Abstract
γ-Herpesviruses (γHVs), including important human pathogens such as Epstein Barr virus, Kaposi’s sarcoma-associated HV, and the murine γHV68, encode homologs of the antiapoptotic, cellular Bcl-2 (cBcl-2) to promote viral replication and pathogenesis. The precise molecular details by which these proteins function in viral infection are poorly understood. Autophagy, a lysosomal degradation pathway, is inhibited by the interaction of cBcl-2s with a key autophagy effector, Beclin 1, and can also be inhibited by γHV Bcl-2s. Here we investigate the γHV68 M11-Beclin 1 interaction in atomic detail, using biochemical and structural approaches. We show that the Beclin 1 BH3 domain is the primary determinant of binding to M11 and other Bcl-2s, and this domain binds in a hydrophobic groove on M11, reminiscent of the binding of different BH3 domains to other Bcl-2s. Unexpectedly, regions outside of, but contiguous with, the Beclin 1 BH3 domain also contribute to this interaction. We find that M11 binds to Beclin 1 more strongly than do KSHV Bcl-2 or cBcl-2. Further, the differential affinity of M11 for different BH3 domains is caused by subtle, yet significant, variations in the atomic details of each interaction. Consistent with our structural analysis, we find that Beclin 1 residues L116 and F123, and M11 residue pairs G86 + R87 and Y60 + L74, are required for M11 to bind to Beclin 1 and downregulate autophagy. Thus, our results suggest that M11 inhibits autophagy through a mechanism that involves the binding of the Beclin 1 BH3 domain in the M11 hydrophobic surface groove.
Keywords: γ-herpesvirus, Beclin 1, Bcl-2, viral Bcl-2, autophagy, apoptosis
Introduction
Murine γ-Herpesvirus (γHV) 68 provides a model system for important human pathogens like Epstein Barr virus and Kaposi’s sarcoma-associated HV (KSHV). γHVs express many cellular protein mimics, including homologs of cellular B-cell lymphoma protein-2 (cBcl-2), the first protein shown to be oncogenic by blocking cell death rather than by increasing cellular proliferation.1–4 γHV Bcl-2s also have an antiapoptotic role, are maximally expressed immediately postinfection,5 are critical for viral reactivation from latency and persistent replication in immunocompromised hosts,1,6–15 and may increase the likelihood of oncogenic transformation of infected cells.5,16 The γHV68 Bcl-2 homolog, M11, inhibits Fas-, TNFα–and Sindbis virus infection-induced apoptosis,17 is important for the induction of atypical lymphoid hyperplasia and lymphoproliferative disease,16 and is the only γHV Bcl-2 known to play a role during infection in vivo.1
It has previously been shown that autophagy is inhibited by cBcl-2s and KSHV Bcl-2.18,19 While this manuscript was in preparation, it was shown that γHV68 M11 also inhibits autophagy.20 Autophagy, the primary intracellular pathway by which long-lived proteins, organelles, and intracellular pathogens undergo lysosomal degradation after sequestration in multi-membrane cytoplasmic vesicles called autophagosomes, is essential for survival during cellular and organismal stress.21,22 Disruption of autophagy is implicated in neurodegenerative disorders, muscular diseases, cardiomyopathy, cancer and infectious diseases.22–24 Many successful intracellular pathogens have evolved to evade and even exploit the autophagic machinery to promote their replication and survival.25–29
In the last decade many conserved autophagy effectors called Atg proteins, as well as major autophagy regulators, have been identified and their roles outlined.21,30 Bcl-2 interacting, coiled-coil protein (Beclin 1), a haploinsufficient tumor suppressor31–35 and one of the first mammalian autophagy proteins identified, shares 30% sequence identity with yeast Atg6 and is an essential component of the Class III phosphatidylinositol 3-kinase (PI3Kc3) complex, responsible for the nucleation of autophagosomes.31–37 In mammalian cells, ER-targeted Bcl-2 inhibits Beclin 1-dependent Class III PI3Kc3 activity, autophagy and autophagic type II programmed cell death caused by excess autophagy.18 Beclin 1 residues 88–140 are sufficient for binding Bcl-2;18,37 within this region, recent structural, mutagenic, and biochemical data show that residues 105–128 constitute a BH3 domain, the primary determinant for binding Bcl-2s.19,38,39 γHV Bcl-2s also directly inhibit Beclin 1-dependent autophagy, which, in theory, might help γHVs escape autophagic degradation and/or also facilitate the malignant transformation of cells.18
Here we use a wide range of biochemical and structural methods to investigate the interaction between γHV68 M11 and Beclin 1 in atomic detail. We find that the Beclin 1 BH3 domain binds in a hydrophobic groove on M11; but unexpectedly, regions outside the BH3 domain also assist in this interaction. Although M11 and other Bcl-2s are all expected to bind BH3 domains in a similar manner, there are subtle, yet significant differences in the interaction of M11 with BH3 domains from different proteins, leading to different binding affinities. We find that consistent with our structural analysis, Beclin 1 residues L116 and F123, as well as M11 residue pairs G86 + R87 and Y60 + L74, are essential for the binding of M11 to Beclin 1, and also for the downregulation of Beclin 1-dependent autophagy by M11. Together, these results show that the Beclin 1 BH3 domain is necessary and sufficient for binding to γHV68 M11, and this interaction results in the inhibition of autophagy, which may be an important physiological function of M11.
Results
The Beclin 1 BH3 domain is sufficient for binding to Bcl-2s
Bcl-2s inhibit autophagy via interaction with the essential autophagy effector protein, Beclin 1.18,19 Human and murine Beclin 1 are 98% identical. Within the first 135 residues of human Beclin 1, there are only two additional residues and four substitutions compared to mouse Beclin 1, while the BH3 domains (residues 105–130 for human Beclin 1) are identical. Hence, residue numbers correspond to human Beclin 1, which was used in this study.
Isothermal titration calorimetry (ITC) was used to quantify the binding affinities of BH3 domain-containing molecules to Bcl-2s [Supplementary Information (SI) Fig. 1– Fig 3]. As expected from their divergent amino acid composition, the different γHV Bcl-2s and cBcl-2s bind each of the BH3 domain-containing molecules with varying affinities (Table 1, SI Table 1).40,41 The Beclin 1 (1–135) fragment binds to γHV68 M11 with approximately 40-fold higher affinity than cBcl-2 and 65-fold higher affinity than KSHV Bcl-2 (Table 1). We find that the isolated Beclin 1 BH3 domain binds to M11 with about ten-fold higher affinity than to KSHV Bcl-2 or cBcl-2 (Table 1), but similar to Bcl-XL which binds with an affinity of 1.1 µM.38 Thus, compared to the Beclin 1 BH3 domain, the 1–135 fragment binds approximately five-fold more strongly to M11, but does not bind significantly better to either cBcl-2 or KSHV Bcl-2. Beclin 1 residues 1–105 may increase affinity for M11 by directly contributing additional interactions with M11, or by increasing the helicity of the BH3 domain, as our circular dichroism results (data not shown) show that the isolated Beclin 1 BH3 domain is unstructured in solution, although, like other BH3 domains, we and others20,38,39 find that it binds as a helix to Bcl-2s.
Table 1.
Binding of different Bcl-2 homologs to BH3 domain-containing fragments from Beclin 1 and two proapoptotic proteins
| Bcl-2 homolog | Beclin 1 (1–135) | BH3 domain from: | ||
|---|---|---|---|---|
| Beclin 1 | BAD | BAD | ||
| γHV68 M11 | 0.2 ± 0.1 | 1.1 ± 0.3 | 33.1 ± 1.4 | 4.0 ± 0.4 |
| KSHV Bcl-2 | 54.2 ± 14.3 | 13.3 ± 2.3 | 3.942 | 0.9842 |
| Bcl-2 | 8.6 ± 1.2 | 8.0 ± 0.2 | 0.01642 | 23.6 ± 5.7 |
Kd are in µM. Results shown are the average of three independent experiments ± s. d.
Autophagy inhibition may be an important function of γHV68 M11
A comparison of the binding of the Beclin 1 BH3 domain to various Bcl-2s, based on our studies with KSHV Bcl-2 and cBcl-2 (Table 1) and a previously published study with Bcl-XL,38 demonstrates that M11 binds more tightly to the Beclin 1 BH3 domain than do other Bcl-2s, with the exception of Bcl-XL. Additionally, as discussed above, residues N-terminal of the BH3 domain further assist in binding to M11 but not to cBcl-2 or KSHV Bcl-2 (Table 1) or Bcl-XL.38 Recent data published while this manuscript was in preparation, show that Beclin 1 residues C-terminal of the BH3 domain also improve binding to M11, but only slightly for cBcl-2,20 while previous results have shown that these residues do not assist in binding to Bcl-XL.38
Our results (Table 1), and other recently published results,20 demonstrate that M11 binds isolated BH3 domains from Beclin 1 and from most proapoptotic proteins within a ten-fold range of sub-micromolar affinities (SI Table 1). In contrast, previous studies have shown that cBcl 2, Bcl-XL, as well as KSHV Bcl-2, bind at least one pro-apoptotic protein BH3 domain substantially more tightly than Beclin 1 (SI Table 1).38, 40, 42, 43 Comparison of these binding data suggests that while many Bcl-2 family members preferentially target apoptosis versus autophagy proteins, M11 likely targets both autophagy and apoptosis proteins equally.
Molecular basis of the M11-Beclin 1 BH3 domain interaction
M11 and other antiapoptotic Bcl-2s share little sequence identity and only the BH1 domain is conserved in M11. However, they have a common three-dimensional structure consisting of a central hydrophobic α-helix surrounded by six amphipathic helices, delineated into four Bcl-2 homology (BH) domains: BH4, BH3, BH1 and BH2.8,44 Proapoptotic Bcl-2 proteins are either “BH3-only proteins” containing a single BH3 domain or “Bax-like proteins” that contain three BH domains, BH4, BH3 and BH1. In both these groups, the BH3 domain is required for interaction with anti-apoptotic Bcl-2 proteins. Typically, BH3 domains of proapoptotic proteins are four-turn amphipathic α-helices, bearing the sequence motif: Hy-X-X-X-Hy-X-X-X-Sm-D/E-X-Hy, where Hy are hydrophobic residues and Sm represents small residues, typically glycine.45 Recently it was shown that Beclin 1 residues 105–127 constitute a BH3 domain.19,38,39
Our 2.5 Å crystal structure of the M11-Beclin 1 BH3 domain complex (Fig. 1) shows that the Beclin 1 BH3 domain binds to a hydrophobic surface groove on M11, similar to the binding of BH3 domains of proapoptotic proteins to other antiapoptotic Bcl-2s.8,42,43,46–48 Although the Beclin 1 BH3 domain binds with similar affinities to M11 (Table 1) and Bcl-XL,38 binding to M11 buries about 1883 Å2 at the interface and involves 12 Beclin 1 residues (Fig. 1A–C), while binding to Bcl-XL buries 2078 Å2 and involves 14 Beclin 1 residues (Fig. 1D). The amphipathic Beclin 1 BH3 domain binds such that the hydrophobic face of the BH3 domain is buried in the hydrophobic binding groove on the M11 surface (Fig. 1C). Both hydrophobic and polar interactions contribute to the M11-Beclin 1 BH3 domain interface (Fig. 1A–C). Beclin 1 residues L116 and G120 are completely buried, while residues L112 and F123 are partially buried. However, due to its larger side chain, F123 probably contributes more than L112 to the interaction with M11. Further, residue L112 is more solvent exposed in the complex with M11 than in the complex with Bcl-XL, suggesting that interactions with this residue may play a more important role in binding to Bcl-XL. Of the polar interactions, perhaps the most significant is a bidentate salt bridge between the Oδ1 and Oδ2 of Beclin 1 D121, a conserved acidic residue in all BH3 domains, and Nε and Nη of M11 R87, which is invariant amongst Bcl-2s (Fig. 1C). Beclin 1 G120 and M11 G86 are packed in an anti-parallel manner against the M11 G86-R87 and G120-D121 backbones, respectively. The absence of a large side chain at the glycine positions is essential to avoid a steric conflict, but superposition of the Beclin 1 BH3 domain-bound M11 and Bcl-XL structures indicates that these interactions are less well packed in the M11-Beclin 1 BH3 domain interface compared to the Bcl-XL-Beclin 1 BH3 domain interface.
Figure 1.
The Beclin 1 BH3 domain bound to M11 or Bcl-XL. The Beclin 1 BH3 domain is rendered in teal, while residues involved in binding to M11 or Bcl-XL are shown in atomic detail. All molecular figures are presented in superimposable orientations and were prepared with the program PYMOL (http://www.pymol.org). Molecular surfaces of M11 and Bcl- XL are colored by atom type—oxygen red, nitrogen blue, sulfur green and carbon grey. (A) M11 residues that interact with the Beclin 1 BH3 domain are labeled. (B) The Beclin 1 BH3 domain. Residues involved in binding to M11 are labeled. (C and D) Interaction of the Beclin 1 BH3 domain with (C) M11 and (D) Bcl-XL. Additional Beclin 1 residues involved in binding Bcl-XL are labeled. (E and F) Conformational changes between the free (magenta) and Beclin 1-bound states (gray) of (E) M11 and (F) Bcl-XL. For M11, secondary structure elements are labeled and large scale changes in conformation are indicated by an arrow.
Conformational changes that widen the M11 hydrophobic surface groove are required to accommodate the Beclin 1 BH3 helix (Fig. 1E). The most significant of these changes involve M11 residues comprising the α2–α3 loop and α3, which may be shifted by up to 10 Å (Fig. 1E). Further, M11 residues 53–55 undergo a secondary structure change from coil to helix, adding an additional turn to α2 (Fig. 1E). Binding of BH3 domains from Beclin 1 and other proteins have also been shown to induce conformational changes in Bcl-XL, which also result in the widening of the BH3-binding groove38,39,46–48 (Fig. 1F).
Comparison of the binding of different BH3 domains to M11
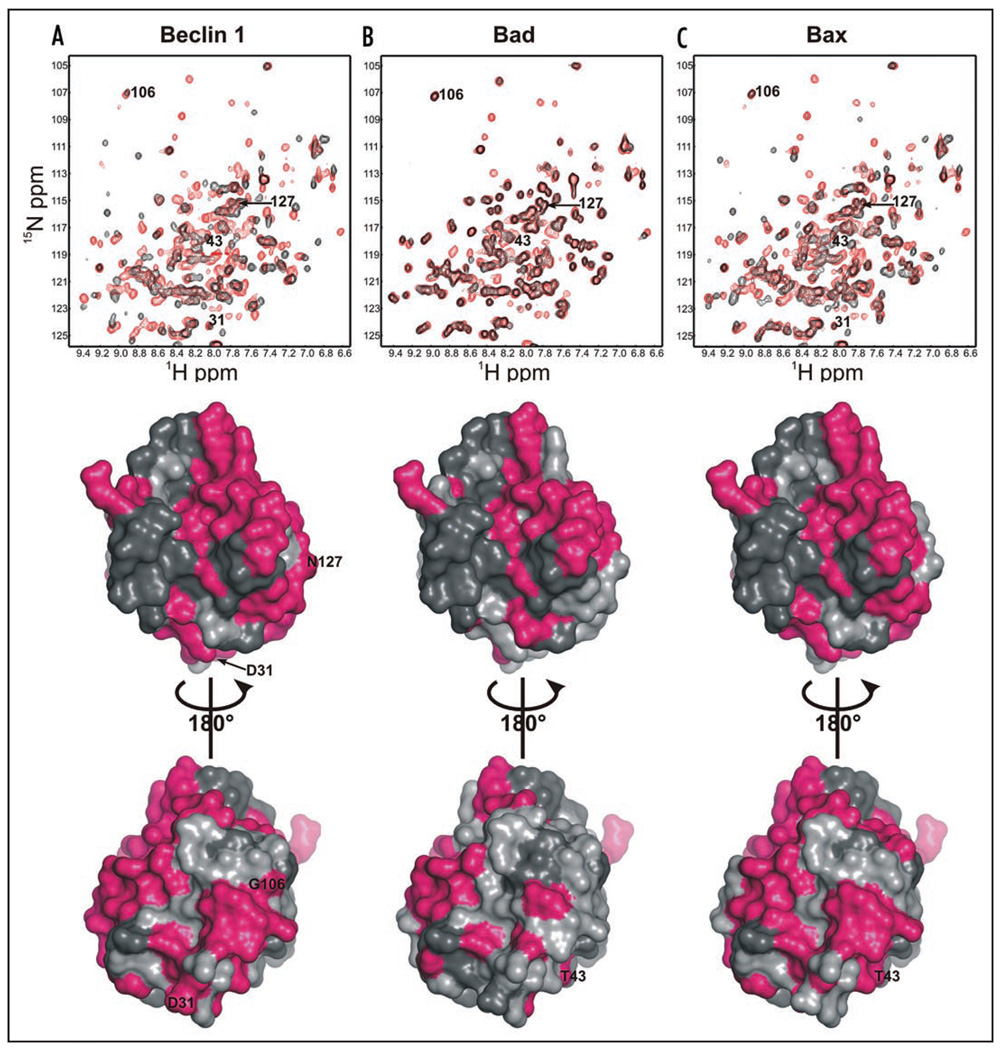
M11 residues affected by binding of BH3 domains from Beclin 1, Bax and Bad were identified by measuring 1H/15N HSQC spectra after titration of peptides corresponding to each BH3 domain (Fig. 2). Analysis of these NMR spectra showed that binding of the Beclin 1 BH3 domain affects extensive regions of M11, including not only residues of the hydrophobic groove and those that undergo conformational changes, but also residues from other regions of M11 (Fig. 2A). Similar widespread effects were seen upon binding of BH3 domains from Bad (Fig. 2B) and Bax (Fig. 2C). However, consistent with the binding affinities of these BH3 peptides to M11 (Table 1), binding of Bad appears to involve the fewest M11 residues as only 46 residues are affected by binding, while binding of Beclin 1 and Bax involves many more M11 residues, with 77 and 76 residues respectively being affected by the interaction (Fig. 2 and SI Table 4).
Figure 2.
M11 residues affected by binding of selected BH3 domains from (A) Beclin 1 (B) Bad and (C) Bax. Each of the upper panels shows superimposed 1H/15N HSQC spectra with red contours corresponding to the spectrum of apo-M11, and black contours corresponding to the spectrum of M11 bound to the BH3 domain indicated. The middle and lower panels display the M11 molecular surface in gray, with dark gray indicating residues with unassigned backbone 1H and 15N chemical shifts, and magenta showing residues affected by binding of the BH3 domain indicated. The middle panels show the molecular surfaces in the same orientation as in Figure 1, while in the lower panels the molecules are rotated by 180° about the Y-axis relative to the middle panels.
The BH3 domains of Beclin 1, Bax and Bad are all expected to bind to the hydrophobic surface groove of M11, and consistent with this, many residues that line this groove, R57, D59, K68, R69, D70, L74, T76, S77, L78, F79, V80, D81, V82, G86, R87, G90 and V94, are affected by the titration of each one of these BH3 domains. Consistent with the measured binding affinities for the BH3 domains of Beclin 1, Bax and Bad (Table 1), binding of the Bad BH3 domain involves the fewest additional residues within the hydrophobic groove of M11 (Fig. 2). Although each BH3 domain has a distinct binding footprint on the M11 molecular surface (Fig. 2), no unique residue within the M11 BH3-binding groove is affected by the binding of only one BH3 domain. However, of residues outside the BH3 domain, D31, G106 and N127, are affected only by the binding of the Beclin 1 BH3 domain, but not by BH3 domains from Bad or Bax, while another residue, T43, is affected by binding of BH3 domains from Bad and Bax, but not from Beclin 1 (Fig. 2). Thus, it is likely that the observed binding affinities and specificities of BH3 domains from different proteins are a consequence of both surface area buried upon binding, as well as shape and electrostatic complementarity of the interacting surfaces.
Mutations at the M11-Beclin 1 BH3 domain interface affect M11-Beclin 1 binding
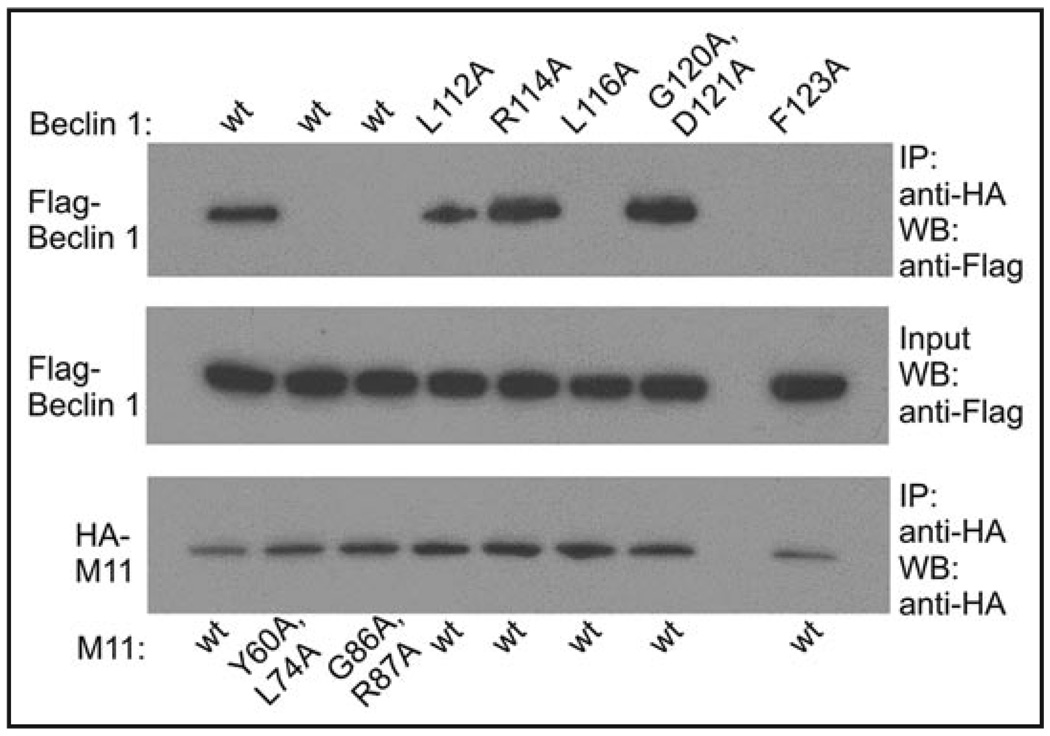
The role of key residues involved in the M11-Beclin 1 interaction identified from our structural analysis was assessed by coimmunprecipitation (CoIP) from mammalian COS7 cells transiently transfected with M11 or Beclin 1 mutants (Fig. 3). Beclin 1 residues tested include L112, L116, G120, D121 and F123. As expected wild-type (wt) M11 did not CoIP significantly different levels of wt Beclin 1 and R114A Beclin 1, a solvent-exposed residue not involved in binding to M11, used as a control (Fig. 3). In contrast, M11 CoIPed undetectable levels of L116A and F123A Beclin 1, but only marginally reduced levels of L112A Beclin 1 (Fig. 3). This confirmed that all three residues are involved in the interaction with M11, but L116 and F123 play more important roles than L112, consistent with our structural analysis. Although a G120E + D121E Beclin 1 double mutant abrogates binding to Bcl-XL, M11 CoIPed a G120A + D121A Beclin 1 double mutant at wt levels (Fig. 3), indicating that these conserved residues were not as important for binding to M11 as to Bcl-XL,38 partly consistent with our structural analysis which suggests that this region is less well-packed in the M11-Beclin 1 interface.
Figure 3.
Co-immunoprecipitation of Flag epitope-tagged Beclin 1 constructs with HA-tagged M11 constructs in COS7 cells.
Interestingly, we find that double mutation of the complementary conserved M11 residues, G86A and R87A, abrogate CoIP of wt Beclin 1 (Fig. 3). A S85A + G86A + R87A triple M11 mutation has been shown to abolish binding to certain proapoptotic BH3-only proteins,8 and more recently to Beclin 1.20 However, our structural analysis suggests that M11 S85 does not directly participate in the interface with Beclin 1, and these CoIPs confirm that mutation of G86 and R87 is sufficient to abolish binding to Beclin 1. Since our CoIP results for the G120A + D121A Beclin 1 double mutant indicate that the bidentate salt bridge between Beclin 1 D121-M11 R87 is not essential for the Beclin 1-M11 interaction, perhaps the abrogation of binding by the G86A + R87A double mutant is largely the effect of the G86A mutation. Finally, we find that a Y60A + L74A M11 double mutant, which affects the BH3 domain-binding M11 groove, also reduces CoIP of wt Beclin 1 to undetectable levels (Fig. 3). Together, the CoIP data suggest that hydrophobic interactions may be more important than electrostatic interactions for binding of M11 to Beclin 1.
M11-Beclin 1 BH3 domain binding is required for autophagy inhibition by M11
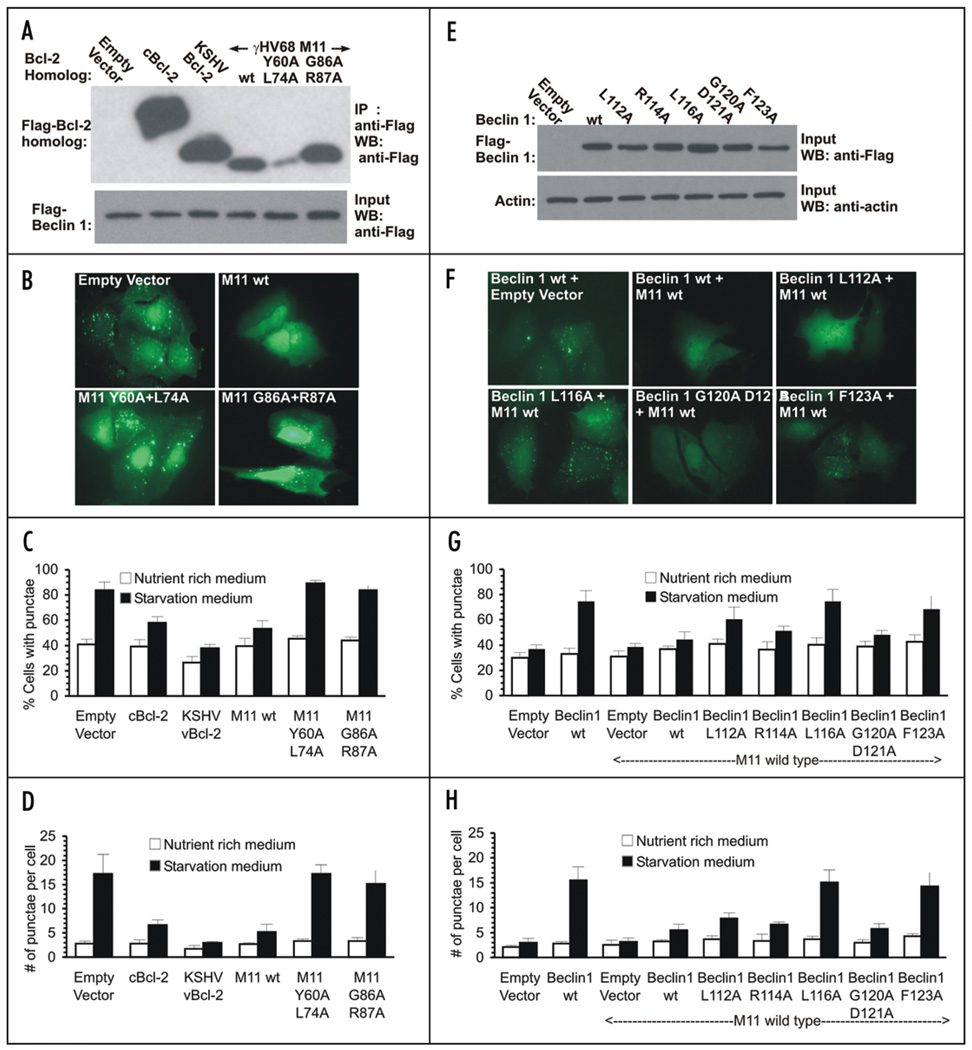
Autophagy levels were monitored by assaying the change in cellular localization of the transiently-expressed specific mammalian autophagy marker, GFP-tagged LC3 (GFP-LC3), from a diffuse cytoplasmic distribution to localized puncta corresponding to autophagosomal structures (Fig. 4, SI Fig. 5). Although GFP-LC3 levels per se do not reflect autophagic flux, only steady-state levels of autophagosomes, previous studies from our laboratory and others have utilized several different methods to demonstrate that Bcl-2s inhibit autophagic flux.18,19 Autophagy was monitored in human MCF7 breast carcinoma cells, which express very low levels of Beclin 1 and do not show starvation-induced increases in autophagy unless Beclin 1 is ectopically expressed.18,31,49,50 The effect of different Bcl-2s and M11 mutants was assayed after transient coexpression of either KSHV Bcl-2, cBcl-2, γHV68 M11 and M11 mutants in MCF7 cells stably transfected with beclin 1 (MCF7.beclin 1 cells) (Fig. 4A), whereas the effect of Beclin 1 mutants was assayed in MCF7 cells after transient coexpression of wt M11, and either wt or mutant Beclin 1 (Fig. 4D).
Figure 4.
The M11-Beclin 1 interaction is essential for the downregulation of autophagy by M11. (A) Expression of cBcl-2, KSHV Bcl-2, γHV68 M11 and M11 mutants in MCF7.beclin 1 cells. (B) Representative images of GFP-LC3 staining in MCF7.beclin 1 cells cotransfected with GFP-LC3 and the indicated plasmid. (C) Light microscopic quantification of the percentage of GFP-positive MCF7.beclin 1 cells with discrete puncta, co-transfected with GFP-LC3 and the Bcl-2 homolog or mutant indicated below the x-axis. (D) Light microscopic quantification of the number of discrete GFP-LC3 puncta per cell in GFP-positive MCF7.beclin 1 cells co-transfected with GFP-LC3 and the Bcl-2 homolog or mutant indicated below the x-axis. (E) Expression of Flag-tagged wt and mutant Beclin 1 constructs in MCF7 cells. Wt M11 was expressed at uniform levels in cells co-transfected with the M11 plasmid. (F) Representative images of GFP-LC3 staining in MCF7 cells cotransfected with GFP-LC3 and the indicated plasmids. (G) Light microscopic quantification of the percentage of GFP-positive MCF7 cells with discrete puncta, co-transfected with GFP-LC3, wt M11 and Beclin 1 or Beclin 1 mutants as indicated below the x-axis. (H) Light microscopic quantification of the number of discrete GFP-LC3 puncta per cell in GFP-positive MCF7 cells co-transfected with GFP-LC3, wt M11 and Beclin 1 or Beclin 1 mutants, as below the x-axis. For (C–F), results shown represent mean ± S.E.M. Similar results were obtained in three independent experiments.
Autophagy levels increase upon starvation of MCF7.beclin 1 cells (p < 0.0003 for starved versus unstarved cells; Fig. 4B–D, SI Fig. 4A). As shown previously,18 Beclin 1-dependent, starvation-induced autophagy levels are downregulated by cBcl-2 (p < 0.001 for cBcl-2 versus empty vector), and even more potently by KSHV Bcl-2 (p < 0.0003 for KSHV Bcl-2 versus empty vector). A clear comparison of downregulation of autophagy by M11 and these other Bcl-2s was not possible, as M11 expresses at very low levels in MCF7 (Fig. 4A) and other cell lines tested (data not shown). However, despite poor expression, we find that M11 inhibits starvation-induced autophagy (p < 0.0004 for M11 versus vector), at least as potently as cBcl-2, although not as much as KSHV Bcl-2 (Fig. 4B–D). Given the low M11 expression in these cells, M11 may actually inhibit autophagy as or more potently than KSHV Bcl-2. As expected, M11 mutants that abolish binding to Beclin 1, i.e., the Y60A + L74A (p = 0.9947 for M11 versus vector) and G86A + R87A (p = 0.3302 for M11 versus vector) double mutants, also abrogate the M11-mediated downregulation of starvation-induced, Beclin 1-dependent autophagy (Fig. 4B–D). However, due to poor expression of the Y60A + L74A double mutant it was not possible to unambiguously demonstrate that the lack of inhibition by that mutant was not simply a consequence of poor expression of that mutant (Fig. 4A–D).
As shown previously,18,31 starvation does not increase autophagy in MCF7 cells that do not express Beclin 1, and ectopic expression of M11 has no effect on autophagy levels in such cells (p = 0.2810 for starved versus unstarved cells; Fig. 4F). Transient expression of Beclin 1 in MCF7 cells leads to a marked increase in autophagy upon starvation (p = 0.0007 for starved versus unstarved cells), and this starvation-induced, Beclin 1-dependent autophagy is significantly downregulated by M11 expression (p = 0.00002 for M11 versus vector; Fig. 4F–H). As expected, the downregulation of starvation-induced autophagy by M11 is abrogated by coexpression of M11 and either L116A Beclin 1 (p = 0.00004 for mutant versus wt Beclin 1) or F123A Beclin 1 (p = 0.0002 for mutant versus wt Beclin 1; Fig. 4F–H); the Beclin 1 mutants that do not bind to M11. In contrast, starvation-induced autophagy levels are unaffected by coexpression of either R114A Beclin 1 (p = 0.0759 for mutant versus wt Beclin 1) or the G120A + D121A Beclin 1 (p = 0.6465 for mutant versus wt Beclin 1) double mutant; mutants which do not display impaired binding to M11 (Fig. 4F–H). Further, starvation-induced autophagy levels in cells expressing L112A Beclin 1 (p = 0.0071 for mutant versus wt Beclin 1), a mutation that weakens, but does not abolish binding to M11, are higher than that observed for wt Beclin 1, yet not as high as that observed for mutants that cannot bind M11 (Fig. 4F–H). Thus, downregulation of starvation-induced autophagy by M11 appears to be directly correlated to its ability to bind Beclin 1.
Discussion
γHV68 M11, a viral homolog of antiapoptotic cBcl-2s, inhibits apoptosis by binding BH3 domains of proapoptotic proteins in a hydrophobic surface groove, like other Bcl-2 homologs.8 Bcl-2s also inhibit autophagy via interactions with the autophagy effector Beclin 1.18,19,31 Recently, Beclin 1 was shown to contain a BH3 domain, which binds in the surface hydrophobic groove of Bcl-XL.19,38,39 While this manuscript was in preparation, another report studying the M11-Beclin 1 interaction was published.20 As discussed below, our results confirm and complement these studies.
Like BH3 domains from most proapoptotic proteins,20 we confirm that the isolated Beclin 1 BH3 domain binds to M11 with sub-micro-molar affinity, showing that the BH3 domain of these proteins is sufficient for binding to M11. We find that Beclin 1 residues N-terminal of the BH3 domain improve binding to M11, complementing recent results showing a similar effect for residues C-terminal of the BH3 domain.20 The molecular mechanism by which these additional residues assist binding of Beclin 1 to M11 is not clearly understood. The N-terminal residues may improve Beclin 1 affinity for Bcl-2s simply by increasing the helicity of the BH3 domain, as the isolated Beclin 1 BH3 domain is unstructured in solution but binds as a helix to M11. However, contrary to this theory, we find that the effect of the Beclin 1 N-terminal residues differs for binding to different Bcl-2s, suggesting that other more selective factors consistent with the variable amino acid composition of the Bcl-2s, such as additional direct interactions between the N-terminal residues and M11, are responsible for this increased affinity. In contrast, it has been suggested that residues C-terminal to the BH3 domain increase affinity of binding primarily by increasing helicity of the BH3 domain,20 although direct interactions between these residues and M11 cannot be ruled out. Thus, our results also raise the interesting possibility that residues outside the BH3-domain of proapoptotic proteins may also play a direct role in binding to different Bcl-2s. This remains to be completely investigated.
We find that the Beclin 1 BH3 domain binds with higher affinity to M11 than to Bcl-2 or KSHV Bcl-2, but with similar affinity to Bcl-XL. Further, regions outside the Beclin 1 BH3 domain dramatically improve binding to M11, but not to other Bcl-2s. Finally, while M11 binds the BH3 domains from the proautophagic protein Beclin 1 and most proapoptotic proteins within a ten-fold affinity range, other Bcl-2s appear to bind BH3 domains of at least one proapoptotic protein significantly more tightly. Thus, our biochemical data suggest that downregulation of autophagy may be as important a function of M11 as inhibition of apoptosis.
As also shown recently,20 our crystal structure of the M11-Beclin 1 BH3 domain complex confirms that the Beclin 1 BH3 helix binds in the hydrophobic surface groove of M11, reminiscent of the binding of other BH3 domains to Bcl-XL. Conformational changes in M11 that result in the widening of this groove, similar to those required for binding of BH3 domains to Bcl-XL, are required to accommodate the Beclin 1 BH3 domain. Both hydrophobic and polar interactions contribute to the M11-Beclin 1 BH3 domain interface. Twelve Beclin 1 BH3 domain residues interact directly with M11. Of these, almost all are conserved amongst vertebrate Beclin 1 homologs, and many are also conserved amongst proapoptotic protein BH3 domains.
Given the divergence in amino acid composition of Bcl-2s, it is not surprising that we find that these homologs bind the Beclin 1 and other BH3 domains with varying affinities, probably translating to widely varying specificity for different BH3 domain-containing proteins. The molecular determinants that enforce these specificities are poorly understood. Although BH3 domains from Beclin 1, Bax and Bad are all expected to bind in the hydrophobic surface groove of M11, the binding footprint of each of these is not identical. Thus, although we expect that functionally important residues conserved amongst all BH3 domains will be involved in interactions with different Bcl-2s, the importance of each residue varies, and depends on the shape and electrostatic complementarity of the interaction partner. Further, even amongst M11 residues affected by the binding of all BH3 domains, it is likely that the contribution of each interacting M11 residue to the total binding energy will differ in the case of each BH3 domain.
We show that M11 downregulates starvation-induced autophagy, and that the binding of M11 to Beclin 1 is essential for this downregulation. However, we find that not all residues at the M11-Beclin 1 interface are equally important. The M11 residue pairs, Y60 + L74, which form part of the hydrophobic surface groove, and G86 + R87, which lie at one end of this groove and are conserved amongst all Bcl-2s, are essential for binding to Beclin 1. Of the five Beclin 1 residues conserved amongst BH3 domains, L112, L116, G120, D121 and F123; L116, G120, D121 and F123 have been shown to be important for binding to Bcl-XL; however, only L116 and F123 appear essential for binding to M11. Thus, we not only identify residues critical for the binding of M11 to Beclin 1, but also prove that the BH3 domain is necessary for binding. Finally, we show that the M11 double mutants Y60A + L74A and G86A + R87A, which do not bind to Beclin 1 cannot downregulate starvation-induced autophagy. Conversely, in the absence of wt Beclin 1, M11 cannot downregulate autophagy if only the Beclin 1 mutants L116A and F123A, which do not bind to M11, are expressed.
Overexpression of Bcl-2s in MCF7.beclin 1 cells indicates that KSHV Bcl-2 and M11 inhibit autophagy more than cBcl-2. Despite this, our ITC data show that the γHV Bcl-2s do not bind the Beclin 1 BH3 domain substantially better than the cBcl-2s. Recently, we have shown that in nutrient deprivation conditions C-Jun N-terminal kinase-mediated Bcl-2 phosphorylation inhibits its interaction with Beclin 1 and increases Beclin 1-dependent autophagy.51 However, as KSHV Bcl-2 lacks equivalent phosphorylation sites, starvation conditions do not similarly downregulate Beclin 1-KSHV Bcl-2 complexes or increase autophagy.51 γHV68 M11 also lacks analogous phosphorylation sites. Therefore, it is likely that the enhanced downregulation of Beclin 1-dependent autophagy by γHV Bcl-2s is chiefly due to the inability of cellular factors to downregulate γHV Bcl-2s, rather than because of an inherently higher affinity of the γHV Bcl-2s for Beclin 1. Thus, γHV Bcl-2s appear to have evolved to constitutively inhibit autophagy, perhaps to enable the γHVs to evade autophagic degradation.
This work elucidates the molecular basis of γHV68 M11 binding to Beclin 1. In mammals, along with Beclin 1, class III PI3Kc3,36,50 UV irradiation resistance-associated gene (UVRAG),52 and Endophilin 1 (Bif-1)53 are essential components of the multiprotein, membrane-associated, autophagosome nucleation complex. Further, both Beclin 1 and Bif-1 have been shown to also form homo-oligomers.53,54 Binding of Beclin 1 to cBcl-2 has been shown to inhibit Beclin 1 interaction with PI3Kc3 or UVRAG, although neither protein appears to interact directly with the Beclin 1 BH3 domain.18,54 Further, it has been suggested that binding of various Bcl-2 homologs may prevent higher order oligomerization of Beclin 1.55 Thus, it appears likely that binding of Bcl-2 homologs to Beclin 1 sterically hinders its interactions with other components of the autophagy nucleation complex, inhibiting the autophagy-inducing function of Beclin 1. The molecular mechanism of Beclin 1 in autophagy and consequently, the exact mechanism by which the Beclin 1-Bcl-2 interaction disrupts the autophagy function of Beclin 1, remains to be understood.
Finally, this study not only furthers our knowledge of the regulation of Beclin 1, and consequently, the vital cellular process of autophagy, but also provides information regarding the subtle differences that constitute specificity determinants of the interaction of M11 with BH3 domains. Information regarding interaction specificity determinants will be useful for constructing mutant viruses to selectively examine the effects of M11 on either host cell autophagy or apoptosis, and consequently on viral pathogenesis and transformation of infected cells. For instance, the highest level of γHV Bcl-2 expression occurs immediately post-infection, abrogating host cell apoptosis, thus promoting virus survival. It is likely that inhibition of virus degradation by autophagy may be another key reason for the early expression γHV Bcl-2. Similarly, it has generally been believed that γHV Bcl-2s cause transformation of infected cells chiefly by inhibiting apoptosis. However, given the established role of Beclin 1 and autophagy in tumor suppression,31–35 it is quite possible, as has been discussed recently,16 that inhibition of autophagy may be another mechanism of malignant transformation of infected cells by viral Bcl-2 family members. Finally, information regarding interaction specificity determinants can also be used for the structure-based design of small molecules that selectively inhibit Beclin 1-γHV Bcl-2 interactions and remove the γHV blockade of autophagy. Such inhibitors would provide a powerful tool to examine the role of autophagy in regulating γHV infection, and ultimately may even form the basis of novel therapeutics to treat γHV infections.
Materials and Methods
Please see Supplementary Information for methods of production of protein and peptide reagents and for details of ITC, X-ray crystallography, CoIP and Western blot analyses.
ITC
For all ITC experiments, the Bcl-2 homolog and either the Beclin 1 BH3 domain or MBP•Beclin 1(1–135)•His6 were loaded into separate dialysis cassettes, and simultaneously dialyzed against three exchanges of 2 L of ITC buffer. ITC was performed in a MicroCal Omega VP-ITC calorimeter (MicroCal Inc., Northampton, Massachusetts), at 14°C with 35 injections of 8 µL each. Data were plotted and analyzed using MicroCal Origin software version 7.0, with a single-site binding model.
NMR spectroscopy
M11 1H/15N HSQC spectra were recorded at 25°C with a Varian Inova 500 MHz spectrometer under conditions similar to those previously reported. Data processing and analysis was performed using the programs NMRPIPE56 and NMRVIEW.57 Backbone 15N and 1H chemical shift assignments were kindly provided by Dr. E. Olejniczak (Abbott Laboratories) and Dr. H. Virgin (Washington University School of Medicine), and applied to the HSQC spectra using NMRVIEW. Backbone chemical shifts affected by binding of different BH3 domains were identified by recording 1H/15N HSQC spectra while sequentially titrating in increasing concentrations of peptide ranging from 60–500 µM into the NMR tube containing 250 µM M11.
X-ray crystallography
The M11-Beclin 1 BH3 domain complex was crystallized by hanging-drop vapor diffusion, and diffraction data from these crystals were collected at the SBC 19ID Advanced Photon Source, Chicago (SI and SI Table 2). The structure was solved by molecular replacement using a single M11 monomer (PDB ID: 2ABO) with residues 52–73 removed as a search model. The final model (SI Table 3) is deposited in the PDB with accession code 3DVU.
Western blot analyses and co-immunoprecipitation assays
Beclin 1/Bcl-2 co-immunoprecipitation assays and Western blot analyses of Beclin 1 or Bcl-2 protein expression were performed as described in detail in the Supplementary Materials and Methods.
Autophagy assays
Autophagy was assayed by light microscopic quantification of fluorescent autophagosomes in either MCF7 or MCF7.beclin 1 cells transfected with GFP-LC3, Bcl-2 homologs and, in the former also with Beclin 1 expression plasmids, as described.50 Cells were either starved for four hours in Earle’s balanced salt solution (EBSS; starvation medium), or cultured in DMEM with 10% fetal calf serum and an addition of 2x amino acid mixture (nutrient rich medium) prior to analysis. The percentage of GFP-LC3 positive cells with GFP-LC3 puncta was assessed by counting a minimum of 100 cells for duplicate samples per condition in three independent experiments, while the number of GPF-LC3 puncta per GFP-LC3 positive cell was assessed by counting a minimum of 50 cells for duplicate samples per condition in three independent experiments. The significance of alterations in autophagy levels were determined by a two-tailed, heteroscedastic student’s t-test.
Supplementary Material
Acknowledgements
This work was funded by NIH grants R21 AI078108 to S. S. and R01 CA109168 to B.L. We thank Johann Deisenhofer for access to facilities for protein purification and structure biology, Michael Rosen for access to the VP-ITC calorimeter, Mischa Machius for assistance with synchrotron data transfer, and Norma Duke for assistance with synchrotron data collection. X-ray diffraction data used in this report are derived from work performed at Argonne National Laboratory, Structural Biology Center at the Advanced Photon Source. Argonne is operated by U. Chicago Argonne, LLC, for the U.S. Department of Energy, Office of Biological and Environmental Research under contract DE-AC02-06CH11357.
Footnotes
Note
Supplementary materials can be found at: www.landesbioscience.com/supplement/SinhaAUTO4-8-Sup.pdf
References
- 1.Gangappa S, van Dyk LF, Jewett TJ, Speck SH, Virgin HW., IV Identification of the in vivo role of a viral Bcl-2. J Exp Med. 2002;195:931–940. doi: 10.1084/jem.20011825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cuconati A, White E. Viral homologs of Bcl-2: role of apoptosis in the regulation of virus infection. Genes Develop. 2002;16:2465–2478. doi: 10.1101/gad.1012702. [DOI] [PubMed] [Google Scholar]
- 3.Vaux D, Cory S, Adams J. Bcl-2 gene promotes haemopoietic cell survival and cooperates with c-myc to immortalize pre-B cells. Nature. 1998;335:440–442. doi: 10.1038/335440a0. [DOI] [PubMed] [Google Scholar]
- 4.McDonnell T, Deane N, Platt F, Nunez G, Jaeger U, McKearn JP, Korsmeyer S. bcl-2-immunoglobulin transgenic mice demonstrate extended B cell survival and follicular lymphoproliferation. Cell. 1989;57:79–88. doi: 10.1016/0092-8674(89)90174-8. [DOI] [PubMed] [Google Scholar]
- 5.Altmann M, Hammerschmidt W. Epstein-Barr virus provides a new paradigm: A requirement for the immediate inhibition of apoptosis. PLoS Biol. 2005;3:404. doi: 10.1371/journal.pbio.0030404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hardwick JM. Viral interference with apoptosis. Semin Cell Dev Biol. 1998;9:339–349. doi: 10.1006/scdb.1998.0243. [DOI] [PubMed] [Google Scholar]
- 7.Benedict CA, Norris PS, Ware CF. To kill or be killed: viral evasion of apoptosis. Nat Immunol. 2002;3:1013–1018. doi: 10.1038/ni1102-1013. [DOI] [PubMed] [Google Scholar]
- 8.Loh J, Huang Q, Petros AM, Nettesheim D, van Dyk LF, Labrada L, Speck SH, Levine B, Olejniczak ET, Virgin HW., IV A Surface groove essential for viral Bcl-2 function during chronic infection in vivo. PLoS Path. 2005;1:80–91. doi: 10.1371/journal.ppat.0010010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sarid R, Sato T, Bohenzky RA, Russo JJ, Chang Y. Kaposi’s sarcoma-associated herpesvirus encodes a functional Bcl-2 homologue. Nat Med. 1997;3:293–298. doi: 10.1038/nm0397-293. [DOI] [PubMed] [Google Scholar]
- 10.Ojala PM, Tiainen P, Salven T, Veikkola E, Castanos-Velez R, Sarid R, Biberfeld P, Makela TP. Kaposi’s sarcoma-associated herpesvirus-encoded v-cyclin triggers apoptosis in cells with high levels of cyclin-dependent kinase 6. Cancer Res. 1999;59:4984–4989. [PubMed] [Google Scholar]
- 11.Henderson S, Huen D, Rowe M, Dawson C, Johnson G, Rickinson A. Epstein-Barr virus-encoded BHRF1 protein, a viral homologue of Bcl-2, protects human B-cells from programmed cell death. Proc Natl Acad Sci USA. 1993;90 doi: 10.1073/pnas.90.18.8479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Foghsgaard L, Jaattela M. The ability of BHRF1 to inhibit apoptosis is dependent on stimulus and cell type. J Virol. 1997;71:7509–7517. doi: 10.1128/jvi.71.10.7509-7517.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Marshall WL, Yim C, Gustafson E, Graf T, Sage DR, Hanify K, Williams L, Fingeroth J, Finberg RW. Epstein-Barr virus encodes a novel homolog of the Bcl-2 oncogene that inhibits apoptosis and associates with Bax and Bak. J Virol. 1999;73:5181–5185. doi: 10.1128/jvi.73.6.5181-5185.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Theodorakis P, D’Sa-Eipper, Subramanian T, Chinnadurai G. Unmasking of a proliferation-restraining activity of the anti-apoptosis protein EBV BHRF1. Oncogene. 1996;12:1707–1713. [PubMed] [Google Scholar]
- 15.Bellows DS, Howell M, Pearson C, Hazlewood SA, Hardwick JM. Epstein-Barr virus BALF1 is a Bcl-2 antagonist of the herpesvirus antiapoptotic Bcl-2 proteins. J Virol. 2002;76:2469–2479. doi: 10.1128/jvi.76.5.2469-2479.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tarakanova VL, Kreisel F, White DW, Virgin HW., IV Murine gammaherpesvirus 68 genes both induce and suppress lymphoproliferative disease. J Virol. 2008;82:1034–1039. doi: 10.1128/JVI.01426-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang GH, Garvey TL, Cohen JI. The murine gammaherpesvirus-68 M11 protein inhibits Fas- and TNF-induced apoptosis. J Gen Virol. 1999;80:2737–2740. doi: 10.1099/0022-1317-80-10-2737. [DOI] [PubMed] [Google Scholar]
- 18.Pattingre S, Tassa A, Qu X, Garuti R, Liang XH, Mizushima N, Packer M, Schneider MD. Levine B. Bcl-2 antiapoptotic proteins inhibit Beclin 1-dependent autophagy. Cell. 2005;122:927–939. doi: 10.1016/j.cell.2005.07.002. [DOI] [PubMed] [Google Scholar]
- 19.Maiuri M, Le Toumelin G, Criollo A, Rain J, Gautier F, Juin P, Tasdemir E, Pierron G, Troulinaki K, Tavernarakis N, Hickman J, Geneste O, Kroemer G. Functional and physical interaction between Bcl-XL and a BH3-like domain in Beclin-1. EMBO J. 2007;26:2527–2539. doi: 10.1038/sj.emboj.7601689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ku B, Woo JS, Liang C, Lee KH, Hong HS, E X, Kim KS, Jung JU, Oh BH. Structural and biochemical bases for the inhibition of autophagy and apoptosis by viral Bcl-2 of murine γ-herpesvirus 68. PLoS Path. 2008;4:25. doi: 10.1371/journal.ppat.0040025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Levine B, Klionsky DJ. Development by self-digestion: Molecular mechanisms and biological functions of autophagy. Dev Cell. 2004;6:463–477. doi: 10.1016/s1534-5807(04)00099-1. [DOI] [PubMed] [Google Scholar]
- 22.Levine B, Kroemer G. Autophagy in the pathogenesis of disease. Cell. 2008;132:27–42. doi: 10.1016/j.cell.2007.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shintani T, Klionsky DJ. Autophagy in health and disease: A double-edged sword. Science. 2004;306:990–995. doi: 10.1126/science.1099993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mizushima N, Levine B, Cuervo A, Klionsky D. Autophagy fights disease through cellular self-digestion. Nature. 2008;451:1069–1075. doi: 10.1038/nature06639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kirkegaard K, Taylor MP, Jackson WT. Cellular autophagy: surrender, avoidance and subversion by microorganisms. Nat Rev Microbiol. 2004;2:301–314. doi: 10.1038/nrmicro865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Deretic V. Autophagy as an immune defense mechanism. Curr Opin Immunol. 2006;18:375–382. doi: 10.1016/j.coi.2006.05.019. [DOI] [PubMed] [Google Scholar]
- 27.Levine B, Deretic V. Unveiling the roles of autophagy in innate and adaptive immunity. Nat Rev Immunol. 2007;7:767–777. doi: 10.1038/nri2161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Orvedahl A, Levine B. Viral evasion of autophagy. Autophagy. 2008;4:280–285. doi: 10.4161/auto.5289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schmid D, Pypaert M, Münz C. Antigen-loading compartments for major histocompatibility complex class II molecules continuously receive input from autophagosomes. Immunity. 2007;1:79–92. doi: 10.1016/j.immuni.2006.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Legakis JE, Klionski DJ. Overview of Autophagy. In: Deretic V, editor. Autophagy in immunity and infection. Weinheim, Germany: Wiley-VCH Verlag GmbH & Co. KGaA; 2006. pp. 3–17. [Google Scholar]
- 31.Liang XH, Jackson S, Seaman M, Brown K, Kempkes B, Hibshoosh H, Levine B. Induction of autophagy and inhibition of tumorigenesis by beclin 1. Nature. 1999;402:672–676. doi: 10.1038/45257. [DOI] [PubMed] [Google Scholar]
- 32.Aita VM, Liang XH, Murty VVVS, Pincus DL, Yu W, Cayanis E, Kalachikov S, Gilliam TC, Levine B. Cloning and genomic organization of beclin 1, a candidate tumor suppressor gene on chromosome 17q21. Genomics. 1999;59:59–65. doi: 10.1006/geno.1999.5851. [DOI] [PubMed] [Google Scholar]
- 33.Qu X, Yu J, Bhagat G, Furuya N, Hibshoosh H, Troxel A, Rosen J, Eskelinen E-L, Mizushima N, Ohsumi Y, Cattoretti G, Levine B. Promotion of tumorigenesis by heterozygous disruption of the beclin 1 autophagy gene. J Clin Invest. 2003;112:1809–1820. doi: 10.1172/JCI20039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yue Z, Jin S, Yang C, Levine AJ, Heintz N. Beclin 1, an autophagy gene essential for early embryonic development, is a haploinsufficient tumor suppressor. Proc Nat Acad Sci. 2003;100:15077–15082. doi: 10.1073/pnas.2436255100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Karantza-Wadsworth V, Patel S, Kravchuk O, Chen G, Mathew R, Jin S, White E. Autophagy mitigates metabolic stress and genome damage in mammary tumorigenesis. Genes Dev. 2007;21:1621–1635. doi: 10.1101/gad.1565707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kihara A, Kabeya Y, Ohsumi Y, Yoshimori T. Beclin-phosphatidylinositol 3-kinase complex functions at the trans-golgi network. EMBO Rep. 2001;2:330–335. doi: 10.1093/embo-reports/kve061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liang XH, Kleeman LK, Jiang HH, Gordon G, Goldman JE, Berry G, Herman B, Levine B. Protection against fatal Sindbis virus encephalitis by Beclin 1, a novel Bcl-2-interacting protein. J Virol. 1998;72:8586–8596. doi: 10.1128/jvi.72.11.8586-8596.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Oberstein A, Jeffrey PD, Shi Y. Crystal structure of the Bcl-XL-Beclin 1 peptide complex: Beclin 1 is a novel BH3 only protein. J Biol Chem. 2007;282:13123–13132. doi: 10.1074/jbc.M700492200. [DOI] [PubMed] [Google Scholar]
- 39.Feng W, Huang S, Wu H, Zhang M. Molecular basis of Bcl-xL’s target recognition versatility revealed by the structure of Bcl-xL in complex with the BH3 domain of Beclin-1. J Mol Biol. 2007;372:223–235. doi: 10.1016/j.jmb.2007.06.069. [DOI] [PubMed] [Google Scholar]
- 40.Chen L, Willis SN, Wei A, Smith BJ, Fletcher JI, Hinds MG, Colman PM, Day CL, Adams JM. Huang DCS. Differential targeting of prosurvival Bcl-2 proteins by their BH3-only ligands allows complementary apoptotic funciton. Mol Cell. 2005;17:393–403. doi: 10.1016/j.molcel.2004.12.030. [DOI] [PubMed] [Google Scholar]
- 41.Flanagan A, Letai A. BH3 domains define selective inhibitory interactions with BHRF-1 and KSHV BCL-2. Cell Death Diff. 2008;15:580–588. doi: 10.1038/sj.cdd.4402292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Huang Q, Petros AM, Virgin HW, Fesik SW, Olejniczak ET. Solution structure of a Bcl-2 homolog from Kaposi sarcoma virus. Proc Nat Acad Sci. 2002;99:3428–3433. doi: 10.1073/pnas.062525799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Huang Q, Petros AM, Virgin HW, Fesik SW, Olejniczak ET. Solution structure of the BHRF1 protein from Epstein-Barr virus, a homolog of human Bcl-2. J Mol Biol. 2003;332:1123–1130. doi: 10.1016/j.jmb.2003.08.007. [DOI] [PubMed] [Google Scholar]
- 44.Muchmore SW, Sattler M, Liang H, Meadows RP, Harlan JE, Yoon HS, Nettesheim D, Chang BS, Thompson CB, Wong CS, Ng SC, Fesik SW. X-ray and NMR structure of homan Bcl-XL, an inhibitor of programmed cell death. Nature. 1996;381:335–341. doi: 10.1038/381335a0. [DOI] [PubMed] [Google Scholar]
- 45.Levine B, Sinha S, Kroemer G. Bcl-2 family members: dual regulators of apoptosis and autophagy. Autophagy. 2008;4:600–606. doi: 10.4161/auto.6260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liu X, Dai SC, Zhu Y, Marrack P, Kappler JW. The structure of a Bcl-XL/Bim fragment complex: implications for Bim function. Immunity. 2003;19:341–352. doi: 10.1016/s1074-7613(03)00234-6. [DOI] [PubMed] [Google Scholar]
- 47.Petros AM, Nettseheim DG, Wang Y, Olejniczak ET, Meadows RP, Mack J, Swift K, Matayoshi ED, Zhang H, Thompson CB, Fesik SW. Rationale for Bcl-XL/Bad peptide complex formation from structure, mutagenesis and biophysical methods. Protein Sci. 2000;9:2528–2534. doi: 10.1110/ps.9.12.2528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sattler M, Liang H, Nettesheim D, Meadows RP, Harlan JE, Eberstadt M, Yoon HS, Shuker SB, Chang BS, Minn AJ, Thompson CB, Fesik SW. Structure of the Bcl-XL-Bak peptide complex: recognition between regulators of apoptosis. Science. 1997;275:983–986. doi: 10.1126/science.275.5302.983. [DOI] [PubMed] [Google Scholar]
- 49.Liang XH, Yu J, Brown K, Levine B. Beclin 1 contains a leucine-rich nuclear export signal that is required for its autophagy and tumor suppressor function. Cancer Res. 2001;61:3443–3449. [PubMed] [Google Scholar]
- 50.Furuya N, Yu J, Byfield M, Pattingre S, Levine B. The evolutionarily conserved domain of Beclin 1 is required for Vps34 binding, autophagy and tumor supppressor function. Autophagy. 2005;1:41–47. doi: 10.4161/auto.1.1.1542. [DOI] [PubMed] [Google Scholar]
- 51.Wei Y, Pattingre S, Sinha S, Bassik M, Levine B. JNK1-mediated phosphorylation of Bcl-2 regulates starvation-induced autophagy. Mol Cell. 2008;30:678–688. doi: 10.1016/j.molcel.2008.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Liang C, Feng P, Ku B, B-H O, Jung JU. UVRAG: A new player in autophagy and tumor cell growth. Autophagy. 2007;3:69–71. doi: 10.4161/auto.3437. [DOI] [PubMed] [Google Scholar]
- 53.Takahashi Y, Coppola D, Matsushita N, Cualing H, Sun M, Sato S, Liang C, Jung J, Cheng J, Mul J, Pledger W, Wang HG. Bif-1 interacts with Beclin 1 through UVRAG and regulates autophagy and tumorigenesis. Nat Cell Biol. 2007;9:1142–1151. doi: 10.1038/ncb1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ku B, Woo JS, Liang C, Lee KH, U JJ, Oh BH. An insight into the mechanistic role of Beclin 1 and its inhibition by prosurvival Bcl-2 family proteins. Autophagy. 2008;4:519–520. doi: 10.4161/auto.5846. [DOI] [PubMed] [Google Scholar]
- 55.Liang C, E X, Jung JU. Downregulation of autophagy by herpesvirus Bcl-2 homologs. Autophagy. 2008;4:268–272. doi: 10.4161/auto.5210. [DOI] [PubMed] [Google Scholar]
- 56.Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J Biomol NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 57.Johnson BA, Blevins RA. NMRView: A computer program for the visualization and analysis of NMR data. J Biomol NMR. 1994;4:603–614. doi: 10.1007/BF00404272. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.