Summary
Nascent transcripts of the Drosophila Ubx gene were detected by in situ hybridization. Following onset of expression, the progress of RNA polymerase (1.4 kb/min) across the gene was visualized as the successive appearance of hybridization signals from different positions within the transcription unit. Nascent transcripts disappeared at mitosis. Hybridization signals reappeared in the next cell cycle first with a 5′ probe, and later, following a delay consistent with the transcription rate, with a 3′ probe. Nascent transcripts were observed continuously in expressing cells of a mutant embryo in which cells are blocked in interphase. We conclude that progression through mitosis causes abortion of nascent transcripts and suggest that periodic abortion of transcription contributes to regulation of expression of large genes.
Introduction
The size of transcription units spans an enormous range, from less than 1 kb to the 2000 kb human dystrophin gene. There is a corresponding variation in the time that RNA polymerase takes to traverse a gene, from about 30 s to 11 hr. In many organisms, embryonic cell cycles are short (8 min in Drosophila, 30 min in frog, and 3 hr in mouse). Indeed, these cell cycle times are shorter than the transcription times of long genes. If DNA synthesis or mitosis disrupts transcription, these rapid cell cycles would preclude expression of large genes. The potential regulatory importance of such a coupling of the rate of cell cycle progression and gene expression led us to examine the influence of cell cycle events on transcription.
Genes governing embryonic pattern formation in Drosophila have transcription units ranging from about 2 kb to 105 kb and would be expected to have transcription times between about 1.5 min and 75 min. During the period when these genes are expressed, cell cycle times change from as short as 8 min to more than 180 min. We have taken advantage of the detailed characterization of cell division times in early Drosophila embryos (Campos-Ortega and Hartenstein, 1985; Foe, 1989) and a powerful in situ hybridization procedure (Tautz and Pfeifle, 1989; O'Farrell et al., 1989) to follow the fates of nascent Ultrabl-thorax (Ubx) transcripts as cells progress through the cell cycle.
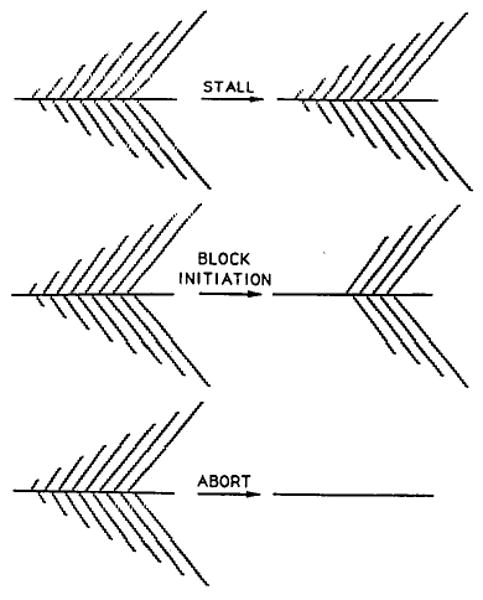
More than 30 years ago, measurement of incorporation of RNA precursors showed that there is little or no RNA synthesis during mitosis (Taylor, 1960; Prescott and Bender, 1962). This inhibition during mitosis appears to be general and occurs in Drosophila embryos (Edgar and Schubiger, 1986). We consider three possible mechanisms of inhibition (Figure 1). First, RNA polymerase might arrest or stall during mitosis and resume synthesis in the next cell cycle. Second, initiation of new RNA chains might be blocked, while engaged polymerases run off the gene during the period of inhibition. Third, RNA polymerase might abort transcription, and nascent transcripts dissociate from the template. Whether mitotic interruption of transcription has a severe or mild impact on expression of large transcripts depends on which of these mechanisms applies. According to the first and second mechanisms, a brief M phase would create only a transient lapse in expression, though according to the second mechanism the lapse would be delayed by the time of RNA polymerase transit of the gene. Only the third mechanism disrupts ongoing transcription and has a long-term consequence. Transcription would have to begin anew, so that a brief M phase will cause a gap in expression proportional to the size of a gene. In rapid cell cycles, mitosis could preclude expression of large genes by erasing nascent transcripts before their completion.
Figure 1. Alternative Models for the Inhibition of Transcription at Mitosis.
The Ubx gene of Drosophila encodes a homeotic function that specifies segmental identity (Lewis, 1978). Ubx is expressed in a pattern that parallels the positions in which it acts; there is predominant and early expression of Ubx in a region called parasegment 6 (PS6) and somewhat later and less extensive expression in the cells of PS5 and PS7–13 (e.g., Akam and Martinez-Arias, 1985). Importantly for this report, the Ubx gene product is expressed from a single promoter, and RNA polymerase requires about 55 min to traverse the 77 kb transcription unit (Figure 2). Additionally, the gene is transcriptionally active during two well-characterized and easily observed embryonic cell cycles (cycles 14 and 15).
Figure 2. Probes for the Ubx Transcription Unit.
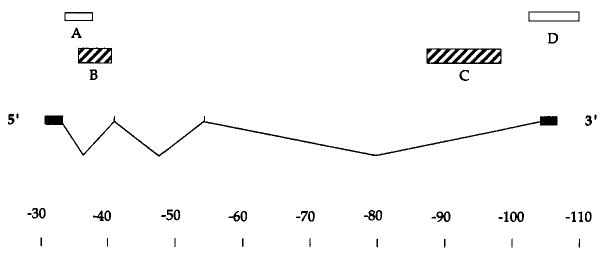
The positions of the four Ubx exons are indicated relative to a scale in kb. Only one of the alternative splicing patterns is indicated. The major mature transcripts of 3.2 and 4.3 kb, while differing in their inclusion of the internal exons, are derived from a roughly 77 kb primary transcript. The boxes above indicate the positions of probes used in this study. Unless indicated otherwise, probes described as 5′ and 3′ refer to probes B and C, respectively. Probe B is recessed from the 5′ end by 3.5 kb, and probe C falls about 10 kb short of the 3′ end. The 5′ ends of these probes are separated by 52 kb. This schematic of the Ubx transcription unit is based on O'Connor et al. (1988).
The first 13 mitotic cycles of Drosophila embryos are exceedingly rapid (as short as 8 min), synchronous, and occur without cell division. The synchronous mitosis 13, whose completion marks the beginning of cell cycle 14, produces about 5000 nuclei arranged in a monolayer at the periphery of the syncytial cytoplasm. Immediately, membranes begin to grow down between the nuclei, producing a cellular layer 50 min later. DNA synthesis begins immediately after mitosis 13 and is completed about 40 min into the cellularization process (McKnight and Miller, 1977; Edgar and Schubiger, 1986; Edgar and O'Farrell, 1989). Programmed variations in the lengths of the subsequent G2 lead to a detailed pattern of mitosis; cells in different locales divide together to create a mosaic of division domains (Foe, 1989; Edgar and O'Farrell, 1989).
Here we examine Ubx expression using a powerful in situ hybridization technique (Tautz and Pfeifle, 1989) that can detect the cluster of nascent transcripts on a gene as a dot of nuclear staining in the light microscope (O'Farrell et al., 1989). We show that Ubx transcription is interrupted by progression through mitosis 14 and mitosis 15 and that this interruption is due to abortion of nascent transcripts.
Results
Detection of Nascent Transcripts
The increased resolution provided by immunohistochemical detection of chemically tagged probes has revolutionized in situ hybridization (Tautz and Pfeifle, 1989). Local accumulation of specific transcripts within the nuclei of both mammalian and Drosophila cells is easily detected (Lawrence et al., 1989; O'Farrell et al., 1989). Several results suggested that dots of nuclear stain observed in Drosophila embryos represent clusters of nascent transcripts on the actively expressed gene. First, like the cytoplasmic signal, the dots were RNAase sensitive and were detected only with probes to sequences within transcription units (data not shown). Thus, the dots are due to RNA. Second, a string probe revealed two dots in normal diploid nuclei and only one dot in the nuclei of embryos heterozygous for a defective string gene (O'Farrell et al., 1989; B. A. Edgar, unpublished data). Thus, the dots are associated with the cognate genetic locus, and there is an independent accumulation of string RNA associated with each homolog. Third, nuclear dots were detected with both intron and exon probes. Thus, the dots include RNA sequences absent in the final processed transcript. Finally and most importantly, as reported here, one can follow the progress of RNA polymerase through a gene by using probes to different sections of a transcription unit. Following initiation of expression of a gene, dots are seen first with a probe to more 5′ sequences and only later, after the expected delay, with probes to more 3′ sequences. This discordance in the time of appearance of 5′ and 3′ signals shows that the dots are not signals from completed transcripts, but include a signal from intermediates in synthesis, i.e., nascent transcripts.
Most of our analysis has been with two Ubx probes, B and C. Although they are somewhat recessed from the ends of the transcription unit (Figure 2), these probes will be referred to as 5′ and 3′ probes, respectively. The 5′ boundaries of these probes are separated by 52 kb and they are complementary to intron sequences. The depth of penetration of cell membranes during cellularization is an accurate developmental clock (Merrill et al., 1988) that allowed us to time the initial detection of Ubx nascent transcripts. As shown by the sequence of embryos depicted in Figure 3A, expression in the region of PS6 was first detected with the 5′ probe about 5 min after the initiation of cell cycle 14. In contrast, expression was detected with the 3′ probe only when embryos had progressed at least 40 min into cycle 14. (Note that, because the images used in Figure 3A were optimized for staging of the embryos, the dots are not visible. At a higher magnification and more superficial focal plane, dots are detected [Figure 3B], and the signal from the dots paralleled the more diffuse signal seen in Figure 3A.)
Figure 3. Progression of Transcription along the Ubx Gene in Early Cycle 14.
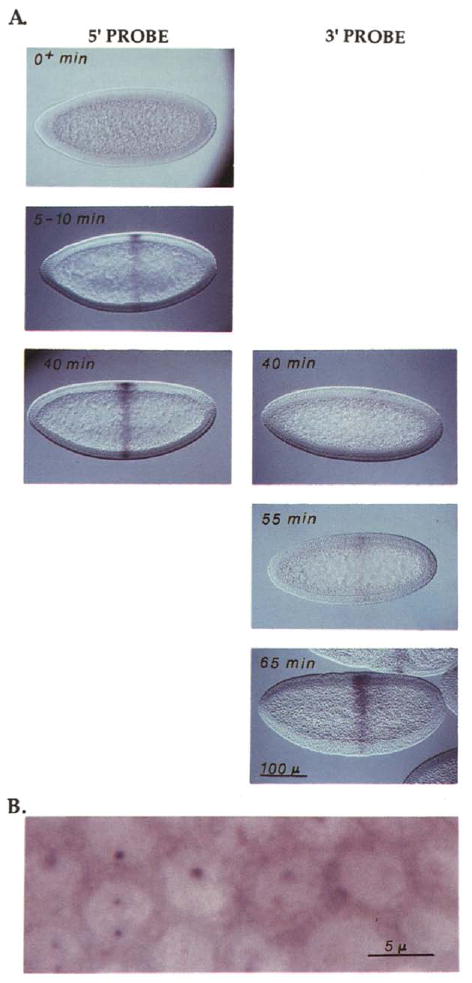
(A) Embryos where hybridized with either a 5′ or 3′ probe, stained, and then staged according to the depth of penetration of the membranes that invaginate between nuclei during cellularization. Based on live time-lapse records, the rate of this penetration is known, allowing translation of the depth of penetration into time after mitosis 13 (note that completion of mitosis 13 occurs at 130 min AED and marks the beginning of cell cycle 14). These times are given in the upper left of each panel, and the panels are arranged in a temporal sequence, top to bottom. Staining is evident earlier with the 5′ probe (left) than with the 3′ probe (right). The signal with the 5′ probe appears about 5 min after mitosis 13, while it is not until 40 min after mitosis 13 that a hint of staining is seen with the 3′ probe. The scale bar indicates about 100 μm.
(B) To visualize the nuclear dots, an early stage 8 embryo, comparable to those in (A), was hybridized with probe A and photographed at a higher magnification and at a superficial focal plane. Each cell with evident Ubx expression has two dots in its nucleus, although not all of these are evident at the focal plane of this picture. Diffuse cytoplasmic signal and a very weak diffuse nuclear signal are also evident. It should be noted that the disposition of the dots is not constant. At some stages only one dot is resolved, and often with Ubx probes the dots are found closely opposed to the nuclear membrane. The bar indicates approximately 5 μm.
Based on the physical distance between these probes and the time of appearance of the signals, we estimate a transcriptional elongation rate of about 1.4 kb/min and 55 min to traverse the entire Ubx transcription unit. Thummel et al. (1990) estimated a transcriptional elongation rate of 1.1 kb/min (± 30%) by measuring the lag time in the induction of sequences representing different intervals within a large ecdysone inducible transcription unit. Irvine et al. (1991) pointed out that the lag between Ubx promoter activation and the beginning of Ubx protein accumulation could be accounted for by a transcriptional elongation rate of 1.3 kb/min. These small differences are probably a reflection of the errors in these estimations, but real differences between genes or stages may exist. In our considerations of transcription times we use our estimate made in the embryo.
Abortion of Ubx Transcripts
Ubx expression becomes more complex as more posterior regions initiate expression (Figure 4d) and as the simple shell of the early embryo is distorted by gastrulation movements (Figures 4a–4c). To simplify the following description, we will focus on PS6 expression (hatched region in Figures 4a–4c and arrows in Figures 4d–4p). This simple stripe of expression crosses the dorsal ectoderm and ventral neurectoderm, which have distinctive times of mitosis. Most of the cells of the dorsal ectoderm initiate mitosis over a 15 min period from 205 to 220 min AED (time after egg deposition; 205 min AED corresponds to 75 min after M13) (Foe, 1989). This group of cells, domain 11 (Figures 4a–4c, stippled area), will exit mitosis about 7 min later (212 to 227 AED). Though the ventral neurectoderm region has a complex division pattern, it can be considered a late domain with respect to the dorsal ectoderm (mitosis occurs between 230 and 295 min AED).
Figure 4. Transient Absence of Ubx Nascent Transcripts in Recently Divided Cells.
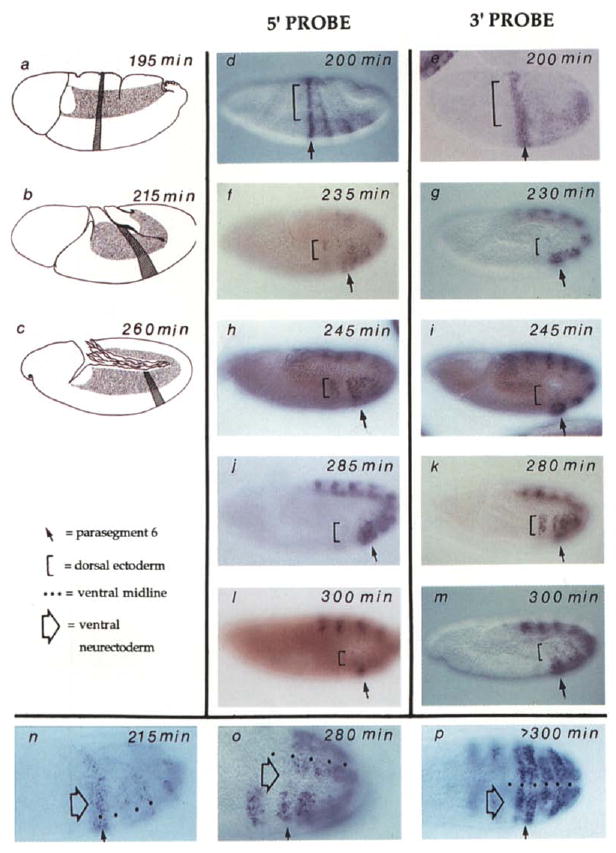
Wild-type embryos at various stages of cell cycle 14 and 15 are shown. In all panels except (n) to (p), anterior is left and ventral is down. The locations of PS6 (hatch stripe) and mitotic domain 11 (stippled region) are shown in the drawings on the left (a–c). These illustrate the shape changes during the movements of germband extension in which cells move ventrally to form the germband, which extends posteriorly, turns, and folds back upon itself. In the photographs, PS6 is indicated by an arrow and the dorsal ectoderm by a bracket (d–m) and a neurectoderm region just lateral to the ventral midline by a large open arrow (n–p). The age of the embryos in min AED was estimated by morphological criteria and progression of cell division domains (Campos-Ortega and Hartenstein, 1985; Foe, 1989) and is indicated on each panel. The embryos that are viewed from the side (d–m) are arranged according to age (top to bottom) so that staining with the 5′ probe (central column) can be compared to embryos at a similar stage stained with the 3′ probe (right column). In (d) and (e) are shown the patterns of Ubx expression as visualized about 10 to 15 min before division 14 begins in the dorsal ectoderm. Note that expression posterior to PS6 initiates later than PS6 expression (compare [d] to Figure 3) and is not yet evident with the 3′ probe in (e). Also, the embryo in (e) was flattened during slide preparation more than the other embryos in this display. In (f) and (g) are shown embryos shortly after completion of mitosis 14 in the dorsal ectoderm. No 3′ signal is seen (g) in this domain, while some heterogeneity (staining and nonstaining cells) is seen in this region with the 5′ probe. As discussed in the text, disappearance of signal is associated with progress through mitosis, and the heterogeneity of expression with the 5′ probe (f) is due to reappearance of signal in cells that have progressed further into cycle 15. Note that the brownish red color of the embryos in (f) and (I) appeared during observation under UV light and is not relevant to the hybridization signal. In (h) and (i) are shown embryos 20 min after mitosis 14 in the dorsal ectoderm. A strong signal is seen in this region with the 5′ probe but not with the 3′ probe. In (j) and (k) are shown embryos about 50 min after mitosis in the dorsal ectoderm. A strong signal is in the dorsal ectoderm with both 5′ and 3′ probes. Mitosis 15 has begun in some regions of the dorsal ectoderm of the embryo shown in (j) (fluorescence of Hoechst 33258–stained DNA allowed visualization of these mitoses; data not shown) and signal with the 5′ probe has been lost in some regions, notably in PSS. In (I) and (m) are shown embryos in which most of the cells of the dorsal ectoderm have progressed through mitosis 15 and signal is again absent from this region with both 5′ and 3′ probes. Note that the faint mottled expression in the dorsal ectoderm of the embryo in (m) is due to staining of cells in a focal plane below the ectoderm. Such nonectodermal cells divide according to a different schedule. In (n), (o), and (p) are shown embryos oriented so that the ventral region is visible. The dots mark the ventral midline. These embryos have been hybridized with the 3′ probe. At stages prior to mitosis 14 (n) and well after mitosis (p) the signal spans the ventral region of the embryo. In (o) there is an interruption in the signal in a region just lateral to the midline (large open arrow). This corresponds to mitotic domain N where mitosis has recently been completed. The signal is mottled in the more ventral region Oust adjacent to the midline marked by dots), which corresponds to mitotic domain M where division is in progress (some cells have divided and others have not).
Expression of Ubx was interrupted by progression through mitosis. This occurred first in the dorsal ectoderm where cells divide earlier (Figure 4g). Figure 4e shows that expression of Ubx in PS6 was detected with the 3′ probe prior to mitosis 14, and Figure 4g shows that this expression was selectively extinguished in dorsal ectoderm upon completion of mitosis in this region. The mitotic loss of signal was also seen with the 5′ probe (compare Figures 4d and 4f), but the signal reappeared more quickly. Spotty signal evident in postmitotic cells (Figure 4f) was the result of the heterogeneity of division times within this domain (about 15 min) and the short lag period (about 10 min) between disappearance of the 5′ signal and its reappearance (Figure 6 and data not shown). In slightly older embryos, strong expression was again evident with the 5′ probe, but not yet with the 3′ probe (compare Figures 4h and 4i). The 3′ signal finally reappeared when dorsal cells had progressed more than 30 min into cell cycle 15 (Figure 4k). The disappearance of signal suggests that nascent transcripts abort at mitosis, and the temporal sequence of reappearance of 5′ and 3′ signals suggests that expression starts anew in each cell cycle.
Figure 6. The Localized Nuclear Hybridization Signal Fades during the Course of Mitosis.
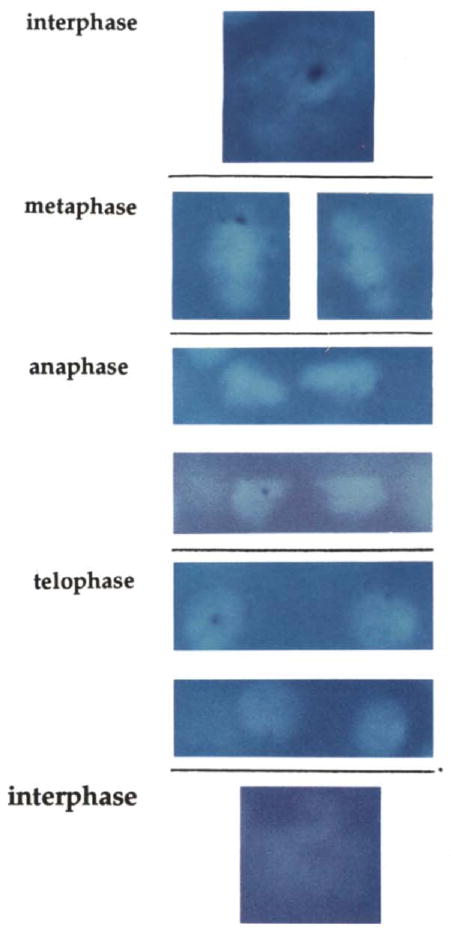
Embryos were hybridized with the 5′ probe and stained with Hoechst 33258. The bright fluorescence of the Hoechst shows the DNA, and the hybridization signal is evident as a dark blue dot. Individual cells from PS6 are shown at different stages of progression through mitosis 14. Although the hybridization signal in metaphase cells is strong, it is reduced in comparison to interphase 14 cells. Anaphase cells have a weak signal, and a signal can sometimes be detected in telophase cells and sometimes not. A pair of telophase nuclei showing a relatively strong signal is shown, as well as two other more representative nuclei that show a signal that is at the limit of detectability. At this level of sensitivity no signal is seen in cells that have just returned to an interphase configuration.
We characterized Ubx expression patterns throughout cycles 14 and 15 and found that nascent Ubx transcripts were lost whenever cells progressed through mitosis. For example, Figures 4l and 4m show the loss of hybridization signal in the dorsal ectoderm at mitosis 15, and Figures 4n–4p show loss of signal in ventral cells at the time of mitosis 14. Furthermore, in all cases reemergence of signal in postmitotic cells occurred first with the 5′, and then with the 3′ probe (data not shown).
A Requirement for Mitosis
To test whether there is a causal connection between progression through mitosis and Ubx transcript abortion, we examined Ubx transcripts in mutant embryos that fail to go through mitosis 14. In the absence of zygotic string function, the cell cycle is arrested in G2 of cycle 14 (Edgar and O'Farrell, 1989). Even though the cell cycle is blocked, other developmental events continue (Edgar and O'Farrell, 1989; Hartenstein and Posakony, 1990). In string mutant embryos the nuclear dots of Ubx hybridization first appear in cycle 14, as they do in wild type, but in the mutant the dots persist through stages when the wild-type embryos show mitotic interruption of expression (Figure 5). Thus, abortion of Ubx transcripts requires progression of the cell cycle beyond G2 of cell cycle 14.
Figure 5. Ubx Nascent Transcripts Persist When the Cell Cycle Is Arrested in Interphase.
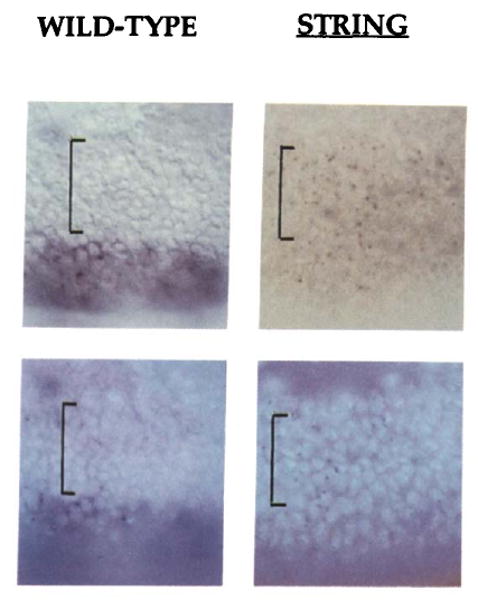
The surface of embryos in the area of PS6 is shown illuminated by transmitted light (top) or by fluorescence of the DNA-specific dye Hoechst 33258 (bottom). The hybridization signal from the 3′ Ubx probe is most easily seen in the upper images. In the wild-type embryo on the left, the dorsal ectoderm (region marked by the bracket) has recently divided and shows no signal, while the larger and as yet undivided cells of the ventral ectoderm show obvious expression. In the string mutant embryo on the right, although at the same stage as the wild-type embryo, there has been no cell division because string blocks the cell cycle in interphase 14. The larger size of the nuclei in the dorsal ectoderm of the string embryo can be seen in the fluorescent image. The dorsal cells of string mutant embryos show a localized nuclear hybridization signal. The low level of diffuse staining of the string mutant embryo is characteristic of later string embryos. At present we do not know why an intron probe should give cytoplasmic background (see text) in wild-type embryos (ventral region of embryo on left), nor why it is reduced in a string mutant embryo.
Nascent Transcripts during Mitosis
Although we had an a priori concern that nascent transcripts might not be detectable on mitotic chromosomes because of restricted accessibility to the condensed chromosomes, this was not the case. Figure 6 shows that hybridization signal was reduced but still detected on condensed mitotic chromosomes. There was a progressive decay in the signal intensity through metaphase, anaphase, and telophase. It should be noted that embryonic mitoses in Drosophila are exceptionally rapid, and the period over which we detected decay of the nuclear signal is about 5 min (early metaphase to late telophase; Minden et al., 1989; Hiraoka et al., 1990), a period during which RNA polymerase would be expected to progress less than 7 kb.
Discussion
We have found that nascent transcripts can be detected as dots of nuclear staining following in situ hybridization, and we suggest that this method may be widely useful in analyses of transcription. We have used this approach to demonstrate that nascent Ubx transcripts are aborted as cells progress through mitosis. Because the transcription of all genes is interrupted during mitosis (Taylor, 1960; Prescott and Bender, 1962), we suggest that this abortion of nascent transcripts is also general and affects all polymerase II transcripts. The mitotic abortion of transcripts is of particular interest because of its potential role in regulation.
Detecting Nascent Transcripts
It appears that in situ hybridization has a high sensitivity for detection of nascent transcripts. When transcripts are detected by Northern blots, heterogeneity in the length of nascent transcripts causes them to be dispersed during separation, while the completed products are concentrated in discrete bands. Consequently, this procedure is relatively insensitive for the detection of nascent transcripts. The situation is reversed for in situ hybridization. Now, the nascent transcripts are concentrated in a compact region around the gene, while the completed RNA products are usually dissipated throughout the cytoplasm.
We initially expected that nascent transcripts would be trace components. However, because of the length of time it takes to transcribe a large gene like Ubx, the levels of nascent intermediates are substantial. Furthermore, the completed Ubx transcript has a very short lifetime (A. W. Shermoen and P. H. O'Farrell, unpublished data), so that the majority of the life span of a Ubx transcript is occupied by its synthesis. In such cases, the nascent transcript can be more abundant than the mature product (see Experimental Procedures for a more quantitative consideration). While the properties of the Ubx transcript facilitate the detection of nascent transcripts, the technique has proven capable of detecting nascent transcripts from every gene that we have studied to date. This includes string, engrailed, Ubx, and Antennapedia, two of which (string and engrailed) have short transcription units. Consequently, we suggest that in situ hyridization may be widely useful for detection of nascent transcripts.
Transcript Abortion
In addition to nascent RNA, nuclear RNA includes intermediates in processing and transport. Since rates of processing, intron sequence degradation, and RNA export are all thought to be fast in comparison to the 55 min required for Ubx RNA synthesis, these intermediates are not likely to make a significant contribution to the nuclear hybridization signal. In any case, the contribution made by nascent transcripts can be identified by the discordance in the time of appearance of signal with probes to different positions in the transcription unit. We show that, upon initial activation of the Ubx gene in cell cycle 14, dots of nuclear hybridization appear first with a 5′ probe and only later, after a lag consistent with transcription rates, with the 3′ probe. Thus, the signal includes intermediates in transcript synthesis.
The disappearance of hybridization signal upon progression through mitosis occurs over a time period (about 5 min) far shorter than the time RNA polymerase takes to transit the gene (55 min). Thus, this disappearance is not due to inhibition of transcription at the promotor. Rather, the disappearance of signal at mitosis suggests that the nascent transcripts are dispersed, masked, or degraded. The sequential reemergence of a 5′ signal followed by a 3′ signal more than 30 min later argues strongly that the nascent transcripts were aborted. In models that do not incorporate abortion, the polymerases distributed throughout the gene would be expected to again synthesize RNA in the postmitotic cell, resulting in simultaneous synthesis of RNA sequences all along the transcription unit. On the other hand, the sequential appearance of the 5′ and 3′ signals is readily explained by a wave of newly initiated polymerases progressing through a gene previously stripped of active polymerase.
Pairing of Genes
The number of dots visualized has interesting implications regarding the organization of DNA in the nucleus. In G2 of cell cycle 14, we resolve at most two dots (observations with string, Ubx, and engrailed), even though these post-replicative cells have four copies of the gene. Since a heterozygote for a string mutation gives only one dot when hybridized with a string probe, each dot is presumed to represent one homolog (O'Farrell et al., 1989). In a G2 cell, there are two copies of each homolog, the sisters of replication. Since these sisters do not give resolvable signals we presume that they are closely juxtaposed. This is consistent with other observations (McKnight and Miller, 1977). However, a genetic interaction between homologs has also suggested that they are paired (Lewis, 1954; Jack and Judd, 1979), which predicts that we should see one dot. Preliminary observations (Y. Hiraoka and J. W. Sedat, personal communication; A. W. Shermoen and P. H. O'Farrell, unpublished data) suggest that homologous loci are not paired in very early embryos but might begin to pair during the postblastoderm divisions.
The Regulatory Consequences of Abortion of Nascent Transcripts
As a consequence of abortion of nascent transcripts at each mitosis, transcription of each gene will have to begin anew in the next cell cycle. Because of the lag involved in the production of complete transcripts, there will be an eclipse period during which no new transcripts will be completed. This eclipse period will be longer for larger genes, being equal to the time required to transcribe the gene.
Transcript abortion provides a means by which progression of the cell cycle can control and coordinate gene expression. Periodic abortion can alter the yield of completed products from a promotor and, in the extreme, can preclude expression of genes having transcription times longer than the cell cycle time. The extremely short cycle times of the cleavage stage Drosophila embryo will severely restrict the size of gene that can be transcribed. These cleavage divisions occur once every 8 min and have an interphase period of about 3.5 min. The maximum size of a gene that could be expressed in this short interphase would be about 5 kb, or even less, if the recovery of transcriptional competence after mitosis is not immediate. Genes such as the gap genes and pair-rule genes that are expressed and function prior to cell cycle 14 are relatively small. The sizes of their transcription units are tightly clustered around 2 kb. As the division cycle gradually gets longer, larger genes can be expressed (Foe and Alberts, 1983; Edgar et al., 1986). Perhaps this gradually changing cutoff serves a timing function that controls the onset of expression of larger transcripts.
In cycle 14, expression of the homeotic genes begins. These tend to be large, extending up to the 105 kb Antennapedia transcription unit, and have transcription times (up to 75 min) approximating the length of the postblastoderm cell cycles (30 to 180 min). Consequently, many of the transcripts that are initiated will be aborted, and the yield of completed transcript will depend on both the time during the cell cycle at which transcription is initiated and the length of the cell cycle. For example, some cells initiate Ubx expression only toward the middle of cell cycle 14 (e.g., in the more posterior parasegments), too late for transcription to reach the end of the gene prior to mitosis 14 in the dorsal ectoderm. As another example, during cell cycles 15 and 16, neuroblasts divide too rapidly (every 30 min) to complete the 77 kb Ubx transcript or the 105 kb Antennapedia transcript (estimated transcription times of 55 and 75 min, respectively). Consequently, abortion of transcription does modify the spatial pattern and level of accumulation of homeotic gene products (A. W. Shermoen and P. H. O'Farrell, unpublished data). It remains to be shown whether or not these modulations in expression influence morphogenesis. Nonetheless, given the importance of the spatial patterns of homeotic gene expression, it would be very surprising if transcript abortion did not influence development.
The Mechanism of Abortion
In string mutants the cell cycle is arrested in G2 of cycle 14, and Ubx transcript abortion does not occur. Thus, abortion requires progression of the cell cycle beyond G2. Furthermore, since some decay of the localized hybridization signals (dots) has occurred by metaphase, an event occurring between G2 and metaphase initiates the decay. The loss of nascent transcripts does not appear to be a direct physical result of chromosome condensation because the dots, though reduced in intensity, are still present in many metaphase cells. The hybridization signal disappeared gradually during metaphase, anaphase, and telophase. There are several possible explanations for this gradual decay of the signal. Perhaps the transcripts are abruptly terminated at a particular time during mitosis, but their dispersal or degradation is gradual. Alternatively, disassembly of a transcription complex may be a multistep process that is slow. Indeed, in normal termination of transcription, a provoking event, polymerase passage through a poly(A) addition site, results in downstream termination.
While we do not yet have any direct data identifying the basis for transcript abortion, it seems reasonable to consider the role of p34cdc2 kinase (or MPF). This kinase is found universally among eukaryotes, and its activation provokes mitosis (Lee and Nurse, 1987; Murray and Kirschner, 1989; Lehner and O'Farrell, 1990). We favor a model for the involvement of cdc2 kinase in which mitotic phosphorylation of either RNA polymerase or an accessory subunit destabilizes the transcription complex so that it becomes much less processive, resulting in a gradual termination of ongoing transcription over the course of a few minutes. The reported phosphorylation of the C-terminal domain of RNA polymerase by p34cdc2 is a candidate for this destabilizing reaction, but we think it is an unlikely candidate because of suggestions that phosphorylation of the C-terminal domain is involved in transcript initiation (Cisek and Corden, 1989; Payne et al., 1989).
Experimental Procedures
Fly Strains and DNA Constructs
Wild-type Drosophila embryos were from the Sevelen strain. The string mutant embryos were homozygous progeny produced by the stg7BI TM3, Ser stock.
Hybridization probes were prepared from cloned fragments for the Ubx gene. Probes A, B, C, and D were prepared from plasmids designated pφ3128, pφ3109, pφ3137, and pφ3144, respectively. These plasmids were kindly provided by Ken Irvine in David Hogness's laboratory and by Welcome Bender. In each case, inserts were isolated, digested with Alul, Haelll, and Sau3AI to reduce the fragment size, and random primed for incorporation of digoxigenin-conjugated dUTP (Boehringer Mannheim) according to Tautz and Pfeifle (1989) as modified by Edgar and O'Farrell (1990).
In Situ Hybridizations
Staining of digoxigenin-tagged hybrids was with alkaline phosphatase–conjugated anti-digoxigenin antibodies (Boehringer Mannheim). The staining procedure, which results in the precipitation of a dye, NBT, was usually for 12 to 14 hr at room temperature. Three hour staining produced lighter dots and none of the additional nuclear and cytoplasmic signal sometimes seen. It should be noted that the practice of observing the progress of the staining reaction in the dissecting microscope can be misleading. In examples where nuclear dots were very successfully and specifically stained with intron probes, the embryos appeared clear and unstained in the dissecting microscope. Apparently, the compact dark dot of staining was not resolved at low magnification. A dispersed signal is required for detection at low resolution. Thus, if staining reactions are terminated only after staining is visible in a dissecting microscope, the nuclear dots can be heavily overstained, and probes completely specific for nuclear dots will be dismissed as negative. For details of the staining procedure see Lehner and O'Farrell (1990).
The ages of embryos were based on a variety of cues: the depth of membrane invagination (Merrill et al., 1988), the domains of mitosis as detected in Hoechst 33258–stained embryos (Foe, 1989), and morphological features (Campos-Ortega and Hartenstein, 1985).
While the results argue that the dots we detect are nascent transcripts, there were two unexpected features of the data. First, contrary to expectations that intron sequences should be exclusively nuclear, probes to intron sequences gave a cytoplasmic signal in addition to the dots (Figure 3B). While it is not currently known what type of RNA product produced this cytoplasmic signal, it is clear that cytoplasmic accumulation is not a general feature of intron sequences. A probe to intron sequences in engrailed did not show substantial cytoplasmic signal (D. Moazed and P. H. O'Farrell, unpublished data). Second, despite the expectation that translocation of Ubx sequences to the cytoplasm would await completion of the transcripts, the 5′ probe, in addition to detecting nuclear dots, detected a faint cytoplasmic signal at stages prior to the appearance of a signal with the 3′ probe. This suggests that a fraction of the nascent transcripts terminate prematurely, or, in the case of intron sequences, that cotranscriptional processing releases sequences from the nascent transcript and from the nucleus (LeMaire and Thummel, 1990). One characterized cDNA includes some sequences from the first Ubx intron and appears to result from a shorter primary transcript (E. Gavis and D. S. Hogness, personal communication). While some of our observations may be explained by the expression of this RNA, we suspect that there may be additional Ubx transcripts.
Calculating Relative Abundance of Nascent Transcripts
To obtain a rough idea of the abundance of nascent transcripts, we used a few simplifying assumptions. We assumed that RNA polymerase progresses through a gene at a uniform rate and that the lifetime of intermediates in processing (known to have low abundance) would make a negligible contribution to the total lifetime of a transcript. Normal release of nascent transcripts by cleavage adjacent to a poly(A) addition site is taken to be the transition between nascent transcripts and completed transcripts. The lifetime of completed transcripts is presumed to be governed by an exponential decay process. The lifetimes of nascent transcripts should be the time required for RNA polymerase to transit the transcription unit, which is given by the length of the transcription unit (L; in kb) divided by the speed of polymerization (S; in kb/min). The average lifetime of the completed transcript is given by the inverse of the first order decay constant (k), which can be calculated from the half-life (k = In2/t½). The ratio of the number of nascent transcripts to completed transcripts (R) at steady state is given by the ratio of the lifetimes of nascent and completed transcripts (R = L × In2/S × t½; see Discussion). Using our estimated rate of polymerization (1.4 kb/min) this relationship is R = (0.5 × L/t½)(min × kb−1). It should be noted that R is a ratio of the number of chains and does not take into account the fact that nascent transcripts are by definition incomplete. In hybridization experiments it should be recalled that all nascent transcripts include the most 5′ sequences but that only a few of the nascent transcripts, those about to be completed, include 3′ sequences. Thus, the relative signal intensity from nascent and completed transcripts will depend on the position of the probe sequence within the transcription unit. Finally, the fraction of nascent transcripts that are detected with intron probes will be reduced if there is cotranscriptional processing (LeMaire and Thummel, 1990), but the signal from completed transcripts is also reduced because intron sequences generally have a shorter t½ than exon sequences. While these parameters are hard to include in calculations, we find that both intron and exon probes detect localized nuclear transcript, but that intron probes give less dispersed signal.
Acknowledgments
We thank Welcome Bender and members of the Hogness laboratory and particularly Ken Irvine for their helpful suggestions and for cloned fragments of the Ubx genes. We are indebted to Bruce Edgar and Danesh Moazed, whose earlier observations stimulated this study. We thank Shelagh Campbell, Jim Jaynes, Danesh Moazed, and Madhu Wahi for their comments on the manuscript and Frances Harrison for the drawings in Figure 4, and Judy Piccini for her assistance in the preparation of this manuscript. This work was supported by NIH grants PO1 HL43821 and RO1 GM37193.
References
- Akam M, Martinez-Arias A. The distribution of Ultrabithorax transcripts in Drosophila embryos. EMBO J. 1985;4:1687–1700. doi: 10.1002/j.1460-2075.1985.tb03838.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campos-Ortega JA, Hartenstein V. The Embryonic Development of Drosophila melanogaster. Berlin, Heidelberg, New York, Tokyo: Springer-Verlag; 1985. [Google Scholar]
- Cisek L, Corden J. Phosphorylation of RNA polymerase by the murine homologue of the cell-cycle control protein cdc2. Nature. 1989;339:679–684. doi: 10.1038/339679a0. [DOI] [PubMed] [Google Scholar]
- Edgar BA, O'Farrell PH. Genetic control of cell division patterns in the Drosophila embryo. Cell. 1989;57:177–187. doi: 10.1016/0092-8674(89)90183-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar BA, O'Farrell PH. The three postblastoderm cell cycles of Drosophila embryogenesis are regulated in G2 by string. Cell. 1990;62:469–480. doi: 10.1016/0092-8674(90)90012-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar BA, Schubiger G. Parameters controlling transcriptional activation during early Drosophila development. Cell. 1986;44:871–877. doi: 10.1016/0092-8674(86)90009-7. [DOI] [PubMed] [Google Scholar]
- Edgar BA, Kiehle CP, Schubiger G. Cell cycle control by the nucleo-cytoplasmic ratio in early Drosophila development. Cell. 1986;44:365–372. doi: 10.1016/0092-8674(86)90771-3. [DOI] [PubMed] [Google Scholar]
- Foe VE. Mitotic domains reveal early commitment of cells in Drosophila embryos. Development. 1989;107:1–22. [PubMed] [Google Scholar]
- Foe VE, Alberts BM. Studies of nuclear and cytoplasmic behavior during the five mitotic cycles that precede gastrulation in Drosophila embryogenesis. J Cell Sci. 1983;61:31–70. doi: 10.1242/jcs.61.1.31. [DOI] [PubMed] [Google Scholar]
- Hartenstein V, Posakony J. Sensillum development in the absence of cell division: the sensillum phenotype of the Drosophila mutant string. Dev Biol. 1990;138:147–158. doi: 10.1016/0012-1606(90)90184-k. [DOI] [PubMed] [Google Scholar]
- Hiraoka Y, Agard DA, Sedat JW. Temporal and spatial coordination of chromosome movement, spindle formation and nuclear envelope breakdown during prometaphase in Drosophila melanogaster embryos. J Cell Biol. 1990;111:2815–2828. doi: 10.1083/jcb.111.6.2815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irvine KD, Helfand SL, Hogness DS. The large upstream control region of the Drosophila homeotic gene Ultrabithorax. Development. 1991;111:407–424. doi: 10.1242/dev.111.2.407. [DOI] [PubMed] [Google Scholar]
- Jack JW, Judd BH. Allelic pairing and gene regulation: a model for the zeste–white interaction in Drosophila melanogaster. Proc Natl Acad Sci USA. 1979;76:1368–1372. doi: 10.1073/pnas.76.3.1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence JB, Singer RH, Marselle LM. Highly localized tracks of specific transcripts within interphase nuclei visualized by in situ hybridization. Cell. 1989;57:493–502. doi: 10.1016/0092-8674(89)90924-0. [DOI] [PubMed] [Google Scholar]
- Lee M, Nurse P. Cell cycle control genes in fission yeast and mammalian cells. Nature. 1987;327:287–290. doi: 10.1016/0168-9525(88)90171-0. [DOI] [PubMed] [Google Scholar]
- Lehner CF, O'Farrell PH. Drosophila cdc2 homologs: a functional homolog is co-expressed with a cognate variant. EMBO J. 1990;9:3573–3581. doi: 10.1002/j.1460-2075.1990.tb07568.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeMaire MF, Thummel CS. Splicing precedes polyadenylation during Drosophila E74A transcription. Mol Cell Biol. 1990;10:6059–6063. doi: 10.1128/mcb.10.11.6059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis EB. The theory and application of a new method of detecting chromosomal rearrangements in Drosophila melanogaster. Am Nat. 1954;88:225–239. [Google Scholar]
- Lewis EB. A gene complex controlling segmentation in Drosophila. Nature. 1978;276:565–570. doi: 10.1038/276565a0. [DOI] [PubMed] [Google Scholar]
- McKnight SL, Miller OJ., Jr Electron microscope analysis of chromatin replication in the cellular blastoderm Drosophila melanogaster embryo. Cell. 1977;12:795–804. doi: 10.1016/0092-8674(77)90278-1. [DOI] [PubMed] [Google Scholar]
- Merrill P, Sweeton D, Wieschaus E. Requirements for autosomal gene activity during precellular stages of Drosophila melanogaster. Development. 1988;104:495–509. doi: 10.1242/dev.104.3.495. [DOI] [PubMed] [Google Scholar]
- Minden JS, Agard DA, Sedat JW, Alberts BM. Direct cell lineage analysis in Drosophila melanogaster by time-lapse, three-dimensional optical microscopy of living embryos. J Cell Biol. 1989;109:505–516. doi: 10.1083/jcb.109.2.505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray A, Kirschner M. Cyclin synthesis drives the early embryonic cell cycle. Nature. 1989;339:280–286. doi: 10.1038/339275a0. [DOI] [PubMed] [Google Scholar]
- O'Connor M, Binari R, Perkins L, Bender W. Alternative RNA products from the Ultrabithorax domain of the bithorax complex. EMBO J. 1988;7:435–445. doi: 10.1002/j.1460-2075.1988.tb02831.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Farrell PH, Edgar BA, Lakich D, Lehner C. Directing cell division during development. Science. 1989;246:635–640. doi: 10.1126/science.2683080. [DOI] [PubMed] [Google Scholar]
- Payne J, Laybourn P, Dahmus M. The transition of RNA polymerase II from initiation to elongation is associated with phosphorylation of the carboxy-terminal domain of subunit IIa. J Biol Chem. 1989;264:19621–19629. [PubMed] [Google Scholar]
- Prescott D, Bender M. Synthesis of RNA and protein during mitosis in mammalian tissue culture cells. Exp Cell Res. 1962;26:260–268. doi: 10.1016/0014-4827(62)90176-3. [DOI] [PubMed] [Google Scholar]
- Tautz D, Pfeifle C. A non-radioactive in situ hybridization method for the localization of specific RNAs in Drosophila embryos reveals translation control of the segmentation gene hunchback. Chromosoma. 1989;98:81–85. doi: 10.1007/BF00291041. [DOI] [PubMed] [Google Scholar]
- Taylor J. Nucleic acid synthesis in relation to the cell division cycle. Ann NY Acad Sci. 1960;90:409–421. doi: 10.1111/j.1749-6632.1960.tb23259.x. [DOI] [PubMed] [Google Scholar]
- Thummel CS, Burtis KC, Hogness DS. Spatial and temporal patterns of E74 transcription during Drosophila development. Cell. 1990;61:101–111. doi: 10.1016/0092-8674(90)90218-4. [DOI] [PubMed] [Google Scholar]


