Abstract
The objectives of this study are as follows: to describe practical implementation challenges of multisite, multivendor quantitative studies; to describe the MRI phantom and analysis software used in the Alzheimer’s Disease Neuroimaging Initiative (ADNI) study, illustrate the utility of the system for measuring scanner performance, the ability to assess gradient field nonlinearity corrections: and to recover human brain images without geometric scaling errors in multisite studies. ADNI is a large multicenter study with each center having its own copy of the phantom. The design of the phantom and analysis software are presented as results from predistribution systematics studies and results from field experience with the phantom at 58 enrolling ADNI sites over a 3 year period. The estimated coefficients of variation intrinsic to measurements of geometry in a single phantom are in the range of 3–5 parts in 104. Phantom measurements accurately detect linear and nonlinear scaling in images. Gradient unwarping methods are readily assessed by phantom nonlinearity measurements. Phantom-based scaling correction reduces observed geometric drift in human images by one-third or more. Repair or replacement of phantoms between scans, however, is a confounding factor. The ADNI phantom can be used to assess both scanner performance and the validity of postprocessing image corrections in order to reduce systematic errors in human images. Reduced measurement errors should decrease measurement bias and increase statistical power for measurements of rates of change in the brain structure in AD treatment trials. Perhaps the greatest practical value of incorporating ADNI phantom measurements in a multisite study is to identify scanner errors through central monitoring. This approach has resulted in identification of system errors including sites misidentification of their own gradient hardware and the disabling of autoshim, and a miscalibrated laser alignment light. If undetected, these errors would have contributed to imprecision in quantitative metrics at over 25% of all enrolling ADNI sites.
Keywords: phantom, multicenter trial, Alzheimer’s Disease Neuroimaging Initiative, ADNI
INTRODUCTION
Alzheimer’s disease (AD) is a progressive neurodegenerative disorder affecting millions of people. The pharmaceutical industry is developing disease modifying treatments with the hope of slowing the rate of neurodegeneration. Longitudinal measurements of brain structure change (typically shrinkage due to neurodegeneration) have been used as a measure of the rate of AD progression. The statistical power to detect brain structure change, including slowing of progression due to effective treatment interventions is limited by measurement errors in addition to biological intrasubject variability. One of the goals of Alzheimer’s Disease Neuroimaging Initiative (ADNI) is to develop improved methods for acquiring and processing MRI and PET data in order to reduce measurement error and thus improve statistical power to detect change in brain structures. A component of that development includes an MRI phantom for tracking and possibly correcting scanner performance.
A natural history study of aging and dementia, ADNI, is jointly funded by the National Institutes of Health and industry via the Foundation for the NIH. The full study will run over five years, following approximately 800 subjects each for 24–36 months. Recruited at 58 clinical enrollment sites, ADNI subjects are scanned on 58 different 1.5 T scanners and 33 3 T scanners. In the ADNI MRI protocol, a phantom is scanned immediately after each subject scan. Each site has an identical copy of the phantom (Phantom Laboratory, Salem, New York) designed for ADNI to measure linear and nonlinear spatial distortion, signal-to-noise ratio (SNR), and image contrast. ADNI images (including those of the phantom and human subjects) are publicly available to researchers via the worldwide web (www.loni.ucla.edu∕ADNI∕).
Previous studies of phantom performance1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21 have been in limited environments, studying small numbers of phantoms and∕or scanners over modest time intervals. The scale of ADNI requires that a fleet of phantoms perform accurately over years. Also, in multicenter trials the selection of enrollment sites is driven by the ability of sites to enroll adequate numbers of subjects that meet clinical inclusion criteria rather than availability of particular MR imaging equipment or access to physics support. Most subjects are scanned on clinical scanners, and locally determined clinical needs drive decisions regarding maintenance and upgrades.
A key assumption when including phantom measurements in this study was that the phantom captures information about the scanner that is applicable to associated human images. Only if that assumption holds can the phantom be used to disentangle instrumental drifts from biological variations and pathological change. Here we assess the utility of phantom-based retrospective data correction for improving intrasubject image consistency.
Designed for scanner calibration, the ADNI phantom can be used both to track scanner changes and to verify that reconstruction operations such as off-line gradient warping corrections are correctly implemented. Phantom-based assessments include (1) geometrical uniformity, (2) SNR, and (3) CNR. This report will focus on the information about scan acquisition geometry obtained through ADNI phantom measurements.
The objectives of this paper are as follows:
-
(1)
to describe the methods used to extract scanner performance data from the phantom scans;
-
(2)
to report the sensitivity of the phantom to changes in scanner performance, calibration, and∕or imaging parameters;
-
(3)
to document the variability of the ADNI phantom fleet at baseline;
-
(4)
to demonstrate the efficacy of the phantom for ensuring that scanner-dependent postprocessing reconstruction is correctly implemented; and
-
(5)
to describe longitudinal tracking information on scanners used in ADNI as well as the range of per-scanner summary statistics.
MATERIALS AND METHODS
Description of ADNI MR image data and study design
ADNI subjects are evaluated at 6–12 month intervals for 2–3 years depending on the clinical diagnosis at baseline. All subjects are scanned at 1.5 T at each time point, half are scanned with FDG PET. Subjects not assigned to the PET arm of the study are eligible for 3 T MRI scanning. The goal is to acquire both 1.5 T and 3 T MRI studies at multiple time points in 25% of the subjects.
Employing an MP-RAGE (Ref. 22) sequence, 3 D T1-weighted structural images are the focus of the ADNI protocol23 to measure rates of brain atrophy. Defined across selected systems from GE Healthcare, Philips Medical Systems and Siemens Medical Solutions with an eye toward minimizing cross-platform differences, the nominal TI∕TR∕TE at 1.5 T for the ADNI MP-RAGE are 1000∕2400∕minimum full ∼5 ms. On systems with birdcage head coils, TR is increased to 3000 ms to compensate for reduced SNR. Platform-specific protocols were distributed digitally through the MRI vendors to minimize inconsistencies expected to arise from building the protocol manually on individual scanners. Detailed platform-specific lists of parameters are provided at http:∕∕www.loni.ucla.edu∕ADNI∕Research∕Cores∕.
In the ADNI data flow, each MP-RAGE scan undergoes 3D gradient unwarping correction during image preprocessing if applicable. Unlike these references,1, 15, 19, 20, 21 the ADNI phantom is not used to correct nonlinearities. Instead, vendor supplied parametrizations of gradient nonlinearity are used to correct image warping off line, and the phantom is used to verify the correction.
Phantom design and analysis
Phantom design
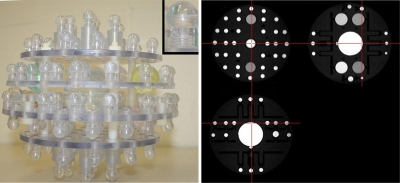
The ADNI phantom consists of spherical inclusions inside a 20 cm diameter water-filled clear urethane shell. Inclusions are copper sulfate filled polycarbonate spherical shells. Approximately 1.4 mm thick, the shells are injection molded polycarbonate with threaded mounts, which, in turn, screw into polycarbonate plates. The lengths of the mounts are varied to follow the curvature of the outer shell as necessary. Plates are positioned by using nylon tubular spacers. Threaded nylon rods pass through the plates and spacers. The assembly is held together by nylon nuts. The interior assembly is shown in Fig. 1. The inclusions are summarized as follows:
Fiducial spheres: Located in an unambiguous pattern, 158 1.0 cm diameter inclusions and 2 1.5 cm diameter inclusions with 3.3 mM copper sulfate solution are used for geometrical measurement.
SNR sphere: A single 6.0 cm diameter sphere that is approximately concentric with the outer shell, containing 3.3 mM copper sulfate solution is used for SNR measurement.
CNR spheres: Four 3.0 cm diameter spheres with copper sulfate concentrations of 0.9, 1.2, 1.7, and 2.4 mM provide contrast information. The T1 relaxation times range from approximately 400 to 1200 ms at 1.5 T to roughly span the range for brain tissue.
Solutions were mixed in large quantities to reduce phantom-to-phantom variability in copper sulfate concentration.
Figure 1.
ADNI phantom. A photograph of the internal components of the ADNI phantom is shown. Each of the spheres is filled with a copper sulfate solution. The colored spheres contain differing solution concentrations. The small inset provides a detailed view of a single sphere and postcomponent. A triplanar view of a phantom image acquired with the MP-RAGE used in the ADNI protocol is also shown.
Analysis software
Fully automatic, the analysis of phantom images uses a hierarchical approach, finding first the large SNR sphere, then the fiducial spheres, and lastly the CNR spheres. Complete containment of all inclusions within the image volume is required. T1-weighted images such as spoiled gradient echo, spoiled FLASH, IR-SPGR, or MP-RAGE are assumed. The analysis software was written in MATLAB (MathWorks Natick, Massachusetts) and executes in 7–12 min on a 3 GHz Pentium IV processor. The software, including a table of relative sphere locations, is available online through the ADNI web site (http:∕∕www.loni.ucla.edu∕ADNI∕Data∕). Throughout the software sanity checks are implemented with the software aborting execution of major problems are detected. Given ADNI images the analysis aborts on less than 0.1% of scans.
Pattern recognition and sphere finding
Although phantom orientation is specified in ADNI scanning protocol, the analysis is orientation insensitive. The SNR sphere is found first, and then the 1.5 cm spheres at expected distances from the SNR sphere center form a unique coordinate system. After establishing a coordinate system, the 1.0 cm spheres are found by searching in the neighborhoods where they are expected.
SNR sphere location and analysis
The SNR sphere is the largest short-T1 object in the phantom. Let the number of voxels contained in the SNR sphere be VSNR. Otsu’s method24 defines an initial threshold, which is then adjusted down until at least 0.8×VSNR voxels are suprathreshold. Clusters of suprathreshold voxels are found. If no single cluster of at least 0.5×VSNR is found, the analysis aborts. The threshold is adjusted until between 98% and 102% of the expected number of voxels around the largest region are suprathreshold, aborting if no suitable threshold is found. The SNR sphere center is taken as the intensity-weighted average position of suprathreshold voxels. The SNR sphere was not designed for precision spatial measurement and the observed spatial manufacturing variability is of the order of a millimeter. Thus, it is used as a rough anchor in finding other navigator spheres.
SNR measurement in the phantom is carried out in three steps. The cluster is eroded by nine passes with “center plus six nearest neighbors” structuring element. A least-squares fit of a smooth quadratic function in 3D is made to the remaining voxels as a bias correction. The noise level is estimated from the standard deviation of the residual intensities after subtracting the smooth function. The signal level is the average of the cluster of voxels after erosion.
Fiducial sphere finding and analysis
The two 1.5 cm fiducial spheres are located 60 and 90 mm from the center of the phantom, respectively. Normalized 2D cross correlation with a 1.5 cm circular template is calculated slice by slice. Correlation maxima in 2 cm thick shells with radii of 60 and 90 mm centered on the SNR sphere center are found. Subregions centered on the locations of the maxima are selected, segmented by Otsu’s method, clusters found, and positions estimated. Given the positions of the 1.5 cm fiducial spheres and the center of the SNR sphere, a provisional linear coordinate transformation with three rotations, translations, and scalings providing nine degrees of freedom (9DOF) is created. The transformation maps sphere positions in a coordinate system local to the phantom to the image coordinate system. Analysis aborts unless both 1.5 cm spheres are found.
Using the provisional transformation, a list of 1.0 cm fiducial marker locations is transformed into the image coordinate system. Working from the center of the phantom outward, the 1.0 cm spheres are found. After three or more 1.0 cm spheres have been found, the provisional coordinate transformation is updated using the 1.5 cm sphere locations, the observed 1.0 cm sphere locations and excluding the SNR sphere location.
The 1.0 cm sphere finding scheme is as follows.
-
(1)
A 3.0×3.0×3.0 cm3 subregion of the image centered on the expected sphere location is extracted. Even in images acquired with most nonlinear gradients for ADNI, this empirically provides a sufficient margin to allow 1.0 cm sphere to be wholly contained in the subregion.
-
(2)
Cross-correlation maps of a 1.0 cm bright sphere on a dark background are calculated over the subregion.
-
(3)
Cross-correlation maps of a 1.0 cm bright sphere surrounded by a 2 voxel thick dark shell on a bright background are calculated over the subregion.
-
(4)
The product of the maps is formed and the location of the maximum is found. If the maximum correlation product occurs because both the individual maps were populated by negative correlations, then the search for this sphere is aborted.
-
(5)
A threshold is estimated which selects voxels occupying 80% of the volume of a 1.0 cm sphere. The subregion is thresholded and clusters of suprathreshold voxels are found. The threshold is lowered by 1% until a cluster is found which overlaps the correlation maximum and contains between 75% and 110% of the expected volume. If the threshold drops below 10% of the maximum intensity in the subimage, the search for this sphere is aborted. The inability to find individual spheres does not stop the search for others.
-
(6)
The final sphere center estimate is found as the intensity-weighted average position of voxels in the sphere.
-
(7)
Dilation is performed on the voxel cluster to include the shell. The mean intensity of a two voxel thick band immediately outside the shell is taken as an estimate of local water background. Tracking the ratio of mean intrasphere intensity to background allows dim sphere detection.
Early production phantoms were found to develop leaks in the small inclusion leading to a loss of contrast. The inclusion of cross correlation in step (3) is to aid in the detection of inclusions which have leaked.
CNR sphere analysis
Given the transformation of fiducial sphere positions, the locations of the CNR spheres are well established. One CNR sphere at time, the 3.8×3.8×3.8 cm3 subregion centered on the expected location is extracted from the image data. Using Otsu’s method, an initial threshold is estimated. Cluster finding is applied to suprathreshold voxels, and the single cluster with a volume between 75% and 110% of the expected volume is found. CNR intensity values are taken as the mean intensity of voxels in the clusters. Local water background is estimated in a fashion analogous to the fiducial sphere finding.
Extracting MRI system geometric information
Comparison of the expected (i.e., nominal) and observed positions of the fiducial spheres provides information on the scanner geometric performance. Two types of geometric information are extracted from the list of observed and nominal positions; a linear component loosely identified with the gradient calibration and an estimate of nonlinearly spatially varying displacements attributed to field nonlinearities.
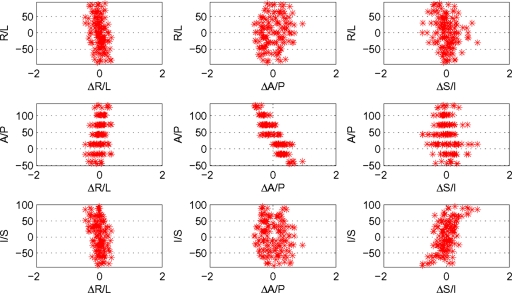
A simple set of plots permits qualitative evaluation of the scanner performance. Viewing of the plots is included in the ADNI data quality control workflow and allows a rapid (<10 s) evaluation by inspection. The expected and observed positions are temporarily registered using only translations and rotations. Define Δαi as the difference between the sphere positions as observed and as designed in the α direction for the ith sphere. Working in the coordinate system of the acquired images (e.g., X, Y, and Z being equivalent to R∕L, A∕P, and S∕I for a human laying head-first-supine in the magnet) plots are made of the locations in each direction versus residuals ΔX, ΔY, and ΔZ. Ideally, the distribution of residuals would be independent of position, centered on zero and narrow. Registration forces the mean residual to be zero. Linear dependence of Δα on coordinate α may be attributed to gradient miscalibration. Non-linear dependence is associated with magnetic field nonideality. Figures 23 present such plots and will be discussed in the Results section.
Figure 2.
Qualitative evaluation of geometric performance. Plots of sphere position (vertical axes) versus displacement (horizontal axes) in each cardinal direction provide qualitative image distortion information. All lengths are measured in mm, and the position origin is MR scanner isocenter.
Figure 3.
Calibration exercise. Sets of plots with data before (left) and after (right) an exercise in scanner calibration are shown. After calibration, the dependence of position residuals (the horizontal axes in the subplots) on position (the vertical axes in the subplots) was greatly reduced. The two obvious outlier points in the A∕P versus ΔA∕P subplots were due to manufacturing defects and subsequently repaired.
Linear (scaling) and nonlinearity measures
After a registration between expected and observed positions with 9DOF, the transformation is decomposed to extract the linear scale factors which would be applied along the axes of the MRI scanner to bring the image into agreement with theory.
To determine nonlinear measures, a vector displacement field parametrized by low-order polynomial functions is fitted that minimizes the distances between observed and nominal positions. For each fiducial marker the residual distance calculated. Summary statistics on the distribution of residual distances are calculated. To assess nonlinearity, the summary statistics after fitting with first-order polynomial function are found.
Studies prior to phantom distribution (systematics)
Prior to distributing phantoms to ADNI scanning sites in the field, we performed several studies to determine the extent to which the phantom can provide meaningful measurements of scanner performance including the following; verify the ability of the system to detect deliberately introduced gradient amplitude scaling, measure stability of serial measurements with a single phantom, measure variability across the fleet of ADNI phantoms ascribed to manufacturing variability, and investigate gradient unwarping by polynomial geometry measures with the phantom.
Longitudinal measures of scaling in ADNI scanners
Variability in linear geometry (i.e., scaling) of the scanners in ADNI was measured over time with serial phantom images. In Sec. 3, we show that measurements of scan scaling change discretely over time with system recalibration. Thus, to estimate the underlying stability of scanners, a pooled variance approach is used. Each axis (R∕L, A∕P, and S∕I) is considered separately. Phantom measures of image scaling are ordered by time and clusters are found as follows: (1) The standard deviations of measurements in a four measurement-wide sliding window are found. (2) The window of data with minimum standard deviation is taken as a cluster seed. (3) A cluster width is defined as the maximum of the cluster standard deviation and 3.5×10−4 (an estimate of the single phantom measurement variability). As the mean scale factors are very close to unity the standard deviation of scale factors, for practical purposes, the same as the coefficient of variation. (5) Adjacent-in-time points are added to the cluster until a point more than 2.58 cluster widths is encountered. That is, points between the first and 99th percentile are added. (6) Points assigned to a cluster are marked as “used” and steps (2)–(5) are repeated until there are no unused strings of data four or more points long. The pooled variance over the clusters is then calculated. Cluster delineation is also visually inspected.
Use of phantom measurements to correct linear scaling changes in human images
In order to assess the efficacy of phantom-based measurements to correct linear scaling changes in human images, we selected image pairs of subjects scanned serially in ADNI. Coregistration was performed using AIR25 allowing rotation, translation, and scaling (9DOF) on pairs of images from the same subjects. Masks including the skull and its contents and excluding the neck were created for each case. The registration targets, each subject’s skull and contents, were therefore spatially invariant over time. Unlike the brain itself, which does change over time, the skull does not. Thus, the anatomic target used for registration here gives an independent measure of change in the gradient scaling over time. No clinical criteria were used in selecting subjects and images used. Two versions of each image, with and without phantom-based scaling correction, were analyzed. From the uncorrected pairs of images sets of coregistration scalings were found. Independently, the phantom-corrected pairs were also coregistered generating another set of scaling parameters. If the phantom captures gradient calibration information which is also applicable to the companion human images, then the distribution of scaling parameters over many pairs of subject images is expected to be narrower and centered closer to unity for the phantom-corrected image pairs than for uncorrected pairs.
RESULTS AND DISCUSSIONS
Qualitative evaluation of geometric performance
Shown in Fig. 2 is a typical result for a representative scanner used in ADNI. The image was acquired sagittally and the scanner performs an in-plane gradient warping correction. 3D gradient warping correction was done in postprocessing. Interpretation of the plots is as follows:
The plot in the upper left corner reveals a slight linear dependence of ΔR∕L on R∕L position. The slope of the distribution is consistent with the image FOV being stretched by approximately 0.1%.
The middle plot in Fig. 2 (A∕P position versus ΔA∕P) reveals a stronger linear dependence. Here the FOV appears to be 0.6% too large. The phantom was placed in the magnet with the plates lying in coronal planes. Mounted at fixed distances from the plates, the spheres cluster at discrete A∕P locations.
An example of a nonlinear distortion is seen in the lower right panel. The curved distribution of the points in the S∕I versus ΔS∕I plot indicates systematically increasing distortion. A linear fit to the data is consistent with the FOV being approximately 0.3% too small.
Off-diagonal plots provide insight into the dependence warping in a given direction on position in another. While the plots on the diagonal can be linked to gradient amplitude, divergences in the off-diagonal plots away from isocenter are indicative of warping which is spatially dependent. Because the fiducial markers are distributed within a sphere, markers at the extreme of any given dimension are closer to the center of the phantom in the other two directions. Hence bulges near the center of a distribution on an off-diagonal plot appear because the sampling is limited.
Little off-diagonal structure is typically found for scanners used in ADNI.
The phantom analysis algorithm has proved to be robust, processing thousands of MR volumes and failing only when the images presented contain gross artifacts and errors such as missing slices, incorrectly ordered slices, and incomplete coverage.
Calibration exercise
A simple exercise was performed in order to verify the ability of the system to detect deliberately introduced gradient amplitude scaling (Fig. 3). The phantom was scanned, the scale factors derived from that scan were multiplicatively applied to the gradient amplifier settings and the phantom was rescanned. Prior to adjustment, the scale factors were (1.0053, 1.0058, and 1.0067)—approximately 1 mm accumulated stretching of a 200 mm diameter phantom in all dimensions. After adjustment, the scale factors were (1.0004, 0.999 98, and 1.0002)—less than 0.1 mm accumulated stretching over 200 mm.
Stability of serial measurements with a single “master” phantom
Nine images acquired back to back on a single scanner at Mayo Clinic were used to estimate variation intrinsic to measurements in a single master phantom. The phantom was repositioned between scans and uncertainty is driven largely by phantom positioning within the scanner. The coefficient of variation was 3–5 parts in 104.
Measurement uncertainty and construction variability across the ADNI phantom fleet
Strict agreement of the phantom construction with design is assumed in the analysis. Acceptance testing was done on all phantoms used in ADNI to evaluate the correctness of assembly and to estimate the variability of construction. Qualitative assessment of analysis output plots to search for manufacturing problems was carried out on all phantoms. Loosely inserted threaded mounts were the most common error in early production units. Phantoms with construction problems were returned to the manufacturer for repair or replacement. Repaired units were reassessed before being accepted for use.
Assessing construction variability over a period of months requires correction for potential drift of the scanner on which acceptance testing was done. A single phantom was selected as a master reference unit. The master phantom was scanned when any other phantom underwent acceptance testing and phantoms under test were normalized to the scanner performance estimated from the master phantom scan. Histograms of the ratios of observed test phantom scaling normalized to master phantom scaling are shown in Fig. 4 for the first 66 ADNI production phantoms. The coefficients of variation in the ratios are 3.5×10−4, 15×10−4, and 4.8×10−4 in the nominal R∕L, A∕P, and S∕I directions, respectively. The R∕L and S∕I directions represent within-plate variability and are consistent with the level of variability introduced by differences in positioning and precise machining. Variability perpendicular to the plates is three to four times larger than within-plate variability and also larger than the estimated measurement variability due to positioning. To maximize longitudinal stability, measurements should ideally be made using only one phantom.
Figure 4.
Construction variability. Histograms of normalized phantom size for the initial 66 production phantoms used in the ADNI study are shown.
Related to construction variability is the issue of manufacturing defects. Early production phantoms commonly had issues with leaking fiducial makers. Manufacturing and analysis software improvements have reduced both the incidence of leakage and the necessity to replace phantoms in which leaking spheres are found. The next most common occurrence necessitating replacement is when the detachment of the large 6 cm SNR sphere from its mounting post. This has occurred four times in 66 phantoms over 3 years, generally when the unit is dropped or rolls off a table. No other issues requiring phantom replacement have been found.
Nonlinearity estimates
As discussed previously, the expected and observed positions are used to fit deformation fields parametrized by low-order polynomial functions. Histograms of the residual radii for different polynomial orders are shown for a phantom scan acquired with on-line 2D [Fig. 5a] and 3D off-line gradient warping correction [Fig. 5b]. In each of Figs. 5a, 5b the distributions are shown for different orders of fitted polynomial (vertical axes). Although the data (radii) are clearly not normally distributed, the standard deviation still serves as a useful summary value. In Fig. 5c the evolution of the standard deviation with polynomial order shows the difference between 2D and 3D correction. Allowing only first-order correction (equivalent to post-hoc adjustment of the image FOV), 3D-corrected data (solid red circles) has a much smaller standard deviation than 2D-corrected data. These data illustrate the value of full 3D correction for gradient nonlinearity. The data also illustrate the fact that the ADNI phantom and analysis method can detect these effects with good sensitivity.
Figure 5.
Nonlinearity estimates. The dependence of the residual radius distribution for different orders of polynomial displacement field is shown for a scan with 2D (left) and 3D (middle) gradient warping corrections. In these plots, each horizontal row contains a histogram of 160 residual radii for a deformation field of given polynomial order (which is indicated on the vertical axes). The sizes of the boxes in the plots are proportional to the density of points. The rightmost plot presents the standard deviation of the distributions for data with 2D warping correction (open stars) and for 3D warping correction (solid red circles).
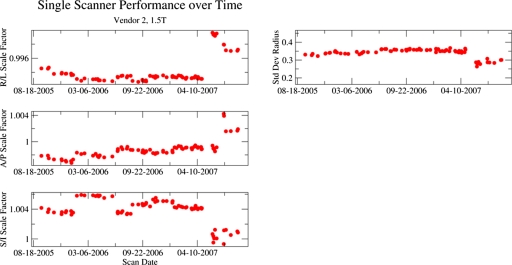
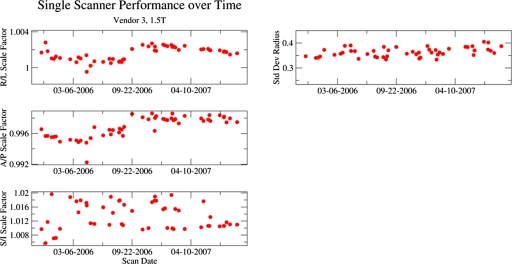
Longitudinal tracking of individual scanners with phantom measurements
Figures 67 contain longitudinal tracking information representative of a scanner from two of the MR system vendors used in ADNI. Each figure presents data acquired on a 1.5 T system from a relatively high-enrolling ADNI site. Each site imaged only a single phantom (i.e., no replacement necessary) in these data. In each figure, the left panel demonstrates scale factors along each cardinal axis. The right panel shows the standard deviation of residual radius (nonlinearity).
Figure 6.
Longitudinal tracking of individual scanner from vendor 2 with phantom measurements. The left panel demonstrates scale factors along each cardinal axis. The right panels show the standard deviation of residual radius (nonlinearity). The system was recalibrated in early and mid-2006 as well as mid-2007 when the system underwent an upgrade. After the upgrade, the standard deviation of residual radius metric for nonlinearity was decreased.
Figure 7.
Longitudinal tracking of individual scanner from vendor 3 with phantom measurements. The left panel demonstrates scale factors along each cardinal axis. The right panels show the standard deviation of residual radius (nonlinearity). Prior to mid-2007, the protocol for this vendor was errantly distributed with autoshimming disabled, a fact reflected in the larger variation in the S∕I scale factors. Note that the vertical range for the S∕I scale factor time course is larger than for other dimensions.
The scanner in Fig. 6 shows four clusters of scale factors. The A∕P scale factor shows drifting between calibrations in 2006. Discrete steps in 2006 were approximately 1∕512. The jump in 2007 occurred with a system upgrade in which the RF equipment was upgraded, the main magnetic field reshimmed and the gradient coils replaced (though with the same basic model as previously present). The nonlinearity measure was reduced by the system upgrade. The reasons for the improvement are not known but could be related to improved production methods for the new gradient coils, reshimming of the main magnetic field as well as other changes in the hardware and software. This highlights one of the challenges of involvement in multicenter studies. It can be difficult to know precisely what has been done to a scanner when changes are observed. Scan timing parameters were unchanged as called for in the ADNI study and accordingly the relative contrast values were unchanged with the upgrade.
In Fig. 7, the R∕L and A∕P scale factors show evidence of slow drifting, but no large discrete jumps. The S∕I scale factor is erratic. Prior to mid-2007 the protocol distributed to sites with 1.5 T scanners from this vendor errantly had autoshimming disabled. The last five points for this scanner were acquired after corrected protocols were distributed.
From Figs. 67, it is apparent that estimating the best case performance of a scanner requires removing the discrete effects of scanner calibration changes and also changes introduced if multiple phantoms were used (e.g., the replacement of a defective phantom). To that end, the clustering algorithm previously outlined was employed. Figure 8 represents the summary of scanner performance for more than 2200 phantom scans. Shown in Fig. 8 are plots of system number (arbitrarily enumerated) versus the mean scale factors with error bars representing the square root of the pooled variance. Scanners perform similarly, with the exception of the S∕I direction for 1.5 T scanners from vendor 3. The mean scale factors differ systematically by vendor. Vendor dependence is less evident in measures of nonlinearity, SNR, and contrast parameters (Figs. 67).
Figure 8.
Summary of scanner performance for more than 2200 phantom scans. A pooled-variance approach is used to estimate the stability of gradient performance factoring out discrete changes generally due to scanner recalibration. Symbols are plotted at the mean scale value over all values, and error bars indicate the square root of the pooled variance. System number is an arbitrary enumeration. R∕L calibration appears less consistent across scanners for vendor 1 than for other vendors. The S∕I per scanner error bars for vendor 3 are much larger than for other vendors and other directions. Scanners from vendor 2 are from two different models and the data are clustered by model in the S∕I mean scale factors.
Use of phantom measurements to correct within-scanner linear scaling changes in human images
An underlying assumption in the ADNI approach is that phantom measurements accurately capture geometric performance information intrinsic to the scanner and that information can be applied to correct human images acquired in the same scanning session as those of the phantom. In ADNI, data sets are made available with the three Cartesian scale factors that govern displayed voxel size modified based on measurements from paired phantom images. If the phantom captures system geometric performance and that performance applies to the accompanying human images, then pairs of images from the same subject acquired at different times should be more compatible than those without phantom scaling. To test the hypothesis that phantom correction of scaling errors in human images is feasible, coregistration of approximately 800 intrasubject image pairs was carried out.
Histograms of relative scale factors from coregistration of 465 intrasubject image pairs are shown in Fig. 9. Histograms are shown for data with and without phantom-based correction. These are “best case” data in that scan pairs were selected such that the same phantom was used for each image pair to eliminate phantom construction variability. Also, the scans were acquired on 1.5 T systems from vendors 1 and 2 and each pair of images was collected on the same scanner with no phantom repair or replacement between scans.
Figure 9.
Use of phantom measurements to correct within-scanner linear scaling changes in human images. Histograms of intrasubject coregistration scale factors from 1.5 T scanners with and without phantom-based voxel size adjustment are shown. The upper (lower) histograms are without (with) correction. Correction reduces the widths of the distributions. The vertical dashed lines are located at 1.00, the ideal intrasubject scale factor.
The means and standard deviations of the scale factor values with and without phantom-based voxel size adjustment are shown in Table 1. The mean values are all quite close to unity, indicating that for this fleet of scanners drift does not appear to be systematic. The standard deviations of the scale factor distributions are reduced with phantom-based voxel size adjustment. Without correction, the frequency-encoded axis has the most variability; with correction, the variabilities are more consistent across all axes.
Table 1.
Summary statistics for the scale factors in each of the cardinal directions are shown under various experimental conditions. AS indicates that data were acquired with autoshim enabled; “no AS” indicates data from vendor 3 with autoshim errantly disabled in the distributed protocol. “Matched phantoms” indicates that each image pair contributing to the underlying distribution was corrected against the same phantom. All 3 T data were acquired with autoshim enabled.
| Data set∕Direction | Mean (SD) scale factors from pairwise coregistration | ||
|---|---|---|---|
| R∕L | A∕P | S∕I | |
| 1.5 T, AS, no correction (N=604) | 0.9998 (0.0026) | 0.9995 (0.0023) | 0.9997 (0.0034) |
| 1.5 T, AS, corrected, matched phantoms (N=465) | 0.9994 (0.0019) | 0.9992 (0.0018) | 0.9999 (0.0020) |
| 1.5 T noAS, matched phantoms (N=62) | 0.9996 (0.0016) | 0.9998 (0.0012) | 1.0000 (0.0051) |
| 1.5 T, AS, corrected, mismatched phantoms (N=139) | 0.9998 (0.0019) | 1.0045 (0.0031) | 1.0001 (0.0020) |
| 3 T AS, corrected, matched phantoms | 1.002 (0.0036) | 1.0004 (0.0020) | 0.9991 (0.0029) |
As previously mentioned, the protocols initially distributed for 1.5 T systems from vendor 3 erroneously had autoshim disabled. Autoshim measurements yield a constant gradient offset in each of the three physical gradient axis directions. These offsets are only expected to affect spatial scaling the frequency-encoded direction for 3D volume scans, because the other two directions are phase encoded. The ADNI MP-RAGE scans are acquired in the sagittal plane, so that the frequency-encoded direction corresponds to the S∕I direction. Summary statistics from 62 pairs of scans on 1.5 T systems from vendor 3 in which the phantom was not repaired or replaced between scans are included in Table 1. R∕L and A∕P coregistration scale factor distributions are narrower than for vendors 1 and 2 (the best case data in the previous paragraph). As expected the S∕I distribution of coregistration scale factors is much broader than for other vendors.
Changing the phantoms within the time series of imaging sessions introduces additional variability. Phantom construction varies most in the A∕P direction and this variability appears in the pairwise scaling when different phantoms were used. From a set of 139 image pairs on ten scanners in which the phantom was replaced between scans, the R∕L and S∕I variability is essentially unchanged; while A∕P variability is larger than with no correction. Moreover, the mean A∕P scale factor is shifted after phantom replacement. Early production phantoms frequently had one or more leaking fiducial markers and initial analysis software versions failed in the presence of one or more undetected fiducial markers. The phantom vendor improved the fiducial marker manufacturing process, reducing the incidence of leaking markers. Additionally, the analysis was rewritten to better find dim fiducial markers and also to be tolerant absent markers. Currently phantoms are not replaced unless absolutely necessary.
Because roughly 25% of the ADNI subjects are scanned at 3 T (in addition to 1.5 T), there are fewer pairs of images from 3 T scanners. Summary statistics are included in Table 1. Performance in the A∕P and S∕I directions are similar to that found for image pairs acquired at 1.5 T. The R∕L variability is worse than at 1.5 T. One particular model of scanner is observed to drive the R∕L variability (SD=0.0082) and when removed from the collection of data the R∕L standard deviation for the remaining scanners is reduced to 0.0022, more consistent with 1.5 T data.
Use of phantom measurements to perform absolute scaling of human images across scanners
Although intrascanner stability is necessary for the success of ADNI, use of the phantom to potentially perform absolute scaling of human images across scanner is of interest too. In the ADNI protocol, each site uses one and only one scanner at each field strength. Thus, the only available cross-system data are also cross field and there is no ready way to disentangle changes related to field strength from changes related to the other system hardware and software. Scale factors from coregistration of 3 T images to 1.5 T images from intrasubject image pairs are shown in Fig. 10. As in Fig. 9, phantoms are invariant and 1.5 T images from scans obtained on vendor 3 machines are excluded yielding 206 pairs of scans. For these distributions the means and standard deviations are 0.9995(0.0047), 0.9986(0.0054), and 1.0005(0.0044) in the R∕L, A∕P, and S∕I directions. There are nine possible pairings of vendors. The pairing of scanners with the minimum standard deviations has R∕L, A∕P, S∕I standard deviations of 0.0019, 0.0009, and 0.0022 indicating performance similar to 1.5 T intrascanner data. The worst combination has standard deviations of 0.0110, 0.0117, and 0.0071.
Figure 10.
Use of phantom measurements to perform absolute scaling of human images across scanner. Histograms of intrasubject coregistration scale factors for image pairs with one scan acquired at 3 T and the other at 1.5 T are shown. The upper (lower) histograms are without (with) correction. Correction reduces the widths of the distributions.
Verifying the correctness of gradient warping corrections
The necessity and availability of full 3D gradient warping correction varies by scanner vendor and model; required corrections range from none to full 3D correction done in postprocessing. Correction coefficients are gradient hardware model specific, and therefore, unwarping algorithms are independent of the image content. The approach neglects B0 inhomogeneity effects. All scans in ADNI are corrected to the level equivalent to full 3D gradient warping correction. Representative measures after first-order polynomial correction for a range of scanners with correct and incorrect correction coefficients are shown in Table 2. These are representative values and not intended to differentiate the various scanners (no vendor-or model-identifying information is presented). Here the requirement is the phantom provides validation that corrections are properly applied. These values are two to four times larger than reported3, 4, 5, 6, 7 after data-driven corrections were made. Detailed information about the gradient hardware may or may not be present (and reliable) in the DICOM headers and incorrect gradient hardware was reported by five ADNI sites when surveyed at the start of the study. The phantom was essential in determining that the geometric corrections were being made properly. As discussed previously, the standard-deviation-of-residual radii provides a measure of image nonlinearity. Empirically, we found that with the correct gradient unwarping the scanners in ADNI had similar standard deviation of residual radii values. Using correction coefficients for the wrong gradient hardware results in distinctly larger values, which was the only way we were able to identify the five sites that had reported incorrect gradient hardware at the beginning of the study. Without the phantom-based system surveillance, incorrect unwarping would have been applied to all human images throughout the duration of the study at these five sites.
Table 2.
Representative values for standard deviation of residuals (mm) for systems used in ADNI are presented. Entries for scanners requiring different levels of gradient warping correction are included. Right and wrong corrections indicate that postprocessing was done using the right and wrong gradient warping coefficients for the actual system.
| Correction required | Standard deviation of residual radii (mm) | ||
|---|---|---|---|
| Uncorrected | Wrong correction | Right correction | |
| None | 0.32 | N∕A | 0.32 |
| 2D on-line+3D post processing | 1.1 | 0.65 | 0.31 |
| 3D post processing | 0.42 | 1.2 | 0.28 |
| 3D on scanner | 0.29 | N∕A | 0.29 |
Detecting system errors with the ADNI phantom
To date, monitoring each MRI system in the ADNI study has resulted in identification of major system errors that can be grouped into three classes. (1) Five sites misreported their own gradient hardware, leading to incorrect 3D distortion correction. When these errors were detected by analysis of the phantom scan, the correct 3D distortion correction was applied. (2) One site’s laser landmark system was misadjusted during an upgrade, leading to geometrical distortion which was detected in one of the off-diagonal scatter plots. The site was unaware of this problem which was uncovered by the phantom measurements. (3) An incorrect protocol parameter (autoshim disabled) was initially distributed to nine sites. Autoshim status is not recorded in the DICOM header in that vendor’s images. Thus without the phantom monitoring, this error would have gone undetected for the duration of the ADNI study. Gone undetected (and hence uncorrected), these problems would have contributed to imprecision in quantitative metrics at over 25% of all enrolling ADNI sites.
CONCLUSION
ADNI is the first large multiyear multicenter MRI trial to employ a phantom scanned with each subject, providing time-locked estimates of scanner performance. The phantom analysis provides precise estimates of linear geometrical scale factors by which the scanner deviates from ideal and which are ascribed to gradient drift and∕or miscalibration. The estimated coefficients of variation intrinsic to measurements in a single phantom are in the range of 3–5 parts in 104 and are driven largely by phantom positioning within the scanner.
Scanner tracking reveals that gradient stability is in many cases disrupted by recalibration, which is often associated with a system hardware or software upgrade. That is, recalibration induces discrete changes which are often larger than observed system drift over periods of months. On the small subset of systems where autoshimming was errantly disabled on the distributed 1.5 T protocols for one vendor, relatively large instability is observed in the frequency encoded (S∕I axis).
In addition to linear fidelity estimates, the analysis produces a summary statistic that captures the spatial nonlinearity in the images. These values are found to be useful in verifying that gradient unwarping corrections made in postprocessing are correctly implemented. With full 3D unwarping, scanners in ADNI perform similarly in this metric. Estimates of residual nonlinearity for the “gradwarp” correction method used in ADNI are two to four times larger than data-driven approaches wherein the deformation field is estimated from phantom images.
The distributions of linear scaling parameters for intrasubject coregistration were narrower after phantom-based voxel size adjustment. This result supports the underlying assumption in the ADNI approach that phantom measurements can accurately capture information about the scanner, which can be applied to correct human images acquired in the same scanning session. However, in situations where the assumptions underlying phantom-based scaling of human images were violated–systems with autoshim disabled and where the phantom was repaired or replaced within the time series—as might be expected, phantom-based scaling of human images was not effective and could introduce more error than simply not scaling the human images.
Based on field experience to date, the greatest practical value of incorporating ADNI phantom measurements in a multisite study is to identify scanner errors through central monitoring. This approach has resulted in identification of three categories of major system errors. Had these gone undetected (and hence uncorrected), these problems would have contributed to imprecision in quantitative metrics at over 25% of all enrolling ADNI sites.
ACKNOWLEDGMENTS
This project was funded by the Alzheimer’s Disease Neuroimaging Initiative (ADNI; Principal Investigator: Michael Weiner; NIH Grant No. U01 AG024904). ADNI is funded by the National Institute of Aging, the National Institute of Biomedical Imaging and Bioengineering (NIBIB), and the Foundation for the National Institutes of Health, through generous contributions from the following companies and organizations: Pfizer Inc., Wyeth Research, Bristol-Myers Squibb, Eli Lilly and Company, Glaxo-SmithKline, Merck and Co. Inc., AstraZeneca AB, Novartis Pharmaceuticals Corporation, the Alzheimer’s Association, Eisai Global Clinical Development, Elan Corporation plc, Forest Laboratories, and the Institute for the Study of Aging (ISOA), with participation from the U.S. Food and Drug Administration. Support was also through National Institute of Aging Grant No. R01 AG11378. Additional infrastructure support was funded through NIH Grant No. C06 RR018898 and AG11378. Data used in the preparation of this article were obtained from the Alzheimer’s Disease Neuroimaging Initiative (ADNI) database (www.loni.ucla.edu∕ADNI). A complete listing of ADNI investigators who contributed to ADNI design, implementation and data collection is available at http:∕∕www.loni.ucla.edu∕ADNI∕Collaboration∕ADNI_Authorship_list.pdf).
References
- Baldwin L. N., Wachowicz K., Thomas S. D., Rivest R., and Fallone B. G., “Characterization, prediction, and correction of geometric distortion in 3 T MR images,” Med. Phys. 34(2), 388–399 (2007). 10.1118/1.2402331 [DOI] [PubMed] [Google Scholar]
- Breeuwer M. M., Holden M., and Zylka W., “Detection and correction of geometric distortion in 3D MR images,” Proc. SPIE 4322, 1120–1120 (2001). [Google Scholar]
- Chen C. C., Wan Y. L., Wai Y. Y., and Liu H. L., “Quality assurance of clinical MRI scanners using ACR MRI phantom: Preliminary results,” J. Digit Imaging 17(4), 279–284 (2004). 10.1007/s10278-004-1023-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colombo P., Baldassarri A., Del Corona M., Mascaro L., and Strocchi S., “Multicenter trial for the set-up of a MRI quality assurance programme,” Magn. Reson. Imaging 22(1), 93–101 (2004). 10.1016/j.mri.2003.04.001 [DOI] [PubMed] [Google Scholar]
- Firbank M. J., Harrison R. M., Williams E. D., Coulthard A., Britson P. J., Gunter J. L., and Ward C. P., “Quality assurance for MRI: Practical experience,” Br. J. Radiol. 73(868), 376–383 (2000). [DOI] [PubMed] [Google Scholar]
- Friedman L. and Glover G. H., “Report on a multicenter fMRI quality assurance protocol,” J. Magn. Reson Imaging 23(6), 827–839 (2006). 10.1002/jmri.20583 [DOI] [PubMed] [Google Scholar]
- Fu L., Fonov V., Pike B., Evans A. C., and Collins D. L., “Automated analysis of multi site MRI phantom data for the NIHPD project,” Medical Image Computing and Computer Assisted Intervention International Conference2006. (unpublished), Vol. 9, pt. 2, pp. 144–151. [DOI] [PubMed]
- Holden M., Breeuwer M. M., McLeish K., Hawkes D. J., Keevil S. F., and Hill D. L., “Sources and correction of higher-order geometrical distortion for serial MR brain imaging,” Proc. SPIE 4322, 69–78 (2001). 10.1117/12.431151 [DOI] [Google Scholar]
- Howe F. A., Canese R., Podo F., Vikhoff B., Slotboom J., Griffiths J. R., Henriksen O., and Bovee W. M., “Quality assessment in in vivo NMR spectroscopy: V. Multicentre evaluation of prototype test objects and protocols for performance assessment in small bore MRS equipment,” Magn. Reson. Imaging 13(1), 159–167 (1995). 10.1016/0730-725X(94)00091-G [DOI] [PubMed] [Google Scholar]
- Ihalainen T., Sipila O., and Savolainen S., “MRI quality control: six imagers studied using eleven unified image quality parameters,” Eur. Radiol. 14(10), 1859–1865 (2004). 10.1007/s00330-004-2278-4 [DOI] [PubMed] [Google Scholar]
- Koch N., Liu H. H., Olsson L. E., and Jackson E. F., “Assessment of geometrical accuracy of magnetic resonance images for radiation therapy of lung cancers,” J. Appl. Clin. Med. Phys. 4(4), 352–364 (2003). 10.1120/1.1617211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemieux L. and Barker G. J., “Measurement of small inter-scan fluctuations in voxel dimensions in magnetic resonance images using registration,” Med. Phys. 25(6), 1049–1054 (1998). 10.1118/1.598288 [DOI] [PubMed] [Google Scholar]
- Lerski R. A. and de Certaines J. D., “Performance assessment and quality control in MRI by Eurospin test objects and protocols,” Magn. Reson. Imaging 11(6), 817–833 (1993). 10.1016/0730-725X(93)90199-N [DOI] [PubMed] [Google Scholar]
- Mascaro L., Strocchi S., Colombo P., Del Corona M., and Baldassarri A. M., “Definition criteria for a magnetic resonance quality assurance program: multicenter study,” Radiol. Med. (Torino) 97(5), 389–397 (1999). [PubMed] [Google Scholar]
- Michiels J., Bosmans H., Pelgrims P., Vandermeulen D., Gybels J., Marchal G., and Suetens P., “On the problem of geometric distortion in magnetic resonance images for stereotactic neurosurgery,” Magn. Reson. Imaging 12(5), 749–765 (1994). 10.1016/0730-725X(94)92200-4 [DOI] [PubMed] [Google Scholar]
- Moore C. S., Liney G. P., and Beavis A. W., “Quality assurance of registration of CT and MRI data sets for treatment planning of radiotherapy for head and neck cancers,” J. Appl. Clin. Med. Phys. 5(1), 25–35 (2004). 10.1120/jacmp.26.147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orth R. C., Sinha P., Madsen E. L., Frank G., Korosec F. R., Mackie T. R., and Mehta M. P., “Development of a unique phantom to assess the geometric accuracy of magnetic resonance imaging for stereotactic localization,” Neurosurgery 45(6), 1423–1429 (1999). 10.1097/00006123-199912000-00030 [DOI] [PubMed] [Google Scholar]
- Tofts P. S., “Standardisation and optimisation of magnetic resonance techniques for multicentre studies,” J. Neurol., Neurosurg. Psychiatry 64, S37–S43 (1998). 10.1136/jnnp.64.1.37 [DOI] [PubMed] [Google Scholar]
- Wang D., Doddrell D. M., and Cowin G., “A novel phantom and method for comprehensive 3-dimensional measurement and correction of geometric distortion in magnetic resonance imaging,” Magn. Reson. Imaging 22(4), 529–542 (2004). 10.1016/j.mri.2004.01.008 [DOI] [PubMed] [Google Scholar]
- Wang D., Strugnell W., Cowin G., Doddrell D. M., and Slaughter R., “Geometric distortion in clinical MRI systems Part I: Evaluation using a 3D phantom,” Magn. Reson. Imaging 22(9), 1211–1221 (2004). 10.1016/j.mri.2004.08.012 [DOI] [PubMed] [Google Scholar]
- Wang D., Strugnell W., Cowin G., Doddrell D. M., and Slaughter R., “Geometric distortion in clinical MRI systems Part II: Correction using a 3D phantom,” Magn. Reson. Imaging 22(9), 1223–1232 (2004). 10.1016/j.mri.2004.08.014 [DOI] [PubMed] [Google Scholar]
- J. P.Mugler, III and Brookeman J. R., “Three-dimensional magnetization-prepared rapid gradient-echo imaging (3D MP RAGE),” Magn. Reson. Med. 15(1), 152–157 (1990). 10.1002/mrm.1910150117 [DOI] [PubMed] [Google Scholar]
- C. R.Jack, Jr., Bernstein M. A., Fox N. C., Thompson P., Alexander G., Harvey D., Borowski B., Britson P. J., Ward L. W. J. C., Dale A. M., Felmlee J. P., Gunter J. L., Hill D. L., Killiany R., Schuff N., Fox-Bosetti S., Lin C., Studholme C., DeCarli C. S., Krueger G., Ward H. A., Metzger G. J., Scott K. T., Mallozzi R., Blezek D., Levy J., Debbins J. P., Fleisher A. S., Albert M., Green R., Bartzokis G., Glover G., Mugler J., and Weiner M. W., “The Alzheimer’s Disease Neuroimaging Initiative (ADNI): MRI methods,” J. Magn. Reson Imaging 27(4), 685–691 (2008). 10.1002/jmri.21049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otsu N., “A threshold selection method from gray-level histograms,” IEEE Trans. Syst. Man Cybern. 9, 62–66 (1979). 10.1109/TSMC.1979.4310076 [DOI] [Google Scholar]
- Woods R. P., Grafton S. T., Holmes C. J., Cherry S. R., and Mazziotta J. C., “Automated image registration: I. General methods and intrasubject, intramodality validation,” J. Comput. Assist. Tomogr. 22(1), 139–152 (1998). 10.1097/00004728-199801000-00027 [DOI] [PubMed] [Google Scholar]