NOTCH1 mutations are found in more than 50 % of patients with T-lineage acute lymphoblastic leukemia (TALL), and have been associated with a favorable outcome in pediatric T-ALL. FBXW7 mutations are found in a small subset of T-ALL, sometimes overlapping with the presence of NOTCH1 mutations. In this study, Baldus and colleagues investigated the prognostic impact of NOTCH1 and FBXW7 mutations in adult T-ALL. See related perspective article on page 1338.
Keywords: acute T-lymphoblastic leukemia, NOTCH1, FBXW7, mutations
Abstract
Background
NOTCH1 mutations have been associated with a favorable outcome in pediatric acute T-lymphoblastic leukemia. However, the results of studies on the prognostic significance of NOTCH1 mutations in adult T-lymphoblastic leukemia remain controversial.
Design and Methods
Here we have investigated the prognostic impact of mutations in the NOTCH1 pathway, in particular, the NOTCH1 and FBXW7 genes, in a large cohort of adult patients with T-lymphoblastic leukemia (n=126). We determined the occurrence of mutations in NOTCH1 and FBXW7 by DNA amplification and direct sequencing of polymerase chain reaction products.
Results
Mutations were identified in 57% and 12% of the NOTCH1 and FBXW7 genes, respectively. The characteristics of patients carrying NOTCH1 and/or FBXW7 (NOTCH1-FBXW7) mutations were similar to those with wild-type genes. Patients with NOTCH1-FBXW7 mutations more often showed a thymic immunophenotype (p=0.001). In the overall cohort, no significant differences were seen in the complete remission or event-free survival rates between patients with mutated or wild-type NOTCH1-FBXW7 (p=0.39).
Conclusions
NOTCH1 and FBXW7 mutations were not predictive of outcome in the overall cohort of adult patients with T-lymphoblastic leukemia, but there was a trend towards a favorable prognostic impact of NOTCH1-FBXW7 mutations in the small subgroup of patients with low-risk ERG/BAALC expression status. Our findings further confirm the high frequency of NOTCH1 mutations in adult T-lymphoblastic leukemia.
Introduction
Acute T-lymphoblastic leukemia (T-ALL) accounts for 25% of all cases of adult acute lymhpoblastic leukemia.1 Using standard chemotherapeutic regimens, long-term disease-free survival in adult T-ALL remains limited with only 32–46% of patients becoming long-term survivors.1,2 Treatment optimization strategies in acute leukemia depend on the accurate assignment of patients at diagnosis to specific risk groups.2,3 In acute myeloid leukemia and B-lineage acute lymphoblastic leukemia molecular risk factors with proven prognostic significance have been identified and guide the development of molecular-based therapies.4–6 In contrast, few molecular risk factors have been established in T-ALL and targeted therapies have yet to be explored.
The NOTCH1 gene was initially found to be involved in the rare t(7;9)(q34;q34.3) in T-ALL.7 Subsequently, activating NOTCH1 mutations were identified in 56% of pediatric T-ALL patients, including mutations in the heterodimerization (HD) domain, the polypeptide enriched proline, glutamate, serine and threonine (PEST) domain, and in the transactivation domain (TAD).8,9 Mutations in the HD domain are proposed to destabilize the protein, likely conferring ligand-independent pathway activation, whereas mutations in the PEST domain result in an increased half-life of the transcriptionally active intracellular NOTCH1 (ICN1) fragment.8,10 Recently, internal duplication insertions leading to expansion in the extracellular juxtamembrane region have been found to be a novel mechanism of NOTCH1 activation in 3.3% of cases of T-ALL.11 While NOTCH1 signaling is essential for the development of normal T-cell progenitors, constitutive activation of NOTCH1 signaling by mutational events has been linked to T-cell leukemogenesis and direct evidence of its pathogenetic role has been provided by murine models that showed the induction of T-cell malignancies in mice carrying NOTCH1 mutations.12,13
The tumor suppressor FBXW7 targets proteins for proteosomal degradation and has been shown to suppress NOTCH1 signaling by ubiquitination of the ICN1 fragment.14,15 Mutations in FBXW7 have been identified in T-lymphoblastic cell lines as well as in primary T-ALL samples and were found to abrogate NOTCH1 binding, thereby enhancing NOTCH1 signaling. Moreover, studies have demonstrated that mutations in FBXW7 mediate resistance to gamma-secretase inhibitors.16,17 In addition, FBXW7 recognizes other substrates for ubiquitination including CYCLIN E, c-MYC, and JUN; thus, in addition to enhancing NOTCH1 signaling, FBXW7 mutations may also affect other oncogenic pathways.
To date, clinical studies have predominately investigated pediatric T-ALL and reported NOTCH1 and FBXW7 mutations in 56% and 12% of cases, respectively.8,16–20 Interestingly, in one study, a mutated NOTCH1 status was associated with a better response to pred-nisone, a lower level of minimal residual disease, and a higher event-free survival rate.18 In contrast, in adult TALL, few data are available on the frequency21 or the prognostic impact of NOTCH1 and FBXW7 mutations, with one study (including only 24 adult T-ALL patients) reporting an inferior outcome for patients with NOTCH1 mutations.20 Since the results from pediatric and adult T-ALL mutation analyses remain controversial, larger studies are necessary to determine the prognostic and therapeutic relevance of NOTCH1 and FBXW7 mutations in adult T-ALL. Thus, we evaluated NOTCH1 and FBXW7 mutation status in a large number of adult T-ALL patients enrolled on German Multicentre Acute Lymphoblastic Leukemia (GMALL) protocols and related the mutation status to clinical characteristics and molecular markers.
Design and Methods
Patients and treatment
We studied 126 consecutive adult patients with newly diagnosed T-ALL, who were registered on the GMALL 05/93 and 06/99 multicenter protocols and had sufficient material available.22,23 The GMALL protocols included intensive chemotherapy as well as autologous or allogeneic stem cell transplantation (SCT).24 All patients gave written informed consent to participation in the study according to the Declaration of Helsinki. The study was approved by the ethics board of the Johann Wolfgang Goethe University Frankfurt/Main, Germany. Genomic DNA from six healthy volunteers was obtained after informed consent and used as control samples. The entry criteria and overall results of patients analyzed in this study did not differ from those of the remaining patients in of the GMALL studies who were not included in this study because of lack of material.
Immunophenotypic analyses
Pre-treatment bone marrow samples were collected centrally. The blast fraction of the samples was enriched by density-gradient centrifugation (Ficoll-Paque Plus, Amersham Biosciences, Uppsala, Sweden) and stored in liquid nitrogen. Immunophenotyping of fresh samples was performed centrally in the GMALL reference laboratory at the Charité, University Hospital Berlin, Campus Benjamin Franklin, Germany. Immuno-phenotyping was carried out as previously described.23,25 T-lineage leukemia was subclassified into pre-T-ALL or early T-ALL (cyCD3+, CD7+, CD5+/−, CD2−, sCD3−, CD4−/+, CD8−/+, CD1a− or cyCD3+, CD7+, CD5−, CD2+, sCD3−, CD4−, CD8−, CD1a−), thymic T-ALL (cyCD3+, CD7+, CD5+/−, CD2+/−, sCD3+/−, CD4+, CD8+, CD1a+), and mature T-ALL (cyCD3+, CD7+, CD5+, CD2+, sCD3+/−, CD4+/−, CD8+/−, CD1a−).
DNA isolation and NOTCH1 and FBXW7 mutation analyses
Genomic DNA from pre-treatment bone marrow samples was isolated using the PureGene kit (Gentra Systems; Plymouth, USA) following the manufacturer’s instructions. NOTCH1 mutations in the N-terminal region of the HD domain (HD-N; hotspot of mutations: bp 4724–4829), the C-terminal region (HD-C; hotspot of mutations: bp 5029–5222), the TAD, and the PEST domain were determined by direct sequencing of polymerase chain reaction (PCR)-amplified products.8 The PEST domain was divided into a N-terminal PEST-1 (hotspot of mutations: bp 7255–7330) region and a C-terminal PEST-2 (hotspot of mutations: bp 7525–7558) region, the latter region includes the FBXW7 binding site. Sufficient material for the analyses of the FBXW7 mutation status was available for 112 of the 126 patients. Sequencing of the amplified PCR products of exons 8 and 9 was carried out as previously described.26 As a control, bone marrow samples from six healthy volunteers were included and no mutations were detected.
Molecular characterization by gene expression analyses
mRNA expression levels of the genes HOX11, HOX11L2, BAALC, and ERG were determined by comparative real-time reverse transcriptase-PCR as previously reported.27,28
Statistical analysis
Comparisons of baseline clinical variables across groups were made using the χ2 Fisher’s exact test for categorical data. The non-parametric Mann-Whitney U test was applied for quantitative variables. A p value of 0.05 or less (two-sided) was considered to indicate a statistically significant difference.
Clinical follow-up data were available from 108 of the 126 T-ALL patients with a median follow-up time of 20.5 months (range, 0.5 to 81.2 months). Complete remission (CR) was assessed after completion of induction chemotherapy and required a granulocyte count of 1.5×109/L or more, a platelet count of 100×109/L or more, without peripheral blood blasts, bone marrow cellularity greater than 20% with maturation of all cell lines, less than 5% bone marrow blasts, and absence of extramedullary leukemia. Relapse was defined as the reappearance of circulating blasts, more than 5% bone marrow blasts, or development of extramedullary leukemia. Event-free survival was calculated using the Kaplan-Meier method and the log-rank test was used to compare differences between survival curves. Event-free survival was measured from the date of starting the study protocol until the date of death of any cause or relapse or failure to achieve complete remission.
In order to identify independent prognostic factors and effect modifiers (i.e. interactions between different factors), Cox proportional hazards models were constructed for event-free survival. The following co-variates were included in the full model: BAALC/ERG expression status (low-risk versus high-risk), HOX11 and HOX11L2 expression (presence versus absence), white blood count, age, immunophenotype (early, thymic, mature), NOTCH1/FBXW7 mutation status (wild-type NOTCH1 and FBXW7 versus mutated NOTCH1 and/or FBXW7), and BAALC/ERG as an effect modifier for NOTCH1/FBXW7. Stepwise forward and backward selection was performed. All calculations were performed using the SPSS software package, version 15 (SPSS Inc., Chicago, IL, USA).
Results
NOTCH1 and FBXW7 mutations in adult T-cell acute lymphoblastic leukemia: frequency and characteristics
NOTCH1 mutations were identified in 72 (57%) of the 126 adult T-ALL patients. Mutations involved the HD domain in 56 patients and the PEST domain in 20 patients. Five patients had mutations in the TAD. Forty-five novel mutations were identified, whereas the remaining mutations had been reported previously, primarily in pediatric T-ALL.8,18–20,28 Twelve patients had more than one mutation, including nine patients with mutations in different domains: three patients had combined HD domain and TAD mutations, and six patients had co-existing HD and PEST domain mutations. In the HD domain, point mutations were predominant. Insertions and/or deletions were present in 16 cases, 14 of which have not been reported previously. These changes included cryptic aberrations with in-frame insertions, duplications and deletions. In the TAD, only new mutations were identified, resulting in a premature stop in four patients. The mutation analyses of the PEST domain revealed the predominance of complex changes with deletions and insertions resulting in truncated proteins in 15 patients (Table 1, Online Supplementary Figure S1).
Table 1.
Frequency of NOTCH1 and FBXW7 mutations in adult T-ALL.
FBXW7 mutations were found in 14 (12%) of the 112 analyzed T-ALL samples. The R465C/H mutation (exon 8) was detected in seven patients, the exon 9 mutations R505C and G517R were identified in six and one patient, respectively. Nine (64%) of the 14 T-ALL patients with FBXW7 mutations had co-existing NOTCH1 mutations, eight in the HD domain and two in the PEST domain, with one patient (ID 88) having three mutations (R465C/H + HD-N + PEST-1; Table 1, Online Supplementary Figure S1).
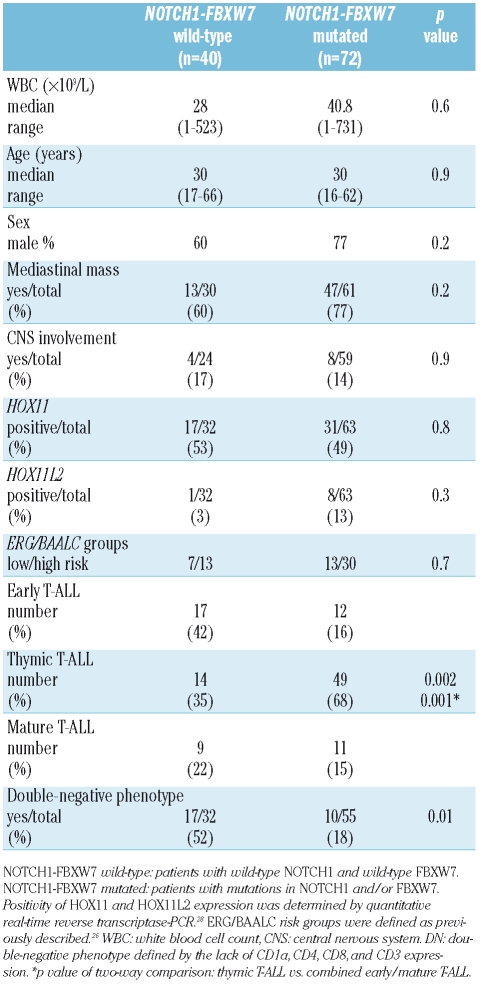
Association of NOTCH1 and FBXW7 mutations with molecular and clinical characteristics
As NOTCH1 and FBXW7 mutations affect the same signaling pathway, patients with mutations in the NOTCH1 and/or FBXW7 gene were grouped into a combined NOTCH1-FBXW7 mutated group. With respect to clinical parameters no significant differences were seen between NOTCH1-FBXW7 wild-type and NOTCH1-FBXW7 mutated patients (Table 2). There was no significant correlation between the mRNA expression of the previously characterized molecular markers with prognostic significance including HOX11, HOX11L2, BAALC and ERG (Table 2) and NOTCH1-FBXW7 mutation status.
Table 2.
Association of NOTCH1-FBXW7 mutation status with clinical and molecular characteristics in adult T-ALL.
Association of NOTCH1 and FBXW7 mutations with mmunological characteristics
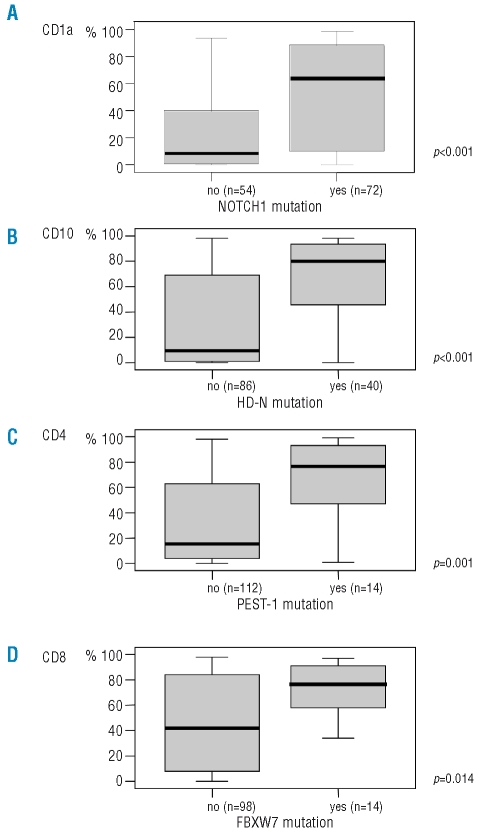
NOTCH1-FBXW7 mutated cases exhibited a thymic immunophenotype with a greater frequency as compared to wild-type cases (68%; p=0.001; Table 2). The association of NOTCH1 and FBXW7 mutations was further investigated by the analyses of surface marker expression reflecting stages of T-cell differation. NOTCH1-FBXW7 wild-type cases predominantly exhibited an immature double-negative phenotype defined by the lack of CD1a, CD4, CD8, and CD3 expression compared to NOTCH1-FBXW7 mutated cases which displayed a more mature phenotype (p=0.01; Table 2). In particular, expression of CD1a was highly indicative of the presence of NOTCH1 mutations (p<0.001; Figure 1A), CD10 expression was associated with mutations in the HD-N, and CD4 expression with mutations in the PEST-1 region (Figure 1B–C). Interestingly, all cases with FBXW7 mutations could be assigned to cell populations double positive for CD8 and CD4 or single positive for CD8 and the FBXW7 mutated status correlated with CD8 expression (Figure 1D).
Figure 1.
Association between mutation status and the expression of surface antigens. Y-axis of the box-plots depicts the expression (in percent) of the surface antigen as determined by flow cytometry.
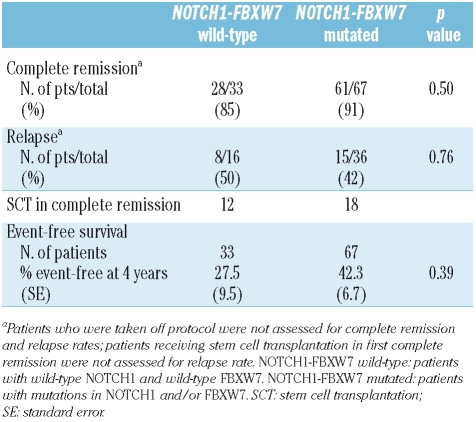
NOTCH1 and FBXW7 mutations and outcome
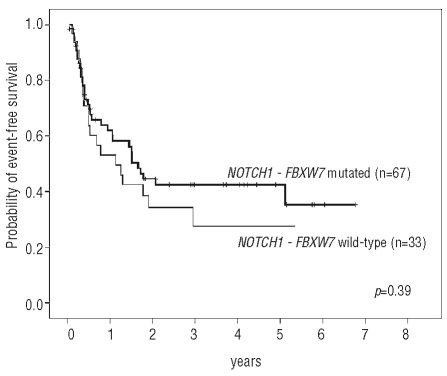
Significant differences were not observed in the complete remission, relapse, or event-free survival rates between NOTCH1-FBXW7 wild-type and mutated cases (Table 3, Figure 2). Similar results were also obtained comparing wild-type versus mutated NOTCH1 cases irrespective of the FBXW7 mutation status. The number of FBXW7 mutated cases was small and thus the prognostic impact of this mutation independent of NOTCH1 mutations was not analyzed. No significant differences were seen at 4 years with respect to the cumulative incidences of relapse (NOTCH1-FBXW7 mutated: 40.2% vs. NOTCH1-FBXW7 wild-type: 57.8%; p=0.63) and overall survival (NOTCH1-FBXW7 mutated: 41.9% vs. NOTCH1-FBXW7 wild-type: 53.0%; p=0.46). Similarily, no significant differences in outcome were observed between NOTCH1-FBXW7 wild-type and NOTCH1-FBXW7 mutated cases within the risk groups defined by immunophenotype (early, thymic, or mature T-ALL), and HOX11L2 and HOX11 expression (data not shown).
Table 3.
Impact of NOTCH1 and FBXW7 mutation status on survival in adult T-ALL.
Figure 2.
Kaplan Meier analyses of event-free survival of patients with mutated NOTCH1-FBXW7 compared to patients with wild-type NOTCH1-FBXW7.
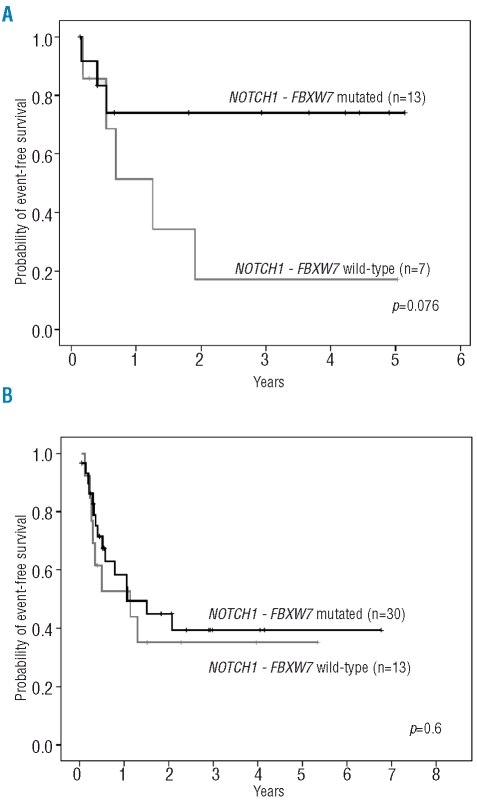
In an exploratory analysis, the prognostic impact of the NOTCH1-FBXW7 mutation status was further investigated within the previously defined ERG/BAALC risk groups. Within the small ERG/BAALC low-risk group (i.e. low expression of ERG and/or BAALC),27 NOTCH1-FBXW7 wild-type patients had an inferior event-free survival (event-free at 4 years: 17%) as compared to NOTCH1-FBXW7 mutated patients (event-free at 4 years: 74%, p=0.076; Figure 3A). The lack of statistical significance is likely due to the small number of patients in each cohort. In contrast, no differences in outcome were seen between NOTCH1-FBXW7 mutated and NOTCH1-FBXW7 wild-type patients within the ERG/BAALC high-risk group (Table 4; Figure 3B). In a multivariate analysis NOTCH1-FBXW7 mutation status was not an independent prognostic significance and no interaction between NOTCH1-FBXW7 and ERG/BAALC could be confirmed. As these analyses were based on a small number of patients, further studies are needed to define the prognostic significance of NOTCH1 and FBXW7 mutations within these molecular risk groups. When patients undergoing allogeneic and autologous stem cell transplantation were excluded from the analyses and censored at the time of their transplant, outcome analyses showed comparable results. In addition, when patients who received consolidation chemotherapy and autologous stem cell transplantation were combined, similar results were obtained.
Figure 3.
Kaplan-Meier analyses of event-free survival within ERG/BAALC expression groups. (A) Within the ERG/BAALC low-risk group, patients with wild-type NOTCH1-FBXW7 had a poorer event-free survival compared to patients with mutated NOTCH1-FBXW7. (B) In the subset of the ERG/BAALC high-risk group no significant difference was seen in event-free survival between NOTCH1-FBXW7 wild-type and NOTCH1-FBXW7 mutated patients.
Table 4.
Impact of NOTCH1 and FBXW7 mutation status on survival in adult T-ALL according to ERG/BAALC expression.
Discussion
The prognostic impact of mutations in the NOTCH1 signaling pathway, including the NOTCH1 and the FBXW7 genes, in pediatric and adult T-ALL remains controversial. In pediatric T-ALL a large study reported by Breit et al. evaluating 157 pediatric T-ALL cases demonstrated a more favorable outcome for patients with a mutated NOTCH1 status.18 Combined mutations of NOTCH1 and FBXW7 have only been analyzed in a small cohort of pediatric patients and also indicated a favorable outcome for patients with mutated NOTCH1.30 However, two smaller studies showed no survival differences with respect to the NOTCH1 mutation status.20,29 In contrast, in adult T-ALL a single, small study investigated the prognostic impact of NOTCH1 mutations and showed an inferior outcome for patients with HOX11L2 expression and mutated NOTCH1.20 Larger studies determining the prognostic impact of NOTCH1 and FBXW7 mutations in adult T-ALL have not yet been reported.
In the current study, we observed no significant prognostic implications of NOTCH1 or FBXW7 mutations with respect to the complete remission rate and event-free survival among the overall cohort of adult T-ALL patients; no impact was seen within defined subgroups based on HOX11 or HOX11L2 expression or immunophenotype. In an exploratory subgroup analysis within the favorable-risk group characterized by low expression of ERG and/or BAALC, patients with wild-type NOTCH1-FBXW7 had a worse outcome than patients with NOTCH1-FBXW7 mutations. However, the analyses remain limited due to the small sample size. In contrast, no outcome differences were seen between NOTCH1-FBXW7 mutated and NOTCH1-FBXW7 wild-type cases in high-risk ERG/BAALC patients.
The discrepancies between pediatric and adult T-ALL may reflect differences in the molecular background of the disease, selection of patients and the treatment protocols used.5 Whereas ERG and BAALC expression was not reported in the pediatric cohort, the distribution of the immunophenotype differed slightly from that in adults, with 79% of pediatric NOTCH1 mutated cases exhibiting the more favorable thymic phenotype as compared to 65% in our adult T-ALL cohort. In accordance with the high prevalence of NOTCH1 mutations in pediatric T-ALL we found mutations in 57% of adult T-ALL cases localized in the known hot spots of the HD and PEST domains and identified novel mutations in the TAD. Indeed, the majority of mutations (62%) described here have not yet been reported, highlighting the wide variation of NOTCH1 mutations and potential molecular differences between pediatric and adult T-ALL.21
The position and type of the new mutations identified in adult T-ALL were similar to those found in childhood T-ALL and likely result in equivalent functional consequences of NOTCH1 activation.
Mutations in the conserved FBXW7-binding pocket (residues R465, R479, and R505) have been shown to abrogate NOTCH1 binding, thereby enhancing NOTCH1 activity.16,17 About 12% of pediatric T-ALL patients had FBXW7 mutations at initial diagnosis.16 We identified FBXW7 mutations in a similar percentage of cases of adult T-ALL (12%). FBXW7 mutations have been primarily associated with the HD domain, resulting in signal amplification, whereas FBXW7 mutations in the PEST domain are less frequent.16,17,31 It has been proposed that mutations in the PEST domain relieve mutational pressure on FBXW7 as a marginal selective advantage would be gained by FBXW7 mutations in the context of existing PEST deletions.15 In contrast to studies in pediatric T-ALL, which found no co-existing mutations in the NOTCH1 PEST domain and the FBXW7 gene,16,17 we identified two patients (ID 88, ID 96) with combined PEST and FBXW7 mutations. In patient 96 the point mutation in the PEST-2 region was C-terminal to the proposed FBXW7-NOTCH1 T2512 interaction residue. In addition to the FBXW7 and PEST-1 mutations, patient 88 carried a mutation in the HD domain.
High expression of ERG and BAALC has been associated with an immature leukemic phenotype.27 Our preliminary data suggesting that NOTCH1-FBXW7 mutated status is associated with a favorable outcome only in the low-risk ERG/BAALC group might indicate a molecularly more differentiated leukemic stage with high susceptibility to anti-proliferative therapy. Leukemic blasts from T-ALL patients with mutated, thus activated, NOTCH1 signaling displayed a more mature phenotype and demonstrated a genotype-phenotype association as distinct surface marker expression correlated with mutations in specific NOTCH1 domains. This genotype-phenotype correlation suggests that a specific cell type might be prone to acquire certain mutational events (HD-N, PEST-1) and this may indicate that mutations preferentially occur in cells at different stages of differentiation. Interestingly, irrespective of the NOTCH1 mutation status, FBXW7 mutations were also associated with a more mature T-lymphoblast phenotype arrested at the CD4 and CD8 double positive and the CD8 single positive state.32 In contrast, patients with high-risk ERG/BAALC as well as wild-type NOTCH1-FBXW7 tended to be in a molecularly undifferentiated stage, which is likely to confer chemotherapy resistance.
NOTCH1 mutations are an ideal target for pharmacological interventions, e. g. gamma-secretase inhibitors that prevent the generation of the ICN1 fragment and thereby suppress NOTCH1 activity.33 Importantly, mutations in FBXW7 have been associated with resistance to gamma-secretase inhibitor treatment, as disruption of FBXW7 function will maintain NOTCH1 signaling.17 Therefore, the identification of FBXW7 mutations in 12% of all patients and in 64% of cases with mutated NOTCH1 signaling needs to be taken into account when choosing targeted therapies. As for other leukemic subtypes, tailoring the therapeutic approaches based on the molecular alteration is critical for treatment optimization in T-ALL. Whether low-risk patients characterized by low ERG/BAALC expression exhibiting a NOTCH1-FBXW7 mutated status might benefit from gamma-secretase inhibitor-based therapy and be spared intensified chemotherapy regimes should be explored in future studies including an even larger cohort of patients.
The complexity of NOTCH1 signaling is further highlighted by the recent identification of juxtamembrane expansion mutations. However, due to the low frequency of these mutations studies including even more TALL patients will be necessary to evaluate their prognostic impact. Moreover, PTEN mutations can be identified in about 8% of patients with newly diagnosed TALL. It has been proposed that resistance to gamma-secretase inhibitor-mediated inhibition of NOTCH1 signaling in T-ALL with PTEN loss of function mutations is a result of uninhibited AKT activation, leading to aberrant proliferation signaling.34 Recent approaches combining gamma-secretase inhibitors with glucocorticoids have been shown to overcome glucocoriticoid resistance in T-ALL and indicate that beneficial effects of gamma-secretase inhibitors can be obtained by concurrent treatment with other antileukemic drugs.35
In summary, molecular characterization of T-ALL should include immunophenotyping, since patients with thymic CD1a-positive T-ALL have a distinctly favorable outcome. Novel molecular markers with prognostic implications including HOX11L2, ERG, and BAALC, once confirmed in prospective studies, may help to filter out patients with high-risk thymic T-ALL who would potentially benefit from more intensive therapy. Mutational analyses of NOTCH1 and FBXW7 are, at present, still exploratory. Recently, Asfani et al. showed a favorable prognostic impact of NOTCH1 and FBXW7 mutations in adult T-ALL.36 Since biological and clinical differences of cases might account, in part, for the differences in results between studies, sequencing of mutations should be considered in future trials to prospectively validate their prognostic impact in adult T-ALL. Furthermore, the future use of specific inhibitors of activated NOTCH1 may be based on the mutational profile within the NOTCH1 signaling pathway.
Acknowledgements
we thank Liliana Mochmann for her critical reading of the manuscript.
Footnotes
The online version of this article contains a supplementary appendix.
Authorship and Disclosures
CDB conducted the study, performed research, analyzed data, and wrote the manuscript. JT performed the laboratory work. NG and DH provided the clinical data and were involved in the data analyses. AS performed the statistical analysis. CS and MM contributed to the laboratory work. TB and SS provided molecular data. CD Bloomfield was involved in the data analysis and writing of the manuscript. ET co-ordinated the research. WKH contributed to the analysis and to the writing of this manuscript.
The authors reported no potential conflicts of interest.
Funding: supported by grants from the Berliner Krebsgesellschaft and the Deutsche Krebshilfe (Max Eder Nachwuchsförderung) to CD Baldus.
References
- 1.Hoelzer D, Gökbuget N, Ottmann O, Pui CH, Relling MV, Appelbaum FR, et al. Acute lymphoblastic leukemia. Hematology (Am Soc Hematol Educ Program) 2002:162–92. doi: 10.1182/asheducation-2002.1.162. [DOI] [PubMed] [Google Scholar]
- 2.Thomas X, Boiron JM, Huguet F, Dombret H, Bradstock K, Vey N, et al. Outcome of treatment in adults with acute lymphoblastic leukemia: analysis of the LALA-94 trial. J Clin Oncol. 2004;22:4075–86. doi: 10.1200/JCO.2004.10.050. [DOI] [PubMed] [Google Scholar]
- 3.Mrozek K, Bloomfield CD. Chromosome aberrations, gene mutations and expression changes, and prognosis in adult acute myeloid leukemia. Hematology (Am Soc Hematol Educ Program) 2006:169–77. doi: 10.1182/asheducation-2006.1.169. [DOI] [PubMed] [Google Scholar]
- 4.Estey E, Döhner H. Acute myeloid leukaemia. Lancet. 2006;368:1894–907. doi: 10.1016/S0140-6736(06)69780-8. [DOI] [PubMed] [Google Scholar]
- 5.Pui CH, Jeha S. New therapeutic strategies for the treatment of acute lymphoblastic leukaemia. Nat Rev Drug Discov. 2007;6:149–65. doi: 10.1038/nrd2240. [DOI] [PubMed] [Google Scholar]
- 6.Armstrong SA, Look AT. Molecular genetics of acute lymphoblastic leukemia. J Clin Oncol. 2005;23:6306–15. doi: 10.1200/JCO.2005.05.047. [DOI] [PubMed] [Google Scholar]
- 7.Ellisen LW, Bird J, West DC, Soreng AL, Reynolds TC, Smith SD, et al. TAN-1, the human homolog of the Drosophila notch gene, is broken by chromosomal translocations in T-lymphoblastic neoplasms. Cell. 1991;66:649–61. doi: 10.1016/0092-8674(91)90111-b. [DOI] [PubMed] [Google Scholar]
- 8.Weng AP, Ferrando AA, Lee W, Morris JP, 4th, Silverman LB, Sanchez-Irizarry C, et al. Activating mutations of NOTCH1 in human T cell acute lymphoblastic leukemia. Science. 2004;306:269–71. doi: 10.1126/science.1102160. [DOI] [PubMed] [Google Scholar]
- 9.Mansour MR, Duke V, Foroni L, Patel B, Allen CG, Ancliff PJ, et al. Notch-1 mutations are secondary events in some patients with T-cell acute lymphoblastic leukemia. Clin Cancer Res. 2007;13:6964–9. doi: 10.1158/1078-0432.CCR-07-1474. [DOI] [PubMed] [Google Scholar]
- 10.Grabher C, von Boehmer H, Look AT. Notch 1 activation in the molecular pathogenesis of T-cell acute lymphoblastic leukaemia. Nat Rev Cancer. 2006;6:347–59. doi: 10.1038/nrc1880. [DOI] [PubMed] [Google Scholar]
- 11.Sulis ML, Williams O, Palomero T, Tosello V, Pallikuppam S, Real PJ, et al. NOTCH1 extracellular juxtamembrane expansion mutations in T-ALL. Blood. 2008;112:733–40. doi: 10.1182/blood-2007-12-130096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Radtke F, Wilson A, Stark G, Bauer M, van Meerwijk J, MacDonald HR, et al. Deficient T cell fate specification in mice with an induced inactivation of Notch1. Immunity. 1999;10:547–58. doi: 10.1016/s1074-7613(00)80054-0. [DOI] [PubMed] [Google Scholar]
- 13.Lin YW, Nichols RA, Letterio JJ, Aplan PD. Notch1 mutations are important for leukemic transformation in murine models of precursor-T leukemia/lymphoma. Blood. 2006;107:2540–54. doi: 10.1182/blood-2005-07-3013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nakayama KI, Nakayama K. Regulation of the cell cycle by SCF-type ubiquitin ligases. Semin Cell Dev Biol. 2005;16:323–33. doi: 10.1016/j.semcdb.2005.02.010. [DOI] [PubMed] [Google Scholar]
- 15.Oberg C, Li J, Pauley A, Wolf E, Gurney M, Lendahl U. The Notch intracellular domain is ubiquitinated and negatively regulated by the mammalian Sel-10 homolog. J Biol Chem. 2001;276:35847–53. doi: 10.1074/jbc.M103992200. [DOI] [PubMed] [Google Scholar]
- 16.Thompson BJ, Buonamici S, Sulis ML, Palomero T, Vilimas T, Basso G, et al. The SCFFBW7 ubiquitin ligase complex as a tumor suppressor in T cell leukemia. J Exp Med. 2007;204:1825–35. doi: 10.1084/jem.20070872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.O'Neil J, Grim J, Strack P, Rao S, Tibbitts D, Winter C, et al. FBW7 mutations in leukemic cells mediate NOTCH pathway activation and resistance to gamma-secretase inhibitors. J Exp Med. 2007;204:1813–24. doi: 10.1084/jem.20070876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Breit S, Stanulla M, Flohr T, Schrappe M, Ludwig WD, Tolle G, et al. Activating NOTCH1 mutations predict favorable early treatment response and long-term outcome in childhood precursor T-cell lym-phoblastic leukemia. Blood. 2006;108:1151–7. doi: 10.1182/blood-2005-12-4956. [DOI] [PubMed] [Google Scholar]
- 19.Lee SY, Kumano K, Masuda S, Hangaishi A, Takita J, Nakazaki K, et al. Mutations of the Notch1 gene in T-cell acute lymphoblastic leukemia: analysis in adults and children. Leukemia. 2005;19:1841–3. doi: 10.1038/sj.leu.2403896. [DOI] [PubMed] [Google Scholar]
- 20.Zhu YM, Zhao WL, Fu JF, Shi JY, Pan Q, Hu J, et al. NOTCH1 mutations in T-cell acute lymphoblastic leukemia: prognostic significance and implication in multifactorial leukemogenesis. Clin Cancer Res. 2006;12:3043–9. doi: 10.1158/1078-0432.CCR-05-2832. [DOI] [PubMed] [Google Scholar]
- 21.Mansour MR, Linch DC, Foroni L, Goldstone AH, Gale RE. High incidence of Notch-1 mutations in adult patients with T-cell acute lymphoblastic leukemia. Leukemia. 2006;20:537–9. doi: 10.1038/sj.leu.2404101. [DOI] [PubMed] [Google Scholar]
- 22.Brüggemann M, Raff T, Flohr T, Gökbuget N, Nakao M, Droese J. Clinical significance of minimal residual disease quantification in adult patients with standard risk acute lymphoblastic leukemia. Blood. 2006;107:1116–23. doi: 10.1182/blood-2005-07-2708. [DOI] [PubMed] [Google Scholar]
- 23.Schwartz S, Rieder H, Schläger B, Burmeister T, Fischer L, Thiel E. Expression of the human homologue of rat NG2 in adult acute lymphoblastic leukemia: close association with MLL rearrangement and a CD10(−)/CD24(−)/CD65s(+)/CD15(+) B-cell phenotype. Leukemia. 2003;17:1589–95. doi: 10.1038/sj.leu.2402989. [DOI] [PubMed] [Google Scholar]
- 24.Baldus CD, Burmeister T, Martus P, Schwartz S, Gökbuget N, Bloomfield CD, et al. High expression of the ETS transcription factor ERG predicts adverse outcome in acute T-lymphoblastic leukemia in adults. J Clin Oncol. 2006;24:4714–20. doi: 10.1200/JCO.2006.06.1580. [DOI] [PubMed] [Google Scholar]
- 25.Bene MC, Castoldi G, Knapp W, Ludwig WD, Matutes E, Orfao A, et al. Proposals for the immunological classification of acute leukemias. Leukemia. 1995;9:1783–6. [PubMed] [Google Scholar]
- 26.Nowak D, Mossner M, Baldus CD, Hopfer O, Thiel E, Hofmann WK. Mutation analysis of hCDC4 in AML cells identifies a new intronic polymorphism. Int J Med Sci. 2006;3:148–51. doi: 10.7150/ijms.3.148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Baldus CD, Martus P, Burmeister T, Schwartz S, Gökbuget N, Bloomfield CD, et al. Low ERG and BAALC expression identifies a new subgroup of adult acute T-lymphoblastic leukemia with a highly favorable outcome. J Clin Oncol. 2007;25:3739–45. doi: 10.1200/JCO.2007.11.5253. [DOI] [PubMed] [Google Scholar]
- 28.Baak U, Gökbuget N, Orawa H, Schwartz S, Hoelzer D, Thiel E, Burmeister T. Thymic adult T-cell acute lymphoblastic leukemia stratified in standard- and high-risk group by aberrant HOX11L2 expression: experience of the German multicenter ALL study group. German Multicenter ALL Study Group. Leukemia. 2008;22:1154–60. doi: 10.1038/leu.2008.52. [DOI] [PubMed] [Google Scholar]
- 29.van Grotel M, Meijerink JP, van Wering ER, Langerak AW, Beverloo HB, Buijs-Gladdines JG, et al. Prognostic significance of molecular-cytogenetic abnormalities in pediatric T-ALL is not explained by immunophenotypic differences. Leukemia. 2008;22:124–31. doi: 10.1038/sj.leu.2404957. [DOI] [PubMed] [Google Scholar]
- 30.Malyukova A, Dohda T, von der Lehr N, Akhoondi S, Corcoran M, Heyman M, et al. The tumor suppressor gene hCDC4 is frequently mutated in human T-cell acute lymphoblastic leukemia with functional consequences for Notch signaling. Cancer Res. 2007;67:5611–6. doi: 10.1158/0008-5472.CAN-06-4381. [DOI] [PubMed] [Google Scholar]
- 31.Maser RS, Choudhury B, Campbell PJ, Feng B, Wong KK, Protopopov A, et al. Chromosomally unstable mouse tumours have genomic alterations similar to diverse human cancers. Nature. 2007;447:966–71. doi: 10.1038/nature05886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Onoyama I, Tsunematsu R, Matsumoto A, Kimura T, de Alborán IM, Nakayama K, et al. Conditional inactivation of Fbxw7 impairs cell-cycle exit during T cell differentiation and results in lymphomatogenesis. J Exp Med. 2007;204:2875–88. doi: 10.1084/jem.20062299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shih IeM, Wang TL. Notch signaling, γ-secretase inhibitors, and cancer therapy. Cancer Res. 2007;67:1879–82. 34. doi: 10.1158/0008-5472.CAN-06-3958. [DOI] [PubMed] [Google Scholar]
- 34.Palomero T, Sulis ML, Cortina M, Real PJ, Barnes K, Ciofani M, et al. Mutational loss of PTEN induces resistance to NOTCH1 inhibition in T-cell leukemia. Nat Med. 2007;13:1203–10. doi: 10.1038/nm1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Real PJ, Tosello V, Palomero T, Castillo M, Hernando E, de Stanchina E, et al. γ-secretase inhibitors reverse glucocorticoid resistance in T cell acute lymphoblastic leukemia. Nat Med. 2009;15:50–8. doi: 10.1038/nm.1900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Asnafi V, Buzyn A, Le Noir S, Baleydier F, Simon A, Beldjord K, et al. NOTCH1/FBXW7 mutation identifies a large subgroup with favourable outcome in adult T-cell acute lymphoblastic leukemia (TALL): a GRAALL study. Blood. 2009;113:3918–24. doi: 10.1182/blood-2008-10-184069. [DOI] [PubMed] [Google Scholar]