Abstract
Stomata, flanked by pairs of guard cells, are small pores on the leaf surfaces of plants and they function to control gas exchange between plants and the atmosphere. Stomata will open when water is available to allow for the uptake of carbon dioxide for photosynthesis. During periods of drought, stomata will close to reduce desiccation stress. As such, optimal functioning of stomata will impact on water use efficiency by plants. The development of an inducible, modular system for robust and targeted gene expression in stomatal guard cells is reported here. It is shown that application of ethanol vapour to activate the gene expression system did not affect the ability of stomata to respond to ABA in bioassays to determine the promotion of stomatal closure and the inhibition of stomatal opening. The system that has been developed allows for robust spatio-temporal control of gene expression in all cells of the stomatal lineage, thereby enabling molecular engineering of stomatal function as well as studies on stomatal development.
Keywords: Guard cells, inducible gene expression, modular system, spatio-temporal gene expression, stomata
Introduction
Stomata are ‘breathing’ pores or ‘gatekeepers’ on the surfaces of leaves that provide for atmospheric continuity between the plant and the atmosphere. About 5% of the total leaf surface area can be attributed to stomatal pores and it is through these pores that water vapour is lost from the leaves and carbon dioxide is gained from the environment (Hetherington and Woodward, 2003). The ability to control water loss through stomata and an impervious leaf cuticle marks a key evolutionary phase in the colonization of land by plants about 400 million years ago (Hetherington and Woodward, 2003). The primary role of stomata is to enable the plant to optimize the gain of carbon dioxide for photosynthesis, and at the same time, minimizing the loss of water during transpiration under changing environmental conditions. Stomata do this by regulating changes in the turgor of the pair of flanking guard cells through modification of ionic fluxes, accumulation of sugars, remodelling of cytoskeletal organization, membrane trafficking, and changes in gene expression (Hetherington, 2001; Schroeder et al., 2001; Hetherington and Woodward, 2003; Fan et al., 2004; Roelfsema and Hedrich, 2005). Because of the fundamental importance of stomata to plant physiology and survival, the control of stomatal pore sizes necessitates the robust co-ordination of multiple cellular processes to bring about changes in guard cell turgor (Hetherington, 2001; Schroeder et al., 2001; Hetherington and Woodward, 2003; Fan et al., 2004; Roelfsema and Hedrich, 2005).
It is becoming increasingly clear that stomatal guard cells employ a diverse array of signalling intermediates to bring about the co-ordinated control of turgor, and hence stomatal pore sizes (Hetherington, 2001; Schroeder et al., 2001; Hetherington and Woodward, 2003; Fan et al., 2004; Roelfsema and Hedrich, 2005). Recent work suggests that the architectural organization of the complex signalling system in stomatal guard cells may take the form of a network comprising modules and hubs, reminiscent of scale-free networks (Hetherington and Woodward, 2003; Li et al., 2006). Such a system would confer the necessary redundancy and robustness to the signalling system present in stomatal guard cells, given their fundamental importance as ‘gatekeepers’ for atmospheric continuity between the plant and the environment. The complexity of the guard cell signalling system makes it a challenging prospect for dissecting its organization. Current understanding of the stomatal guard cell signalling system can largely be attributed to the detailed analysis of mutants harbouring genetic lesions that affected stomatal behaviour (Hetherington, 2001; Schroeder et al., 2001; Hetherington and Woodward, 2003; Fan et al., 2004; Roelfsema and Hedrich, 2005). However, it is noteworthy that these mutants often exhibit pleiotropic effects throughout the plant. In order to dissect the organization of the guard cell signalling system, tools are needed for the spatio-temporal regulation of gene expression, i.e. inducible and targeted expression only in stomatal guard cells.
Several promoters have been cloned that confer guard cell-specific gene expression or enhanced gene expression in guard cells and these include the KAT1 promoter (Nakamura et al., 1995), the pGC1 promoter (Yang et al., 2008), and the gcPEPC promoter (Kopka et al., 1997). These promoters are useful for the spatial regulation of gene expression in stomatal guard cells. However, these promoters do not allow for temporal regulation of gene expression. There are several chemical-inducible systems available for temporal regulation of gene expression in plants and these include the use of inducers like tetracycline, dexamethasone, 17-β-oestradiol, and ethanol (Moore et al., 2006). In order to achieve spatio-temporal control of gene expression in stomatal guard cells, a system was developed that incorporates the use of a guard cell-specific promoter (gcPEPC) (Kopka et al., 1997) and the ethanol-inducible gene switch (AlcR/alcA) (Caddick et al., 1998) in a modular system (pSATn/pPZP-RCS2) (Chung et al., 2005; Tzfira et al., 2005) that will enable the expression of multiple genes from a single binary plasmid.
Materials and methods
Plant growth
Arabidopsis thaliana Col-0 plants were grown in a controlled atmosphere room under the following conditions: 8/16 h and 22/18 °C light/dark; PPFD of 250 μmol m−2 s−1. A. thaliana seeds were sown in Petri dishes (9 cm diameter) containing half-strength Murashige and Skoog medium (Sigma, UK) supplemented with 1% (w/v) sucrose (Sigma, UK) and 0.6% plant cell culture-tested agar (Sigma, UK). Seeds were then stratified at 4 °C for 3 d in the dark before being placed in the controlled atmosphere room under the conditions listed above. 10-d-old seedlings were then planted into Arabaskets (BetaTech, Belgium) in a compost:vermiculite (3:1 v/v) mix (Shamrock multipurpose compost, Shamrock Horticulture, UK) and allowed to grow. Plants were watered by fine misting daily. Plant transformation by the floral-dip method was carried out as described by Logemann et al. (2006). Three-week-old transgenic plants were placed in Magenta GA-7 containers with a microfuge tube each containing 1 ml of ethanol (4% or 25%) or water (0% ethanol). For expression analyses, youngest, fully-expanded leaves were harvested at various times (0, 2, and 10 h) post-induction and snap frozen for subsequent RNA extraction. For fluorescence microscopy, the youngest, fully expanded or developing leaves were examined under the fluorescence stereomicroscope or confocal laser scanning microscope 10 h post-induction with 25% ethanol vapour.
Engineering of expression cassettes and assembly into the binary vector
The coding sequence of the AlcR transcription factor was amplified from vector pBin-ΔalcR (Syngenta, UK) with the following primers containing the HindIII (forward) and SalI (reverse) endonuclease restriction sites for subsequent insertion into the MCS of the pSAT2 vector by restriction cloning (see Supplementary Fig. S1 at JXB online).
AlcR: forward primer: 5′-TTT AAG CTT ATG GCA GAT ACG CGC CGA-3′; reverse primer: 5′-GGG GTC GAC CTA CAA AAA GCT GTC AAC-3′
The alcA promoter fragment was amplified from vector pUC-del-alcAN (Syngenta, UK) with the following primers containing the AgeI (forward) and NcoI (reverse) endonuclease restriction sites for replacing the constitutively active promoters in vectors, pSAT3, pSAT4, pSAT6, and pSAT7 (see Supplementary Fig. S1 at JXB online).
alcA: forward primer: 5′-GGG ACC GGT GGG ATA GTT CCG ACC TAG GAT TGG-3′; reverse primer: 5′-GGG CCA TGG GTC GTC CTC TCC AAA TGA AAT GAA C-3′
The coding sequence of Citrine was amplified from vector pRSET B citrine (kindly provided by Professor Roger Tsien, University of California, San Diego, USA) with the following primers containing the ApaI (forward) and BamHI (reverse) endonuclease restriction sites for subsequent insertion into the MCS of pSAT3 by restriction cloning (see Supplementary Fig. S1 at JXB online).
Citrine: forward primer: 5′-TTT GGG CCC ATG GTG AGC AAG GGC GAG-3′; reverse primer: 5′-TTT GGA TCC CTT GTA CAG CTC GTC CAT-3′
The coding sequence of DsRed2 was amplified from vector pGDR (Goodin et al., 2002) (kindly provided by Dr Michael Goodin, University of Kentucky, USA) with the following primers containing the ApaI (forward) and BamHI (reverse) endonuclease restriction sites for subsequent insertion into the MCS of pSAT4 by restriction cloning (see Supplementary Fig. S1 at JXB online).
DsRed2: forward primer: 5′-AAA GGG CCC ATG GCC TCC TCC GAG AAC GT-3′; reverse primer: 5′-TTT GGA TCC CAG GAA CAG GTG GTG GCG-3′
The individual expression cassettes were subcloned into the binary vector, pPZP-RCS2 using homing endonucleases before being transformed into Agrobacterium tumefaciens strain GV3101 (GV3101 carries a disarmed Ti plasmid that possesses the vir genes needed for T-DNA transfer, but has no functional T-DNA region of its own).
All sequences assembled into the individual vectors were sent to Eurofins Biotech (Germany) for sequencing and sequence verification.
Fluorescence microscopy
Fluorescence stereomicroscopy was carried out using an Olympus SZX16 Zoom Stereomicroscope equipped with a 2× Plan Apochromat objective coupled with a DP71 digital colour CCD camera. Citrine was visualized with a blue excitation long pass filter (BP460-495) and a barrier filter BA510IF (Interference filter). DsRed2 was visualized with a narrow green excitation filter (BP530-550) and a barrier filter BA575IF (Olympus, UK). Confocal microscopy was carried out with a Leica TCS-SL laser scanning microscope (Leica Microsystems, Germany). Citrine and DsRed2 fluorescent proteins were imaged using an excitation wavelength of 514 nm and 563 nm, respectively.
Stomatal bioassay
Four–five-week-old Arabidopsis plants were pre-exposed to 25% ethanol vapour overnight. The youngest, fully-expanded leaves were harvested at the end of the night period and expression of the Citrine and DsRed2 fluorescent reporter proteins were determined using an epifluorescence microscope. Inhibition of stomatal opening and promotion of stomatal closure bioassays were conducted to determine the effects of ethanol vapour treatment and fluorescent reporter protein expression on stomatal function. Peels from lower epidermes which had been exposed to ethanol vapour or water vapour were incubated in Petri dishes containing 10 ml of opening buffer (50 mM KCl, 10 mM MES-KOH, pH 6.15) at 22 °C. For inhibition of stomatal opening, epidermal peels were incubated for 2 h in the dark to ensure that stomatal apertures are closed. Initial stomatal apertures (closed) were determined by measuring after the 2 h dark incubation. The epidermal peels were then transferred to fresh opening buffer containing ABA (0, 1, 10, 50 μM) at 22 °C under illumination at 200 μmol m−2 s−1 for 3 h. Pictures of stomata were acquired at 40× magnification for each treatment and analysed with ImageJ to determine the stomatal apertures. For the promotion of stomatal closure, epidermal peels were placed in Petri dishes containing 10 ml of opening buffer at 22 °C for 2 h under illumination at 200 μmol m−2 s−1. Epidermal peels were then incubated for a further 3 h in the presence of ABA (0, 1, 10, 50 μM). Pictures were taken and stomatal apertures were measured as described for the inhibition of stomatal opening experiments.
Expression analysis
The youngest fully-expanded leaves of 3-week-old plants were harvested at 0, 2, and 10 h post-induction with ethanol vapour (4% or 25%) or water vapour (0% ethanol) and snap frozen in liquid nitrogen. Total RNA was isolated using the Qiagen RNeasy kit (Qiagen, UK) according to the manufacturer's instructions. The quality and quantity of the total RNA was determined using a NanoDrop 1000 spectrophotometer (Thermo Scientific, UK). 950 ng of total RNA was treated with 1 U of DNaseI (Invitrogen, UK) before being used for cDNA synthesis using 200 U of M-MLV Reverse Transcriptase (Invitrogen, UK). The resultant cDNA was used for PCR amplification using 0.5 U of Go-Taq™ DNA polymerase (Promega, UK). The sequences of the primers used for amplification of EF1α, Citrine, and DsRed2 are listed below.
EF1α: forward primer: 5′-ATG CCC CAG GAC ATC GTG ATT TCA t-3′; reverse primer: 5′-TTG GCG GCA CCC TTA GCT GGA TCA-3′
Citrine: forward primer: 5′-ATG GTG AGC AAG GGC GAG GA-3′; reverse primer: 5′-CTT GTA CAG CTC GTC CAT GCC G-3′
DsRed2: forward primer: 5′-ATG GCC TCC TCC GAG AAC GT-3′; reverse primer: 5′-CAG GAA CAG GTG GTG GCG-3′
Results and discussion
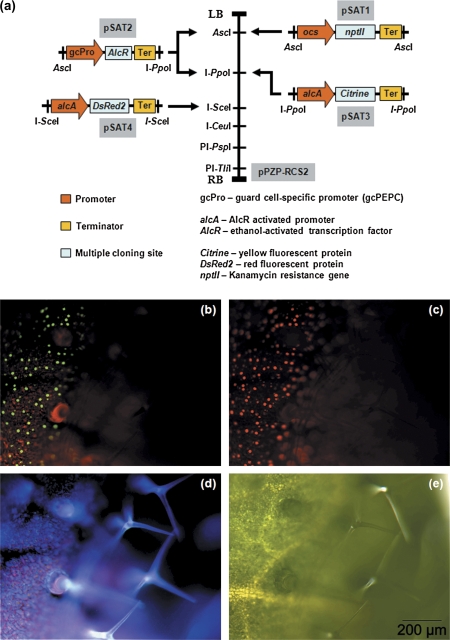
The strategy for developing the system to achieve spatio-temporal control of gene expression in stomatal guard cells is illustrated in Fig. 1a. As proof-of-concept, the constitutively active promoters were replaced in the modular satellite vectors (pSATn) (Chung et al., 2005; Tzfira et al., 2005) with either the guard cell-specific, gcPEPC promoter (Kopka et al., 1997) or the alcA promoter of the ethanol gene switch (Caddick et al., 1998). The rationale for using the gcPEPC promoter is because it is guard cell-specific (von Groll and Altmann, 2001), and it has been used successfully to knockdown expression of phospholipase C (PLC) in stomatal guard cells (Hunt et al., 2003). Because the alcA promoter is specifically activated by the AlcR transcription factor in the presence of ethanol (Caddick et al., 1998), the AlcR gene was cloned downstream of the gcPEPC promoter so that AlcR expression is spatially restricted to stomatal guard cells. In addition, the reporter genes for Citrine fluorescent protein (Citrine) and Discosoma sp. Red fluorescent protein 2 (DsRed2) was cloned downstream of the alcA promoter in two separate pSAT vectors (Fig. 1a). The pSAT expression cassettes were then subcloned, as discrete modules, into the binary plasmid, pPZP-RCS2 using homing endonucleases (Fig. 1a). The completed binary plasmid was then transformed into Agrobacterium tumefaciens and transgenic Arabidopsis thaliana plants were engineered following floral-dip transformation (Logemann et al., 2006) and stable transformants were selected on the basis of resistance to the antibiotic, kanamycin.
Fig. 1.
Schematic representation of the strategy for the development of an inducible, modular system for targeted and robust gene expression in stomatal guard cells and fluorescence stereomicroscopy for detecting the expression of the Citrine and DsRed2 fluorescent proteins in stomatal guard cells. (a) Salient features of the inducible, modular vector system for targeted and robust gene expression in stomatal guard cells. The modular nature of the pSAT vectors allows the assembly of multiple gene expression cassettes within a single binary vector (pPZP-RCS2) using homing endonucleases (AscI, I-PpoI, I-SceI, I-CeuI, PI-PspI, and PI-TliI). (b–e) Bioimaging of ethanol-inducible expression (10 h post-induction with 25% ethanol vapour) of Citrine and DsRed2 using fluorescence stereomicroscopy. (b) Expression of Citrine (green) and (c) DsRed2 (red) fluorescent proteins only in stomatal guard cells. (d) Autofluorescence from cell walls showing the presence of trichomes on the surface of the same leaf and (e) bright field image of the leaf. Representative scale bar (200 μm) is shown in (e). (This figure is available in colour at JXB online.)
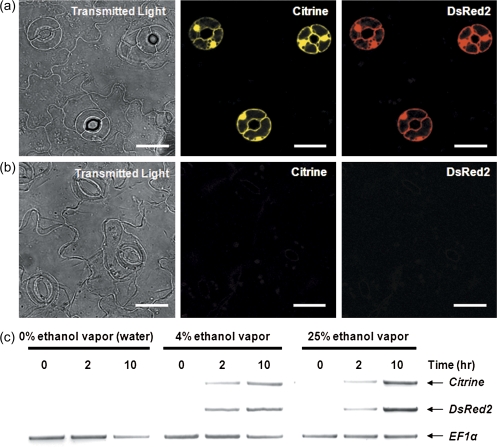
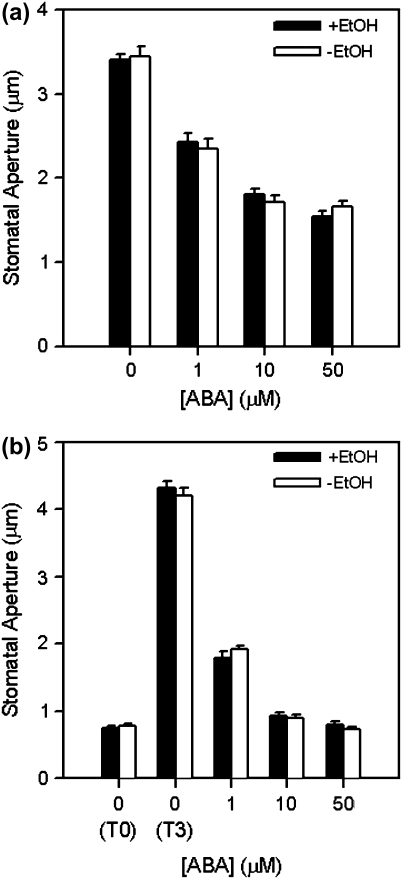
To test the effectiveness of the system that has been developed, stable transformants harbouring the construct shown in Fig. 1a were exposed to ethanol vapour. Stable transformants were exposed to water vapour as a negative control. The expression of the Citrine and DsRed2 fluorescent proteins in stomatal guard cells was visualised using fluorescence stereomicroscopy (Fig. 1b–e) and confocal laser scanning microscopy (Fig. 2a, b). It was demonstrated conclusively that expression of Citrine and DsRed2 was induced only in stomatal guard cells following exposure to ethanol vapour (Figs 1b, c, 2a) and that these two fluorescent proteins were not expressed in the absence of ethanol (Fig. 2b). Importantly, the expression of the fluorescent reporter proteins was not observed in any other cell types (data not shown). Semi-quantitative RT-PCR was used to demonstrate detectable levels of Citrine and DsRed2 transcripts as early as 2 h after exposure to 4% ethanol vapour (Fig. 2c). Interestingly, similar steady-state levels of transcripts were observed when expression levels were examined following induction with 4% or 25% ethanol vapour, indicating that the system that had been developed is highly sensitive to the inducer (Fig. 2c). Previous studies reporting the utilization, in Arabidopsis, of ethanol vapour as the inducer of the ethanol-inducible gene expression system used concentrations of ethanol ≥95% (Roslan et al., 2001; Laufs et al., 2003; Maizel and Weigel, 2004). It is shown here that a much lower concentration (4%) of ethanol vapour is sufficient to induce robust expression of the reporter genes in stomatal guard cells. It is also shown that ethanol exposure and the subsequent expression of Citrine and DsRed2 fluorescent reporter proteins did not affect the ability of ABA (1, 10, and 50 μM) to promote stomatal closure (Fig. 3a) or inhibit light-induced stomatal opening (Fig. 3b). Our data clearly demonstrated that the ethanol-inducible system for the control of gene expression is non-toxic, highly sensitive, and robust in stomatal guard cells.
Fig. 2.
Confocal laser scanning microscopy of isolated epidermes from fully-expanded leaves of transgenic Arabidopsis thaliana plants harbouring the ethanol-inducible construct shown in Fig. 1a and semi-quantitative RT-PCR analysis of the kinetics of expression induction by ethanol. (a) Confocal scanning micrographs were acquired 10 h post-induction with 25% ethanol vapour. Exposure to ethanol vapour induced the expression of Citrine (yellow) and DsRed2 (red) only in stomatal guard cells. Images are representative of leaf epidermes from three independent experiments. Scale bars=20 μm. (b) No expression of Citrine or DsRed2 was observed in the absence of ethanol vapour. Images are representative of leaf epidermes from three independent experiments. Scale bars=20 μm. (c) Steady-state levels of reporter gene transcripts (Citrine and DsRed2) at 0, 2, and 10 h post-induction with 4% or 25% ethanol vapour. Plants exposed to water (0% ethanol vapour) were used as negative controls. The results are shown as reversed gel images (dark bands on white background) and are representative of 2–3 independent experiments. (This figure is available in colour at JXB online.)
Fig. 3.
Effects of ethanol exposure on stomatal function. (a) Effects of exposure to 25% ethanol vapour on (a) promotion of stomatal closure and (b) inhibition of stomatal opening by ABA (1, 10, and 50 μM). Values are means ±SE of 90–200 stomata.
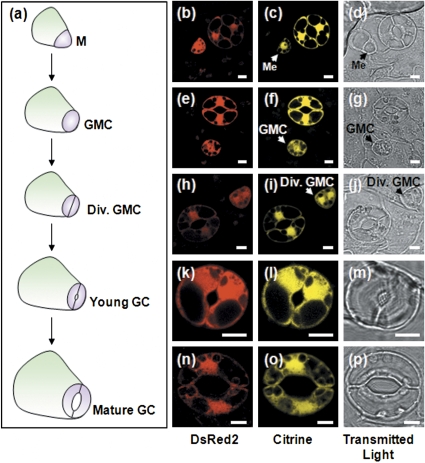
Interestingly, it was also observed that expression of the Citrine and DsRed2 fluorescent proteins was not restricted only to mature guard cells 10 h post-induction by 25% ethanol vapour (Fig. 4a–p). It was possible to visualize Citrine and DsRed2 fluorescent proteins in cells of the stomatal lineage: meristemoids (Fig. 4a–c), guard mother cells (Fig. 4d–f), dividing guard mother cells (Fig. 4h–j), young guard cells (Fig. 4k–m), and mature guard cells (Fig. 4n–p).
Fig. 4.
Confocal laser scanning microscopy showing ethanol-inducible expression of Citrine and DsRed2 in all cell-types of the stomatal lineage 10 h post-induction with 25% ethanol vapour. (a) Schematic representation of stomatal development. (b–d) Meristemoid (Me), (e–g) guard mother cell (GMC), (h–j) dividing guard mother cell (Div. GMC), (k–m) developing guard cell, and (n–p) mature guard cell. Images are representative of 50–60 cells. Scale bars=5 μm. (This figure is available in colour at JXB online.)
The system described has the potential to be used for engineering the functional responses of mature guard cells as it was observed that expression of the reporter genes are spatially restricted only to stomatal guard cells. In addition, the system that has been developed here is highly sensitive to the inducer (ethanol), as concentrations as low as 4% ethanol supplied as a vapour is sufficient to induce expression of the reporter genes. The ability for the spatio-temporal control of expression of multiple genes in stomatal guard cells will be an important tool for dissecting the complex signalling system utilized by stomatal guard cells to sense and respond to changes in the prevailing environmental conditions.
Also, robust activity was observed of the system developed for spatio-temporal control of gene expression in cells of the stomatal lineage. Stomatal development begins with the asymmetric division of the meristemoid mother cell to give rise to a triangular-shaped meristemoid cell. The meristemoids will differentiate into the guard mother cell, and the guard mother cell will divide symmetrically to give rise to the pair of guard cells that flank a single stoma (Bergmann and Sack, 2007; Pillitteri and Torri, 2007; Casson and Gray, 2008). We are beginning to understand the underlying molecular mechanisms controlling stomatal cell state transitions. For example, three closely related basic-helix-loop-helix (bHLH) transcription factor genes, SPEECHLESS (SPCH), MUTE, and FAMA have been shown to be involved in controlling guard cell development. These three bHLH transcription factors have been shown to function sequentially to affect stomatal cell state transition. SPCH regulates the transition from meristemoid mother cells to meristemoid cells, MUTE regulates the transition from meristemoid to guard mother cell, and FAMA regulates the transition from guard mother cell to guard cells (Bergmann and Sack, 2007; Pillitteri and Torri, 2007, Casson and Gray, 2008). The spatio-temporal system that has been developed for controlling expression of multiple genes will be a particularly useful tool for manipulating the expression of the trio of bHLH transcription factors (SPCH, MUTE, and FAMA) and other factors like SCREAM/ICE1 and SCREAM2 (Kanaoka et al., 2008) to further our understanding of the molecular mechanisms underpinning stomatal development.
In summary, the system developed here represents a significant advance to current methods for spatio-temporal control of gene expression in plants. The system described has the potential to be used for engineering the functional responses of mature guard cells. In addition, because of the robust activity of the system in cells of the stomatal lineage, it can also be used in studies on developmental processes determining stomatal cell state transitions. Because of the modular nature of the vector system, it can easily be adapted or modified for use in different cell types through the use of cell-type specific promoters.
Supplementary data
Supplementary data are available at JXB online.
Supplementary Fig. S1. Schematic representation of the structural features of the pSAT vectors showing the restriction endonuclease sites within the multipe cloning site (MC) and the restricition endo nuclease sites (Age1 and Nco1) that can be used for promoter (Pro) substitution.
Acknowledgments
This research was funded by a Science Foundation Ireland (SFI) grant RF/06/GEN034 to CK-YN, an Irish Research Council for Science, Engineering, and Technology (IRCSET) Postdoctoral Fellowship to TCX, IRCSET Postgraduate Scholarships to CMH and JPC, and a SFI UREKA-Phytotechnology Undergraduate Research Bursary to MS.
References
- Bergmann DC, Sack FD. Stomatal development. Annual Review of Plant Biology. 2007;58:163–181. doi: 10.1146/annurev.arplant.58.032806.104023. [DOI] [PubMed] [Google Scholar]
- Caddick MX, Greenland AJ, Jepson I, Krause KP, Qu N, Riddell KV, Salter MG, Schuch W, Sonnewald U, Tomsett AB. An ethanol inducible gene switch for plants used to manipulate carbon metabolism. Nature Biotechnology. 1998;16:177–180. doi: 10.1038/nbt0298-177. [DOI] [PubMed] [Google Scholar]
- Casson S, Gray JE. Influence of environmental factors on stomatal development. New Phytologist. 2008;178:9–23. doi: 10.1111/j.1469-8137.2007.02351.x. [DOI] [PubMed] [Google Scholar]
- Chung S-M, Frankman EL, Tzfira T. A versatile vector system for multiple gene expression in plants. Trends in Plant Science. 2005;10:357–361. doi: 10.1016/j.tplants.2005.06.001. [DOI] [PubMed] [Google Scholar]
- Fan L-M, Zhao Z, Assmann SM. Guard cells: a dynamic signalling model. Current Opinion in Plant Biology. 2004;7:537–546. doi: 10.1016/j.pbi.2004.07.009. [DOI] [PubMed] [Google Scholar]
- Goodin MM, Dietzgen RG, Schichnes D, Ruzin S, Jackson AO. GD vectors: versatile tools for the expression of green and red fluorescent protein fusions in agroinfiltrated plant leaves. The Plant Journal. 2002;31:375–383. doi: 10.1046/j.1365-313x.2002.01360.x. [DOI] [PubMed] [Google Scholar]
- Hetherington AM. Guard cell signalling. Cell. 2001;107:711–714. doi: 10.1016/s0092-8674(01)00606-7. [DOI] [PubMed] [Google Scholar]
- Hetherington AM, Woodward IF. The role of stomata in sensing and driving environmental change. Nature. 2003;424:901–908. doi: 10.1038/nature01843. [DOI] [PubMed] [Google Scholar]
- Hunt L, Mills LN, Pical C, Leckie CP, Aitken FL, Kopka J, Mueller-Roeber B, McAinsh MR, Hetherington AM, Gray JE. Phospholiase C is required for the control of stomatal apertureby ABA. The Plant Journal. 2003;34:47–55. doi: 10.1046/j.1365-313x.2003.01698.x. [DOI] [PubMed] [Google Scholar]
- Kanaoka MM, Pillitteri LJ, Fujii H, Yoshida Y, Bogenschutz NL, Takabayashi J, Zhu J-K, Torii KU. SCREAM/ICE1 and SCREAM2 specify three cell-state transitional steps leading to Arabidopsis stomatal differentiation. The Plant Cell. 2008;20:1775–1785. doi: 10.1105/tpc.108.060848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopka J, Provart NJ, Müller-Rober B. Potato guard cells respond to drying soil by a complex change in the expression of genes related to carbon metabolism and turgor regulation. The Plant Journal. 1997;11:871–882. doi: 10.1046/j.1365-313x.1997.11040871.x. [DOI] [PubMed] [Google Scholar]
- Laufs P, Coen E, Kronenberger J, Traas J, Doonan J. Separable roles of UFO during floral development revealed by conditional restoration of gene function. Development. 2003;130:785–796. doi: 10.1242/dev.00295. [DOI] [PubMed] [Google Scholar]
- Li S, Assmann SM, Albert R. Predicting essential components of signal transduction networks: a dynamic model of guard cell abscisic acid signalling. PLoS Biology. 2006;4:e312. doi: 10.1371/journal.pbio.0040312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logemann E, Birjenbihl RP, Ülker B, Somssich IE. An improved method for preparing Agrobacterium cells that simplifies the Arabidopsis transformation. Plant Methods. 2006;2:16. doi: 10.1186/1746-4811-2-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maizel A, Weigel D. Temporally and spatially controlled induction of gene expression in Arabidopsis thaliana. The Plant Journal. 2004;38:167–171. doi: 10.1111/j.1365-313X.2004.02027.x. [DOI] [PubMed] [Google Scholar]
- Moore I, Samalova M, Kurup S. Transactivated and chemically inducible gene expression in plants. The Plant Journal. 2006;45:651–683. doi: 10.1111/j.1365-313X.2006.02660.x. [DOI] [PubMed] [Google Scholar]
- Nakamura RL, McKendree WL, Jr, Hirsch RE, Sedbrook JC, Gaber RF, Sussman MR. Expression of an Arabidopsis potassium channel gene in guard cells. Plant Physiology. 1995;109:371–374. doi: 10.1104/pp.109.2.371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pillitteri LJ, Torii KU. Breaking the silence: three bHLH proteins direct cell-fate decisions during stomatal development. BioEssays. 2007;29:861–870. doi: 10.1002/bies.20625. [DOI] [PubMed] [Google Scholar]
- Roelfsema MR, Hedrich R. In the light of stomatal opening: new insights into ‘the Watergate’. New Phytologist. 2005;167:665–691. doi: 10.1111/j.1469-8137.2005.01460.x. [DOI] [PubMed] [Google Scholar]
- Roslan HA, Salter MG, Wood CD, et al. Characterization of the ethanol-inducible alc gene-expression system in Arabidopsis thaliana. The Plant Journal. 2001;28:225–235. doi: 10.1046/j.1365-313x.2001.01146.x. [DOI] [PubMed] [Google Scholar]
- Schroeder JI, Kwak JM, Allen GJ. Guard cell abscisic acid signalling and engineering drought hardiness in plants. Nature. 2001;410:327–330. doi: 10.1038/35066500. [DOI] [PubMed] [Google Scholar]
- Tzfira T, Tian GW, Lacroix B, Vyas S, Li J, Leitner-Dagan Y, Krichevsjy A, Taylor T, Vainstein A, Citovsky V. pSAT vectors: a modular series of plasmids for autofluorescent protein tagging and expression of multiple genes in plants. Plant Molecular Biology. 2005;57:503–516. doi: 10.1007/s11103-005-0340-5. [DOI] [PubMed] [Google Scholar]
- von Groll U, Altmann T. Stomatal cell biology. Current Opinion in Plant Biology. 2001;4:555–560. doi: 10.1016/s1369-5266(00)00215-6. [DOI] [PubMed] [Google Scholar]
- Yang Y, Costa A, Leonhardt N, Siegel RS, Schroeder JI. Isolation of a strong Arabidopsis guard cell promoter and its potential as a research tool. Plant Methods. 2008;4:6. doi: 10.1186/1746-4811-4-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.