Abstract
A recent analysis of the hexokinase (HXK) gene family from Arabidopsis revealed that three hexokinase-like (HKL) proteins lack catalytic activity, but share about 50% identity with the primary glucose (glc) sensor/transducer protein AtHXK1. Since the AtHKL1 protein is predicted to bind glc, although with a relatively decreased affinity, a reverse genetics approach was used to test whether HKL1 might have a related regulatory function in plant growth. By comparing phenotypes of an HKL1 mutant (hkl1-1), an HXK1 mutant (gin2-1), and transgenic lines that overexpress HKL1 in either wild-type or gin2-1 genetic backgrounds, it is shown that HKL1 is a negative effector of plant growth. Interestingly, phenotypes of HKL1 overexpression lines are generally very similar to those of gin2-1. These are quantified, in part, as reduced seedling sensitivity to high glc concentrations and reduced seedling sensitivity to auxin-induced lateral root formation. However, commonly recognized targets of glc signalling are not apparently altered in any of the HKL1 mutant or transgenic lines. In fact, most, but not all, of the observed phenotypes associated with HKL1 overexpression occur independently of the presence of HXK1 protein. The data indicate that HKL1 mediates cross-talk between glc and other plant hormone response pathways. It is also considered Whether a possibly decreased glc binding affinity of HKL1 could possibly be a feedback mechanism to limit plant growth in the presence of excessive carbohydrate availability is further considered.
Keywords: Auxin, glucose signalling, growth regulation, GUS staining, hexokinase, hexokinase-like, hypocotyl elongation, plant hormones
Introduction
All living organisms have complex regulatory networks that enable them to sense their nutrient status and to adjust their growth and development accordingly. Glucose (glc) is an important metabolic nutrient, which also functions as a signalling molecule that regulates gene expression in a variety of organisms (Towle, 2005; Rolland et al., 2006; Gancedo, 2008). In plants, glc affects the expression of more than 1000 genes involved in a diverse array of biological processes (Price et al., 2004; Osuna et al., 2007). Many of the glc-regulated genes are involved in phytohormone biosynthesis and response pathways which control plant growth (Gibson, 2004). Furthermore, genetic studies indicate that many mutants of plant glc signalling are alleles of genes with defined roles in ABA or ethylene biosynthesis, or their signalling networks (Leon and Sheen, 2003; Rognoni et al., 2007).
Genetic and biochemical evidence indicates that two Arabidopsis proteins, hexokinase1 (HXK1) and the regulator of G-protein signalling1 (RGS1), have independent roles in glc sensing and phytohormone responses (Rolland et al., 2006). As a glc sensor, AtHXK1 modulates plant growth at many different developmental stages (Moore et al., 2003). A null mutant of AtHXK1, gin2-1, has reduced shoot and root growth, increased apical dominance, delayed flowering, and altered sensitivities to auxin, cytokinin, and glc (Moore et al., 2003). On the other hand, AtRGS1 has been suggested to function as a glc binding protein that can attenuate cell division in primary root apical meristems through its interaction with GPA1, a heterotrimeric G-protein subunit (Chen et al., 2006; Johnston et al., 2007). Similar to HXK1, plant heterotrimeric G-proteins also affect a diverse array of developmental and hormone responses (Perfus-Barbeoch et al., 2004). However, even though seedlings of null mutants of both AtHXK1 and AtRGS1 fail to undergo normal glc-dependent cell cycle arrest, their hypocotyl elongation responses at low light are opposite to each other (Chen et al., 2003; Moore et al., 2003).
Plant HXKs are encoded by a modest family of about 5–10 genes (Claeyssen and Rivoal, 2007). HXK proteins are reported to occur in the cytosol, mitochondria, plastids, nuclei, and Golgi (Balasubramanian et al., 2007, and references therein). AtHXK1 is predominantly associated with the mitochondria, but also reportedly can occur in the nucleus (Cho et al., 2006). There is evidence that from both locations, AtHXK1 can regulate gene and/or protein expression, but there are questions regarding both scenarios (see Balasubramanian et al., 2008). In rice, OsHXK5 and OsHXK6 have been shown recently to act as glc sensors and similarly to have a predominantly mitochondrial association, but possible nuclear function (Cho et al., 2009). However, in contrast to AtHXK1, both OsHXK5 and OsHXK6 do contain a predicted nuclear localization signal.
A recent analysis of the Arabidopsis HXK gene family revealed that three of the six members lack catalytic activity when assayed with varying concentrations of glc or fructose (Karve et al., 2008). These were designated as hexokinase-like (HKL) proteins since they also are about 50% identical to AtHXK1. The basis for the lack of catalytic activity in the HKL proteins was attributed to a number of changes throughout the primary sequences and not to any specific single amino acid change. Known functional domains and key residues are reasonably well conserved in AtHKL1 (At1g50460) and AtHKL2 (At3g20040), and both proteins can probably bind glc (Karve et al., 2008). However, sequence divergence in AtHKL3 (At4g37840) is so extensive that the protein might not bind either glc or ATP. Interestingly, all three Arabidopsis HKL proteins have a mitochondrial targeting peptide which is very similar to that of AtHXK1. Experimental evidence for their mitochondrial association has been shown by using a proteomics approach (Heazlewood et al., 2004) and by examining the cellular expression of C-terminal GFP fusion proteins (Karve et al., 2008).
Non-catalytic HXKs have been reported in fungi and possibly occur commonly among higher plants (A Virnig and Bd Moore, unpublished data). The fungal HKL proteins have divergent roles including one as a meiosis-specific transcription factor in Saccharomyces cerevisiae (Daniel, 2005) and others as regulators of a carbon starvation response in Aspergillus nidulans (Bernardo et al., 2007). Despite the reports of the presence of HKL proteins in evolutionarily diverse species, their lack of catalytic activity has made it challenging to define their functions. In this study, a reverse genetics approach was used to determine whether AtHKL1 might have a role in plant growth, perhaps as an effector of glc signalling. Analyses of phenotypes from gain-of-function Arabidopsis plants and from an identified mutant line with a T-DNA insertion in HKL1, show that HKL1 is a negative regulator of plant growth and that it affects seedling growth responses to glc and auxin. However, HKL1 does not affect glc signalling, as shown in protoplast transient expression assays and by seedling candidate gene expression assays. These data indicate that AtHKL1 has an important role in plant growth and development, perhaps by mediating cross-talk between glc and hormone response pathways.
Materials and methods
Plant material and growth conditions
Seeds of Arabidopsis thaliana ecotype Columbia (Col-0), ecotype Landsberg erecta (Ler), and a Col line with a T-DNA insertion within the HKL1 locus (At1g50460; line WISCDSLOX383A5; hereafter designated hkl1-1) were obtained from the Arabidopsis Biological Resource Center (Ohio State University). Seeds of maize (Zea mays L.) were purchased (Seed Genetics, Lafayette, IN). Lines for gin2-1, tir1, and transgenic lines expressing HXK1-HA or HXK1-FLAG were as previously described (Moore et al., 2003). A homozygous line containing the T-DNA insertion in the HKL1 gene was identified by PCR genotyping using the following primers: p745 (5′-AACGTCCGCAATGTGTTATTAAGTTG-3′) and HKL1A5RP (5′-CCGTGTTATCTGAGCCTTACG-3′) for the T-DNA insertion allele; and, HKL1A5LP (5′-TGCAAACAAATTTAACGGCTC-3′) and HKL1A5RP for the WT allele. The insertion position in the hkl1-1 mutant was mapped by sequencing the PCR product obtained by the primers L1WLP (5′-TGCAAACAAATTTAACGGCTC-3′) and L1WRP (5′-CCGTGTTATCTGAGCCTTACG-3′), using hkl1-1 genomic DNA as template.
Arabidopsis seeds were surface-sterilized and stratified for 2 d at 4 °C as in Jang et al. (1997). Plants grown in soil were in a growth chamber (125 μmol m−2 s−1, 22 /20 °C day/night temperature) at either a 12 h photoperiod (normal), an 8 h photoperiod (short day, SD), or a 16 h photoperiod (long day, LD). Plants were also grown for some assays on 1× MS agar plates (modified basal medium with Gamborg vitamins; PhytoTechnology Laboratories, Shawnee Mission, KS) at pH 5.7, normally with 0.5% sucrose, and under constant light (30 μmol m−2 s−1). For glc repression assays, seedlings were grown on 1× MS plates with a substituted carbon source as 3–7% glc or 3–7% mannitol, for 7 d under constant light. Hypocotyl elongation assays were done at reduced light and nutrients as described before (Moore et al., 2003). For the assay of auxin-induced lateral root formation, Arabidopsis seeds were grown on 1× MS plates with 0.5% sucrose plus 5 μM 1-naphthylphthalamic acid (NPA) for 5 d and then were transferred to sucrose plates with or without 0.1 μM naphthalene acetic acid (NAA) for 5 d (Chen et al., 2003).
To perform glc signalling assays by candidate gene expression, 15–20 seedlings were grown in 125 ml flasks containing 50 ml of half-strength MS medium supplemented with 1% sucrose. Seedlings were grown on a rotary shaker at 250 rpm under constant light (70 μmol m−2 s−1) at 22 °C for 7 d. Seedlings were then washed with sugar-free half-strength MS medium for 24 h in the dark while shaking, and subsequently transferred to the light in fresh sugar-free medium (control) or in medium plus 2% glc. Seedlings were treated under constant light with shaking for 8 h, and were then harvested by quickly blotting with filter paper before freezing in liquid N2.
Plasmid constructs
RBCS-LUC, PPDK-LUC, and UBQ10-GUS constructs have been described previously (Schaffner and Sheen, 1991; Balasubramanian et al., 2007). An available clone of HKL1 with a double haemagglutinin (HA) tag (Karve et al., 2008) was subcloned with a substituted C-terminal FLAG tag in the HBT vector (Moore et al., 2003). Each fusion gene was then transferred into the pCB302 binary vector (bar selection marker; Xiang et al., 1999), using BamHI and PstI cloning sites. For cloning the HKL1 promoter, a 3098 bp fragment upstream of the start codon was PCR amplified using the following primers: L1PGUSFP (5′-CCCAAGCCTGGGCAGCGAGCTGTCAAACTGGGGA-3′) and L1PGUSRP (5′-GCTCTAGATGCCCCAAAACAGAACCAAAAAGACA-3′). The promoter was cloned into the binary vector pSMAB704 (bar selection marker; Igasaki et al., 2002), using HindIII and SmaI cloning sites upstream of the β-glucuronidase (GUS) gene. The identities of all clones were verified by DNA sequencing.
Binary constructs were introduced into Agrobacterium tumefaciens GV3101 by electroporation. Arabidopsis plants of Col-0, Ler or gin2-1 were transformed using the floral dip method (Clough and Bent, 1998). Transformants were selected for herbicide resistance (200 μM glufosinate ammonium; Rely 200, Bayer Crop Science, Kansas City, MO). Seeds of transgenic lines segregating 3:1 for herbicide resistance in the T2 generation were selected for isolating homozygous lines. Seeds from two or more T3 lines homozygous for the single insert were used for experiments.
RT- PCR analysis
Total RNA was isolated from whole seedlings of different lines using the RNeasy plant kit (Qiagen, Germantown, MD). One μg of total RNA was converted to cDNA using the Protoscript II RT-PCR kit (New England BioLabs, Ipswich, MA). PCR primer sequences for HXK1, HKL1, and UBQ5 were described previously (Karve et al., 2008). The expression of a variety of candidate genes was assessed in preliminary experiments by semi-quantitative RT-PCR, based on published data from glc transcript profiling studies (Price et al., 2004). Selected glc regulated genes are a subset which responded most robustly under the current treatment conditions. The PCR primer sequences for the candidate genes were generated using the AtRTPrimer public database (Han and Kim, 2006): ASN1 (asparagine synthase1, At3g47340; 5′-TGATTCTCAGGCCAAGAGAGTTCGT-3′, 5′-CCCAACCAATGTAGAGCGAAGTGAC-3′, expected size=413 bp), T6P (trehalose 6-phosphate synthase8, At1g70290; 5′-AGCTCCATTGTTCAAGATCCAAGCA-3′, 5′-GCTCCCCGCGTTCTACCATTTCTC-3′, expected size=626 bp), and GLYK (glycerate kinase, At1g80380; 5′-TTGGTGCGAAGATCAGATTGCTTTG-3′, 5′-GGAGACAGCATCGCATTAGTTTGC-3′, expected size=544 bp). All the primers were designed to span one or more introns such that the amplicon size from cDNA would be different than that from genomic DNA. The template amounts were first titrated to balance the UBQ5 expression in different samples (using densitometry), and corresponding template amounts were used thereafter, while varying PCR cycle numbers.
Immunoblots and gluokinase activity assays
Total soluble proteins were extracted as described by Karve et al. (2008). The protein concentration in the leaf extracts was measured by Coomassie Blue (Bio-Rad, Hercules, CA). Equal amounts of proteins were electrophoresed by SDS-PAGE and transferred onto Immobilon-P membrane (Millipore, Bedford, MA). The membranes were probed with monoclonal anti-HA (Roche, Indianapolis, IN) or anti-FLAG M2 (Sigma-Aldrich, St Louis, MO) antibodies, then incubated with HRP conjugated secondary antibody, followed by chemiluminescence reagents (SuperSignal West Pico, Pierce Biotechnology, Rockford, IL) and detection by film (Blue X-ray, Phenix Research Products, Candler, NC). Glucokinase activity was measured directly from leaf extracts or from lysates of maize protoplasts transfected with the indicated plasmids (Karve et al., 2008).
Protoplast transient expression assays
Leaves of greening maize seedlings or Arabidopsis plants (Col-0 or hkl1-1) were used as a source of protoplasts for protein expression and signalling assays (Jang and Sheen, 1994; Hwang and Sheen, 2001). Protoplasts were transfected (Yoo et al., 2007) with promoter constructs for RBCS-LUC (4 μg) or PPDK-LUC (6 μg), and with UBQ10-GUS (2 μg) as an internal control (Balasubramanian et al., 2007). Protoplasts were co-transfected as indicated with effectors HXK1-HA (6 μg) and/or HKL1-HA (8 μg). Transfection efficiencies were routinely >60%, as determined using WRKY-GFP (Balasubramanian et al., 2007). An empty vector was included to maintain a balanced concentration of DNA during transfections. Following transfection, protoplasts were incubated in the dark for 90 min, then treated with 2 mM glc and incubated in the light for 6–8 h at 30 μmol m−2 s−1. Protoplasts were collected by low speed centrifugation. After resuspending in lysis buffer, GUS and LUC activities were measured as described previously (Balasubramanian et al., 2007). Promoter activities are expressed as relative LUC/GUS values, normalized to control samples, which had no added glc.
Histochemical GUS staining and fluorometric GUS assays
Histochemical staining of transgenic Arabidopsis plants expressing the pHKL1-GUS fusion construct was performed as described by Crone et al. (2001). The plant tissue was incubated in GUS staining buffer containing 25 mg ml−1 of X-Glc (Gold BioTechnology, St Louis, MO) for 2–4 h and destained with 95% ethanol for 6–8 h. For measuring total extractable GUS activity, seedlings were extracted in buffer containing 50 mM NaH2PO4 (pH 7.0), 10 mM EDTA, 0.1% Triton X-100, 0.1% sodium lauryl sarcosine, and 10 mM β-mercaptoethanol. The enzymatic reaction was carried out in 100 μl of extraction buffer plus 1 mM 4-methyl umbelliferyl glucuronide (MUG, Sigma-Aldrich) at 37 °C for the indicated times, before stopping with 300 μl of 0.2 M Na2CO3. Fluorescence was measured in a 96-well microtitre plate format using a GENios spectrophotometer (Phenix Research Products) at a 360 nm excitation wavelength and a 465 nm emission wavelength. Sample GUS activities were calculated from a standard curve made using 0.1–1 μM 4-methyl-umbelliferone (Sigma-Aldrich).
In one experiment, transgenic seeds expressing pHKL1-GUS were grown on 1× MS plates plus 0.5% sucrose for 7 d, then transferred to liquid MS medium for 4 h with 10 μM indoleacetic acid (IAA), 1 μM abscisic acid (ABA), 50 μM 1-aminocylcopropane-1-carboxylic acid (ACC), or 10 μM zeatin (all from Sigma-Aldrich). Both treated and control seedlings were analysed for GUS staining and extractable GUS activity as described above.
Light microscopy
Light microscopy was used to view and capture images for routine seedling pictures, as well as for the GUS-stained seedlings or tissues, using a Nikon SMZ1500 stereo microscope (Nikon Instruments Inc., Melville, NY) with a MicroPublisher CCD cooled colour camera and Image Pro Plus v5.0 software (Media Cybernetics, Bethesda, MD). For measuring the hypocotyl lengths, the stereomicroscope was calibrated throughout the magnification range, using a stage micrometer.
Results
Molecular characterization of HKL1 knockout and increased expression lines
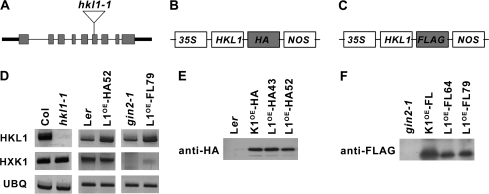
To understand the biological role of AtHKL1, a functional genomics approach was used by examining phenotypes of mutant and transgenic lines with altered HKL1 protein expression level. Seeds of a T-DNA insertion line for AtHKL1, generated by the University of Wisconsin knockout facility, were obtained through ABRC. Homozygous knockout plants with a possible single insert were identified by PCR screening. The T-DNA insertion was shown using real-time PCR and the 2–ΔΔCt method of relative quantification (Ingham et al., 2001) to be present as a single copy (see Supplementary Fig. S1 at JXB online), as shown by the dilution series values close to 1. The insertion site was mapped to exon VI of HKL1 (Fig. 1A). Using semi-quantitative RT-PCR, the mutant line was found to have no detectable HKL1 transcript (Fig. 1D). This line was designated hkl1-1.
Fig. 1.
Molecular characterization of Arabidopsis HKL1 mutant and transgenic lines. (A) Schematic diagram showing the gene structure of HKL1 (At1g50460). Exons are indicated by grey rectangles, introns are indicated by the thinner lines. The location of the T-DNA insertion in hkl1-1 is shown with the open triangle. (B, C) Design of plasmid constructs used to transform Arabidopsis lines. HKL1-HA was used to transform Ler and HKL1-FLAG was used to transform gin2-1. Boxes are not drawn to scale. 35S, CaMV promoter; NOS, nopaline synthetase terminator; HA, 2 copies of the 10 amino acid haemagglutinin tag; FLAG, 1 copy of the 8 amino acid FLAG tag. (D) Transcript expression of HKL1 and HXK1 by semi-quantitative RT-PCR: Col and hkl1-1; Ler and HKL1-HA line 52; and gin2-1 and HKL1-FLAG line 79. AtUBQ5 mRNA was used as a control for the amount of template. PCR cycle numbers for HKL1, HXK1, and UBQ were 33, 30, and 30, respectively. L1OE, HKL1 overexpression. (E) Immunoblot analysis using anti-HA antibody and 1 μg protein from leaf extracts of Ler, HXK1-HA transgenic (K1OE-HA), and two HKL1-HA lines. (F) Immunoblot analysis using anti-FLAG antibody and 1 μg protein from leaf extracts of gin2-1, HXK1-FLAG transgenic (K1OE-FL), and two HKL1-FLAG lines.
Transgenic Arabidopsis plants that constitutively express HKL1 in different genetic backgrounds were made: HKL1-HA expressed in Ler or HKL1-FLAG expressed in gin2-1 (Fig. 1B, C). This was done in order to distinguish possible HKL1 dependent phenotypes in relation to HXK1 expression. Three independent homozygous lines were obtained for the HKL1-HA transformants and seven lines for the HKL1-FLAG transformants. Representative transformed lines had substantially increased HKL1 transcripts, relative to each respective parental line (Fig. 1D). Notably, the HXK1 mRNA abundance was not altered in hkl1-1 or in transformed Ler lines which expressed HKL1-HA. The transformed lines with HKL1-FLAG did not have HXK1 transcripts, consistent with their parental background being gin2-1.
Western blot analysis of leaf extracts was carried out using antibodies to the introduced epitope tags (Fig. 1E, F). All of the transgenic lines expressed the corresponding tagged protein, while the parental lines did not. Positive controls included transgenic lines that expressed either HA or FLAG-tagged forms of HXK1 protein. From these assays, the two indicated lines expressing each construct were selected for further phenotypic analyses, with data presented for HKL1-HA line 52 and for HKL1-FLAG line 79.
Growth phenotypes of HKL1 knockout and increased expression lines
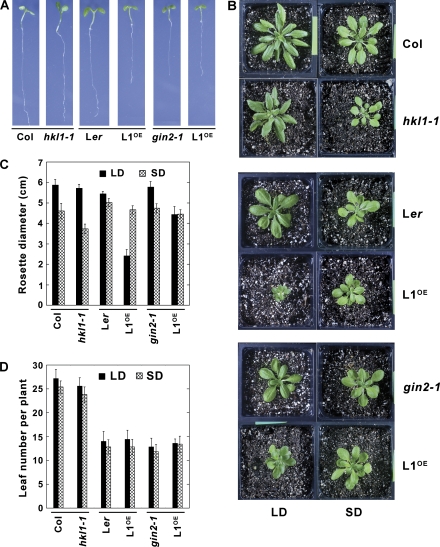
To test whether the HKL1 protein has a discernible function in plant growth, the different experimental lines were grown under different conditions. When grown on agar plates with 0.5% sucrose, the HKL1-HA seedlings were distinctly smaller than were the parental Ler seedlings, as were the gin2-1 seedlings (Fig. 2A). However, expression of HKL1 in the gin2-1 background had no apparent affect on seedling growth. Growth of hkl1-1 seedlings on sucrose plates resembled growth of the parental Col-0 seedlings. These results indicated that HKL1 might be a negative regulator of plant growth when overexpressed in the Ler background.
Fig. 2.
Growth phenotypes of HKL1 mutant and transgenic lines. Dark bars indicate corresponding parental controls and modified lines. (A) Seven-day-old seedlings on 1× MS plates+0.5% sucrose. (B) Plants grown 30 d in a growth chamber under 8 h (short day, SD) or 16 h (long day, LD) photoperiods. (C) Average rosette diameter (cm) after 30 d ±SD, n=10. The difference in average diameters of Col and hkl1-1 plants under SD conditions is statistically significant by a 2-tailed T test at P >0.95. (D) Average leaf number per plant at the time of bolting ±SD, n=10.
Transgenic and mutant lines also were grown in soil under different light conditions. When grown under SD conditions, both HKL1 overexpression lines had normal growth, when compared with control plants (Fig. 2). However, the hkl1-1 plants under SD conditions were somewhat smaller than control plants, with a rosette diameter reduced by about 20% (Fig. 2B, C). Growth of the hkl1-1 plants under LD conditions was similar to Col-0. By contrast, growth of the overexpression lines in either Ler or gin2-1 backgrounds was considerably reduced under LD conditions. For example, the rosette diameter for HKL1-HA plants was 50% smaller than for Ler plants. This resulted in mature plants of the transgenic line being about 4-fold smaller. Also, the diameter of HKL1-FLAG plants was reduced by 25% compared to gin2-1 plants, resulting in almost a 2-fold decrease in plant size. The reduced rosette sizes were not associated with a change in leaf numbers at the time of flowering for the different transformants relative to control lines (Fig. 2D), or with a change in the time to flowering (data not shown). These observations indicate that the intrinsic developmental programme was not changed due to increased HKL1 protein expression. However, seed yield from the small plants was greatly reduced, but not seed viability. Notably then, HKL1 overexpression in the Ler background resulted in an even smaller plant than when overexpressed in the absence of HXK1 protein in the gin2-1 background (see Supplementary Fig. S2 at JXB online).
Seedling hypocotyl growth among different HKL1 expression lines
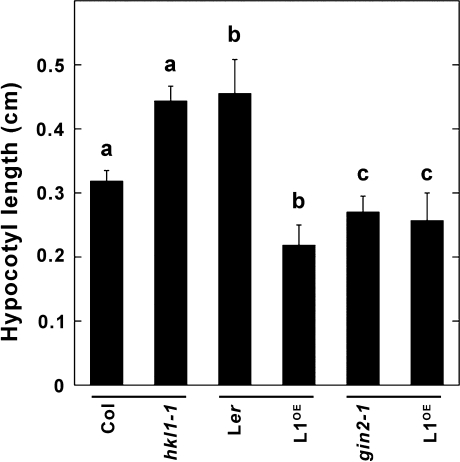
Since Arabidopsis hypocotyl growth is sensitive to many endogenous factors that regulate plant cell elongation (Salchert et al., 1998), the hypocotyl growth of 7-d-old seedlings grown vertically on plates under constant low light conditions was measured (Fig. 3). The average hypocotyl length of HKL1-HA seedlings was about 50% less than of the parental Ler seedlings. On the other hand, hkl1-1 seedlings had a 40% increase in hypocotyl length relative to Col-0 seedlings. The average hypocotyl length of gin2-1 seedlings was about 45% less than that for Ler seedlings. However, HKL1-FLAG seedlings did not show any significant change in hypocotyl growth when compared with the parental genotype, gin2-1. By this assay, HKL1 again was a negative regulator of seedling growth when expressed in a WT background, which contains HXK1.
Fig. 3.
Average seedling hypocotyl length of HKL1 mutant and transgenic lines. Seedlings were grown vertically for 7 d on 1/5× MS plates under constant light (15 μmol m−2 s−1) at 22 °C. Values are means ±SD, n=15. a, b, by 2-tailed T tests, values are statistically different at P >0.95; c, values are not different at P >0.95.
Auxin-induced lateral root formation among different Arabidopsis lines
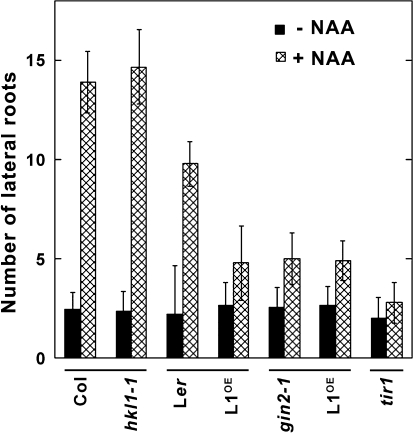
The reduced hypocotyl growth of gin2-1 seedlings was previously linked to its being relatively insensitive to auxin (Moore et al., 2003). Therefore, the different transgenic and mutant lines were also tested by an auxin assay for lateral root formation. In this assay, seedling growth in the presence of the auxin transport inhibitor NPA greatly reduces the number of lateral roots (Himanen et al., 2002). Lateral root formation can then be initiated after seedling transfer to plates with NAA. With these treatments, both Col-0 and Ler seedlings showed a robust induction of lateral root formation, increasing 5-fold and 4-fold, respectively, after transfer to plates with NAA (Fig. 4). Seedlings of hkl1-1 showed a similar increase in their number of lateral roots relative to Col-0 seedlings. However, auxin treatment induced relatively fewer lateral roots in gin2-1, HKL1-HA, and HKL1-FLAG seedlings, about a 2-fold increase. As a control for this assay, the same treatments of the auxin receptor mutant tir1 (Col background) did not appreciably induce any lateral roots, with the tir1 mutant having fewer roots even than gin2-1 or the two HKL1 overexpression lines. These data indicate that HXK1 has a significant role in the auxin induction of lateral roots and that HKL1 blocks this induction response to a level comparable with that observed in the absence of HXK1.
Fig. 4.
Auxin-induced lateral root formation in seedlings of HKL1 mutant and transgenic lines. The number of lateral roots were counted 5 d after seedling transfer from plates with 5 μM NPA to plates with or without 0.1 μM NAA. Values are average lateral root numbers ±SD, n=10.
Glucokinase activities and glc signalling assays using different expression lines
The growth phenotypes of the HKL1 transgenic and mutant lines that have been described could be due to an influence of HKL1 protein on HXK1 protein catalytic activity, on HXK1 signalling functions, and/or on the function of an unknown protein. To test for the possible influence of HKL1 protein on glucokinase activity, rate measurements were carried out using leaf extracts from the different lines. There was no significant difference for enzyme activities between the transgenic lines and their respective control lines (Fig. 5A). As reported previously, HXK enzyme activity in gin2-1 is about one-half of that in Ler (Moore et al., 2003) and HKL1-HA did not have any glc phosphorylation activity (Karve et al., 2008). The possible inhibition of HXK1 by HKL1 was also tested after transiently expressing HXK1-HA and HKL1-HA in maize protoplasts. However, HKL1 protein did not affect the measured glucokinase activity (Fig. 5B).
Fig. 5.
Glucokinase activity of HKL1 mutant and transgenic lines. (A) Clarified leaf extracts of greenhouse-grown plants were assayed directly for enzyme activity. Values are means ±SD, n=3. (B) Maize protoplast extracts were assayed for enzyme activity after expression of plasmids with HXK1-HA and/or HKL1-HA. Protein expression was routinely monitored by labelling with [35S]-methionine (data not shown; as in Karve et al., 2008). Values are means ±SD, n=3, expressed relative to control protoplasts with empty vector DNA only.
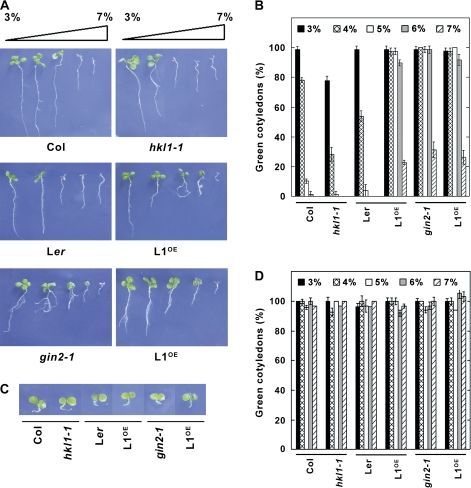
Since HKL1 lacks glucokinase activity, but has a largely conserved glc binding domain, it is possible that, instead, the protein affects glc signalling activities. A widely used screen to identify mutants in glc signalling is based on the ability of some mutants to develop normally on otherwise inhibitory concentrations of exogenous glc (Rolland et al., 2006). Therefore, seedling growth of the different lines was assessed in the presence of varying glc concentrations (Fig. 6A, B; see Supplementary Fig. S3 at JXB online). At relatively high glc levels, Col-0 and Ler seedlings underwent developmental arrest, with much reduced root and shoot growth, and did not accumulate chlorophyll. The hkl1-1 seedlings were hypersensitive to developmental arrest, showing substantial repression even on 4% glc. By contrast, the HKL1-HA seedlings were glc-insensitive relative to the Ler control line. When grown on 6% glc, >90% of the HKL1-HA seedlings have green cotyledons versus 0% of the Ler seedlings. The responses of HKL1-FLAG seedlings were comparable with those of gin2-1 seedlings. As an osmotic control, all lines were shown to have a similar phenotype on MS plates with 6% mannitol (Fig. 6C). Also, mannitol did not repress cotyledon greening in any of the lines (Fig. 6D). The observed glc-dependent phenotype suggested that HKL1 could be a negative regulator of glc signalling.
Fig. 6.
Phenotypes of HKL1 mutant and transgenic lines grown on agar plates with 3–7% glc. (A) Images are representative 7-d-old seedlings. (B) Percentage of seedlings in (A) at corresponding glc concentrations which had green cotyledons. Values are expressed relative to the total number of germinated seedlings (30–40), as means ±SD, n=3. (C) Images of representative 7-d-old seedlings grown on agar plates with 6% mannitol. (D) Percentage of seedlings in (C) at corresponding mannitol concentrations which had green cotyledons. Values are expressed relative to the total number of germinated seedlings, as means ±SD, n=3.
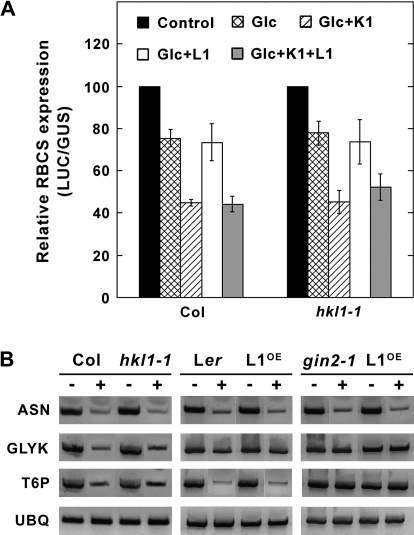
To test whether HKL1 might have a role in glc signalling, protoplast transient expression assays were carried out using pRBCS-LUC and pPPDK-LUC as established reporters of HXK1 signalling (Balasubramanian et al., 2007). Leaf protoplasts of Col-0 and hkl1-1 plants were used in independent assays. Relative RBCS-driven LUC activities expressed in protoplasts of either genotype was reduced by 25% with 2 mM glc (Fig. 7A). In both cases, co-transfection with HXK1 plus treatment with 2 mM glc reduced the reporter activity by about 55%. By contrast, transfected HKL1 did not affect the relative expressed RBCS-driven LUC activity with glc alone or with HXK1 plus glc, using protoplasts from either wild-type or mutant leaves. Similar results were obtained using pPPDK-LUC (data not shown). Notably, in all cases the expression of pUBQ10-GUS was not affected by co-transfection of HXK1, HKL1 and/or by addition of 2 mM glc.
Fig. 7.
Glc signalling assays. (A) Transient expression assays using leaf protoplasts from WT Col or hkl1-1. Protoplasts were co-transfected with pRBCS-LUC and an internal control, pUBQ10-GUS, plus or minus effectors HXK1-HA and/or HKL1-HA. Protoplast treatments include without glc or effectors (Control), with 2 mM glc (Glc), with 2 mM glc+HXK1-HA (Glc+K1), with 2 mM glc+HKL1-HA (Glc+L1), and with 2 mM glc+HXK1-HA+HKL1-HA (Glc+K1+L1). Values are means ±SD of the relative LUC units to GUS activities for replicated assays normalized to the control. GUS activity was not affected by the presence of glc or either effector. (B) Expression of glc regulated genes in HKL1 transgenic lines and mutants. Semi-quantitative RT-PCR was used to determine the transcript levels of asparagine synthase (ASN), glycerate kinase (GLYK), trehalose 6-phosphate synthase (T6P), and ubiquitin (UBQ). Seedlings grown in liquid medium were challenged without (–) or with (+) 2% glc for 8 h (see Materials and methods for further details). The number of PCR cycles was varied in each case, but for the presented data are as follows: ASN, 32 cycles, GLYK, 32 cycles, T6P, 33 cycles, UBQ, 31 cycles.
To complement the transient expression assays, an alternate assay of glc signalling was carried out using seedlings grown in liquid culture and treated with or without 2% glc for 8 h. The selected GLYK and T6P genes are thought to be regulated by HXK1-dependent glc signalling, and ASN by a glycolysis-dependent glc signalling pathway (Price et al., 2004). Supporting this interpretation, transcripts of ASN, GLYK, and T6P were all repressed by glc treatment of Col-0 and Ler seedlings, while GLYK and T6P mRNA abundance were not affected by treatment of gin2-1 seedlings (Fig. 7B). The response of these transcripts was not differentially affected in any of the tested mutant or transgenic lines, relative to the corresponding control lines. These data indicate that HKL1 probably does not affect the commonly recognized transcriptional targets of glc signalling, whether by a HXK1-dependent or a glycolysis-dependent pathway.
HKL1 promoter expression and activity assays
To improve our understanding of possible HKL1 functions, transgenic Arabidopsis plants were made that express an HKL1 promoter–GUS fusion construct (pHKL1-GUS). At the early stages of seedling development, GUS staining was detected mainly in the root, particularly towards the root tip (Fig. 8A). With increased seedling growth, GUS staining was progressively localized to the vascular tissues of cotyledons (Fig. 8B), was relatively strong in the root and shoot meristems, but not in leaf primordia (Fig. 8C). In adult plants, GUS expression was highest in the root and leaf vascular tissue, and in the emerging lateral roots (Fig. 8D, E, F). In stem cross-sections, GUS staining was observed in phloem tissue. In flowers, GUS staining was observed in anther filaments, but not in the pistils (Fig. 8G, H). Staining was also observed broadly in developing siliques, becoming localized apparently to the funiculi of more mature seeds (Fig. 8I).
Fig. 8.
Organ and tissue expression of pHKL1-GUS. (A) Seedlings grown for 3 d on MS plates. (B) Seedlings grown for 7 d on MS plates. (C) Shoot of a 5-d-old seedling, with the arrowhead pointing to specific stain in the meristem. (D) Leaf from a 21-d-old plant. (E) Stem cross-section, with the arrowhead pointing to staining of phloem. (F) Root of a 10-d-old seedling, with the arrowhead pointing to enhanced staining at the site of lateral root initiation. (G) Opened flower. (H) Anthers and filaments. (I) Developing silique, with insert showing a mature silique and an arrowhead pointing to the funiculus of a developing seed.
Since HKL1 overexpression reduced the sensitivity of seedlings to auxin-dependent lateral root formation (Fig. 4) and reduced the sensitivity of seedlings to glc repression of development (Fig. 6), the influence of short-term treatment of seedlings with different hormones was examined on the expression of pHKL1-GUS activity (Fig. 9). The effect of the hormone treatments was determined visually and also quantitatively after extraction and assay of GUS activity. For the latter, initial assays in the absence of added stimuli established that a 2 h reaction time with seedling extracts was within the linear range of activity (Fig. 9B). IAA treatment (or GA3 treatment, data not shown) did not induce pHKL1-GUS expression or enzyme activity (Fig. 9A, C). However, ABA treatment greatly reduced seedling GUS staining and reduced the extractable GUS activity by 50%. On the other hand, zeatin or ACC treatments induced GUS expression throughout the seedling and not just in the vascular tissue. Correspondingly, the extracted GUS activities following these treatments increased up to 2-fold relative to the control treatment. The results of the GUS assays indicate that AtHKL1 might be regulated by multiple plant hormones and, thereby, could have a regulatory role in plant growth and development.
Fig. 9.
Effect of different plant hormones on pHKL1-GUS expression. Seedlings of pHKL1-GUS lines were grown for 7 d on MS plates, then transferred to liquid MS medium for 4 h with different plant hormones: control (no additions), 10 μM IAA, 1 μM ABA, 50 μM ACC, and 10 μM zeatin. (A) Seedlings stained for GUS activity. (B) Reaction time-course for GUS activity assayed from control seedlings. (C) GUS activity of seedlings after a 2 h reaction. Values are means ±SD, n=3.
Discussion
Non-catalytic HXKs have been identified in fungi including S. cerevisiae and A. nidulans (Daniel, 2005; Bernardo et al., 2007) and also in Arabidopsis (Karve et al., 2008). Whether non-catalytic homologues of known enzymes are commonly present in other protein families is not known. The Arabidopsis glutathione transferase family does include both non-catalytic as well as catalytic forms, although their relative distribution between the groups apparently has not been strictly determined (Dixon et al., 2003). Recently, β-amylase4 (BAM4) of Arabidopsis was shown to lack apparent catalytic activity, yet somehow to facilitate starch breakdown (Fulton et al., 2008). BAM4 is one of perhaps four chloroplastic isoforms within Arabidopsis. Also, the plant shikimate kinase gene family includes two non-catalytic homologues which have been present in all major plant lineages for over 400 million years (Fucile et al., 2008). In Arabidopsis, these express novel functions, one of which is required for chloroplast biogenesis (Fucile et al., 2008). Non-catalytic enzyme homologues might occur somewhat more often among plant gene families than what is currently appreciated, since sequence divergence levels within families are often >25%. That is, in order to transfer all four digits of an EC number at an error rate below 10%, the estimated level of sequence identity needs to be >75% (Rost et al., 2003). It is suggested that when non-catalytic homologues of known enzymes do occur, they are likely to have important regulatory functions. For example, several catalytically inactive homologues of phosphoinositide 3-phosphatases have been linked to specific human diseases (Robinson and Dixon, 2006).
As one general approach to understand protein function, the tissue expression pattern and regulation of gene expression can provide an important physiological context. The AtHKL1 transcript was previously shown to be expressed in the principal plant organs (Karve et al. 2008). These observations have been extended in this study by demonstrating that pHKL1-GUS activity occurs predominantly in the vascular tissues of different sink organs such as roots, stems, and anthers (Fig. 8). In stem cross-sections, the vascular staining was associated with phloem tissue. While we are not aware of any HXK family members having been reported in surveys of the phloem proteome, nonetheless many phytohormones and a number of regulatory proteins have been detected in phloem sap (Giavalisco et al., 2006). The HKL1 promoter activity was also found to be influenced by several phytohormones, including being repressed by ABA and induced by both ACC and cytokinin. Hormone induction of the HKL1 promoter occurred in both vascular and non-vascular tissues (Fig. 8). Our analysis of the HKL1 promoter sequence for known regulatory elements (Higo et al., 1999; Molina and Grotewold, 2005; Obayashi et al., 2007) indicates that the promoter does have motifs proposed to be regulated by several hormones (data not shown).
Phenotypes of the AtHKL1 overexpression lines provide evidence that the HKL1 protein is a negative regulator of plant growth. HKL1 overexpression in Ler (HKL1-HA) resulted in reduced seedling growth on sucrose plates (Fig. 2), reduced hypocotyl elongation under low light conditions (Fig. 3), severely reduced rosette size under LD conditions (Fig. 2), and a decreased sensitivity to auxin-induced lateral root formation (Fig. 4). In a recent initial report, some rice HXK family members also were considered to be possible negative regulators of seedling growth (Yu and Chiang, 2008). The status of the glc binding domain in possible regulatory HXKs needs to be evaluated experimentally. It has previously been shown that AtHXK1-G173A has a 90% decrease in glc phosphorylation activity (Karve et al., 2008). Since AtHKL1 has the same recognized glc binding domain as does this mutated protein, we speculate that glc binding affinity is reduced in AtHKL1, but not eliminated. Thus, for a negative regulator, decreased glc binding affinity could be a feedback mechanism to limit plant growth in the presence of excessive carbohydrate availability.
The HKL1 protein might function as a negative regulator of cell expansion, based on reduced hypocotyl growth of HKL1-HA seedlings and on increased hypocotyl growth of the hkl1-1 seedlings (Fig. 3). Seedling hypocotyl growth by cell elongation integrates diverse signals including light, temperature, nutrients, and most plant hormones (Collett et al., 2000; De Grauwe et al., 2005). In gin2-1, reduced hypocotyl growth has been attributed to the possible insensitivity of seedlings to auxin signalling (Moore et al., 2003). However, ethylene also can repress hypocotyl elongation in seedlings grown under conditions similar to those in our experiment (Smalle et al., 1997). Thus, it is possible that HKL1 expression promotes ethylene sensitivity instead of attenuating auxin sensitivity. Consistent with this possibility, while lateral root formation does require auxin synthesis, transport, and/or signalling (Casimiro et al., 2003), enhanced ethylene signalling has more recently been shown to repress lateral root formation by modulating auxin transport (Negi et al., 2008). Thus, the observed HKL1 repression phenotype for auxin-induced root formation (Fig. 4) might instead be associated with an altered ethylene response. Further experiments are needed to clarify the mechanisms involved.
The mode of action of AtHKL1 is not known, but does merit further consideration. On the one hand, since both HXK1 and HKL1 are targeted to mitochondria (Heazlewood et al., 2004; Karve et al., 2008), the two proteins have the potential to interact such that HKL1 could act as a dominant negative effector. In this case, the overexpression of HKL1 in the gin2-1 background might not result in a novel phenotype relative to its overexpression in the presence of HXK1. Assay results for hypocotyl growth (Fig. 3), for auxin induction of lateral root growth (Fig. 4), and for glc tolerance (Fig. 6) are consistent with this possibility. Furthermore, the contrasting phenotypes observed by these assays with the hkl1-1 mutant also support this interpretation. On the other hand, the overexpression of HKL1 in WT did result in a much more diminutive plant under LD conditions than was observed in gin2-1 (Fig. 2). This implies that HKL1 could have a more complicated mode of action by also independently affecting one or more targets possibly involved in mediating phytohormone responses.
In summary, the present results indicate that the non-catalytic AtHKL1 protein can negatively influence plant growth, possibly by somehow influencing cross-talk between glc and other plant hormone response pathways. Elucidating the functions of non-catalytic proteins will be an ongoing challenge for contemporary biologists.
Supplementary data
Supplementary data are available at JXB online.
Supplementary Fig. 1. The number of T-DNA insertions in hkl1-1 as determined by real time PCR.
Supplementary Fig. S2. Growth under 16 h (long day) photoperiod conditions of transgenic Arabidopsis expressing HKL1 protein in different genetic backgrounds.
Supplementary Fig. S3. Growth response on agar plates with varying glc concentrations for the transgenic HKL1-Flag line 43.
Supplementary Fig. S4. Transcript abundance by semi-quantitative RT-PCR of HXK1 and HKL1 from Ler seedlings grown on plates with 0.5% sucrose (–) or with 6% glucose (+).
Acknowledgments
We very much appreciate technical help from Ms Xiaoxia Xia. We thank Dr Zhou Li and Dr Jen Sheen for sharing their initial observations on the relative auxin insensitivity for lateral root induction in gin2-1. We also thank Dr J-C Jang for useful discussions on seedling glc signalling assays. This paper is technical contribution no. 5642 of the Clemson University Experiment Station. This material is based upon work supported by the CSREES/USDA, under project number SC1700190. Any opinions, findings, conclusions or recommendations expressed in this publication are those of the author(s) and do not necessarily reflect the view of the USDA.
Glossary
Abbreviations
- GFP
green fluorescent protein
- GUS
glucuronidase
- HA
haemagglutinin
- HKL
hexokinase-like
- HXK
hexokinase
- LD
long day
- LUC
luciferase
- NAA
naphthalene acetic acid
- NPA
1-naphthylphthalamic acid
- PPDK
pyruvate orthophosphate dikinase
- RBCS
ribulose-1,5-bisphosphate carboxylase small subunit
- RT-PCR
reverse transcriptase polymerase chain reaction
- SD
short day
- UBQ
ubiquitin
References
- Balasubramanian R, Karve A, Kandasamy M, Meagher RB, Moore B. A role for F-actin in hexokinase-mediated glucose signaling. Plant Physiology. 2007;145:1423–1434. doi: 10.1104/pp.107.108704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balasubramanian R, Karve A, Moore B. Actin-based framework for cellular glucose signalling by Arabidopsis hexokinase1. Plant Signaling and Behavior. 2008;3:322–324. doi: 10.4161/psb.3.5.5319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernardo SM, Gray KA, Todd RB, Cheetham BF, Katz ME. Characterization of regulatory non-catalytic hexokinases in Aspergillus nidulans. Molecular Genetics and Genomics. 2007;277:519–532. doi: 10.1007/s00438-006-0203-z. [DOI] [PubMed] [Google Scholar]
- Casimiro I, Beekman T, Graham N, Bhalerao R, Zhang H, Casero P, Sandberg G, Bennett MJ. Dissecting Arabidopsis lateral root development. Trends in Plant Science. 2003;8:165–171. doi: 10.1016/S1360-1385(03)00051-7. [DOI] [PubMed] [Google Scholar]
- Chen J-G, Willard FS, Huang J, Liang J, Chasse SA, Jones AM, Siderovski DP. A seven-transmembrane RGS protein that modulates plant cell proliferation. Science. 2003;301:1728–1731. doi: 10.1126/science.1087790. [DOI] [PubMed] [Google Scholar]
- Chen Y, Ji F, Xie H, Liang J, Zhang J. The regulator of G-protein signalling proteins involved in sugar and abscisic acid signalling in Arabidopsis seed germination. Plant Physiology. 2006;140:302–310. doi: 10.1104/pp.105.069872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho JI, Ryoo N, Eom JS, et al. Role of the rice hexokinases OsHXK5 and OsHXK6 as glucose sensors. Plant Physiology. 2009;149:745–759. doi: 10.1104/pp.108.131227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho YH, Yoo SD, Sheen J. Regulatory functions of nuclear hexokinase1 complex in glucose signalling. Cell. 2006;127:579–589. doi: 10.1016/j.cell.2006.09.028. [DOI] [PubMed] [Google Scholar]
- Claeyssen E, Rivoal J. Isozymes of plant hexokinase: occurrence, properties and functions. Phytochemistry. 2007;68:709–731. doi: 10.1016/j.phytochem.2006.12.001. [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. The Plant Journal. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- Collett CE, Harberd NP, Leyser O. Hormonal interactions in the control of Arabidopsis hypocotyl elongation. Plant Physiology. 2000;124:553–562. doi: 10.1104/pp.124.2.553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crone D, Rueda J, Martin KL, Hamilton DA, Mascarenhas JP. The differential expression of a heat shock promoter in floral and reproductive tissues. Plant, Cell and Environment. 2001;24:869–874. [Google Scholar]
- Daniel J. Sir-dependent downregulation of various aging processes. Molecular Genetics and Genomics. 2005;274:539–547. doi: 10.1007/s00438-005-0040-5. [DOI] [PubMed] [Google Scholar]
- De Grauwe L, Vandenbussche F, Tietz O, Palme K, Van Der Straeten D. Auxin, ethylene and brassinosteroids: tripartite control of growth in the Arabidopsis hypocotyl. Plant and Cell Physiology. 2005;46:827–836. doi: 10.1093/pcp/pci111. [DOI] [PubMed] [Google Scholar]
- Dixon DP, McEwen AG, Lapthorn AJ, Edwards R. Forced evolution of a herbicide detoxifying glutathione transferase. Journal of Biological Chemistry. 2003;278:23930–23935. doi: 10.1074/jbc.M303620200. [DOI] [PubMed] [Google Scholar]
- Fucile G, Falconer S, Christendat D. Evolutionary diversification of plant shikimate kinase gene duplicates. PLoS Genetics. 2008;4:e1000292. doi: 10.1371/journal.pgen.1000292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fulton DC, Stettler M, Mettler T, et al. β-AMYLASE4, a noncatalytic protein required for starch breakdown, acts upstream of three active β-amylases in Arabidopsis chloroplasts. The Plant Cell. 2008;20:1040–1058. doi: 10.1105/tpc.107.056507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gancedo JM. Early steps of glucose signalling in yeast. FEMS Microbiology Review. 2008;32:673–704. doi: 10.1111/j.1574-6976.2008.00117.x. [DOI] [PubMed] [Google Scholar]
- Giavalisco P, Kapitza K, Kolasa A, Buhtz A, Kehr J. Towards the proteome of Brassica napus phloem sap. Proteomics. 2006;6:896–909. doi: 10.1002/pmic.200500155. [DOI] [PubMed] [Google Scholar]
- Gibson SI. Sugar and phytohormone response pathways: navigating a signalling network. Journal of Experimental Botany. 2004;55:253–264. doi: 10.1093/jxb/erh048. [DOI] [PubMed] [Google Scholar]
- Han S, Kim D. AtRTPrimer: database for Arabidopsis genome-wide homogeneous and specific RT-PCR primer-pairs. BMC Bioinformatics. 2006;7:179. doi: 10.1186/1471-2105-7-179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heazlewood JL, Tonti-Filippini JS, Gout AM, Day DA, Whelan J, Millar AH. Experimental analysis of the Arabidopsis mitochondrial proteome highlights signalling and regulatory components, provides assessment of targeting prediction programs, and indicates plant-specific mitochondrial proteins. The Plant Cell. 2004;16:241–256. doi: 10.1105/tpc.016055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higo KY, Ugawa M, Iwamoto Korenaga T. Plant cis-acting regulatory DNA elements (PLACE) database. Nucleic Acids Research. 1999;27:297–300. doi: 10.1093/nar/27.1.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Himanen K, Boucheron E, Vanneste S, de Almeida Engler J, Inzé D, Beeckman T. Auxin-mediated cell cycle activation during early lateral root initiation. The Plant Cell. 2002;14:2339–2351. doi: 10.1105/tpc.004960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang I, Sheen J. Two-component circuitry in Arabidopsis cytokinin signal transduction. Nature. 2001;413:383–389. doi: 10.1038/35096500. [DOI] [PubMed] [Google Scholar]
- Igasaki T, Ishida Y, Mohri T, Ichikawa H, Shnohara K. Transformation of Populus alba and direct selection of transformants with the herbicide bialaphos. Bulletin of Forestry and Forest Products Research Institute. 2002;1:235–240. [Google Scholar]
- Ingham D, Beer S, Money S, Hansen G. Quantitative real-time PCR assay for determining copy number in transformed plants. BioTechniques. 2001;31:132–140. doi: 10.2144/01311rr04. [DOI] [PubMed] [Google Scholar]
- Jang JC, Leon P, Zhou L, Sheen J. Hexokinase as a sugar sensor in higher plants. The Plant Cell. 1997;9:5–19. doi: 10.1105/tpc.9.1.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang JC, Sheen J. Sugar sensing in higher plants. The Plant Cell. 1994;6:1665–1679. doi: 10.1105/tpc.6.11.1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston CA, Taylor JP, Gao Y, Kimple AJ, Grigston JC, Chen JG, Siderovski DP, Jones AM, Willard FS. GTPase acceleration as the rate-limiting step in Arabidopsis G protein-coupled sugar signalling. Proceedings of the National Academy of Sciences, USA. 2007;104:17317–17322. doi: 10.1073/pnas.0704751104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karve A, Rauh BL, Xia X, Kandasamy M, Meagher RB, Sheen J, Moore BD. Expression and evolutionary features of the hexokinase gene family in Arabidopsis. Planta. 2008;228:411–425. doi: 10.1007/s00425-008-0746-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leon P, Sheen J. Sugar and hormone connections. Trends in Plant Science. 2003;8:110–116. doi: 10.1016/S1360-1385(03)00011-6. [DOI] [PubMed] [Google Scholar]
- Molina C, Grotewold E. Genome wide analysis of Arabidopsis core promoters. BMC Genomics. 2005;6:25. doi: 10.1186/1471-2164-6-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore B, Zhou L, Rolland F, Hall Q, Cheng WH, Liu YX, Hwang I, Jones T, Sheen J. Role of the Arabidopsis glucose sensor HXK1 in nutrient, light, and hormonal signalling. Science. 2003;300:332–336. doi: 10.1126/science.1080585. [DOI] [PubMed] [Google Scholar]
- Negi S, Ivanchenko MG, Muday GK. Ethylene regulates lateral root formation and auxin transport in Arabidopsis thaliana. The Plant Journal. 2008;55:175–187. doi: 10.1111/j.1365-313X.2008.03495.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obayashi T, Kinoshita K, Nakai K, Shibaoka M, Hayashi S, Saeki M, Shibata D, Saito K, Ohta H. ATTED-II: a database of co-expressed genes and cis elements for identifying co-regulated gene groups in Arabidopsis. Nucleic Acids Research. 2007;35:D863–D869. doi: 10.1093/nar/gkl783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osuna D, Usadel B, Morcuende R, et al. Temporal responses of transcripts, enzyme activities and metabolites after adding sucrose to carbon-deprived Arabidopsis seedlings. The Plant Journal. 2007;49:463–491. doi: 10.1111/j.1365-313X.2006.02979.x. [DOI] [PubMed] [Google Scholar]
- Perfus-Barbeoch L, Jones AM, Assmann SM. Plant heterotrimeric G protein function: insights from Arabidopsis and rice mutants. Current Opinion in Plant Biology. 2004;7:719–731. doi: 10.1016/j.pbi.2004.09.013. [DOI] [PubMed] [Google Scholar]
- Price J, Laxmi A, St Martin SK, Jang JC. Global transcription profiling reveals multiple sugar signal transduction mechanisms in Arabidopsis. The Plant Cell. 2004;16:2128–2150. doi: 10.1105/tpc.104.022616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson FL, Dixon JE. Myotubularin phosphatases: policing 3-phosphoinositides. Trends in Cell Biology. 2006;16:403–412. doi: 10.1016/j.tcb.2006.06.001. [DOI] [PubMed] [Google Scholar]
- Rognoni S, Teng S, Arru L, Smeekens S, Perata P. Sugar effects on early seedling development in Arabidopsis. Plant Growth Regulation. 2007;52:217–228. [Google Scholar]
- Rolland F, Baena-Gonzalez E, Sheen J. Sugar sensing and signalling in plants: conserved and novel mechanisms. Annual Review of Plant Biology. 2006;57:675–709. doi: 10.1146/annurev.arplant.57.032905.105441. [DOI] [PubMed] [Google Scholar]
- Rost B, Liu J, Nair R, Wrzeszczynski KO, Ofran Y. Automatic prediction of protein function. Cellular and Molecular Life Sciences. 2003;60:2637–2650. doi: 10.1007/s00018-003-3114-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salchert K, Bhalerao R, Koncz-Kálmán Z, Koncz C. Control of cell elongation and stress responses by steroid hormones and carbon catabolic repression in plants. Philosophical Transactions of the Royal Society of London, Biological Sciences. 1998;353:1517–1520. doi: 10.1098/rstb.1998.0307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaffner AR, Sheen J. Maize rbcS promoter activity depends on sequence elements not found in dicot rbcS promoters. The Plant Cell. 1991;3:997–1012. doi: 10.1105/tpc.3.9.997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smalle J, Haegman M, Kurepa J, Van Montagu M, Straeten DV. Ethylene can stimulate Arabidopsis hypocotyl elongation in the light. Proceedings of the National Academy of Sciences, USA. 1997;94:2756–2761. doi: 10.1073/pnas.94.6.2756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Towle HC. Glucose as a regulator of eukaryotic gene transcription. Trends in Endocrinology and Metabolism. 2005;16:489–494. doi: 10.1016/j.tem.2005.10.003. [DOI] [PubMed] [Google Scholar]
- Xiang C, Han P, Lutziger I, Wang K, Oliver D. A mini binary vector series for plant transformation. Plant Molecular Biology. 1999;40:711–717. doi: 10.1023/a:1006201910593. [DOI] [PubMed] [Google Scholar]
- Yoo SD, Cho YH, Sheen J. Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nature Protocols. 2007;2:1565–1572. doi: 10.1038/nprot.2007.199. [DOI] [PubMed] [Google Scholar]
- Yu SM, Chiang CM. Distinct hexokinases (HXKs) act as positive and negative regulators in sugar signalling pathways. BMC Plant Biology. 2008:60–61. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.