This paper shows that the expression of the ribosomal protein S14 (RPS14) is reduced in about two thirds of patients with non-5q- myelodysplastic syndrome. See related perspective article on page 1336.
Keywords: low RPS14, myelodysplastic syndromes, prolonged survival, 5q- aberration
Abstract
To further clarify the role of ribosomal protein S14 (RPS14) in myelodysplastic syndrome, we examined RPS14 transcription in bone marrow derived CD34+ cells from patients with non-5q- myelodysplastic syndrome and found a reduced expression of RPS14 in 51 of 72 (71%) patients. MDS patients with an intermediate-1 risk (INT-1) score according to the international prognostic scoring system and low RPS14 expression had a superior median overall survival of not reached versus 25 months compared to INT-1 patients with high RPS14 expression (p=0.0249). Using multivariate analysis, the RPS14 expression status was confirmed as an independent predictor for survival in INT-1 patients.
Introduction
The 5q- syndrome is a hematologic malignancy characterized by a distinct phenotype and interstitial deletion of the long arm of chromosome 5.1,2 One particular clinical feature of patients with 5q- syndrome is a longer survival compared to patients with other types of myelodysplastic syndrome (MDS).3 Despite many candidate genes within the commonly deleted region, RPS14, which is an integral part of the 40S ribosomal subunit, could be identified as a primary candidate gene and its loss may be causative for the 5q- syndrome.4 Recently, deletions within the RPS14 coding region were found in MDS patients without concurrent cytogenetic aberrations on chromosome 5 (non-5q- MDS).5 To further clarify the role of RPS14 in non-5q- MDS, we examined bone marrow derived CD34+ selected cells from 72 patients with various types of non-5q- MDS with regard to their level of RPS14 gene expression and its possible prognostic relevance.
Design and Methods
Patient and donor characteristics
All patients examined were diagnosed with MDS at our department and included in the Duesseldorf MDS Registry. Bone marrow samples were obtained between 2001 and 2006 according to institutional guidelines following approval by the local ethics committee. All 83 patients and 11 normal donors gave written informed consent. The median age was 70 years (range 38–86 years) and 76 years (range 28–85) for patients and normal donors, respectively. Fifty patients (61%) were male. MDS types diagnosed according to WHO classification were as follows: 4 5q- syndrome, one refractory anemia (RA), one RA with ringsideroblasts (RARS), 17 refractory cytopenia with multilineage dysplasia (RCMD), 16 ringsideroblastic RCMD (RSRCMD), 13 RA with excess of blasts (RAEB) I, 15 RAEB II and 16 secondary acute myeloid leukemia following MDS (sAML). Overall, 11 patients had deletions on the long arm of chromosome 5 of which 4 had 5q- syndrome and 7 had additional cytogenetic aberrations (5q- MDS). For prognosis and survival analyses, only the 72 patients with non-5q- MDS were evaluated. Of these 72 patients, 2 had low, 33 intermediate-1 (INT-1), 13 intermediate-2 (INT-2) and 24 high-risk MDS according to the IPSS. In December 2008 the median follow-up was 24 months (range 1–91 months).
Sample preparation and quantitative RT-PCR
Bone marrow CD34+ cells were selected using immunomagnetic beads as published.6 RNA was purified as described6 and samples were stored at −80°C. For detection and quantification of mRNA levels of RPS14 and GAPDH the LightCycler 1.2 technology (Roche, Mannheim, Germany) was used. PCR reactions were performed using the LightCycler-FastStart DNA Master SYBR Green I kit (Roche). The PCR was carried out in a final volume of 20 μL using 0.5 μM of each primer, 4 mM MgCl2, 2 μL supplied enzyme mix containing the reaction buffer, FastStart Taq DNA polymerase and DNA double-strand specific SYBR Green I dye for the detection of PCR products were used. The PCR protocol was as follows: 480 s preincubation at 95°C; 45 cycles of 15 s at 95°C, 5 s at 63°C and 20 s at 72°C. To test the specifity of the PCR, the reaction products were subjected to melting curve analysis with the LightCycler system and to conventional agarose gel electrophoresis to rule out synthesis of unspecific products. Primer sequences were as follows: RPS14 forward: 5′-ATG TTG GCT GCC CAG-3′; RPS14 reverse: 5′-GGT CTT GGT CCTATT TCC TC3′-; GAPDH forward: 5′-TCC ATG ACA ACT TTG GTA TCG-3′; GAPDH reverse: 5′-GTC GCT GTT GAA GTC AGA GGA-3′. Relative gene expression levels were calculated as the difference of CT values of RPS14 and the housekeeping gene GAPDH as a control (ΔCT). For each individual MDS patient sample, the respective RPS14 fold of expression was calculated against the mean expression level of the 11 normal donors (Δ-ΔCT). To analyze the prognostic relevance of RPS14 expression levels in non-5q- MDS, we dichotomized the patient samples at the median ΔCT values of the respective IPSS groups and divided them into two RPS14 expression groups: a RPS14 low expression group with ΔCT values above the median value and an RPS14 high expression group with deltaCT values below the median value. Online Supplementary Figure 1 shows the individual deltaCT values of all 94 samples analyzed.
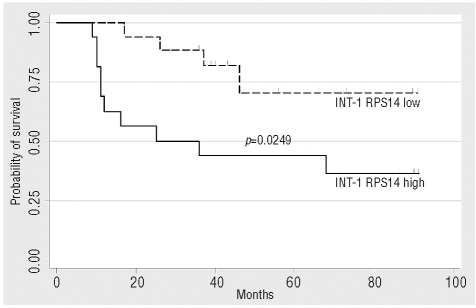
Figure 1.
Unadjusted survival of all 33 patients with INT-1 according to their RPS14 gene expression calculated with the Kaplan Maier method. Patients with INT-1 and a low expression of RPS14 have a superior median overall survival of not reached versus 25 months in the patients with INT-1 and high RPS14 expression. The log-rank test was used for testing significance of the survival functions (p=0.0249).
Statistical methods
Overall survival was calculated from the day of sample collection. Overall survival was estimated according to the Kaplan-Meier method. The equality of survival between different groups was tested using the log-rank test. Prognostic factor analysis was performed with Cox’s multivariate regression method.
Results and Discussion
To test the accuracy of our assay to detect a reduced RPS14 gene expression level, we first compared RPS14 mRNA levels in CD34+ cells of 11 normal donors and 11 patients with deletions on the long arm of chromosome 5 (5q- MDS). Here, RPS14 expression in 5q- MDS patients was reduced by a median fold change of −1.69 (range 3.9 to −7.75) as compared to normal donors (Online Supplementary Figure 2A). This fold of reduction is perfectly in line with previous studies analyzing the gene expression profile of CD34+ cells from patients with 5q- syndrome in comparison to normal donors, where a −1.46 fold of reduction of RPS14 expression was reported.7 We continued our analysis with bone marrow derived CD34+ cells from 72 patients with various types of non-5q- MDS and found a reduced RPS14 expression by at least −1.5-fold in 51 patients (71%). While 13 of 31 (42%) patients with early MDS (RA, RARS, RCMD, RS-RCMD) had low RPS14 expression (Online Supplementary Figure 2B), the respective proportion in patients with advanced MDS (RAEB I, RAEB II, sAML) was 93% (38 out of 41 patients, Online Supplementary Figure 2C). This high number of patients with advanced MDS and decreased RPS14 expression might be the result of clonal proliferation with an increased proportion of presumably RPS14 low expressing CD34+ blast cells within the whole CD34+ population as, to this date, no deletion or mutation of both alleles or pathological hypermethylation of any of the genes within the commonly deleted region, including RPS14, could be detected.8,9 One potential mechanism leading to the reduced RPS14 expression in patients with non-5q- MDS without blast excess, may involve small deletions in the gene encoding RPS14 as recently demonstrated in such patients.5 A specific genomic event is the cause for the reduced RPS14 expression in patients with 5q- syndrome and considering the superior survival of these patients, we wondered if the RPS14 expression status may also have prognostic relevance in non-5q- MDS patients. Therefore, we continued our analysis according to the patients’ IPSS risk score, which is a well established prognostic tool for patients with MDS.10 When using the respective median deltaCT values to discriminate RPS14 low and high expressing patients, we did not find any differences with regard to overall survival when the analysis was confined to patients with IPSS INT-2 or high-risk MDS. Interestingly, as far as the 33 INT-1 patients are concerned, the group of INT-1 patients with low RPS14 expression (INT-1 low) had not yet reached their median overall survival in contrast to those INT-1 patients with high RPS14 expression (INT-1 high) whose median overall survival was 25 months (p=0.0249). With a median follow-up of 40 months (range 9–91 months, Figure 1), a significant survival advantage for patients with low RPS14 expression and an INT-1 risk score was observed. On multivariate analysis the RPS14 expression status was identified as a significant independent predictor for survival in the 33 INT-1 risk patients (Table 1) analyzed here, indicating that the RPS14 expression status may be used to separate two distinct INT-1 risk groups with different prognosis. As it has been shown that lenalidomide-responsive non-5q- patients have a gene expression signature, which resembles a defect in erythroid differentiation11 and that loss of RPS14 expression in CD34+ cells causes such a phenotype,2 one may speculate that the individual RPS14 expression status might help to identify patients with IPSS intermediate risk who will most likely respond to treatment with lenalidomide. Taken together, we confirmed a low RPS14 expression in MDS patients with 5q- aberrations and found that low RPS14 expression levels can also be commonly observed in patients without 5q- aberrations. Of clinical relevance, a low RPS14 expression level is apparently associated with a favorable prognosis in patients with INT-1 risk MDS.
Table 1.
Cox’s multivariate regression to determine predictors for survival of INT-1 patients.
Footnotes
The online version of this article contains a supplementary appendix.
Authorship and Disclosures
AC was the principal investigator and takes primary responsibility for the paper; AC, IB, and UG wrote the paper; BJ and RS performed the laboratory work for this study; AC, IB and RH coordinated the research; GK, AC and UG carried out the statistical analysis; RH reviewed the manuscript.
The authors reported no potential conflicts of interest.
References
- 1.Boultwood J, Lewis S, Wainscoat JS. The 5q-syndrome. Blood. 1994;84:3253–60. [PubMed] [Google Scholar]
- 2.Giagounidis AA, Germing U, Haase S, Hildebrandt B, Schlegelberger B, Schoch C, et al. Clinical, morphological, cytogenetic, and prognostic features of patients with myelodysplastic syndromes and del(5q) including band q31. Leukemia. 2004;18:113–9. doi: 10.1038/sj.leu.2403189. [DOI] [PubMed] [Google Scholar]
- 3.Haase D, Germing U, Schanz J, Pfeilstocker M, Nosslinger T, Hildebrandt B, et al. New insights into the prognostic impact of the karyotype in MDS and correlation with subtypes: evidence from a core dataset of 2124 patients. Blood. 2007;110:4385–95. doi: 10.1182/blood-2007-03-082404. [DOI] [PubMed] [Google Scholar]
- 4.Ebert BL, Pretz J, Bosco J, Chang CY, Tamayo P, Galili N, et al. Identification of RPS14 as a 5q- syndrome gene by RNA interference screen. Nature. 2008;451:335–9. doi: 10.1038/nature06494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sohal DS, Pellagatti A, Zhou L, Mo Y, Opalinska JB, Alencar C, et al. Downregulation of ribosomal proteins is seen in non-5q- MDS Blood 2008(ASH Annual Meeting Abstracts)112.[Abstract 854]17890457 [Google Scholar]
- 6.Bruns I, Steidl U, Fischer JC, Czibere A, Kobbe G, Raschke S, et al. Pegylated granulocyte colony-stimulating factor mobilizes CD34+ cells with different stem and progenitor subsets and distinct functional properties in comparison with unconjugated granulocyte colony-stimulating factor. Haematologica. 2008;93:347–55. doi: 10.3324/haematol.12081. [DOI] [PubMed] [Google Scholar]
- 7.Boultwood J, Pellagatti A, Cattan H, Lawrie CH, Giagounidis A, Malcovati L, et al. Gene expression profiling of CD34+ cells in patients with the 5q- syndrome. Br J Haematol. 2007;139:578–89. doi: 10.1111/j.1365-2141.2007.06833.x. [DOI] [PubMed] [Google Scholar]
- 8.Valencia A, Cervera J, Such E, Sanz MA, Sanz GF. Lack of RPS14 promoter aberrant methylation supports the haploinsufficiency model for the 5q- syndrome. Blood. 2008;112:918. doi: 10.1182/blood-2008-05-159707. [DOI] [PubMed] [Google Scholar]
- 9.Boultwood J, Fidler C, Strickson AJ, Watkins F, Gama S, Kearney L, et al. Narrowing and genomic annotation of the commonly deleted region of the 5q- syndrome. Blood. 2002;99:4638–41. doi: 10.1182/blood.v99.12.4638. [DOI] [PubMed] [Google Scholar]
- 10.Greenberg P, Cox C, LeBeau MM, Fenaux P, Morel P, Sanz G, et al. International scoring system for evaluating prognosis in myelodysplastic syndromes. Blood. 1997;89:2079–88. [PubMed] [Google Scholar]
- 11.Ebert BL, Galili N, Tamayo P, Bosco J, Mak R, Pretz J, et al. An erythroid differentiation signature predicts response to lenalidomide in myelodysplastic syndrome. PLoS Med. 2008;5:e35. doi: 10.1371/journal.pmed.0050035. [DOI] [PMC free article] [PubMed] [Google Scholar]