Abstract
The shoot apical meristem (SAM) is responsible for the development of all the above-ground parts of a plant. Our understanding of the SAM at the molecular level is incomplete. This study investigates the gene expression repertoire of SAMs in the garden pea (Pisum sativum). To this end, 10 346 EST sequences representing 7610 unique genes were generated from SAM cDNA libraries. These sequences, together with previously reported pea ESTs, were used to construct a 12K oligonucleotide array to identify genes with differential SAM expression, as compared to axillary meristems, root apical meristems, or non-meristematic tissues. A number of genes were identified, predominantly expressed in specific cell layers or domains of the SAM and thus are likely components of the gene networks involved in stem cell maintenance or the initiation of lateral organs. Further in situ hybridization analysis confirmed the spatial localization of some of these genes within the SAM. Our data also indicate the diversification of some gene expression patterns and hence functions in legume crop plants. A number of transcripts highly expressed in all three meristems have also been uncovered and these candidates may provide valuable insight into molecular networks that underpin the maintenance of meristematic functionality.
Keywords: Garden pea, meristem, Pisum sativum, transcript profiling
Introduction
The development of the shoot apical meristem (SAM) has attracted widespread attention during recent years as an excellent model for understanding stem cell maintenance during plant development. Meristems are considered to be a plant's equivalent of stem cell niches which are the cellular microenvironments that provide signals to maintain stem cells in a slowly dividing and undifferentiated state (Bhalla and Singh, 2006; Singh and Bhalla, 2006; Scheres, 2007). Stem cells in the central zone of the SAM provide cells for continuous development of the aerial parts of the plant. Dividing cells, that leave the central zone, enter the flanking peripheral zone and can be recruited for lateral organ primordia, which involves rapid division and differentiation.
The SAM is located at the tip of the growing shoot. Histological analysis of the SAM shows that it is highly-patterned and well-organized; one of the most obvious properties of the SAM is that it is made up of superimposed cell layers designated as L1, L2, and L3 (Satina et al., 1940). In general, L1-derived cell lineages form the epidermis, L2-derived cells develop into sub-epidermis cells like photosynthesizing and gametal cells, and L3-derived cells make up the internal tissues (Stewart and Burk, 1970; Szymkowiak and Sussex, 1996). Different plant species may organize their cell layers differently; for example, the SAM only contains two layers—the single-layered tunica (L1) and the corpus (L2) (Steffensen, 1968)—in maize, but four superimposed cell layers are present in birch (Rinne and van der Schoot, 1998).
Extensive genetic investigations, especially in the model angiosperm Arabidopsis thaliana, have led to the identification of several key meristem regulatory genes. The regulatory interactions between these genes have been elucidated through genetic and molecular analyses. These key regulatory genes include CLAVATA3 (CLV3), WUSCHEL (WUS), and SHOOTMERISTEMLESS (STM) (Long et al., 1996; Mayer et al., 1998; Fletcher et al., 1999). WUS and CLV3 form a feedback regulatory pathway that sustains the stem cell pools (Schoof et al., 2000). The functions of CLV3, CLV1, and STM orthologues appear to be well conserved in monocot and dicot systems (Vollbrecht et al., 1991; Sato et al., 1996; Suzaki et al., 2004, 2006; Chu et al., 2006). At the same time, the STM-leading pathways maintain cells in the meristem indeterminate, which is analogous to the role of the CLV–WUS pathway (Clark et al., 1996; Lenhard et al., 2002). However, the CLV–WUS pathway appears to be less conserved than the STM pathway. The maize and rice WUS orthologues show obvious differences with Arabidopsis regarding their expression patterns (Nardmann and Werr, 2006). Another gene, ROSULATA in Antirrhinum, which is considered to be the counterpart of Arabidopsis WUS, also demonstrates different functions due to mutations, resulting in abnormalities in leaf arrangement and branching patterns (Kieffer et al., 2006). These results show that observations for Arabidopsis cannot always be extended to other species, including other dicot species. We are particularly interested in exploring the molecular basis of meristem functionality in legumes, which diverged from Arabidopsis approximately 92 million years ago (Eckardt, 2001). Moreover, legumes are the second most important family of crop plants—they are widely used as a food and feed source (Graham and Vance, 2003). Legumes are particularly important for sustainable agriculture because of their ability to fix atmospheric nitrogen. In this study, the focus was on the garden pea (Pisum sativum), a classic model species for crop and plant development studies. In addition, the availability of various developmental pea mutants (Singer et al., 2002; Foucher et al., 2003) makes this system amenable to genomics studies.
In recent work, isolated SAM was used to generate cDNA libraries enriched in genes expressed in the SAM (Wong et al., 2008). In the present study, sequencing performed using these cDNA libraries allowed us to discover apical meristem-expressed genes. These sequences were used to make a 12K microarray to investigate the transcriptional repertoire of the pea SAM. Furthermore, the spatial expression pattern within the SAM was investigated for selected genes by in situ hybridization. Our results show that spatially limited gene activation or the repression of genes underpins meristem development and functionality. Our data also provide a catalogue of target genes that can be used for both reverse-genetics approaches and meristem cell-type specific markers.
Materials and methods
Plant materials and RNA isolation
Garden pea (Pisum sativum) cultivar Torsdag was grown under greenhouse conditions. 10-day-old pea seedlings at the vegetative stage were used. Four different samples were harvested and these included the shoot apical meristem (SAM), dormant axillary meristem (AM), root apical meristem (RAM), and non-meristem tissue (NM; consisting of tendril, leaf, stem, and mature root tissues). Total RNA was extracted from each sample using the Qiagen RNeasy Mini Kit according to manufacturer's instructions. Three independent tissue collections and RNA extractions were performed for each of the microarray hybridization experiment.
EST sequencing and the construction of a 12K custom array
Normalized library derived from dissected SAMs as described in Wong et al. (2008) was used for the generation of a further 10 000 ESTs. These ESTs were sequenced at the Australian Genome Research Facility (AGRF), Australia using T7 primer and the resulting data were cleaned and assembled using TIGR Gene Indices clustering tools (TGICL, Pertea et al., 2003). These sequences have been deposited in GenBank under the accession numbers FG528667 to FG539012.
In addition to in-house ESTs, approximately 2000 publically available ESTs were also downloaded from NCBI and the IPK (the Leibniz Institute of Plant Genetics and Crop Plant Research) Crop EST database (http://pgrc.ipk-gatersleben.de/cr-est/index.php; ESTs with accession numbers that begin with CL). A number of transcripts (n=1237) were also randomly selected to be represented twice on the array for the subsequent construction of a 12K Combimatrix pea array. The array was made based on CombiMatrix technology (hyperlink) using in situ synthesis as outlined at www.combimatrix.com.
Microarray data acquisition and analysis
RNA samples were hybridized to two-colour Combimatrix arrays manufactured with a custom library of 11 958 probes. Target preparation and hybridization to Combimatrix 12K arrays were performed at the Australian Genome Research Facility Ltd (AGRF), according to the standard CombiMatrix (www.combimatrix.com). Images were scanned and quantified using GenePix Pro 4.0. The limma software package for R (Smyth, 2005) was used in the statistical analysis of the data generated. The microarray data has been submitted to GEO (www.ncbi.nlm.nih.gov/geo/) under the accession GSE13451.
On examination of the data during quality control assessment, it became evident that the Cy3 channel showed little dynamic range and background correction and normalization could not correct for this. Therefore only the Cy5 channel was used in the subsequent analysis. This resulted in somewhat unbalanced sample sizes with six replicates of SAM and one each of RAM, AM, and NM. Although this reduces the degrees of freedom for error to 5, there are well-established statistical methods that can handle this situation (Smyth, 2004).
The red channel intensities were pre-processed and normalized using standard methods for single-channel microarray data (Bolstad et al., 2003). The intensities were background subtracted using local median background estimates, then quantile normalized and log2 transformed.
Differential expression was assessed using linear modelling and empirical Bayes moderated t-statistics (Smyth, 2004). This approach allows variance information to be borrowed across both samples and genes, resulting in reliable statistical inference when the number of replicates is small. The residual variance is estimated from the available replicates, allowing statistical tests to be performed even for the groups with only one sample. The false discovery rate (FDR) was controlled at less than 1% for each comparison using the method of Benjamini and Hochberg (1995).
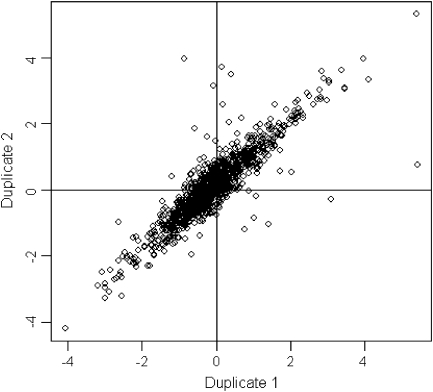
The normalization procedure improved consistency between the replicate arrays, as evidenced by our ability to detect a large number of statistically significant differentially expressed transcripts at a very stringent FDR. For example, 1350 transcripts were detected as differentially expressed between SAM and non-meristematic tissue at 1% FDR after normalization, whereas only 17 transcripts are detected for this comparison using un-normalized log2 transformed data. This highlights the importance of performing appropriate normalization of microarray data. As a further quality control check, the consistency of estimated log2 fold changes was examined for the 1237 transcripts which had duplicate probes on the arrays. The duplicates showed strong agreement after normalization (Fig. 1).
Fig. 1.
Consistency of log2 fold changes for duplicate spots. The plot compares log2 fold changes for the SAM versus a non-meristematic tissue comparison for the 1237 probes with duplicate spots. The first duplicate of each probe is on the x-axis and the second duplicate of the same probe is on the y-axis. Duplicates show a very high level of concordance.
RT- and real time-PCR
For reverse transcription (RT)-PCR studies, 1 μg of DNA-free RNA extract was converted into first-strand cDNA using the SuperScript III system for first-strand cDNA synthesis (Invitrogen) and oligo(dT)12–18. 2 μl of cDNA samples were amplified in a 25 μl PCR mixture. Quantitative-PCR was performed using SYBR® GreenER™ qPCR SuperMix Universal (Invitrogen) on Mx3000P instrument (Stratagene). ROX at a final concentration of 50 nM was used as a reference dye. All reactions were run in duplicate and a melting curve analysis was performed to check for specific product amplification. Relative expression was calculated using MxPro QPCR software v. 1.00 (Stratagene) according to the 2–ΔΔCT method (Livak and Schmittgen, 2001).
In situ hybridization
Non-radioactive in situ hybridization was performed as described previously (Haerizadeh et al., 2009; Wong et al., 2009).
Results and discussion
Features of generated ESTs
Sequences obtained from 10 346 ESTs were assembled and clustered, resulting in 5754 singlets and 1856 contigs. Close to 97% of these 7610 unique sequences (hereafter referred to as unigenes) could be designated as genes with known or putative functions, or as genes encoding hypothetical proteins based on sequence similarity (see the Materials and methods). Using BLAST2GO, 5124 of the unigenes were classified in terms of their GO biological process (data not shown). The successfully classified unigenes cover a broad range of GO functional categories, ranging from genes involved in cell surface receptor-linked signal transduction to cell differentiation for genes associated with transcription and protein metabolic processes (see Supplementary Table 1 at JXB online). Closer inspection of the unigenes grouped in the category ‘Cell differentiation’ revealed a number of sequences with orthologues known to be essential for regulating developmental activities in the SAM. These orthologues include sequences predicted to encode WUS (FG529966), Homeobox protein KNOTTED-1-like 2 (FG538349), YABBY2-like protein (FG532702), FASCIATA2 (FG529452), ARGONAUTE (FG529161), and Homeobox protein SBH1 (FG536622). The occurrence of these sequences demonstrates the utility of our EST collection and supports the potential of our experimental approach in providing insight into the processes underlying SAM function and maintenance.
Functional domains of pea SAM
Base on sequence similarity (at E ≤10−10), transcripts have been identified that are putative orthologues to Arabidopsis genes known to be essential in the functioning and maintenance of the SAM among our EST collection. The spatial expression of these genes was explored in pea to investigate whether their expression was conserved with their putative Arabidopsis orthologues and in doing so distinguishing the different functional domains of SAM in the garden pea.
Maize KNOTTED1 (KN1) and its Arabidopsis counterpart (STM) are mainly expressed in the inner layers (L2 and L3) of the SAM (Smith et al., 1992; Jackson et al., 1994; Long et al., 1996; Long and Barton, 2000). A similar gene has been uncovered in pea (Hofer et al., 2001) and our in situ hybridization analysis revealed the expression of PsKN1 in the pea's inner layers (Fig. 2B) consistent with the expression pattern previously reported by Hofer et al. (2001); this is probably an indicator of undifferentiated cells in the SAM region, as reported for KN1 or STM in maize or Arabidopsis, respectively. Similar to Arabidopsis and maize, the expression of PsKN1 is down-regulated at leaf primordium initiation sites and this has also been shown by Hofer et al. (2001) (Fig. 2B). These results imply the possible conserved functions of PsKN1 with that of the Arabidopsis STM.
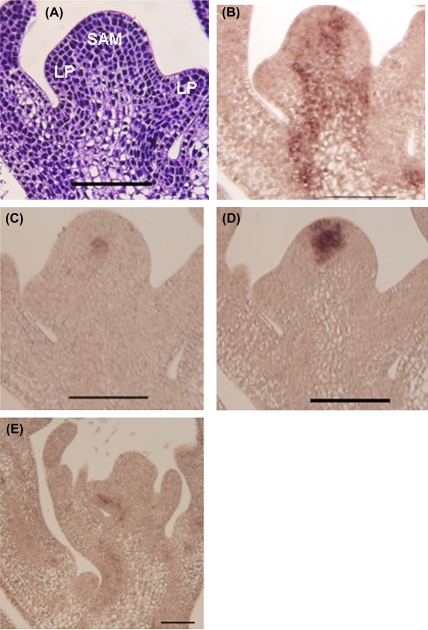
Fig. 2.
Expression of key meristem genes in the pea SAM. (A) A longitudinal section of a paraffin-embedded SAM from a 10-d-old seedling stained with toluidine blue. (B) PsKN1 is expressed throughout the meristem. (C) PsWUS expression is found in the stem cell niche. (D) PsCLV3 is expressed in the stem cell region. (E) PsCLV1 is expressed mainly in the vascular system. Scale bar: 100 μm.
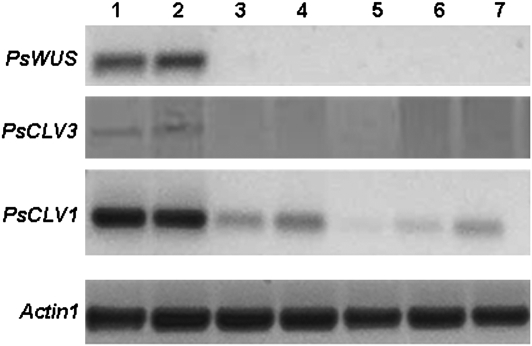
In Arabidopsis, the domain of WUS expression spans the lower part of the central zone (CZ) and the upper part of the rib zone (RZ), defining a functional domain in the CZ called the organizing centre; this centre confers stem cell fates to neighbouring cells (Mayer et al., 1998). Among our EST collection, a likely pea orthologue of WUS was identified (PsWUS; FG529966; see Supplementary Fig. 1A at JXB online). Although PsWUS only shows 48% sequence similarity to petunia WUS at the amino acid level and 33% to Arabidopsis (data not shown), a saturated RT-PCR confirmed the SAM-specific expression of PsWUS (Fig. 3). Further in situ analysis of this gene revealed the expression of PsWUS in a small round area of about two or three cells in diameter in the SAM, just beneath the fourth layer (tunica layer) and in part of the L5 layer (corpus layer; Fig. 2C), which is probably the organizing centre as WUS has been identified to be a marker for the organizing centre (Mayer et al., 1998). This expression pattern strongly suggests that, like the Arabidopsis WUS, the PsWUS also plays key role in stem cell maintenance in pea.
Fig. 3.
RT-PCR analysis of key stem cell maintenance genes in pea. Lanes 1, 2, 3, 4, 5, 6, and 7 contain samples from SAM, AM, RAM, tendril, leaf, stem, and mature root, respectively.
WUS expression in the organizing centre has been shown to activate CLV genes that encode important signalling molecules. While CLV1 encodes a leucine-rich repeat (LRR) receptor-like protein kinase (Clark et al., 1997), CLV3 encodes a secreted protein that can be used to indicate stem cell identity (Fletcher et al., 1999). Since no CLV3 orthologue could be found in our EST collection, the CLV3 gene was cloned using a degenerate PCR method based on the alignment of the reported CLV3 genes of Medicago, Arabidopsis, rice, and castor beans. In addition, saturated RT-PCR of the cloned PsCLV3 showed that it is expressed exclusively in the SAM (Fig. 3). Spatial localization analysis indicated its activity in the four outer layers of the central zone (Fig. 2D), an expression pattern similar to that of Arabidopsis CLV3 (Fletcher et al., 1999).
Although the WUS–CLV3 pathway appears to be conserved in pea, based on their similar spatial expression patterns to that of Arabidopsis, PsCLV1 (FG529170) shows a different expression pattern in pea compared with Arabidopsis CLV1. In contrast to the reported expression of Arabidopsis CLV1 in the SAM (Clark et al., 1997), PsCLV1 is predominantly expressed in the vascular system (Fig. 2E). This result suggests that the receptor kinase activity of PsCLV1 has been replaced by other genes or that PsCLV1 has been recruited to function within different pathways. The latter theory is not unlikely in view of the fact that soybean GmNark shares high sequence homology with CLV1 and is involved in the process of symbiosis (Searle et al., 2003). In addition, the soybean CLV1-like receptor kinase acts predominantly in the vascular tissue (Beveridge et al., 2007). Since CLV3 is expressed in the stem cell region of the pea meristem, the nature of its receptor remains to be resolved.
Detection of differentially expressed genes using the 12K pea oligonucleotide array
The 7610 unigenes from this study were combined with approximately 3000 publically available pea EST sequences to construct a 12K oligonucleotide array (see Materials and methods) that is representative of the pea genome and is enriched for genes exhibiting SAM-associated expression. This microarray was used to perform an extensive analysis of the transcript profile of the pea SAM in comparison to other types of meristems, namely the axillary meristem (AM) and the root apical meristem (RAM), as well as to non-meristem (NM) tissues.
The microarray results showed that the most significant differential expression occurs between non-meristematic tissue and the shoot apical meristem (see Supplementary Table 2 at JXB online), with 1350 transcripts or 10% of the total number of transcripts (11 335) showing differential expression at 1% FDR. As expected, the majority of the SAM up-regulated genes are represented in our collection of SAM-enriched clones, highlighting the significance of constructing an SAM library.
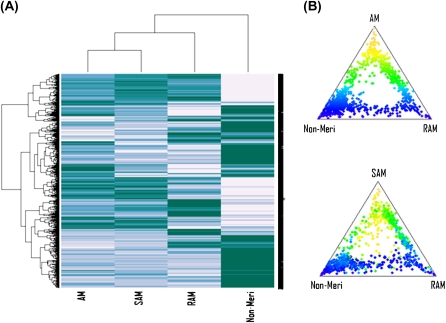
When hierarchical clustering was performed for the 1000 genes exhibiting the most variable expression across the four groups, the resulting heat map illustrated the similarity of the AM's transcript profile to that of SAM (Fig. 4A). Ternary plots of relative expression also showed strong consistency between AM and SAM. Transcripts generally show the same pattern of expression in SAM as in AM relative to the other two tissues (Fig. 4B). Meanwhile, the non-meristematic tissue revealed the most dissimilar profile in comparison to all three meristems (Fig. 4) which is also reflected by the small number of differentially expressed genes in the AM and SAM comparison (Table 1). To verify the results of our microarray analysis, RT-PCR analysis was performed for 25 genes that were selected on the basis of their diverse range of differential expression levels in the SAM and NM comparison. In general, the outcomes are in agreement with the microarray data (Fig. 5).
Fig. 4.
Expression profiles of the 1000 transcripts with the most variable expression across the four tissues used. (A) Hierarchical clustering and heatmap and (B) ternary plots of relative expression. The ternary plots position each transcript according to its relative expression in three different tissues. The distance of each point to each corner is inversely proportional to its expression level in that tissue. Points near each corner of the ternary plot correspond to transcripts expressed exclusively in that tissue whereas transcripts near the centre of the plot are expressed equally in all three tissues. Transcripts are colour-coded according to the first ternary plot, i.e. according to relative expression in AM versus RAM and non-meristem, and the same colour coding is used in the second plot. Both heatmap and ternary plots show strong consistency between AM and SAM.
Table 1.
Number of transcripts with significant differential expression in the comparison of tissues as indicated (FDR ≤0.01)
| Direction of regulation | SAM versus NM | SAM versus AM | RAM versus NM |
| Up | 538 | 2 | 218 |
| Down | 812 | 25 | 404 |
| Total | 1350 | 27 | 622 |
Fig. 5.
RT-PCR analysis of differentially expressed genes. RT-PCR analysis was carried out for 25 selected transcripts as indicated. The ACTIN gene was used as an internal control. FG531364, FG532117, EX570080, FG536909, and FG538772 were identified as those that were significantly down-regulated in the SAM in comparison to the NM. Negative control consisted of samples subjected to PCR analysis with ACTIN primers to examine the absence of any genomic DNA. Lanes 1, 2, 3, and 4 contain samples from SAM, AM, RAM, and non-meristematic tissues (a mixture of tendril, leaf, stem, and mature root tissues), respectively.
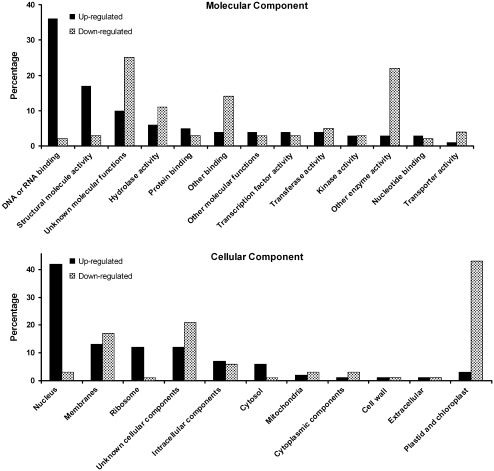
A survey of the top 300 genes up-regulated in the pea SAM in comparison to non-meristematic tissues showed that about one-third of the genes can be classified as encoding gene products with DNA or RNA-binding activities. On the other hand, genes in this class represent only 2% of the top 300 genes that are down-regulated in the SAM in comparison to non-meristematic tissues. Based on cellular component classifications, approximately 40% of the genes up-regulated in the SAM resided in the ‘nucleus’ category (Fig. 6). Among those genes down-regulated in the SAM, 43% belonged to the ‘plastid and chloroplast’ class consistent with the fact that SAMs are heterotrophic.
Fig. 6.
Gene ontology (GO) functional categorization of transcripts differentially regulated in SAM in comparison to non-meristematic tissues. The matching Arabidopsis thaliana gene index (AtGI) for the top 300 transcripts was used in the automated GO categorization available at www.arabidopsis.org.
Comparison of differentially expressed AM and SAM transcripts
AM is the meristem responsible for the secondary axes of plant growth. Many pea orthologues of key meristem identity genes such as PsWUS, PsCLV3, and PsKN1 show expression in the SAM as well as in the AM (Fig. 2). This is not surprising as studies have indicated the formation process of AM resembles that of the SAM (reviewed by Bennett and Leyser, 2006). Despite the similarity in transcript profiles of the AM and SAM, a total of 27 transcripts have been successfully identified that potentially distinguish the AM gene expression profile from that of SAM (at FDR ≤0.01; Table 2). Among them is a transcript known as DRM4 and reported to be associated with AM dormancy (Stafstrom et al., 1998). Further investigation of transcripts in Table 2 should reveal novel dormancy associated genes.
Table 2.
Transcripts that are differentially regulated in the SAM in comparison to the AM (FDR ≤0.01)
| Accession no. | Annotation | Fold change (log2) |
| FG531608 | Cytochrome P450-like protein | 3.0 |
| BV165610 | WD-40 repeat protein MSI4 | 2.1 |
| FG536909 | AP2 domain-containing protein | −2.1 |
| EX569019 | No BLASTX match | −2.2 |
| EX569165 | No BLASTX match | −2.2 |
| FG531433 | F-box/LRR-repeat protein 20–like | −2.3 |
| FG528742 | No BLASTX match | −2.5 |
| FG536535 | No BLASTX match | −2.5 |
| FG534876 | No BLASTX match | −2.5 |
| EX569623 | F-box protein family-like protein | −2.5 |
| FG538233 | Albumin-2 (PA2) | −2.5 |
| FG530490 | Hypothetical protein | −2.6 |
| FG537964 | No BLASTX match | −2.6 |
| FG531181 | Ethylene–responsive transcription factor 5 | −2.6 |
| X65154.1 | GA regulated mRNA | −2.7 |
| FG531688 | Histidine kinase-like protein | −2.7 |
| EX569457 | No BLASTX match | −2.8 |
| AF515796.1 | Drm4 mRNA | −2.8 |
| FG535658 | Acetylglucosaminyltransferase SPINDLY | −3.0 |
| FG534193 | 18 kDa seed maturation protein | −3.0 |
| FG530878 | No BLASTX match | −3.0 |
| FG535973 | Peripheral-type benzodiazepine receptor | −3.1 |
| FG530812 | Cold-acclimation specific protein 15 | −3.4 |
| FG529829 | Hypothetical protein | −3.4 |
| FG538772 | Sex determination protein tasselseed-2 | −3.6 |
| FG529247 | Hypothetical protein | −4.0 |
| FG538246 | Hypothetical Cys-3-His zinc finger protein | −4.0 |
| FG530800 | Isoflavone reductase | −4.4 |
Intriguingly, one transcript that showed lower expression in the AM is one that is predicted to encode the WD-40 repeat MSI4-like protein (BV165610). Members of the MSI-like protein family are known to interact directly or indirectly with histones and hence may target chromatin assembly factors and histone deacetylases and are found to control cell proliferation and differentiation in animals (reviewed by Hennig et al., 2005). The observation that a similar transcript is expressed at a lower level in AMs in comparison to SAMs suggests that it may play roles in regulating activities that are specific to the SAM. Recently, strigolactone has been identified as a hormonal signal that represses axillary bud outgrowth but which does not appear to affect apical growth in pea (Gomez-Roldan et al., 2008; Umehara et al., 2008). It will be interesting to determine how it affects the expression of these genes during the transition of the AM to a SAM that occurs during axillary bud outgrowth.
Shared transcriptional signatures of meristematic activity
Hierarchical clustering of the top 1000 significantly differentially expressed genes revealed a subset of sequences whose up-regulation relative to non-meristematic tissues is shared among all meristem samples, i.e. SAM, AM, and RAM (Fig. 4; see Supplementary Table 3 at JXB online). These genes provide a valuable insight into molecular networks that underpin the maintenance of meristematic functionality. As expected, a significant number of these encode the core histones H1, H2, H3, and H4 (see Supplementary Table 3 at JXB online). Other genes whose preferential expression is shared among the three meristems examined include those that encode the WD-40 repeat protein MSI3, nucleolar protein Nop56 (Nucleolar protein 5A), non-specific lipid transfer protein, palmitoyltransferase ZDHHC3, and LIM domain only protein 7. The WD-40 repeat protein MSI3 is a retinoblastoma-binding protein that plays an important role in controlling cell cycle progression (Dennis, 2007). Nucleolar protein Nop56 activity highlights the increasing relevance of the nucleolus in regulating mitosis and cell cycle progression (Boisvert et al., 2007). Shared up-regulation of palmitoyltransferase ZDHHC3 in meristematic tissues is of particular relevance as this enzyme mediates palmitoylation, a post-translational modification that can have multiple effects on protein function and localization. For example, palmitoylation can influence membrane binding and membrane targeting, and palmitoylation of many proteins is required for efficient signal transduction (Resh, 2006). Our findings indicate that the role of post-translational protein palmitoylation in cell signalling is required to maintain meristematic competence in plants.
A survey of the top 1000 significantly differentially expressed genes also showed that gene repression is a more widespread feature of meristematic activity than up-regulation (see Supplementary Table 4 at JXB online). The number of genes sharing repression in the SAM, AM, and RAM is nearly five times higher than those undergoing up-regulation. Understandably, a majority of the genes repressed in the meristem are involved in glycolytic and photosynthetic metabolism processes. Significant development-related genes that are repressed in all three meristems include the homeobox protein SBH1, late embryogenesis abundant protein 76 (LEA 76), Pisum sativum PsAD2 mRNA, Pisum sativum GA 20-oxidase mRNA, and zinc finger protein CONSTANS-LIKE 16. SBH1 belongs to the Knotted-1 family of homeodomain proteins that are strongly expressed in differentiated cell types (Tioni et al., 2003). GA-20 oxidases regulate the final rate-limiting steps in GA biosynthesis (Desgagne-Penix and Sponsel, 2008). CONSTANS-LIKE 16 CO belongs to a family of 17 putative transcription factors defined by two conserved domains (Robson et al., 2001). The first is a zinc finger region near the amino terminus that resembles B-boxes, which regulate protein–protein interactions in several animal transcription factors. Our data thus highlight candidate genes whose spatially limited gene activation or repression probably underpins meristematic competence in plants.
Apical meristem-specific genes
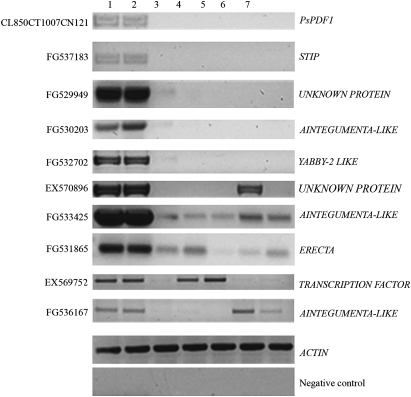
As previously mentioned, saturated RT-PCR analysis confirmed the exclusive expression of key SAM regulatory genes such as PsWUS and PsCLV3 in SAMs or AMs (Fig. 2). In order to identify transcripts that demonstrate similar expression profiles, candidates were selected that were up-regulated when the transcript profile of SAM was compared with that of RAM or NM for further saturated RT-PCR analysis. Among 10 selected candidates, five SAM-specific genes were found (Fig. 7). These genes were expressed only in the SAM or AM and not in any other tissues: tendril, leaf, stem, and root. To validate these observations, further real-time PCR analysis was also carried out for a subset of these candidates (Fig. 8) and the results are in line with that of the RT-PCR analysis.
Fig. 7.
Saturated PCR showing the gene expression of differentially expressed ESTs. Lanes 1, 2, 3, 4, 5, 6, and 7 contain samples from SAM, AM, RAM, tendril, leaf, stem, and mature root, respectively.
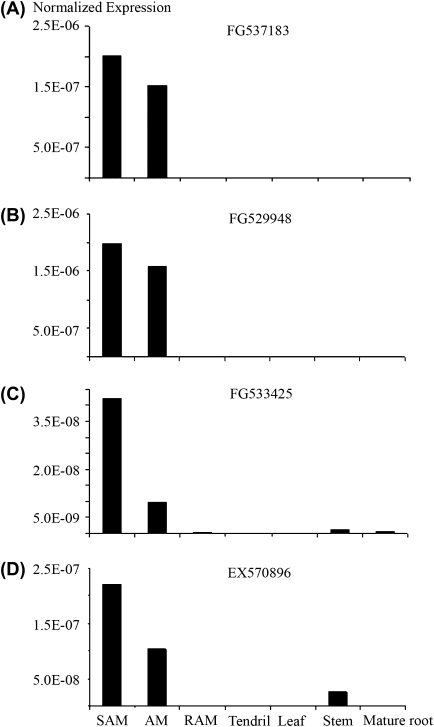
Fig. 8.
Real-time PCR analyses of transcripts with specific (A, B) or enriched (C, D) gene expression in apical meristems.
The SAM-specific genes identified include transcripts predicted to encode PROTODERMAL FACTOR1 (PDF1; CL850CT1007CN1219), a WUS-related product, STIP (FG537183), YABBY2 (FG532702), an unknown protein (FG529949), and AINTEGUMENTA (FG530203) (Fig. 7). As the expression domains of these within the SAM are essential for understanding their functional significance, their spatial expression patterns were investigated.
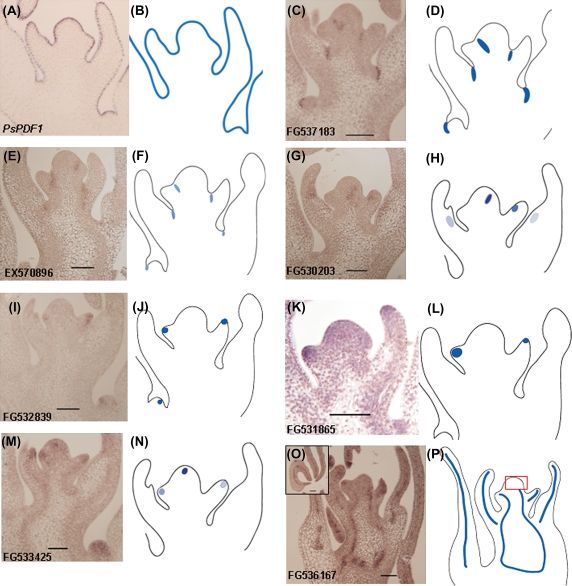
PDF1 is a known marker gene for the L1 layer in Arabidopsis (Abe et al., 1999). The pea orthologue PsPDF1 (CL850CT1007CN1219) shows high sequence similarity to Arabidopsis PDF1 (Wong et al., 2008) with 60% identity at the amino acid level (data not shown). In situ hybridization revealed that the PsPDF1 is also exclusively expressed in the L1 layer (Fig. 9A) indicating the conserved function of this gene in pea as in Arabidopsis. Meanwhile, the expression of STIP (FG537183) was detected in the SAM and leaf primordia (Fig. 9C) and more specifically it is only expressed in the boundary region corresponding to the leaf axil (Fig. 9C), which initiates the axillary meristem. In the SAM region, the expression of STIP is restricted to a small domain that probably separates the leaf anlage and the stem cells (Fig. 9C). After a new axillary meristem is initiated, its expression is again restricted to the leaf axil (Fig. 9C). This expression pattern indicates that this gene is recruited to mark the boundary region separating organ domains. Key genes for axillary meristem initiation reported to date share a common expression feature: the boundary expression domain in the SAM (Schmitz et al., 2002; Keller et al., 2006; Muller et al., 2006). Intriguingly, another transcript annotated to encode an unknown protein (EX570896) with enriched expression in the SAM shows similar spatial expression profile to that of STIP (Fig. 9E) and thus they may be involved in regulating AM initiation as well as in SAM maintenance. This is not unlikely as there are reports suggesting that boundary genes are required for SAM and AM development (Hibara et al., 2006; Borghi et al., 2007).
Fig. 9.
In situ localization of transcripts with specific or enriched expression in the apical meristems. Gene expression patterns in the pea shoot apex [(A), (C), (E), (G), (I), (K), (M) and (O)] with the corresponding modelled expression as shown in (B), (D), (F), (H), (J), (L), (N) and (P). (A, B): Pea PsPDF1(CL850CT1007CN1219) expression was detected in the outermost layer of the meristem. (C, D) and (E, F): Expressions of STIP (FG537183) and an unknown gene (EX570896) were found in the boundary region corresponding to the axillary meristem initiation area. (G, H): ANT-like gene (FG530203) was expressed in a region likely to be the presumptive organ primordium as well as in the central and marginal region of the primordium and developing leaf. (I, J) and (K, L): YABBY (FG532839; I, J) and ERECTA (FG531865; K, L) were mainly expressed in the organ primordia. (M, N): The expression of FG533425, a putative member of the ANT-like genes, is limited to the central zone, an area whereby the expression of PsWUS was detected as well as at the tip of leaf primordial (Fig. 2C). (O, P): Another putative ANT-like gene (FG536167) is expressed in the provascular region in the stem and leaf primordia suggesting its likely roles in vascular differentiation. All are longitudinal sections of paraffin-embedded shoot apices probed with the antisense strand of digoxigenin-labelled RNA.
When the expression of the transcripts annotated as AINTEGUMENTA-LIKE (FG530203) was examined in situ, signals were detected related to its expression in the presumptive organ as well as localized to the central and marginal regions of the primordium and developing leaf (Fig. 9G). This is consistent with what has been reported for a similar Arabidopsis gene (Elliott et al., 1996), suggesting a likely conserved function of this gene in pea.
Transcripts with high expression in the SAM
In addition to investigating the spatial expression of SAM-specific genes, the in situ expression of five other candidates that have enriched expression in the SAM, in comparison to other tissues, has also been examined.
Genes involved in the initiation of lateral organs:
Genes involved in leaf primordium initiation are represented in our array data. YABBY genes in Arabidopsis and maize have been shown to play key roles in leaf primordium initiation and expansion (Bowman, 2000; Eshed et al., 2004; Juarez et al., 2004). FG532839, a putative orthologue of the Arabidopsis YABBY5 gene based on a BLASTX match, is highly expressed in the SAM (Fig. 7). In situ localization showed that this gene accumulates mainly in the leaf primordium (Fig. 9I) indicating the conserved role of pea YABBY genes in lateral organ formation. The discrepancy between the RT-PCR results and the in situ data is due to the fact that, while it was attempted to exclude as much leaf primorium from the dissected SAM as possible, the youngest developing leaf primordium often could not be eliminated as it was difficult to identify on the SAM under the dissecting microscope.
Genes important for cell-to-cell signalling in lateral organ development were also identified in this experiment. The ERECTA gene was characterized as a receptor-like kinase signalling receptor, a component of cell-to-cell signalling (Torii et al., 1996). A similar pea sequence predicted to encode ERECTA-like kinase (FG531865) is highly expressed in the lateral primordium (Fig. 9K) and is consistent with the expression pattern described for Arabidopsis ERECTA (Shpak et al., 2004; Yokoyama et al., 1998).
AINTEGUMENTA (ANT)-LIKE genes:
A total of three ANT-LIKE genes (see Supplementary Fig. 1B at JXB online)—FG533425, FG530203, and FG536167—were highly expressed in the SAM in comparison to the NM (see Supplementary Table 2 at JXB online). In addition to the SAM-specific sequence (FG530203), two other sequences are meristem-enriched in their expression (FG533425 and FG536167) (Fig. 7). FG533425 is expressed not only in the leaf primordium, but also in the centre of the SAM (Fig. 9M). The latter expression pattern has not been reported for the Arabidopsis ANT that is exclusively expressed in the organ primordia and is used to mark the shoot organ primordia (Long and Barton, 1998, 2000). Another ANT-like gene, FG536167, shows a completely different expression pattern compared to the known ANT genes. It is expressed in the provascular region throughout the meristem (Fig. 9O) and the expression is confined to the middle vascular system in the developing leaflet or tendril (inset picture in Fig. 9O). This unique expression pattern indicates that ANT-like genes could be involved in the initiation and formation of the provascular system.
Despite the rapid growth in the understanding and identification of key genes controlling meristem development, a comprehensive view of the temporal and spatial gene expression pattern involved in SAM functionality remains elusive. The development of nearly 12 000 ESTs, representing genes transcribed in the pea SAM, and the identification of gene subsets that are up-regulated or specifically-expressed in SAM cell layers/zones represents a significant advance towards identifying the regulatory networks involved in specifying cell types and providing positional information within the meristem. Our data indicate the usefulness of the transcriptome approach to identifying meristem regulatory genes in legumes and offers the possibility of transferring information from pea to other agronomically important legume species. Moreover, analysis of genes in the legume class contributes to the evolutionary context of plant development and should highlight conserved and divergent mechanisms.
Supplementary data
Supplementary data is available at JXB online.
Supplementary Fig. 1. Phylogenetic analysis of (A) PsWUS and (B) PsANT-LIKE genes.
Supplementary Table 1. Functional classification of unigenes according to GO biological processes using BLAST2GO (Conesa et al., 2005).
Supplementary Table 2. Differentially expressed transcripts in the pea SAM in comparison to NM (at adjusted P-value ≤0.05).
Supplementary Table 3. Up-regulated transcripts in meristems (SAM, AM, and RAM) in comparison to the NM (at adjusted P-value ≤0.01).
Supplementary Table 4. Down-regulated transcripts in meristems (SAM, AM and RAM) in comparison to the NM (at adjusted P-value ≤0.01).
Acknowledgments
Our sincere thanks to Ms Lin Zhang for her assistance in growing the pea plants, and to the Australian Research Council for financially supporting this project as a part of the ARC Centre of Excellence grant.
References
- Abe M, Takahashi T, Komeda Y. Cloning and characterization of an L1 layer-specific gene in Arabidopsis thaliana. Plant and Cell Physiology. 1999;40:571–580. doi: 10.1093/oxfordjournals.pcp.a029579. [DOI] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. Journal of the Royal Statistical Society B. 1995;57:289–300. [Google Scholar]
- Bennett T, Leyser O. Something on the side: axillary meristems and plant development. Plant Molecular Biology. 2006;60:843–854. doi: 10.1007/s11103-005-2763-4. [DOI] [PubMed] [Google Scholar]
- Beveridge CA, Mathesius U, Rose RJ, Gresshoff PM. Common regulatory themes in meristem development and whole-plant homeostasis. Current Opinion in Plant Biology. 2007;10:44–51. doi: 10.1016/j.pbi.2006.11.011. [DOI] [PubMed] [Google Scholar]
- Bhalla PL, Singh MB. Molecular control of stem cell maintenance in shoot apical meristem. Plant Cell Reports. 2006;25:249–256. doi: 10.1007/s00299-005-0071-8. [DOI] [PubMed] [Google Scholar]
- Boisvert F-M, van Koningsbruggen S, Navascues J, Lamond AI. The multifunctional nucleolus. Nature Reviews Molecular and Cell Biology. 2007;8:574–585. doi: 10.1038/nrm2184. [DOI] [PubMed] [Google Scholar]
- Bolstad BM, Irizarry RA, Astrand M, Speed TP. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics. 2003;19:185–193. doi: 10.1093/bioinformatics/19.2.185. [DOI] [PubMed] [Google Scholar]
- Borghi L, Bureau M, Simon R. Arabidopsis JAGGED LATERAL ORGANS is expressed in boundaries and coordinates KNOX and PIN activity. Plant Cell. 2007;19:1795–1808. doi: 10.1105/tpc.106.047159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman JL. The YABBY gene family and abaxial cell fate. Current Opinion in Plant Biology. 2000;3:17–22. doi: 10.1016/s1369-5266(99)00035-7. [DOI] [PubMed] [Google Scholar]
- Chu H, Qian Q, Liang W, et al. The floral organ number4 gene encoding a putative ortholog of Arabidopsis CLAVATA3 regulates apical meristem size in rice. Plant Physiology. 2006;142:1039–1052. doi: 10.1104/pp.106.086736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark SE, Jacobsen SE, Levin JZ, Meyerowitz EM. The CLAVATA and SHOOT MERISTEMLESS loci competitively regulate meristem activity in Arabidopsis. Development. 1996;122:1567–1575. doi: 10.1242/dev.122.5.1567. [DOI] [PubMed] [Google Scholar]
- Clark SE, Williams RW, Meyerowitz EM. The CLAVATA1 gene encodes a putative receptor kinase that controls shoot and floral meristem size in Arabidopsis. Cell. 1997;89:575–585. doi: 10.1016/s0092-8674(00)80239-1. [DOI] [PubMed] [Google Scholar]
- Conesa A, Gotz S, Garcia-Gomez JM, Terol J, Talon M, Robles M. Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics. 2005;21:3674–3676. doi: 10.1093/bioinformatics/bti610. [DOI] [PubMed] [Google Scholar]
- Dennis F. The plant cell cycle–15 years on. New Phytologist. 2007;174:261–278. doi: 10.1111/j.1469-8137.2007.02038.x. [DOI] [PubMed] [Google Scholar]
- Desgagne-Penix I, Sponsel VM. Expression of gibberellin 20-oxidase1 (AtGA20ox1) in Arabidopsis seedlings with altered auxin status is regulated at multiple levels. Journal of Experimental Botany. 2008;59:2057–2070. doi: 10.1093/jxb/ern063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckardt NA. Everything in its place: conservation of gene order among distantly related plant species. The Plant Cell. 2001;13:723–725. doi: 10.1105/tpc.13.4.723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elliott RC, Betzner AS, Huttner E, Oakes MP, Tucker WQJ, Gerentes D, Perez P, Smyth DR. AINTEGUMENTA, an APETALA2-like gene of arabidopsis with pleiotropic roles in ovule development and floral organ growth. The Plant Cell. 1996;8:155–168. doi: 10.1105/tpc.8.2.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eshed Y, Izhaki A, Baum SF, Floyd SK, Bowman JL. Asymmetric leaf development and blade expansion in Arabidopsis are mediated by KANADI and YABBY activities. Development. 2004;131:2997–3006. doi: 10.1242/dev.01186. [DOI] [PubMed] [Google Scholar]
- Fletcher LC, Brand U, Running MP, Simon R, Meyerowitz EM. Signaling of cell fate decisions by CLAVATA3 in Arabidopsis shoot meristems. Science. 1999;283:1911–1914. doi: 10.1126/science.283.5409.1911. [DOI] [PubMed] [Google Scholar]
- Foucher F, Morin J, Courtiade J, Cadioux S, Ellis N, Banfield MJ, Rameau C. DETERMINATE and LATE FLOWERING are two TERMINAL FLOWER1/CENTRORADIALIS homologs that control two distinct phases of flowering initiation and development in pea. The Plant Cell. 2003;15:2742–2754. doi: 10.1105/tpc.015701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Roldan V, Fermas S, Brewer PB, et al. Strigolactone inhibition of shoot branching. Nature. 2008;455:189–194. doi: 10.1038/nature07271. [DOI] [PubMed] [Google Scholar]
- Graham PH, Vance CP. Legumes: importance and constraints to greater use. Plant Physiology. 2003;131:872–877. doi: 10.1104/pp.017004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haerizadeh F, Wong CE, Singh MB, Bhalla PL. Genome-wide analysis of gene expression in soybean shoot apical meristem. Plant Molecular Biology. 2009;69:711–727. doi: 10.1007/s11103-008-9450-1. [DOI] [PubMed] [Google Scholar]
- Hennig L, Bouveret R, Gruissem W. MSI1-like proteins: an escort service for chromatin assembly and remodeling complexes. Trends in Cell Biology. 2005;15:295–302. doi: 10.1016/j.tcb.2005.04.004. [DOI] [PubMed] [Google Scholar]
- Hibara K, Karim MR, Takada S, Taoka K, Furutani M, Aida M, Tasaka M. Arabidopsis CUP-SHAPED COTYLEDON3 regulates postembryonic shoot meristem and organ boundary formation. The Plant Cell. 2006;18:2946–2957. doi: 10.1105/tpc.106.045716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofer J, Gourlay C, Michael A, Ellis THN. Expression of a class 1 knotted1-like homeobox gene is down-regulated in pea compound leaf primordia. Plant Molecular Biology. 2001;45:387–398. doi: 10.1023/a:1010739812836. [DOI] [PubMed] [Google Scholar]
- Jackson D, Veit B, Hake S. Expression of maize KNOTTED1 related homeobox genes in the shoot apical meristem predicts patterns of morphogenesis in the vegetative shoot. Development. 1994;120:405–413. [Google Scholar]
- Juarez MT, Twigg RW, Timmermans MCP. Specification of adaxial cell fate during maize leaf development. Development. 2004;131:4533–4544. doi: 10.1242/dev.01328. [DOI] [PubMed] [Google Scholar]
- Keller T, Abbott J, Moritz T, Doerner P. Arabidopsis REGULATOR OF AXILLARY MERISTEMS1 controls a leaf axil stem cell niche and modulates vegetative development. The Plant Cell. 2006;18:598–611. doi: 10.1105/tpc.105.038588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kieffer M, Stern Y, Cook H, Clerici E, Maulbetsch C, Laux T, Davies B. Analysis of the transcription factor WUSCHEL and its functional homologue in Antirrhinum reveals a potential mechanism for their roles in meristem maintenance. The Plant Cell. 2006;18:560–573. doi: 10.1105/tpc.105.039107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenhard M, Jurgens G, Laux T. The WUSCHEL and SHOOTMERISTEMLESS genes fulfil complementary roles in Arabidopsis shoot meristem regulation. Development. 2002;129:3195–3206. doi: 10.1242/dev.129.13.3195. [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Long J, Barton MK. Initiation of axillary and floral meristems in Arabidopsis. Developmental Biology. 2000;218:341–353. doi: 10.1006/dbio.1999.9572. [DOI] [PubMed] [Google Scholar]
- Long JA, Barton MK. The development of apical embryonic pattern in Arabidopsis. Development. 1998;125:3027–3035. doi: 10.1242/dev.125.16.3027. [DOI] [PubMed] [Google Scholar]
- Long JA, Moan EI, Medford JI, Barton MK. A member of the KNOTTED class of homeodomain proteins encoded by the STM gene of Arabidopsis. Nature. 1996;379:66–69. doi: 10.1038/379066a0. [DOI] [PubMed] [Google Scholar]
- Mayer KFX, Schoof H, Haecker A, Lenhard M, Jurgens G, Laux T. Role of WUSCHEL in regulating stem cell fate in the Arabidopsis shoot meristem. Cell. 1998;95:805–815. doi: 10.1016/s0092-8674(00)81703-1. [DOI] [PubMed] [Google Scholar]
- Muller D, Schmitz G, Theres K. Blind homologous R2R3 Myb genes control the pattern of lateral meristem initiation in Arabidopsis. The Plant Cell. 2006;18:586–597. doi: 10.1105/tpc.105.038745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nardmann J, Werr W. The shoot stem cell niche in angiosperms: expression patterns of WUS orthologues in rice and maize imply major modifications in the course of mono- and dicot evolution. Molecular Biology and Evolution. 2006;23:2492–2504. doi: 10.1093/molbev/msl125. [DOI] [PubMed] [Google Scholar]
- Pertea G, Huang X, Liang F, et al. TIGR gene indices clustering tools (TGICL): a software system for fast clustering of large EST datasets. Bioinformatics. 2003;19:651–652. doi: 10.1093/bioinformatics/btg034. [DOI] [PubMed] [Google Scholar]
- Resh MD. Palmitoylation of ligands, receptors, and intracellular signaling molecules. Science STKE. 2006 doi: 10.1126/stke.3592006re14. 2006, re14. [DOI] [PubMed] [Google Scholar]
- Rinne PL, van der Schoot C. Symplasmic fields in the tunica of the shoot apical meristem coordinate morphogenetic events. Development. 1998;125:1477–1485. doi: 10.1242/dev.125.8.1477. [DOI] [PubMed] [Google Scholar]
- Robson F, Costa MMR, Hepworth SR, Vizir I, Pineiro M, Reeves PH, Putterill J, Coupland G. Functional importance of conserved domains in the flowering-time gene CONSTANS demonstrated by analysis of mutant alleles and transgenic plants. The Plant Journal. 2001;28:619–631. doi: 10.1046/j.1365-313x.2001.01163.x. [DOI] [PubMed] [Google Scholar]
- Satina S, Blakeslee AF, Avery AG. Demonstration of the three germ layers in the shoot apex of Datura by means of induced polyploidy in periclinal chimeras. American Journal of Botany. 1940;27:895–905. [Google Scholar]
- Sato Y, Hong SK, Tagiri A, Kitano H, Yamamoto N, Nagato Y, Matsuoka M. A rice homeobox gene, OSH1, is expressed before organ differentiation in a specific region during early embryogenesis. Proceedings of the National Academy of Sciences, USA. 1996;93:8117–8122. doi: 10.1073/pnas.93.15.8117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheres B. Stem-cell niches: nursery rhymes across kingdoms. Nature Reviews Molecular and Cell Biology. 2007;8:345–354. doi: 10.1038/nrm2164. [DOI] [PubMed] [Google Scholar]
- Schmitz G, Tillmann E, Carriero F, Fiore C, Cellini F, Theres K. The tomato Blind gene encodes a MYB transcription factor that controls the formation of lateral meristems. Proceedings of the National Academy of Sciences, USA. 2002;99:1064–1069. doi: 10.1073/pnas.022516199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoof H, Lenhard M, Haecker A, Mayer KF, Jurgens G, Laux T. The stem cell population of Arabidopsis shoot meristems in maintained by a regulatory loop between the CLAVATA and WUSCHEL genes. Cell. 2000;100:635–644. doi: 10.1016/s0092-8674(00)80700-x. [DOI] [PubMed] [Google Scholar]
- Searle IR, Men AE, Laniya TS, Buzas DM, Iturbe-Ormaetxe I, Carroll BJ, Gresshoff PM. Long-distance signaling in nodulation directed by a CLAVATA1-like receptor kinase. Science. 2003;299:109–112. doi: 10.1126/science.1077937. [DOI] [PubMed] [Google Scholar]
- Shpak ED, Berthiaume CT, Hill EJ, Torii KU. Synergistic interaction of three ERECTA-family receptor-like kinases controls Arabidopsis organ growth and flower development by promoting cell proliferation. Development. 2004;131:1491–1501. doi: 10.1242/dev.01028. [DOI] [PubMed] [Google Scholar]
- Singer S, Maki S, Sollinger J, Plotz J, Fitzgerald K, Fishbach J, Mullen H. Flower development in pea: role of proliferating inflorescence meristem (PIM), an AP1 homolog. Developmental Biology. 2002;247:518–518. [Google Scholar]
- Singh MB, Bhalla PL. Plant stem cells carve their own niche. Trends in Plant Science. 2006;11:241–246. doi: 10.1016/j.tplants.2006.03.004. [DOI] [PubMed] [Google Scholar]
- Smith LG, Greene B, Veit B, Hake S. A dominant mutation in the maize homeobox gene, Knotted-1, causes its ectopic expression in leaf cells with altered fates. Development. 1992;116:21–30. doi: 10.1242/dev.116.1.21. [DOI] [PubMed] [Google Scholar]
- Smyth GK. Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Statistical Applications in Genetics and Molecular Biology. 2004 doi: 10.2202/1544-6115.1027. 3, Article 3. [DOI] [PubMed] [Google Scholar]
- Smyth GK. limma: linear models for microarray data. In: Gentleman R, Carey VJ, Huber W, Irizarry RA, Dudoit S, editors. Bioinformatics and computational biology solutions using R and bioconductor. New York: Springer; 2005. pp. 397–420. [Google Scholar]
- Stafstrom JP, Ripley BD, Devitt ML, Drake B. Dormancy-associated gene expression in pea axillary buds. Planta. 1998;205:547–552. doi: 10.1007/s004250050354. [DOI] [PubMed] [Google Scholar]
- Steffensen DM. A reconstruction of cell development in the shoot apex of maize. American Journal of Botany. 1968;55:354–369. [Google Scholar]
- Stewart RN, Burk LG. Independence of tissues derived from apical layers in ontogeny of the tobacco leaf and ovary. American Journal of Botany. 1970;57:1010–1016. [Google Scholar]
- Suzaki T, Sato M, Ashikari M, Miyoshi M, Nagato Y, Hirano HY. The gene FLORAL ORGAN NUMBER1 regulates floral meristem size in rice and encodes a leucine-rich repeat receptor kinase orthologous to Arabidopsis CLAVATA1. Development. 2004;131:5649–5657. doi: 10.1242/dev.01441. [DOI] [PubMed] [Google Scholar]
- Suzaki T, Toriba T, Fujimoto M, Tsutsumi N, Kitano H, Hirano HY. Conservation and diversification of meristem maintenance mechanism in Oryza sativa: function of the FLORAL ORGAN NUMBER2 gene. Plant and Cell Physiology. 2006;47:1591–1602. doi: 10.1093/pcp/pcl025. [DOI] [PubMed] [Google Scholar]
- Szymkowiak EJ, Sussex IM. What chimeras can tell us about plant development. Annual Review of Plant Physiology and Plant Molecular Biology. 1996;47:351–376. doi: 10.1146/annurev.arplant.47.1.351. [DOI] [PubMed] [Google Scholar]
- Tioni MF, Gonzalez DH, Chan RL. Knotted1-like genes are strongly expressed in differentiated cell types in sunflower. Journal of Experimental Botany. 2003;54:681–690. doi: 10.1093/jxb/erg077. [DOI] [PubMed] [Google Scholar]
- Torii KU, Mitsukawa N, Oosumi T, Matsuura Y, Yokoyama R, Whittier RF, Komeda Y. The Arabidopsis ERECTA gene encodes a putative receptor protein kinase with extracellular leucine-rich repeats. The Plant Cell. 1996;8:735–746. doi: 10.1105/tpc.8.4.735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umehara M, Hanada A, Yoshida S, et al. Inhibition of shoot branching by new terpenoid plant hormones. Nature. 2008;455:195–U129. doi: 10.1038/nature07272. [DOI] [PubMed] [Google Scholar]
- Vollbrecht E, Veit B, Sinha N, Hake S. The developmental gene Knotted-1 is a member of a maize homeobox gene family. Nature. 1991;350:241. doi: 10.1038/350241a0. [DOI] [PubMed] [Google Scholar]
- Wong CE, Bhalla PL, Ottenhof H, Singh MB. Transcriptional profiling of the pea shoot apical meristem reveals processes underlying its function and maintenance. BMC Plant Biology. 2008;8:73–84. doi: 10.1186/1471-2229-8-73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong CE, Singh MB, Bhalla PL. Molecular processes underlying the floral transition in the soybean shoot apical meristem. The Plant Journal. 2009;57:832–845. doi: 10.1111/j.1365-313X.2008.03730.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoyama R, Takahashi T, Kato A, Torii KU, Komeda Y. The Arabidopsis ERECTA gene is expressed in the shoot apical meristem and organ primordia. The Plant Journal. 1998;15:301–310. doi: 10.1046/j.1365-313x.1998.00203.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.