaphase.
The kinetochore is assembled at a unique locus on each natural chromosome. However, for organisms with regional centromeres (Pluta et al., 1995), this reflects only a preference¤, and not an absolute requirement for particular DNA sequences. Kinetochores can form on a wide range of single-copy and repeated DNA sequences, leading to the conclusion that the ultimate determinants of kinetochore assembly are epigenetic. The long-term purpose of our studies is to determine the epigenetic “landscape” that promotes kinetochore assembly and its maintenance during cell divisions.
Experiments including yeast genetics, RNA interference (RNAi) studies in mammalian cells, and gene knockout analysis in mouse and chicken DT40 cells have revealed that kinetochores assemble on a foundation of specialized chromatin containing the kinetochore-specific histone H3 variant CENP-A (Earnshaw and Rothfield, 1985). CENP-A is upstream of almost all other known components in the kinetochore assembly pathway. However, that pathway is multiplex, as recent studies in chicken and Drosophila show that inner kinetochore proteins CENP-H and -C are required for normal CENP-A loading or retention (Okada et al., 2006; Goshima et al., 2007; Erhardt et al., 2008).
Our work was inspired by an approach first developed a number of years ago in which cloned fragments of human centromeric DNA were used to form human artificial chromosomes (HACs) in HT1080 fibrosarcoma cells (Harrington et al., 1997; Ikeno et al., 1998). Originally, HAC formation was only achieved with regular repeated arrays of α-satellite DNA containing CENP-B boxes (Masumoto et al., 1998; Ohzeki et al., 2002; Okada et al., 2007). We extended those studies by developing HACs based on a synthetic alphoidtetO DNA array that resembles centromeric repeats, but contains a tetracycline operator in every second alphoid monomer (Nakano et al., 2008). This allows the targeting of a wide range of proteins into the functional kinetochore as fusions to tetracycline repressor. The power of this system is that it allows the specific manipulation of the protein complement of a single kinetochore in vivo, while leaving all other kinetochores unperturbed.
We previously showed that targeting of either a transcriptional activator or repressor (the tTA and tTS, respectively) could inactivate the synthetic kinetochore (Nakano et al., 2008). Surprisingly, targeting of factors promoting the formation of heterochromatin associated with HP1α strongly disrupted the conditional kinetochore, as judged by loss of centromeric proteins, alteration of the pattern of chromatin modifications within the centromere, and destabilisation of the HAC (Nakano et al., 2008).
The alphoidtetO HAC system has important differences from the only other conditional centromere system, which was developed for the point centromere of budding yeast (Hill and Bloom, 1987). That centromere is inactivated by being bombarded by transcription from an adjacent strong promoter. Thus, it is not suited for the analysis of the effects of transcriptional repressors on centromere function, a key question, given the demonstrated role of transcription in centromere function in Schizosaccharomyces pombe (Volpe et al., 2002, 2003; Chen et al., 2008). Furthermore, in its inactive state, the budding yeast conditional centromere was reported to be fully occupied with CENP-A/Cse4 (Collins et al., 2005), although this was disputed (Mythreye and Bloom, 2003). In the case of the human conditional centromere, inactivation induced by the tTS transcriptional repressor is accompanied by loss of CENP-A (Nakano et al., 2008).
Here, we have used the alphoidtetO HAC system to further investigate the process of HAC disruption by the transcriptional corepressor KAP1, a downstream effector of the tTS. We show that KAP1 inactivates the HAC in a complex manner that appears to involve a hierarchy of loss of inner and outer kinetochore components that precedes loss of CENP-A from centromeric nucleosomes.
MATERIALS AND METHODS
Construction of a HeLa-HT1080 Cell Line Containing an AlphoidtetO HAC
A HT1080 cell line carrying 1 copy/cell of a HAC obtained from a synthetic centromeric α-satellite DNA incorporating a tetO sequence every second alphoid monomer (AB2.2.18.21; Nakano et al., 2008) was fused to a HeLa cell line resistant to the drug neomycin. AB2.2.18.21 cells, resistant to blasticidin (the resistance marker present on the HAC DNA), were plated with neomycin-resistant HeLa cells in a 50%-to-50% ratio and left to grow overnight. The next day, growth medium was washed, and cells were incubated for 3 min with a solution of 50% polyethyleneglycol (PEG; Roche, Indianapolis, IN) in 1× PBS. PEG was then removed by washing with warm medium, and cells were grown in the presence of blasticidin (8 μg/ml) for HAC selection and geneticin (800 ng/ml) for HeLa selection. Several hybrid clones were obtained from single cells and analyzed by fluorescence in situ hybridization (FISH) with a BAC-probe (the HAC vector backbone). 1C7 cells selected for further study were cultured in RPMI medium (Invitrogen, Carlsbad, CA) supplemented with 10% of fetal bovine serum (Invitrogen) and with 500 μg/ml geneticin and 8 μg/ml blasticidin.
Construction of TetR:EYFP-tagged Targeting Constructs
KAP1 constructs were inserted at the C-terminus of TetR:EYFP in the plasmid pEYFP-C1 (Nakano et al., 2008). KAP1 deletion mutants were amplified by PCR using the following oligonucleotides as primers: 5′- TTCGGCCGCAGCCTCGGCCT-3′ (RBCC_1_Fwd), 5′- GAGGGGCCATGGGTGCAGGG-3′ (RBCC_1_Rev), 5′- ATGGCCCCTCCAAGAGCCCC-3′ (HPBD_1_Fwd), 5′- CCCTCCGCAAGAGCCATAAGC-3′ (HPBD_1_Rev), 5′- GGTGGCCCGGGAACCCTGGA-3′ (PHD_1_Fwd), 5′- GGGGCCATCACCAGGGCCAC-3′ (PHD_1_Rev). PCR products were cloned in pGEMTeasy, cut and cloned blunt into the BglII site of pEYFP-C1 (TetR:EYFP). TetR:EYFP:KAP1[20–559] was obtained by inserting a stop codon at residue 559 by site-directed mutagenesis of KAP1Δ20.
Generation of the 1C7-KAP1 Cell Line and Washout Experiments
For constructs allowing stable, puromycin-dependent expression in 1C7 cells, the BsaHI/BspEI fragment of TetR:EYFP containing the coding sequence for tet repressor–NLS–EYFP was ligated into the ClaI/BspEI-digested pIRESpuro2 (Clontech, Palo Alto, CA) generating tYIP. The 2.5-kb BspEI/BclI fragment of TetR:EYFP:KAP1 was cloned into the BspEI/BamHI-digested tYIP backbone, generating tYIP-KAP1.
Stable transfections were carried out in the presence of 1 μg/ml doxcycline, using Fugene HD (Roche) in a transfection mix of 15 μl Fugene HD reagent and 5 μg plasmid in 250 μl OptiMEM (Invitrogen), essentially according to the manufacturer's instructions. Twenty-four hours after transfection, puromycin and blasticidin were added to final concentrations of 2 μg/ml and 4 μg/ml, respectively. From this point onward, doxycycline was added fresh every 2 d. Clonal lines of drug resistant 1C7 cells were isolated by limiting dilution, and a clone showing medium-to-high levels of the fluorescent construct, as determined by fluorescence microscopy, was selected for experiments.
The doxycycline washout time course was started with a subconfluent culture of 1C7-KAP1 cells growing in the presence of all drugs. The cell layer was rinsed twice with excess D-PBS, followed by incubation in cRPMI without drugs for 30 min at 37°C. This procedure was repeated once, and cells were subsequently incubated over night in cRPMI with 2 μg/ml puromycin. The following morning, the cell layer was washed in excess D-PBS followed by PBS/EDTA, cells were harvested in TrypLE Express (Invitrogen) and centrifuged at 1000 rpm for 3 min. The cell pellet was resuspended in cRPMI with 2 μg/ml Puromycin and seeded on coverslips for fixation and antibody staining as described at the relevant time points.
Immunostaining and Cytological Analysis
Immunostaining and cytological analyses were performed as described in (Nakano et al., 2008) except that anti-CENP-A was as described in (Valdivia et al., 1998). A DeltaVision (Applied Precision, Issaquah, WA) system based on an Olympus IX-70 microscope stand (Melville, NY) coupled to a CH350 CCD camera (Photometrics, Tucson, AZ) and controlled through SoftWorx (Applied Precision) was used for image acquisition of transient transfection experiments. Images were taken using an Olympus S-Plan apocromat 100× 1.40 NA oil immersion objective. Imaging of the stable 1C7-KAP1 cells was performed on an Olympus IX-71 microscope stand coupled to a Photometrics Cool Snap HQ camera, using an Olympus UPlanSApo 100× 1.40 NA oil immersion objective. All images were deconvolved with SoftWorx before processing. Subsequently, SoftWorx or Image ProPlus (Media Cybernetics, Silver Spring, MD) was used for image analysis.
For fluorescence quantification of transient transfection experiments, cells expressing similar amounts of exogenous protein (judged by the similar fluorescence background) were chosen and line-profile measurements were taken on sum projections of deconvolved images. For quantification of ACA staining at HAC and endogenous centromeres, a selected region of interest (ROI) was defined on image projections as above. The line-profile algorithm was also used to measure interkinetochore distances on mitotic HACs.
Fluorescence quantification of stable 1C7-KAP1 cells showing a single EYFP spot was performed on deconvolved images with a z section spacing of 0.2 μm (or 0.3 μm for mitotic cells). Quantification was performed in Image Pro, applying a custom-written macro (details available upon request). In brief, HAC enhanced yellow flourescent protein (EYFP)-associated fluorescent spot signals of the Texas Red–conjugated secondary antibodies within a nine-pixel diameter, circular ROI were measured in the relevant sections. For each section, the background over a nuclear area not showing any CENP staining in an identical ROI was subtracted and the net values were summed. These sum values were used to calculate the average fluorescent intensity per section quantified (arbitrary fluorescent units per unit area).
Chromatin immunoprecipitation and Real-Time PCR
For the HAC loss assay, 1C7 cells transfected with plasmids expressing the indicated targeting constructs were collected and genomic DNA prepared using the DNeasy Blood and Tissue Kit (Qiagen, Chatsworth, CA) according to the manufacturer's instructions, including a treatment with RNase A. Quantitative real-time PCR using a SYBR green mastermix (JumpStart, Sigma, St. Louis, MO) was subsequently preformed using the following oligonucleotides: 5SDNA-F1/-R1 for 5S ribosomal DNA, tet-1/tet-3 for the alphoidtetO array, and bsr-F/-R for the bsr marker gene (Nakano et al., 2008). For each sample, the average ratio of the amounts of [alphoidtetO] and [bsr] to [5S rDNA] was calculated. To determine the relative HAC copy number per cell, this ratio was then normalized to the ratio derived from 1C7 cells transfected with TetR:EYFP that were grown in the presence of doxycycline for the course of the assay (arbitrary HAC copy number per cell = 1).
Chromatin immunoprecipitation (ChIP) to analyze HAC chromatin was performed essentially as described in Nakano et al. (2008). Monoclonal antibodies used in ChIP were as described in Kimura et al. (2008).
RESULTS
Isolation of a HeLa Hybrid Cell Line Bearing the AlphoidtetO Synthetic HAC
To study the alphoidtetO HAC in a cell line with favorable growth and transfection properties, HAC-containing HT1080 cell line AB2.2.18.21 (Nakano et al., 2008) was fused to neomycin-resistant HeLa cells. Clone 1C7, with 1 HAC per nucleus in >85% of cells (Figure 1A and I) was selected for further characterization. 1C7 cells had a morphology typical of HeLa cells, and a doubling time of ∼18–20 h.
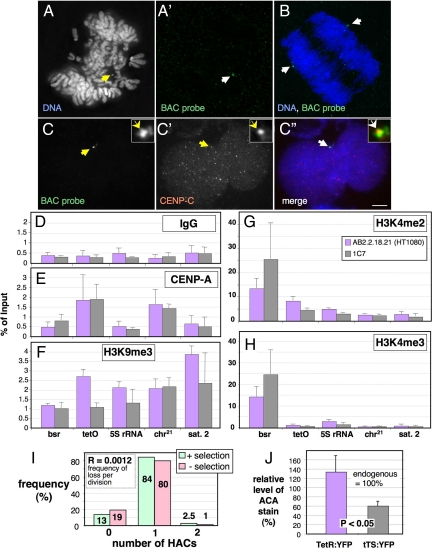
Figure 1.
Isolation and characterization of a Hela-HT1080 hybrid cell line carrying a stable alphoidtetO HAC. (A) FISH with a BAC probe (green in A′) on mitotic chromosomes from the 1C7 cell line with 1 alphoidtetO HAC per nucleus (arrow in A). (B) FISH on anaphase 1C7 cell. The HAC (green, arrows in B) segregates with the endogenous chromosomes. Blue, DAPI staining for DNA. (C) Immuno-FISH with a BAC probe (C, green in the merged image in C″) and antibody against CENP-C (C', red in the merged image in C″) on an interphase 1C7 cell. CENP-C colocalizes with the BAC-probe on the HAC (arrows). Scale bar, 5 μm. (D–H) ChIP with control IgG antibody (D) and antibodies specific for CENP-A (E), trimethylated Lysine 9 on histone H3 (F), dimethylated Lysine 4 on histone H3 (G) and trimethylated Lysine 4 on histone H3 (H). Real-time PCR on the purified DNA was performed with primers specific for sequences in the actively transcribed Bsr (blasticidin resistance) gene on the BAC vector and the alphoidtetO array (tetO) forming the HAC centromere. As controls, primers specific for the DNA coding for rRNA (5S rDNA), endogenous chromosome 21 centromeric alphoid DNA (chr.21) and endogenous satellite 2 DNA (Sat 2), which is located in the pericentromeric heterochromatin of chromosomes 1 and 16, were used. Error bars, SEM of three independent ChIP experiments. (I) Analysis of HAC mitotic stability in 1C7 cells grown for 40 generations in the absence of blasticidin selection. The number of HACs per interphase nucleus was determined by FISH with a BAC probe. (J) Quantification of the amount of ACA human autoantibody (Earnshaw and Rothfield, 1985) staining associated with the alphoidtetO HAC in 1C7 cells fixed 48 h after transfection with TetR:EYFP and tTS:EYFP. Values are normalized to the average ACA staining of endogenous centromeres (100%). Error bars, SD.
The synthetic kinetochore of the alphoidtetO HAC assembled a normal complement of kinetochore proteins in 1C7 cells, as shown for CENP-C in Figure 1C. ChIP with specific antibodies revealed that the HAC alphoidtetO sequences bound similar levels of CENP-A and H3K4me2 in AB2.2.18.21 and 1C7 cells, while having appreciable levels of H3K4me3 only on the active bsr marker gene (Figure 1, D–H). The alphoidtetO HAC was mitotically stable in 1C7 cells, with a loss rate of 0.0012 per division after growth without selection for 40 generations (Figure 1I).
The synthetic kinetochore retained its conditional activity in 1C7 cells. Expressed TetR:EYFP showed a diffuse nuclear localization plus one bright spot that colocalized with CENP-A and -C (Supplemental Figure S1A). TetR:EYFP binding did not affect the localization of CENP-A, -B and -C recognized by anti-centromere antibodies (ACA; Earnshaw and Rothfield, 1985) at the synthetic kinetochore. In contrast, targeting the transcriptional repressor tTS:EYFP to the HAC kinetochore reduced the ACA signal to <50% (Figure 1J, Supplemental Figure S1B). We therefore decided to explore in more detail the events that occur during kinetochore disruption mediated by the tTS.
Inactivation of the Synthetic Kinetochore by KAP1
The transcriptional silencing function of the tTS is mediated by a protein domain called the Kruppel-associated box (KRAB). One principal downstream effector of this class of transcriptional repressors is the scaffolding protein KAP1 (Kruppel-associated protein 1, also known as TIF1β and Trim28 (Friedman et al., 1996; Le Douarin et al., 1996; Shojaei et al., 2004). KAP1 is proposed to recruit chromatin-remodelling factors to loci targeted by KRAB-family repressors (Schultz et al., 2001). To determine whether KAP1 could be responsible for HAC kinetochore inactivation downstream of tTS binding, TetR:EYFP was fused to a KAP1 construct lacking the first 20 residues (KAP1Δ20 ; Figure 2A), which has been reported to function identically to the full-length protein in in vivo gene silencing assays (Sripathy et al., 2006).
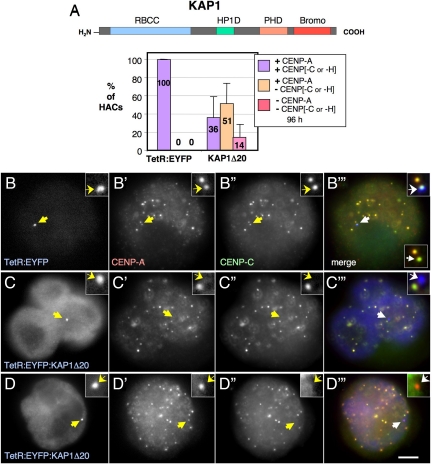
Figure 2.
Disruption of the HAC kinetochore by KAP1Δ20. (A) Schematic of KAP1 and its domains (top). Bottom, the frequency of the HACs targeted with KAP1Δ20 positive or not for CENP-A and -C or -H is plotted. For this experiment, 1C7 cells were transfected with control TetR:EYFP and TetR:EYFP:KAP1Δ20, fixed 96 h after transfection and costained with the indicated antibodies. Error bars, SEM. (B–D) Representative images of the staining of transfected 1C7 cells analyzed in B. Arrows point to the alphoidtetO HAC. Colors in merge: blue, EYFP; green, CENP-C; red, CENP-A. Merged inset, bottom right of C‴ shows only red and green channels. Scale bar, 5 μm.
In 1C7 cells transfected with constructs encoding TetR:EYFP:KAP1Δ20, we observed complete loss of CENP-A and -C from 14% of centromeres at 96 h after transfection (Figure 2, A and C). About half of centromeres with bound TetR:EYFP:KAP1Δ20 lacked detectable CENP-C or CENP-H but still had CENP-A (Figure 2, A and D). Just over a third of centromeres with bound TetR:EYFP:KAP1Δ20 appeared structurally normal in these experiments (Figure 2A). This variability could reflect cell cycle differences or differences in the levels of the expressed proteins.
To test the hypothesis that kinetochore inactivation by KAP1 requires loss of CENP-A from the kinetochore chromatin, we used quantitative fluorescence analysis to determine the relationship between KAP1-induced loss of CENP-A and the loss of CENP-C/CENP-H from the alphoidtetO kinetochore. Most HACs had a relatively narrow range of CENP-A antibody binding (intensity values ranging between 100 and 240 arbitrary units in these experiments; left of the vertical dotted line in Figure 3A), but a much wider range of associated CENP-C (330–2400 arbitrary units). On only 4 of 19 HACs with bound TetR:EYFP was a higher CENP-A signal accompanied by a greater CENP-C signal.
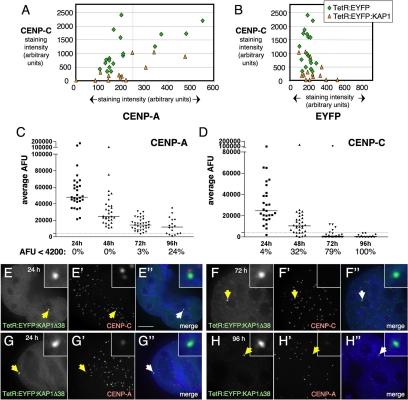
Figure 3.
CENP-C is lost more rapidly than CENP-A from KAP1 targeted HAC kinetochores. (A and B) Measurement of the amounts of CENP-C and -A (A) or -C and EYFP (B) associated with alphoidtetO HAC in1C7 cells 48h after transient transfection with TetR:EYFP or TetR:EYFP:KAP1Δ20 (n = 13–19). (C and D) 1C7-KAP1 cells were fixed and stained for either CENP-A or -C at the indicated time points after washing out of doxycycline to allow binding of TetR:EYFP:KAP1Δ38 to the HAC. Background-subtracted average arbitrary fluorescent units (AFU) of the HAC associated antibody staining are plotted. The median AFU is indicated for each antibody and time point as solid line in the graph. The percentage of analyzed HACs showing an AFU value of <4200 (dotted line in the graph) is indicated. Targeting of KAP1 causes levels of CENP-C to diminish more rapidly than levels of CENP-A. (E–H) Maximum intensity projections of cells stained for CENP-C 24 (E) and 72 (F) h after doxycycline washout, with HAC staining corresponding approximately to the median AFU in D. By 72 h, most HACs lack a detectable CENP-C staining. For CENP-A, time points at 24 (G) and 96 (H) h are shown. Staining corresponds to the median AFU in C. Even at the 96-h time point, a weak HAC associated CENP-A signal can be detected in the majority of cells. Scale bar, 5 μm.
Line-profile analysis of CENP-A and CENP-C signals at the disrupted HAC kinetochores with bound TetR:EYFP:KAP1Δ20 suggested that loss of CENP-C does not require a prior loss or decrease in CENP-A levels at the kinetochore. Indeed, in this experiment, the levels of CENP-A found on HACs with very little or no detectable CENP-C were similar to the levels found on HACs targeted with control TetR:EYFP (Figure 3A).
When levels of HAC-associated EYFP signals were compared, the amount of bound TetR:EYFP:KAP1Δ20 was similar to that of TetR:EYFP, although CENP-C levels were much reduced with the former (Figure 3B). Thus, the differential effects of TetR:EYFP and TetR:EYFP:KAP1Δ20 on kinetochore stability cannot be explained by differences in the efficiency of binding of the two EYFP fusion proteins to the synthetic kinetochore.
To confirm that the loss of CENP-C occurs independently of CENP-A loss, and to obtain a better understanding of the dynamics of KAP1-mediated kinetochore disruption, we generated a 1C7 derived cell line (1C7-KAP1) stably expressing TetR:EYFP:KAP1Δ38. These cells were maintained in the presence of 1 μg/ml doxycycline to prevent binding of TetR:EYFP:KAP1Δ38 to the HAC (Materials and Methods). To examine the time course of events at the HAC kinetochore after KAP1 binding, we washed out the doxycycline and subsequently fixed cells for staining with antibodies directed against either CENP-A or -C after 24, 48, 72, and 96 h. We have previously shown that stable expression of TetR:EYFP in AB2.2.18.21 cells for up to 30 d does not affect HAC stability in most cells (Nakano et al., 2008).
In 1C7-KAP1 cells however, quantification of the HAC-associated signals revealed a rapid, time-dependent loss of CENP-C. After only 48 h, 33% of cells failed to display levels of HAC-associated CENP-C above the background level of 4200 fluorescent units per unit area in these experiments (Figure 3D). By 72 h, almost 80% of cells lacked detectable CENP-C staining at the HAC (Figure 3, E and F), and after 96 h, none of the HACs examined were positive for CENP-C. In the same cells, levels of HAC-associated CENP-A declined much more slowly, with more than 75% of cells retaining weak but detectable staining after 96 h (Figure 3, C, G, and H). Consistently, the average fold decrease in median fluorescent intensity for CENP-C was almost twice that for CENP-A over a 24-h interval within the first 72 h after doxycycline washout. As a control, average levels of CENP-A or -C staining at endogenous centromeres did not change over the course of the experiment (data not shown).
These experiments demonstrate that KAP1 fused to TetR:EYFP can effectively disrupt the HAC inner kinetochore as judged by loss of CENP-C or -H and that this occurs independently of effects on CENP-A localization.
Functional Dissection of the Role of KAP1 in Centromere Inactivation
KAP1 is a scaffold protein that recruits an array of adapter proteins and chromatin modifiers to loci where it binds. KAP1 has a modular organization with several well-characterized functional domains (Figure 4A), including a KRAB-binding N-terminal RBCC (RING finger, B-Box, coiled coil) region, a HP1-binding region, and a PHD-finger/Bromodomain. The PHD-finger promotes sumoylation of the Bromodomain, which then provides a specific binding interface for chromatin modifiers SETDB1 and NuRD (Ivanov et al., 2007; Zeng et al., 2008), leading to chromatin-mediated gene silencing (Schultz et al., 2002; Ivanov et al., 2007).
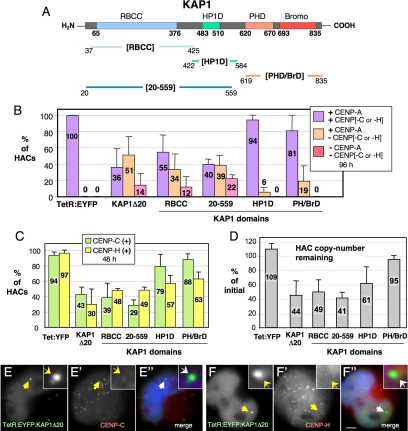
Figure 4.
Disruption of CENP-H and CENP-C kinetochore association. (A) Detailed schematic of the KAP1 subdomains used for targeting. (B) Frequency of the HACs targeted with KAP1-derived constructs positive or not for CENP-A and -C or CENP-H (n = 50–95). 1C7 cells were transfected with TetR:EYFP or KAP1-derived constructs, fixed 96 h after transfection and costained with the indicated antibodies. (C) Frequency of HACs targeted with the indicated constructs and positive for CENP-C or -H (n = 55–120). (D) Real-time PCR analysis of HAC retention after transfection with the indicated constructs as described for Figure 3B. Error, SEM. (E and F) HACs targeted with TetR:EYFP:KAP1Δ20 display complete loss of CENP-C (E′) and -H (F′) staining. Merge: blue, DAPI; green, EYFP; red, CENP-C/H. Scale bar, 5 μm.
To identify functional domain(s) of KAP1 capable of kinetochore inactivation, several KAP1Δ20 truncation mutants were fused to TetR:EYFP for HAC targeting. These included (Figure 4A): the RBCC region (KAP1[RBCC], aa 37–425); the HP1-binding domain (KAP1[HP1D], aa 422–584); a region starting with residue 20 and including the RBCC and HP1-binding motif (KAP1[20–559], aa 20–559); and the PHD-finger and Bromodomain (KAP1[PH/BrD], aa 619–835).
Hierarchical disruption of the kinetochore was observed after the targeting of the various KAP1 subdomains to the synthetic kinetochore in 1C7 cells. When screened at 96 h after transfection, the two constructs from the C-terminal half of KAP1 (TetR:EYFP:KAP1[HP1D] and TetR:EYFP:KAP1[PH/BrD]) had relatively mild effects. Both failed to induce the loss of CENP-A from kinetochores, causing the loss of CENP-C or -H in 6 or 19% of kinetochores, respectively (Figure 4, B and C; Supplemental Figure 2, C and D).
TetR:EYFP:KAP1[RBCC], which contains the N-terminal half of KAP1, was much more effective at disrupting the synthetic kinetochore. This protein induced CENP-A loss at 12% of targeted kinetochores, and the loss of either CENP-C or -H from 34% of the remaining CENP-A-positive kinetochores (Figure 4B). All kinetochores that were missing CENP-A were also missing CENP-C or -H.
Addition of the HP1-binding domain to give TetR:EYFP:KAP1[20–559] yielded a protein that was even more potent at disruption of the synthetic kinetochore. In this case CENP-A was lost from 22% of targeted kinetochores and 39% of the remaining CENP-A–positive kinetochores were missing CENP-C or -H (Figure 4B).
Given our previous finding that HP1α targeting was sufficient to inactivate the alphoidtetO synthetic kinetochore (Nakano et al., 2008), we were initially surprised that the KAP1 HP1-binding domain on its own was not a strong disrupter of the kinetochore. This was potentially explained by the observation that TetR:EYFP:KAP1[HP1D] was much less efficient at recruiting HP1α to the alphoidtetO kinetochore than TetR:EYFP:KAP1Δ20 (Supplemental Figure S4D). KAP1 oligomerization mediated by the RBCC region may be necessary for optimal KAP1-HP1 interactions. Indeed, it has been reported that the KAP1 HP1-binding region requires oligomerization for optimal transcriptional silencing activity and localization at pericentromeric heterochromatin (Matsuda et al., 2001).
To better understand the effect of KAP1 constructs on CENP-C/H kinetochore association, 1C7 cells were transfected and fixed 48 h later for immunostaining. All fusion proteins tested caused CENP-H loss from 37 to ∼70% of kinetochores (Figure 4, C and F; Supplemental Figure S3, B–D). The same fusion constructs were more variable in their effects on CENP-C localization at kinetochores. Chimeras containing the N-terminal half of KAP1 were most efficient at disrupting CENP-C binding to the synthetic kinetochore: only 39% of TetR:EYFP:KAP1[RBCC]-positive kinetochores and 29% of TetR:EYFP:KAP1[20–559]-positive kinetochores retained detectible CENP-C (Figure 4, C and E; Supplemental Figure S2, A and B). In contrast the C-terminal PHD/Bromodomain and HP1-binding domains (TetR:EYFP:KAP1[PH/BrD] and TetR:EYFP:KAP1[HP1D]) had little effect on CENP-C localization, with 88 and 79% of targeted HACs retaining CENP-C (Figure 4C, Supplemental Figure S4, B–D). We could observe an apparent tendency of a given domain to preferentially displace one of the two assessed centromere proteins over the other, with TetR:EYFP:KAP1[20–559] showing a higher frequency of loss of CENP-C relative to CENP-H, and the two C-terminal domains more efficiently displacing CENP-H relative to CENP-C (Figure 4C), however, this difference was not statistically significant.
Measurements of the effect of KAP1 domains on HAC stability in vivo were consistent with the above results on CENP-C and -H localization. When the various subdomains were expressed in cells and the levels of HAC DNA determined by quantitative PCR 12 d later, TetR:EYFP:KAP1Δ20, TetR:EYFP:KAP1[RBCC] and TetR:EYFP:KAP1[20–559] caused significant loss of the HAC, whereas TetR:EYFP:KAP1[PH/BrD] was much less effective (Figure 4D). In this assay, tetR:EYFP:KAP1[HP1D] yielded an intermediate level of HAC destabilization. Thus, the loss of kinetochore proteins detected in immunofluorescence experiments corresponds to inactivation of the HAC kinetochore.
These results suggest that there is a hierarchy of kinetochore disruption by KAP1, with CENP-C and -H being lost more readily, whereas CENP-A is the more resistant of these three proteins.
KAP1 Also Disrupts the Outer Kinetochore
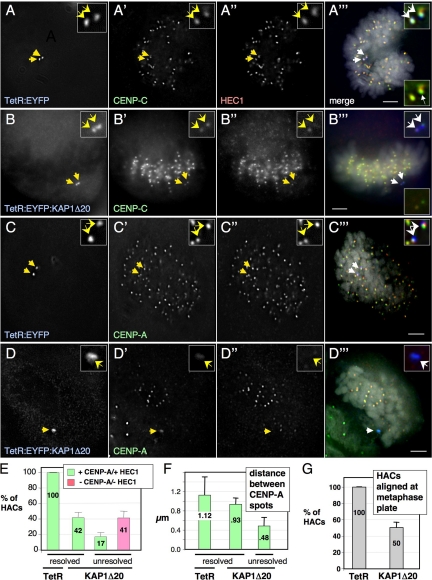
To determine whether disruption of the inner kinetochore by KAP1 also affects other kinetochore regions, we determined the effect of TetR:EYFP:KAP1Δ20 targeting on HEC1/Ndc80 in the outer kinetochore. At 96 h after transfection, inner CENP proteins and HEC1/Ndc80 were readily detected on HACs targeted with TetR:EYFP, appearing as two spots colocalizing with the paired EYFP-dots of the HAC, which was aligned with the other congressed mitotic chromosomes (Figure 5, A and C). The paired EYFP-dots indicate that the targeted HAC had replicated, and the distance between CENP-A spots (∼1.12 μm) suggests that the sister kinetochores had attached to the mitotic spindle and were under tension.
Figure 5.
KAP1Δ20 mediates outer kinetochore disruption. (A–D) Imaging of the alphoidtetO HAC in cells transiently expressing TetR:EYFP (A and C) or TetR:EYFP:KAP1Δ20 (B and D) for 96 h. Fixed cells were costained with an anti-CENP-A (C′ and D′) or anti-CENP-C antibody (A′ and B′) and an anti-Ndc80/HEC antibody (A″–D″). Colors in merge: blue, EYFP; green, CENP-A/C; red, Ndc80/HEC1. Arrows indicate targeted HACs. Scale bar, 5 μm. (E) Frequency of targeted EYFP-positive resolved HAC sisters (as in A and C). (F) Distance between CENP-A spots (when distinct) on resolved (as in B) or unresolved (as in D) HAC sisters. (G) Frequency of targeted HACs aligned across chromosomes congressed on the metaphase plate (as in C). Error bars, (E–G) SEM.
TetR:EYFP:KAP1Δ20 targeting to the HAC resulted in a markedly different outcome. In a significant fraction of mitotic HACs observed (∼41%) both inner and outer kinetochore proteins were absent (Figure 5E), and the HAC appeared as unresolved EYFP spots lying at the edge of the congressed chromosomes (see below, Figure 5D). Rarely, on HACs targeted with TetR:EYFP:KAP1Δ20, a HEC1/Ndc80 signal was found only on 1 EYFP-positive HAC sister kinetochore (Figure 5B). To investigate this intriguing observation in more detail, we quantified fluorescence staining of CENP-C at mitotic HAC sister kinetochores in the stable 1C7-KAP1 cell line 48 h after washing out of doxycycline. Strikingly, in a large proportion of cells, the two targeted HAC sister kinetochores displayed markedly unequal levels of CENP-C staining (Supplemental Figure S5, A and B). This trend toward biased kinetochore protein levels was also observed in cells stained for the outer kinetochore components hMis12 and KNL1 (data not shown). Importantly, unequal levels of CENP-C did not correlate with the amount of the KAP1 fusion construct bound, as this showed largely comparable levels on both HAC sister chromatids (Supplemental Figure S5B).
Targeting of TetR:EYFP:KAP1Δ20 to the synthetic kinetochore caused a reduction in the spacing between sister kinetochores to 0.93 μm in HACs where the two sister chromatids were clearly resolved. More commonly (∼60%), HACs targeted with TetR:EYFP:KAP1Δ20 showed two unresolved EYFP dots (Figure 5, B and D), suggesting that they were experiencing reduced tension across the centromere. In cases with two unresolved EYFP dots where two CENP-A spots could be observed, the later were closer together than normally seen for unattached kinetochores (0.48 μm), suggesting that not only were they not under tension, but that their centromeric heterochromatin was abnormally compacted. These observations could reflect a proposed role of KAP1 in promoting chromosome condensation (Ziv et al., 2006).
HACs with disrupted kinetochores (as judged by the absence of detectible HEC1/Ndc80) were typically found at the edge of the cluster of congressed chromosomes during mitosis (Figure 5D). Overall, the number of TetR:EYFP:KAP1Δ20-targeted HACs with a normal metaphase alignment was reduced from ∼100% of HACs with bound TetR:EYFP to ∼50% (Figure 5G).
We also assessed HAC alignment defects in the stable 1C7-KAP cell line at 48 and 72 h after removal of doxycycline. Consistent with the observation that HACs in most interphase nuclei retained readily detectable levels of CENP-A and -C at the 48-h time point (see Figure 3, C and D), only 6% (34/36) of HACs failed to show a loss of correct metaphase alignment in late prometaphase and metaphase cells (Supplemental Figure S6, A and C). However, by 72 h, 48% (22/42) of HACs were misaligned, and the sister chromatids of these unaligned HACs were generally not resolved (Supplemental Figure S6, B and C). This is despite that fact that CENP-A was still detectable in 97% of interphase cells analyzed (Figure 3C).
Combined, these analyses confirmed the hierarchical disruption of the kinetochore by KAP1, suggesting that KAP1-induced perturbation of the inner-kinetochore affects the structure of the outer kinetochore.
DISCUSSION
We previously described the isolation in HT1080 human fibrosacroma cells of a HAC whose kinetochore is based on a synthetic DNA sequence containing tetracycline operators (Nakano et al., 2008). After transfer of this chromosome into the 1C7 cell hybrid, which retains the growth and transfection properties of HeLa cells, the HAC retained the chromatin composition previously described in the HT1080 background with one exception. The synthetic centromere appeared to contain less of the heterochromatin-associated modification H3K9me3 associated with alphoidtetO sequences in the more rapidly growing 1C7 hybrid cells. Importantly, the alphoidtetO HAC was extremely stable in 1C7 cells, with a loss rate per division of 0.0012. This is within the range of mitotic stabilities reported for normal chromosomes in cultured cells (Burns et al., 1999). Thus, this HAC is a relevant model for the study of the epigenetic regulation of centromere function.
The alphoidtetO HAC is the first experimental system in which the targeting of a transcriptional repressor into the kinetochore has been shown to result in kinetochore inactivation (Nakano et al., 2008). This is consistent with studies of the role of transcription and RNAi in centromere activity in S. pombe (Volpe et al., 2002, 2003; Chen et al., 2008); however, the role for transcription in metazoan kinetochore activity has been little studied (Saffery et al., 2003; Fukagawa et al., 2004; Wong et al., 2007; Chueh et al., 2009). Our previous studies found that alphoidtetO kinetochore inactivation by the tTS correlated with the recruitment of heterochromatin protein HP1α and that direct recruitment of HP1α as a TetR:EYFP fusion protein could also inactivate the kinetochore (Nakano et al., 2008). This suggested that repressive chromatin in general might be incompatible with kinetochore stability, possibly because of a requirement for transcription within the centromere DNA array.
To understand how heterochromatin inactivates the kinetochore, we examined kinetochore disassembly by the multidomain scaffolding protein KAP1, a downstream effector of the tTS. As expected, KAP1 targeting inactivates the synthetic alphoidtetO centromere. However, we could not identify any single KAP1 domain with a dominant effect on the integrity of the synthetic kinetochore. Rather, combinatorial effects appeared to lead to a hierarchical disruption of the kinetochore. CENP-A is the most robust kinetochore component, because KAP1 induces the loss of CENP-C more rapidly than CENP-A. Furthermore, certain KAP1 domains showed a mild, though not statistically significant, tendency to induce the loss of CENP-H more efficiently than CENP-C. Ultimately the kinetochore was inactivated, as shown by the loss of the microtubule-binding component Hec1/Ndc80. These observations suggest that this approach may in the future permit a controlled dissection of the kinetochore in vivo.
Our finding that some mitotic HAC sister kinetochores targeted by KAP1 display unequal levels of CENP-C is intriguing. After targeting different constructs, we occasionally observed paired EYFP spots in interphase cells, presumably reflecting tetR fusion constructs binding to replicated HAC sister chromatids. In some of these cases, we detected a single spot for any kinetochore component that was associated with only one of the two EYFP spots (J.H.B., S.C., and W.C.E., unpublished data). A comparable observation was previously reported based on immuno-FISH experiments showing duplex α-satellite signals associated with only a single ACA spot (Haaf and Ward, 1994).
At present, it is unknown how pre-kinetochore components assembled in G1 are distributed onto sister centromeres during or after S phase. Our results together with those of Haaf and Ward (1994) raise the question whether at least some components of the prekinetochore are seeded on the newly synthesized centromere DNA after replication rather than concomitantly. Chromatin modifications introduced by KAP1 might prevent efficient reassembly of these components onto the new daughter centromere. Although more detailed analysis of this idea is beyond the scope of the present manuscript, our HAC system should allow us to address this question in future experiments, possibly providing new insights into the replication of the interphase prekinetochore structure.
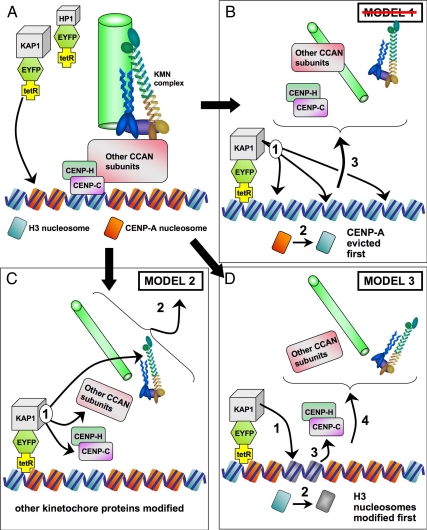
There have been numerous studies of kinetochore assembly pathways, but to our knowledge, this is the first study of single kinetochore inactivation in vivo. We have considered three models to explain the hierarchical disruption of the kinetochore induced by targeting of tet-R fusion proteins. All three models are subject to the caveat that they do not distinguish between KAP1-mediated disassembly of intact kinetochores and interference with kinetochore assembly.
In model 1 (Figure 6B), the kinetochore is built on a chromatin foundation of CENP-A nucleosomes (Palmer and Margolis, 1985; Palmer et al., 1991; Yoda et al., 2000; Black et al., 2007; Dalal et al., 2007; reviewed in Carroll and Straight, 2006; Cheeseman and Desai, 2008; Vagnarelli et al., 2008). In this model, eviction of CENP-A or prevention of its targeting caused by chromatin modifiers would disrupt the foundation for the kinetochore as a primary effect, and the structure would subsequently fall apart. This model is not consistent with our observations. We saw many kinetochores targeted by KAP1 that lacked detectable CENP-C, but had wild-type levels of CENP-A. In our previous study, binding of the tTS transcriptional repressor resulted in recruitment of high levels of H3K9me3 and the reduction of CENP-A chromatin levels at the alphoidtetO array (Nakano et al., 2008). Our present data reveal that the loss of CENP-A may follow, rather than precede, the loss of CENP-C.
Figure 6.
Models for the disruption of the alphoidtetO HAC kinetochore mediated by KAP1 targeting. (A) The alphoidtetO kinetochore, with H3 and CENP-A containing nucleosomes (blue and orange, respectively). CENP-C is associated with H3-containing nucleosomes (Hori et al., 2008). A microtubule (green) is bound by the KMN complex (Cheeseman et al., 2006) anchored through the CCAN (constitutive centromere-associated network) complex (Cheeseman and Desai, 2008). (B) Model 1, chromatin modifiers evict CENP-A and causing the kinetochore to fall apart. (C) Model 2, chromatin modifiers modify nonchromatin components, causing the kinetochore to fall apart. (D) Model 3, chromatin modifiers alter H3-containing nucleosomes, resulting in dissociation of CENP-C and dissolution of the kinetochore.
Model 2 (Figure 6C) suggests that chromatin modifiers recruited to the kinetochore by KAP1 may covalently modify inner or outer kinetochore components other than histones, leading to kinetochore destabilization. One variant of this model could involve HP1α recruited by the action of chromatin modifiers disrupting Mis12 complex assembly or function. HP1α can associate with the Mis12 complex in vivo and is required for its association with the kinetochore (Obuse et al., 2004a). Alternatively, the chromatin modifiers could act on CENP-A chromatin, weakening its links with associated subunits such as CENP-C and -H, but not causing dissociation of CENP-A.
A third model (Figure 6D) postulates that the kinetochore contains both CENP-A and H3-containing nucleosomes. CENP-C can bind DNA in vitro, and this does not require its association with CENP-A (Yang et al., 1996), and in Drosophila, CENP-C is required for efficient CENP-A localization to kinetochores (Goshima et al., 2007; Erhardt et al., 2008). Furthermore, a recent study has shown nucleosomes containing CENP-A and H3 to be locally interspersed in the kinetochore and CENP-C to be associated with H3-containing nucleosomes (Hori et al., 2008). This is consistent with an earlier study in which extensive digestion of chromatin with micrococcal nuclease resulted in a decline in the amount of CENP-B and -C recovered in CENP-A immunoprecipitates (Obuse et al., 2004b). According to model 3, chromatin modifiers recruited by KAP1 would preferentially affect H3 nucleosomes, causing loss of CENP-C without affecting the properties of CENP-A nucleosomes.
According to either of the latter two models, chromatin modifiers could cause the loss of kinetochore components without necessarily requiring the initial loss of CENP-A. Thus both models are consistent with our observations, and further experiments are required to determine the mechanism of kinetochore inactivation.
These studies represent a first description of a novel approach to the study of kinetochore function, and clearly there is much more to be done before we will fully understand the epigenetic requirements for kinetochore function. Nonetheless, the present work raises some interesting questions. How do chromatin modifiers appear to disrupt the kinetochore from the outside in, rather than from the CENP-A nucleosome outward? Is it possible that the modification of kinetochore components by chromatin modifiers could possibly have a role in observed cell cycle variations in kinetochore composition and structure? These questions can be addressed in future studies with the synthetic kinetochore.
Supplementary Material
ACKNOWLEDGMENTS
We thank David Schultz (Wistar Institute, Philadelphia, PA) for the gift of KAP1 wild-type and mutant clones. S.C. and J.H.B. were funded by Ph.D. studentships from The Darwin Trust of Edinburgh and The Wellcome Trust, respectively. This research was supported by the intramural research program of the National Institutes of Health, National Cancer Institute, Center for Cancer Research (V.L.); a grant-in-aid from the Ministry of Education, Science, Sports and Culture of Japan (H.M.); the Ministerio de Ciencia y Tecnología (SAF2007-61863; M.M.V.); and the Genome Network project and Grants-in-aid from the MEXT of Japan (H.K.). Work in the W.C.E. lab is funded by The Wellcome Trust, of which he is a Principal Research Fellow.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E09-06-0489) on August 5, 2009.
REFERENCES
- Black B. E., Jansen L. E., Maddox P. S., Foltz D. R., Desai A. B., Shah J. V., Cleveland D. W. Centromere identity maintained by nucleosomes assembled with histone H3 containing the CENP-A targeting domain. Mol. Cell. 2007;25:309–322. doi: 10.1016/j.molcel.2006.12.018. [DOI] [PubMed] [Google Scholar]
- Burns E. M., Christopoulou L., Corish P., Tyler-Smith C. Quantitative measurement of mammalian chromosome mitotic loss rates using the green fluorescent protein. J. Cell Sci. 1999;112:2705–2714. doi: 10.1242/jcs.112.16.2705. [DOI] [PubMed] [Google Scholar]
- Carroll C. W., Straight A. F. Centromere formation: from epigenetics to self-assembly. Trends Cell Biol. 2006;16:70–78. doi: 10.1016/j.tcb.2005.12.008. [DOI] [PubMed] [Google Scholar]
- Cheeseman I. M., Chappie J. S., Wilson-Kubalek E. M., Desai A. The conserved KMN network constitutes the core microtubule-binding site of the kinetochore. Cell. 2006;127:983–997. doi: 10.1016/j.cell.2006.09.039. [DOI] [PubMed] [Google Scholar]
- Cheeseman I. M., Desai A. Molecular architecture of the kinetochore-microtubule interface. Nat. Rev. Mol. Cell. Biol. 2008;9:33–46. doi: 10.1038/nrm2310. [DOI] [PubMed] [Google Scholar]
- Chen E. S., Zhang K., Nicolas E., Cam H. P., Zofall M., Grewal S. I. Cell cycle control of centromeric repeat transcription and heterochromatin assembly. Nature. 2008;451:734–737. doi: 10.1038/nature06561. [DOI] [PubMed] [Google Scholar]
- Chueh A. C., Northrop E. L., Brettingham-Moore K. H., Choo K. H., Wong L. H. LINE retrotransposon RNA is an essential structural and functional epigenetic component of a core neocentromeric chromatin. PLoS Genet. 2009;5 doi: 10.1371/journal.pgen.1000354. e1000354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins K. A., Castillo A. R., Tatsutani S. Y., Biggins S. De novo kinetochore assembly requires the centromeric histone H3 variant. Mol. Biol. Cell. 2005;16:5649–5660. doi: 10.1091/mbc.E05-08-0771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalal Y., Furuyama T., Vermaak D., Henikoff S. Structure, dynamics, and evolution of centromeric nucleosomes. Proc. Natl. Acad. Sci. USA. 2007;104:15974–15981. doi: 10.1073/pnas.0707648104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earnshaw W. C., Rothfield N. Identification of a family of human centromere proteins using autoimmune sera from patients with scleroderma. Chromosoma. 1985;91:313–321. doi: 10.1007/BF00328227. [DOI] [PubMed] [Google Scholar]
- Erhardt S., Mellone B. G., Betts C. M., Zhang W., Karpen G. H., Straight A. F. Genome-wide analysis reveals a cell cycle-dependent mechanism controlling centromere propagation. J. Cell Biol. 2008;183:805–818. doi: 10.1083/jcb.200806038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman J. R., Fredericks W. J., Jensen D. E., Speicher D. W., Huang X. P., Neilson E. G., Rauscher F. J., 3rd. KAP-1, a novel corepressor for the highly conserved KRAB repression domain. Genes Dev. 1996;10:2067–2078. doi: 10.1101/gad.10.16.2067. [DOI] [PubMed] [Google Scholar]
- Fukagawa T. The kinetochore and spindle checkpoint in vertebrate cells. Front Biosci. 2008;13:2705–2713. doi: 10.2741/2877. [DOI] [PubMed] [Google Scholar]
- Fukagawa T., Nogami M., Yoshikawa M., Ikeno M., Okazaki T., Takami Y., Nakayama T., Oshimura M. Dicer is essential for formation of the heterochromatin structure in vertebrate cells. Nat. Cell Biol. 2004;6:784–791. doi: 10.1038/ncb1155. [DOI] [PubMed] [Google Scholar]
- Goshima G., Wollman R., Goodwin S. S., Zhang N., Scholey J. M., Vale R. D., Stuurman N. Genes required for mitotic spindle assembly in Drosophila S2 cells. Science. 2007;316:417–421. doi: 10.1126/science.1141314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haaf T., Ward D. C. Structural analysis of alpha-satellite DNA and centromere proteins using extended chromatin and chromosomes. Hum. Mol. Genet. 1994;3:697–709. doi: 10.1093/hmg/3.5.697. [DOI] [PubMed] [Google Scholar]
- Harrington J. J., Van Bokkelen G., Mays R. W., Gustashaw K., Willard H. F. Formation of de novo centromeres and construction of first-generation human artificial microchromosomes. Nat. Genet. 1997;15:345–355. doi: 10.1038/ng0497-345. [DOI] [PubMed] [Google Scholar]
- Hill A., Bloom K. Genetic manipulation of centromere function. Mol. Cell. Biol. 1987;7:2397–2405. doi: 10.1128/mcb.7.7.2397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hori T., Okada M., Maenaka K., Fukagawa T. CENP-O class proteins form a stable complex and are required for proper kinetochore function. Mol. Biol. Cell. 2008;19:843–854. doi: 10.1091/mbc.E07-06-0556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeno M., Grimes B., Okazaki T., Nakano M., Saitoh K., Hoshino H., McGill N. I., Cooke H., Masumoto H. Construction of YAC-based mammalian artificial chromosomes. Nat. Biotechnol. 1998;16:431–439. doi: 10.1038/nbt0598-431. [DOI] [PubMed] [Google Scholar]
- Ivanov A. V., et al. PHD domain-mediated E3 ligase activity directs intramolecular sumoylation of an adjacent bromodomain required for gene silencing. Mol. Cell. 2007;28:823–837. doi: 10.1016/j.molcel.2007.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura H., Hayashi-Takanaka Y., Goto Y., Takizawa N., Nozaki N. The organization of histone H3 modifications as revealed by a panel of specific monoclonal antibodies. Cell Struct. Funct. 2008;33:61–73. doi: 10.1247/csf.07035. [DOI] [PubMed] [Google Scholar]
- Le Douarin B., Nielsen A. L., Garnier J. M., Ichinose H., Jeanmougin F., Losson R., Chambon P. A possible involvement of TIF1 alpha and TIF1 beta in the epigenetic control of transcription by nuclear receptors. EMBO J. 1996;15:6701–6715. [PMC free article] [PubMed] [Google Scholar]
- Masumoto H., Ikeno M., Nakano M., Okazaki T., Grimes B., Cooke H., Suzuki N. Assay of centromere function using a human artificial chromosome. Chromosoma. 1998;107:406–416. doi: 10.1007/s004120050324. [DOI] [PubMed] [Google Scholar]
- Matsuda E., Agata Y., Sugai M., Katakai T., Gonda H., Shimizu A. Targeting of Kruppel-associated box-containing zinc finger proteins to centromeric heterochromatin. Implication for the gene silencing mechanisms. J. Biol. Chem. 2001;276:14222–14229. doi: 10.1074/jbc.M010663200. [DOI] [PubMed] [Google Scholar]
- Mythreye K., Bloom K. S. Differential kinetochore protein requirements for establishment versus propagation of centromere activity in Saccharomyces cerevisiae. J. Cell Biol. 2003;160:833–843. doi: 10.1083/jcb.200211116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano M., Cardinale S., Noskov V. N., Gassmann R., Vagnarelli P., Kandels-Lewis S., Larionov V., Earnshaw W. C., Masumoto H. Inactivation of a human kinetochore by specific targeting of chromatin modifiers. Dev. Cell. 2008;14:507–522. doi: 10.1016/j.devcel.2008.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obuse C., Iwasaki O., Kiyomitsu T., Goshima G., Toyoda Y., Yanagida M. A conserved Mis12 centromere complex is linked to heterochromatic HP1 and outer kinetochore protein Zwint-1. Nat. Cell Biol. 2004a;6:1135–1141. doi: 10.1038/ncb1187. [DOI] [PubMed] [Google Scholar]
- Obuse C., Yang H., Nozaki N., Goto S., Okazaki T., Yoda K. Proteomics analysis of the centromere complex from HeLa interphase cells: UV-damaged DNA binding protein 1 (DDB-1) is a component of the CEN-complex, while BMI-1 is transiently co-localized with the centromeric region in interphase. Genes Cells. 2004b;9:105–120. doi: 10.1111/j.1365-2443.2004.00705.x. [DOI] [PubMed] [Google Scholar]
- Ohzeki J., Nakano M., Okada T., Masumoto H. CENP-B box is required for de novo centromere chromatin assembly on human alphoid DNA. J. Cell Biol. 2002;159:765–775. doi: 10.1083/jcb.200207112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada M., Cheeseman I. M., Hori T., Okawa K., McLeod I. X., Yates J. R., Desai A., Fukagawa T. The CENP-H-I complex is required for the efficient incorporation of newly synthesized CENP-A into centromeres. Nat. Cell Biol. 2006;8:446–457. doi: 10.1038/ncb1396. [DOI] [PubMed] [Google Scholar]
- Okada T., Ohzeki J., Nakano M., Yoda K., Brinkley W. R., Larionov V., Masumoto H. CENP-B controls centromere formation depending on the chromatin context. Cell. 2007;131:1287–1300. doi: 10.1016/j.cell.2007.10.045. [DOI] [PubMed] [Google Scholar]
- Palmer D. K., Margolis R. L. Kinetochore components recognized by human autoantibodies are present on mononucleosomes. Mol. Cell. Biol. 1985;5:173–186. doi: 10.1128/mcb.5.1.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer D. K., O'Day K., Le Trong H., Charbonneau H., Margolis R. L. Purification of the centromeric protein CENP-A and demonstration that it is a centromere specific histone. Proc. Nat. Acad. Sci. USA. 1991;88:3734–3738. doi: 10.1073/pnas.88.9.3734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pluta A. F., Mackay A. M., Ainsztein A. M., Goldberg I. G., Earnshaw W. C. The centromere: Hub of chromosomal activities. Science. 1995;270:1591–1594. doi: 10.1126/science.270.5242.1591. [DOI] [PubMed] [Google Scholar]
- Saffery R., Sumer H., Hassan S., Wong L. H., Craig J. M., Todokoro K., Anderson M., Stafford A., Choo K. H. Transcription within a functional human centromere. Mol. Cell. 2003;12:509–516. doi: 10.1016/s1097-2765(03)00279-x. [DOI] [PubMed] [Google Scholar]
- Schultz D. C., Ayyanathan K., Negorev D., Maul G. G., Rauscher F. J., 3rd. SETDB1, a novel KAP-1-associated histone H3, lysine 9-specific methyltransferase that contributes to HP1-mediated silencing of euchromatic genes by KRAB zinc-finger proteins. Genes Dev. 2002;16:919–932. doi: 10.1101/gad.973302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz D. C., Friedman J. R., Rauscher F. J., 3rd. Targeting histone deacetylase complexes via KRAB-zinc finger proteins: the PHD and bromodomains of KAP-1 form a cooperative unit that recruits a novel isoform of the Mi-2alpha subunit of NuRD. Genes Dev. 2001;15:428–443. doi: 10.1101/gad.869501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shojaei F., Gallacher L., Bhatia M. Differential gene expression of human stem progenitor cells derived from early stages of in utero human hematopoiesis. Blood. 2004;103:2530–2540. doi: 10.1182/blood-2003-09-3209. [DOI] [PubMed] [Google Scholar]
- Sripathy S. P., Stevens J., Schultz D. C. The KAP1 corepressor functions to coordinate the assembly of de novo HP1-demarcated microenvironments of heterochromatin required for KRAB zinc finger protein-mediated transcriptional repression. Mol. Cell. Biol. 2006;26:8623–8638. doi: 10.1128/MCB.00487-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vagnarelli P., Ribeiro S. A., Earnshaw W. C. Centromeres: old tales and new tools. FEBS Lett. 2008;582:1950–1959. doi: 10.1016/j.febslet.2008.04.014. [DOI] [PubMed] [Google Scholar]
- Valdivia M. M., Figueroa J., Iglesias C., Ortiz M. A novel centromere monospecific serum to a human autoepitope on the histone H3-like protein CENP-A. FEBS Lett. 1998;422:5–9. doi: 10.1016/s0014-5793(97)01583-4. [DOI] [PubMed] [Google Scholar]
- Volpe T., Schramke V., Hamilton G. L., White S. A., Teng G., Martienssen R. A., Allshire R. C. RNA interference is required for normal centromere function in fission yeast. Chromosome Res. 2003;11:137–146. doi: 10.1023/a:1022815931524. [DOI] [PubMed] [Google Scholar]
- Volpe T. A., Kidner C., Hall I. M., Teng G., Grewal S. I., Martienssen R. A. Regulation of heterochromatic silencing and histone H3 lysine-9 methylation by RNAi. Science. 2002;297:1833–1837. doi: 10.1126/science.1074973. [DOI] [PubMed] [Google Scholar]
- Wong L. H., et al. Centromere RNA is a key component for the assembly of nucleoproteins at the nucleolus and centromere. Genome Res. 2007;17:1146–1160. doi: 10.1101/gr.6022807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C. H., Tomkiel J., Saitoh H., Johnson D. H., Earnshaw W. C. Identification of overlapping DNA-binding and centromere-targeting domains in the human kinetochore protein CENP-C. Mol. Cell. Biol. 1996;16:3576–3586. doi: 10.1128/mcb.16.7.3576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoda K., Ando S., Morishita S., Houmura K., Hashimoto K., Takeyasu K., Okazaki T. Human centromere protein A (CENP-A) can replace histone H3 in nucleosome reconstitution in vitro. Proc. Natl. Acad. Sci. USA. 2000;97:7266–7271. doi: 10.1073/pnas.130189697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng L., Yap K. L., Ivanov A. V., Wang X., Mujtaba S., Plotnikova O., Rauscher F. J., 3rd,, Zhou M. M. Structural insights into human KAP1 PHD finger-bromodomain and its role in gene silencing. Nat. Struct. Mol. Biol. 2008;15:626–633. doi: 10.1038/nsmb.1416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziv Y., Bielopolski D., Galanty Y., Lukas C., Taya Y., Schultz D. C., Lukas J., Bekker-Jensen S., Bartek J., Shiloh Y. Chromatin relaxation in response to DNA double-strand breaks is modulated by a novel ATM- and KAP-1 dependent pathway. Nat. Cell Biol. 2006;8:870–876. doi: 10.1038/ncb1446. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.