This paper shows for the first time that patients with X-linked neutropenia, caused by mutations in the Wiskott Aldrich syndrome gene, developed myelodysplasia/acute myeloid leukemia with acquisition of mutations in the CSF3R gene and loss of chromosome 7. See related perspective article on page 1333.
Keywords: G-CSF, X-linked neutropenia, acute myeloid leukemia
Abstract
X-linked neutropenia (XLN) is a rare form of Congenital Neutropenia (CN) caused by inherited gain-of-function mutations of WAS. Here we report 2 cases of the original L270P X-linked neutropenia kindred that evolved to MDS or AML, with acquisition of G-CSFR (CSF3R) mutations and monosomy 7. Thus, leukemic transformation with acquisition of CSF3R mutations and monosomy 7 is not restricted to classical congenital neutropenia with autosomal inheritance, but can also occur in other genotypes of inherited neutropenia.
Introduction
In 2001, we described X-linked neutropenia (XLN) as a novel subtype of congenital neutropenia (CN) in a three-generation family with 5 affected males and with a g.810T>C WAS mutation, resulting in a p.L270P gain-of-function mutation in the GTPase binding domain (GBD) of the Wiskott-Aldrich Syndrome protein (WASP).1,4 In 2006, 2 more XLN cases were described, one with a p.S272P and one with a p.I294T mutation, the latter reportedly presenting with myelodysplasia (MDS).2 In 2008, we reported a large XLN kindred with 10 affected males and 8 female carriers with a p.I294T WASP mutation, which also results in a gain-of function. CN is a heterogeneous disease with mostly autosomal inheritance patterns. Autosomal dominant CN is caused by ELA2 mutations in 60% of cases.5 Autosomal recessive CN or Kostmann’s disease is caused by HAX1 mutations.6 Other rare subtypes of the disease include autosomal dominant mutations of GFI17 and autosomal recessive mutations of G6PC3.8 The diagnosis of CN is usually made at birth or in the first months of life with recurrent, severe infections and neutrophil counts below 0.5 109/L. Monocyte and eosinophil counts are elevated in CN.7,9 In contrast, XLN has an X-linked inheritance pattern and although also characterized by severe neutropenia, it is often diagnosed at a later age, due to the infectious phenotype which is relatively mild considering the degree of neutropenia. Monocytopenia and low NK cell numbers are typical hematologic features of XLN. Low B-cell counts, low-normal platelets, inversion of the CD4/CD8 ratio and low-normal IgA levels can be present.3
Autosomal dominant and recessive CN carry an increased risk of myeloid transformation with a cumulative incidence of up to 25% at 20 years.9 This leukemic conversion is often associated with the development of monosomy 7 and mutations in the G-CSF receptor gene.6,9–14 Whether a risk of malignant transformation in XLN exists has not yet been reported to date. Here we report for the first time 2 cases with XLN, who developed MDS-RAEB and AML, both with monosomy 7 and CSF3R mutations in leukemic samples.
Design and Methods
Patient material
Blood and bone marrow samples were obtained according to the guidelines of the local IRB at the University Hospitals Leuven.
Detection and quantification of CSF3R mutations in XLN patients
DNA was extracted from viably frozen blood, bone marrow samples or from fixed cells. Exon 17 of CSF3R11,13,15 was amplified using the primers: CSF3R-F 5’-accctttgtgttccaccagt-3’ and CSF3R-R 5’-ttggtcctttcttcctccct-3 (GeneAmp PCR system 2400, Applied Biosystems, Foster City, California). PCR products were verified with 6% PAGE-gels and sequenced in both directions, using BigDye® Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems). Sequencing results were analyzed using Sequence Scanner (Applied Biosystems).
For the detection of the g.2425T>G (p.Y729) mutation by qPCR, we used primers DF 5’-ccagcgatcaggtcctttag-3’ as allele specific forward primer together with 5’-gggcac-tatctccgctgtga-3’ as reverse primer. QPCR was performed in a Real Time PCR quantification AB7000 using p53 as a control.
Results and Discussion
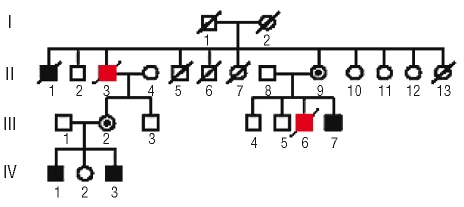
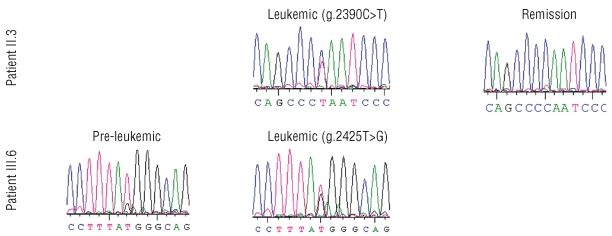
Case II.3 (Figure 1) was diagnosed with neutropenia with neutrophil counts between 0.15–0.9 109/L in his late twenties. Before that time his medical history consisted of frequent staphylococcal skin infections, recurrent bronchitis, pulmonary tuberculosis, a sacral fistula and peritonitis of unknown origin (possibly TBC). The bone marrow karyotype was normal. At age 63, a marrow aspirate showed a poorly represented myeloid lineage with neutrophil maturation arrest and discrete dysplasia in the red lineage and a normal karyotype. From age 63 onwards, G-CSF (filgrastim) was initiated at 0.3 mg/day subcutaneously. At age 65, the diagnosis of MDS type RAEB16 was made and monosomy 7 was found in 10/10 evaluated metaphases.17–18 At age 65, he was admitted with fever and a leukocyte count of 17.9 109/L of which 31% were blasts. After discontinuation of G-CSF, the leukocyte number and circulating blasts decreased and the cytological diagnosis was revised to MDS-RAEB. A stable clinical and hematologic remission was achieved after 4 courses of decitabine. Although the bone marrow blastosis remained at 10%, this was interpreted as evidence of maturation arrest and not of MDS, as repeated marrow karyotypes remained normal. A truncating CSF3R mutation g.2390C>T (p.Q718), previously described by other authors,11,13,15 was identified in a heterozygous pattern in a marrow sample obtained at the time of RAEB (Figure 2). The same mutation could also be documented at lower levels in a sample 2.5 years after treatment, during stable remission (Figure 2). He died at age 68, due to an unrelated cause.
Figure 1.
Updated L270P Pedigree. Filled squares: males with documented neutropenia; circles with a black dot: carrier females; open symbols: unaffected individuals; patients with MDS/AML are indicated in red squares.
Figure 2.
Sequencing profile of patients II.3 and III.6 at different time points.
In case III.6 (Figure 1), neutropenia was first diagnosed at age 27, with a neutrophil count of 0.3 109/L and a medical history consisting of frequent herpes labialis and anal fistulas. He reportedly had a hypocellular bone marrow aspirate with low numbers of granulocyte precursors. At age 31, on the occasion of neutropenic fever and a staphylococcal abscess in the right thigh, a bone marrow aspirate showed a maturation arrest in the myeloid lineage and monosomy 7 in 2 of 11 metaphases. He was not on G-CSF at that time. Interfase FISH was also consistent with monosomy 7 in a minor subpopulation (7%). However, two subsequent marrows six months and five years later revealed a normal karyotype. At age 33, supportive treatment with G-CSF was started at 0.3 mg SC 3 x/wk. At age 37, monosomy 7 reappeared in the bone marrow (7/10 metaphases)19. One year later he developed a refractory AML with monosomy 7 (10/10) as sole karyotypic abnormality. CSF3R mutation screening in a bone marrow sample at this time showed a heterozygous g.2425T>G (p.Y729) mutation in approximately all cells. The same mutation was also identified by qPCR in ~34% of cells in a pre-leukemic sample obtained at age 37 (Figure 2).
In this article we describe 2 cases of XLN with evolution to AML/MDS with acquired monosomy 7 and CSF3R mutations. Monosomy 7 is among the most common chromosomal aberrations in AML and MDS(20). Acquired loss of 7 or of 7q is associated with an array of congenital bone marrow failure syndromes, including CN, Shwachman-Diamond Syndrome and Fanconi anemia and is considered a poor prognostic factor.17,20
CN is well known as a pre-leukemic condition.6,9,13,21–23 Evolution to acute myeloid leukemia has been best documented in autosomal dominant CN (with ELA-2 mutations).9,13 The cumulative incidence of malignant transformation in CN is reportedly more than 25% after 20 years of observation.9 In CN, development of MDS/AML is significantly associated with the acquisition of truncating CSF3R mutations. The frequency of CSF3R mutations in CN patients with leukemic transformation is approximately 80% in both ELA2 and HAX1 positive patients, whereas in CN patients without leukemia, the frequency of CSF3R mutations is only 30%.9 CSF3R mutations have also been described in one of 4 members of the original Kostmann family with a presumable HAX1 mutation without clear malignant evolution.24 In addition, 3 other patients with a HAX1-mutation have been described with a CSF3R mutation, one of whom developed myelodysplasia.6 Moreover, Ward et al. described a young patient with an ELA2 mutation and a constitutive extracellular G-CSF-R mutation, leading to hyporesponsiveness to G-CSF treatment. This patient later developed consecutive acquired (intracellular) CSF3R mutations, leading to restoration of CSF3R responsiveness, but so far without malignant conversion.25
The g.2390C>T (p.Q718) and g.2425T>G (p.Y729) CSF3R mutations that were observed here, have already been reported in CN and were found to be associated with malignancy.11,13,15 Thus, XLN also encompasses an increased risk of conversion to a myeloid malignancy, as does classical CN. At the time of overt leukemia, both patients exhibited a heterozygous CSF3R mutation in approximately 100% of analyzed cells. In case III.6 we could show that the CSF3R mutation preceded the leukemic transformation by approximately 21 months. In patient II.3, no antecedent samples were available but the mutational burden was decreased in two remission bone marrow samples 1.5 and 2.5 years later. Thus, the CSF3R mutational burden was found to correlate with the leukemic burden, as was also described in CN.13 Given the context of congenital neutropenia and administration of G-CSF in both cases, it is unclear to what extent the chronic neutropenia and the administration of G-CSF contributed to the malignant transformation. Patient II.3 had received high doses of G-CSF for two years and patient III.6 had received only supportive G-CSF during the five years preceding the development of AML. Although the duration of G-CSF is not a risk factor for leukemia, a G-CSF dose-effect has been suggested in CN patients requiring high doses of G-CSF. Whether this is due to the severity of the disease, the apparent resistance to G-CSF or the high doses of G-CSF is still under debate.14
CSF3R mutations often precede the leukemic transformation and can be present for years in patients who never develop leukemia. Therefore, CSF3R mutations are considered an early event in leukemogenesis, occurring prior to malignant transformation, as an adaptive response of a defective stem cell pool in CN and leading to activation of anti-apoptotic signaling cascades.13 In line with this is the observation that mutant ELA2 leads to increased apoptosis of myeloid progenitors and that this pro-apoptotic effect could be overridden by CSF3R mutations, leading to enhanced survival and proliferation of hematopoietic progenitor cells.26 According to this model, G-CSF therapy may cause genomic instability of the progenitor cells due to increased pressure on proliferation. Long-term G-CSF therapy could lead to preferential clonal expansion of CSF3R mutant cells that may already be more susceptible to additional aberrations, possibly leading to leukemia.9,27
Non-truncating CSF3R mutations have also been documented in de novo MDS and AML and aberrant expression of G-CSF and its receptor have been observed in solid tumors like ovarian cancer, bladder cancer and squamous cell carcinoma.28 Although G-CSF is frequently administered in these malignancies to overcome chemotherapy-induced neutropenia, this could be a reason for concern.
So far, no cases of hematologic malignancies have been diagnosed in the large Irish p.I294T kindred that we recently reported, nor have we detected major sub-populations with CSF3R mutations using direct sequencing. However, most cases in the latter family are younger than in the first family; the oldest p.I294T XLN case in the Irish family is now 48 years old (range 10–48; mean 35 years) and G-CSF is used more sparingly in this family.
We conclude that XLN is a pre-leukemic condition, similar to autosomally inherited CN. Acquisition of monosomy 7 and CSF3R mutations appear to be a final common pathway associated with leukemic transformation, irrespective of the genetic defect underlying the neutropenia. The benefits of G-CSF treatment should, therefore, be carefully weighed against the possible effects in leukemogenesis in pre-leukemic diseases, such as congenital neutropenias. Based on our findings and on the mild infectious phenotype of XLN, we recommend that G-CSF be used sparingly in XLN and restricted to infectious episodes.
Acknowledgments
for clinical data gathering, we are grateful to Dr Isabelle Meyts, Emiel Baeten, Koen Devriendt and Hilde Demuynck.
Footnotes
Authorship and Disclosures
KB designed research, performed experiments, collected and analyzed data, and wrote the article; PV designed research, analyzed data and wrote the article.
This text presents research results of the Belgian program of Interuniversity Poles of Attraction initiated by the Belgian State, Prime Minister’s Office, Science Policy Programming. The authors assume complete scientific responsibility.
The authors reported no potential conflicts of interest.
Funding: fund for Scientific Research-Flanders (Belgium) (FWO Vlaanderen). KB is an aspirant researcher and PV is a senior clinical investigator of the Fund for Scientific Research-Flanders (Belgium) (FWO Vlaanderen).
References
- 1.Devriendt K, Kim AS, Mathijs G, Frints SG, Schwartz M, Van Den Oord JJ, et al. Constitutively activating mutation in WASP causes X-linked severe congenital neutropenia. Nat Genet. 2001;27:313–7. doi: 10.1038/85886. [DOI] [PubMed] [Google Scholar]
- 2.Ancliff PJ, Blundell MP, Cory GO, Calle Y, Worth A, Kempski H, et al. Two novel activating mutations in the Wiskott-Aldrich syndrome protein result in congenital neutropenia. Blood. 2006;108:2182–9. doi: 10.1182/blood-2006-01-010249. [DOI] [PubMed] [Google Scholar]
- 3.Beel K, Cotter MM, Blatny J, Bond J, Lucas G, Green F, et al. A large kindred with X-linked neutropenia with an I294T mutation of the Wiskott-Aldrich syndrome gene. Br J Haematol. 2009;144:120–6. doi: 10.1111/j.1365-2141.2008.07416.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.den Dunnen JT, Antonarakis SE. Mutation nomenclature extensions and suggestions to describe complex mutations: a discussion. Hum Mutat. 2000;15:7–12. doi: 10.1002/(SICI)1098-1004(200001)15:1<7::AID-HUMU4>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- 5.Dale DC, Link DC. The many causes of severe congenital neutropenia. N Engl J Med. 2009;360:3–5. doi: 10.1056/NEJMp0806821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Klein C, Grudzien M, Appaswamy G, Germeshausen M, Sandrock I, Schaffer AA, et al. HAX1 deficiency causes autosomal recessive severe congenital neutropenia (Kostmann disease) Nat Genet. 2007;39:86–92. doi: 10.1038/ng1940. [DOI] [PubMed] [Google Scholar]
- 7.Horwitz MS, Duan Z, Korkmaz B, Lee HH, Mealiffe ME, Salipante SJ. Neutrophil elastase in cyclic and severe congenital neutropenia. Blood. 2007;109:1817–24. doi: 10.1182/blood-2006-08-019166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boztug K, Appaswamy G, Ashikov A, Schaffer AA, Salzer U, Diestelhorst J, et al. A syndrome with congenital neutropenia and mutations in G6PC3. N Engl J Med. 2009;360:32–43. doi: 10.1056/NEJMoa0805051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zeidler C, Germeshausen M, Klein C, Welte K. Clinical implications of ELA2-, HAX1-, and G-CSF-receptor (CSF3R) mutations in severe congenital neutropenia. Br J Haematol. 2009;144:459–67. doi: 10.1111/j.1365-2141.2008.07425.x. [DOI] [PubMed] [Google Scholar]
- 10.Donadieu J, Leblanc T, Bader Meunier B, Barkaoui M, Fenneteau O, Bertrand Y, et al. Analysis of risk factors for myelodysplasias, leukemias and death from infection among patients with congenital neutropenia. Experience of the French Severe Chronic Neutropenia Study Group. Haematologica. 2005;90:45–53. [PubMed] [Google Scholar]
- 11.Dong F, Dale DC, Bonilla MA, Freedman M, Fasth A, Neijens HJ, et al. Mutations in the granulocyte colony-stimulating factor receptor gene in patients with severe congenital neutropenia. Leukemia. 1997;11:120–5. doi: 10.1038/sj.leu.2400537. [DOI] [PubMed] [Google Scholar]
- 12.Freedman MH, Bonilla MA, Fier C, Bolyard AA, Scarlata D, Boxer LA, et al. Myelodysplasia syndrome and acute myeloid leukemia in patients with congenital neutropenia receiving G-CSF therapy. Blood. 2000;96:429–36. [PubMed] [Google Scholar]
- 13.Germeshausen M, Ballmaier M, Welte K. Incidence of CSF3R mutations in severe congenital neutropenia and relevance for leukemogenesis: Results of a long-term survey. Blood. 2007;109:93–9. doi: 10.1182/blood-2006-02-004275. [DOI] [PubMed] [Google Scholar]
- 14.Rosenberg PS, Alter BP, Bolyard AA, Bonilla MA, Boxer LA, Cham B, et al. The incidence of leukemia and mortality from sepsis in patients with severe congenital neutropenia receiving long-term G-CSF therapy. Blood. 2006;107:4628–35. doi: 10.1182/blood-2005-11-4370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dong F, Brynes RK, Tidow N, Welte K, Lowenberg B, Touw IP. Mutations in the gene for the granulocyte colony-stimulating-factor receptor in patients with acute myeloid leukemia preceded by severe congenital neutropenia. N Engl J Med. 1995;333:487–93. doi: 10.1056/NEJM199508243330804. [DOI] [PubMed] [Google Scholar]
- 16.Bennett JM, Catovsky D, Daniel MT, Flandrin G, Galton DA, Gralnick HR, et al. Proposals for the classification of the myelodysplastic syndromes. Br J Haematol. 1982;51:189–99. [PubMed] [Google Scholar]
- 17.Luna-Fineman S, Shannon KM, Lange BJ. Childhood monosomy 7: epidemiology, biology, and mechanistic implications. Blood. 1995;85:1985–99. [PubMed] [Google Scholar]
- 18.Brozek I, Babinska M, Kardas I, Wozniak A, Balcerska A, Hellmann A, et al. Cytogenetic analysis and clinical significance of chromosome 7 aberrations in acute leukaemia. J Appl Genet. 2003;44:401–12. [PubMed] [Google Scholar]
- 19.Barrios NJ, Kirkpatrick DV, Levin ML, Varela M. Transient expression of trisomy 21 and monosomy 7 following cyclosporin A in a patient with aplastic anemia. Leuk Res. 1991;15:531–3. doi: 10.1016/0145-2126(91)90065-2. [DOI] [PubMed] [Google Scholar]
- 20.Perkins D, Brennan S, Carstairs K, Bailey D, Pantalony D, Poon A, et al. Regional cancer cytogenetics: a report on 1,143 diagnostic cases. Cancer Genet Cytogenet. 1997;96:64–80. doi: 10.1016/s0165-4608(96)00363-9. [DOI] [PubMed] [Google Scholar]
- 21.Rosenberg PS, Alter BP, Link DC, Stein S, Rodger E, Bolyard AA, et al. Neutrophil elastase mutations and risk of leukaemia in severe congenital neutropenia. Br J Haematol. 2008;140:210–3. doi: 10.1111/j.1365-2141.2007.06897.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yetgin S, Germeshausen M, Touw I, Koc A, Olcay L. Acute lymphoblastic leukemia in a patient with congenital neutropenia without G-CSF-R and ELA2 mutations. Leukemia. 2005;19:1710–1. doi: 10.1038/sj.leu.2403850. [DOI] [PubMed] [Google Scholar]
- 23.Badolato R, Fontana S, Notarangelo LD, Savoldi G. Congenital neutropenia: advances in diagnosis and treatment. Curr Opin Allergy Clin Immunol. 2004;4:513–21. doi: 10.1097/00130832-200412000-00007. [DOI] [PubMed] [Google Scholar]
- 24.Carlsson G, Aprikyan AA, Ericson KG, Stein S, Makaryan V, Dale DC, et al. Neutrophil elastase and granulocyte colony-stimulating factor receptor mutation analyses and leukemia evolution in severe congenital neutropenia patients belonging to the original Kostmann family in northern Sweden. Haematologica. 2006;91:589–95. [PubMed] [Google Scholar]
- 25.Ward AC, Gits J, Majeed F, Aprikyan AA, Lewis RS, O’Sullivan LA, et al. Functional interaction between mutations in the granulocyte colony-stimulating factor receptor in severe congenital neutropenia. Br J Haematol. 2008;142:653–6. doi: 10.1111/j.1365-2141.2008.07224.x. [DOI] [PubMed] [Google Scholar]
- 26.Aprikyan AA, Kutyavin T, Stein S, Aprikian P, Rodger E, Liles WC, et al. Cellular and molecular abnormalities in severe congenital neutropenia predisposing to leukemia. Exp Hematol. 2003;31:372–81. doi: 10.1016/s0301-472x(03)00048-1. [DOI] [PubMed] [Google Scholar]
- 27.Germeshausen M, Welte K, Ballmaier M. In vivo expansion of cells expressing acquired CSF3R mutations in patients with severe congenital neutropenia. Blood. 2009;113:668–70. doi: 10.1182/blood-2008-09-178087. [DOI] [PubMed] [Google Scholar]
- 28.Ward AC. The role of granulocyte colony-stimulating factor receptor (G-CSF-R) in disease. Frontiers in Bioscience. 2007;12:608–18. doi: 10.2741/2086. [DOI] [PubMed] [Google Scholar]