Abstract
Interferon gamma (IFN-γ), being the hallmark of the T cell TH1 response, has been extensively studied with respect to its expression and regulation of immune function. This gene has been extensively characterized in many mammalian species, making it one of the most widely cloned immunoregulatory genes. Recently, the gene has been identified in avian and piscine species and we have identified the gene in the frog genome. Based on these identified DNA sequences, we have constructed an evolutionary history of IFN-γ that shows this molecule can be traced back more than 450 million years ago. Our analysis shows that type II interferon (IFN-γ) function evolved before the tetrapod-fish split, a finding that contrasts earlier studies showing its origins in tetrapods. The IFN-γ gene has undergone a further duplication event in teleosts after the tetrapod-fish split suggesting a specific-evolutionary adaptation in fish. The analyses of IFN-γ, IL-22 and IL-26 genomic region in mammals, chicken, frog and fish reveal an evolutionary conservation of the loci and several regulatory elements controlling IFN-γ gene transcription. Furthermore, across the vertebrata, the first intron of IFN-γ gene contains a polymorphic microsatellite that has been closely correlated with disease susceptibility. Comparative-modeling of IFN-γ structure revealed differences among the representative species but with an overall conservation of the fold, dimer interface and some interactions with the receptor. The structural and functional conservation of IFN-γ suggests the presence of an innate, natural killer (NK) like response or even an adaptive TH1 immune response in lower vertebrates.
Keywords: Interferon gamma, vertebrates, evolution, structure
1.0 Introduction
Interferon gamma (IFN-γ) is a major cytokine regulating both innate and cell-mediated immune responses. The bio-activity of IFN-γ has been studied extensively in human and mouse and the major producers of this gene are activated T-cells, natural killer (NK) and NKT-cells. This gene has been demonstrated to play a key role in pathogen clearance and tumor surveillance[1,2]. It induces immunoglobulin class switching in B cells and along with IL (interleukin)-12, polarizes T-cells to a T-helper 1 (Th1) phenotype. In the genomic context, IFN-γ is located along with IL-22 and IL-26 on human chromosome 12q14+3 and mouse chromosome 10. The IFN-γ gene has been widely cloned and characterized from mammals and other vertebrates (Supplementary Table 1)[3,4]. The mammalian IFN-γ gene shares very low sequence similarity to non-mammalian species making it difficult to identify gene homologs in lower vertebrates. However, through genomics using sequence comparisons, the non-mammalian vertebrate homologs of cytokine-genes are now being identified[5,6,7]. Among non-mammals, IFN-γ has been cloned in birds and recently from fish[8,9,10]. In fish, the initial identifications were based on the genome sequence analysis aided by genomic annotation of the pufferfish (Takifugu rubripes & Tetraodon nigirovirdis) and zebrafish (Danio rerio) genomes[9,10]. The structural investigation of the interactions in the IFN-γ and the receptor (IFN-γR) complex has been widely reported[11,12,13].
The structural information obtained from X-ray crystallography and Nuclear Magnetic Resonance (NMR) methods show that IFN-γ is a homo-dimer and its activity arises as a consequence of binding to a receptor complex, which is composed of a four chain bundle of IFN-γR1 and IFN-γR2 genes. These receptors are classified as class II cytokine receptors, harboring two conserved fibronectin type III domains and showing strict species specificity. The IFN-γR1 and IFN-γR2 receptors are located on chromosomes 6q23-q24 and 21q22.11 in human[14,15] and chromosomes 10 and 16 in mouse[16,17], respectively. IFN-γ homo-dimer binds to the two IFN-γR1 chains but does not directly interact with IFN-γR2. However, IFN-γR2 has been shown to be essential for downstream signaling events; binding of the IFN-γ homo-dimer to the pre-assembled receptor triggers downstream JAK-STAT events that activate IFN-γ regulated genes[18].
Since, IFN-γ is a hallmark of the TH1 T-cell phenotype, studying the evolution of this gene will shed light on the origins of a TH1 like response. Furthermore, the transcription and translation of IFN-γ is regulated by several factors targeting the promoter and the un-translated regions. Although, non-coding regions are not under the same constraints as coding sequences, strong conservation of non-coding sequence across species might suggest a gene-regulatory function. The evolution of the IFN-γ structure, ligand-receptor interaction and downstream signaling events, makes us appreciate its distinct role in immune function and responses provides information on the regulation of this gene. During this study, we compared the IFN-γ gene across divergent species to find genomic similarities in order to draw conclusions on their functional unity. Moreover, species like fish, chicken and frog have not only become vertebrate models to study infectious diseases and tumors, but also, these animals are present at key positions in evolution and contribute significantly for studies related to development and comparative analysis of immune system function and development. Furthermore, vertebrate models like zebrafish and frog are amenable for both reverse and forward genetics and could be a valuable tool for dissecting the role of IFN-γ in the host response to infection and other immune challenges.
Genes involved in immunity evolve rapidly as they are subjected to stringent selection to combat rapidly evolving or emerging pathogens. These evolutionary pressures necessarily lower sequence conservation of immune genes, greatly hindering progress in finding gene orthologs in disparate organisms. During this study, an extensive database analysis was carried out to find orthologs of IFN-γ from vertebrates and invertebrates by exploiting conserved genome synteny. In the current study, we report, for the first time the identification of IFN-γ from an amphibian by using the genomic database of western clawed frog (Xenopus tropicalis). Evidence of structural and functional similarity is required to assign orthology to the identified genes. As the IFN-γ in lower vertebrates shows low sequence similarity with mammalian counterparts, and the fact that no structural data existed for non-mammalian vertebrates, we have used the mammalian IFN-γ structures from previous X-ray crystallography data, together with secondary structure predictions, to derive unique structural features which are common for these divergent species. Taken together, these data give important insights on the origin and structural evolution of IFN-γ.
2.0 Origin of IFN-γ dates back to 450 million years ago (mya)
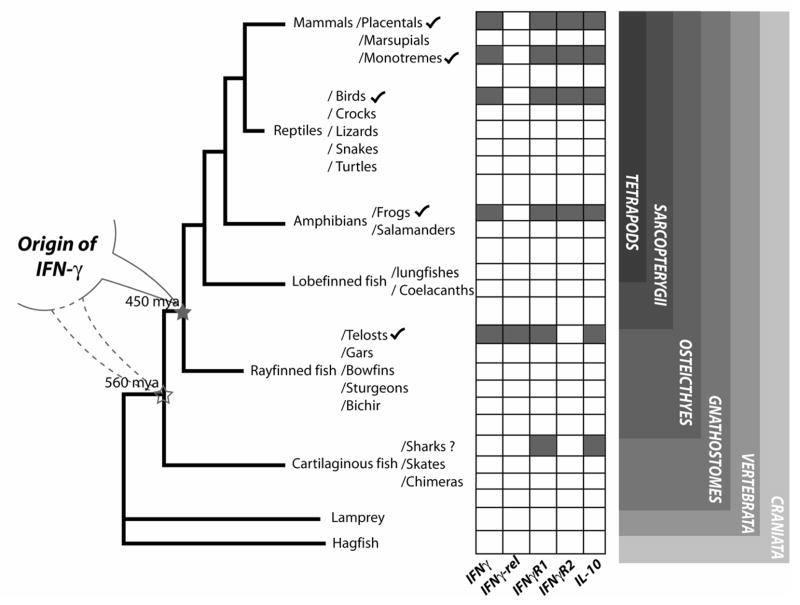
The immune related genes are rapidly evolving and often present great difficulties in finding orthologous genes by conventional “BLAST” (The Basic Local Alignment Search Tool) searches[19]. However, non-immune genes are typically conserved making it easy to search for genes flanking those related to immunity. Among the sub-phylum vertebrata (which consists of 53,000 species), Actinoptergians, Amphibians, Avians, and Mammals have been represented in our analyses. Computational screening of the genome databases of vertebrates and invertebrates for the search for mammalian IFNγ genes orthologs was conducted in these analyses. Agnathan fishes, Chondricthyan fishes (sharks and rays), Sarcopterygian fishes (Coelacanths and Lungfishes) and reptiles have not been represented in our studies because of the non-availability of the genomic sequences or sufficient expressed sequence tags (EST) databases for such analyses. Initially, DYRK2, MDM, RAP1B, NUP107, SLC35E3, CNTN1B, OPN1SW1, CALU harboring scaffolds or contigs were identified. These contigs were then searched for IFN-γ gene using FGENESH+ software and curated manually (http://www.softberry.com/berry.phtml). With the recent release of several draft sequences of vertebrate genomes, it is possible to analyze chicken, frog and fish genomes leading to the identification of the IFN-γ gene. Along with IFN-γ, IL-26 and IL-22 genes have also been identified in fish and chicken (Fig. 1) [9,6]. In this study, we have identified IFN-γ in the frog genome using the sequences from the genome database. The study is important as, in vertebrates, amphibians are an important evolutionary landmark and they depict an evolutionary-interface from aquatic to terrestrial environment and the emergence of tetrapods. However, the identification of IL-10 and IFNγR1 genes in shark [IL-10, Spiny dog fish, Squalus acanthias, (CX789095); bony fish, IFNγR1 (CAD67765) and shark IFNγR1, (EB158140) is suggestive of the presence of IFN-γ gene in cartilaginous fishes. Although, there is no direct evidence of the IFN-γ gene in reptiles, we predict its existence since amphibians and birds, which are close to reptiles, possess this gene. Previous studies have shown that invertebrates do not harbor any of the mammalian cytokine orthologs except for tumor necrosis factor like gene, which is implicated in apoptosis related functions[20]. We searched for IFN-γ gene orthologs in asidians, sea-urchin and fly genome databases. Neither the cytokines nor the genes flanking them (MDM1/RAP1B/DYRK2) were identified from these invertebrates. These analyses suggest that this gene originated in vertebrates rather than from tetrapods as originally described (Fig. 2).
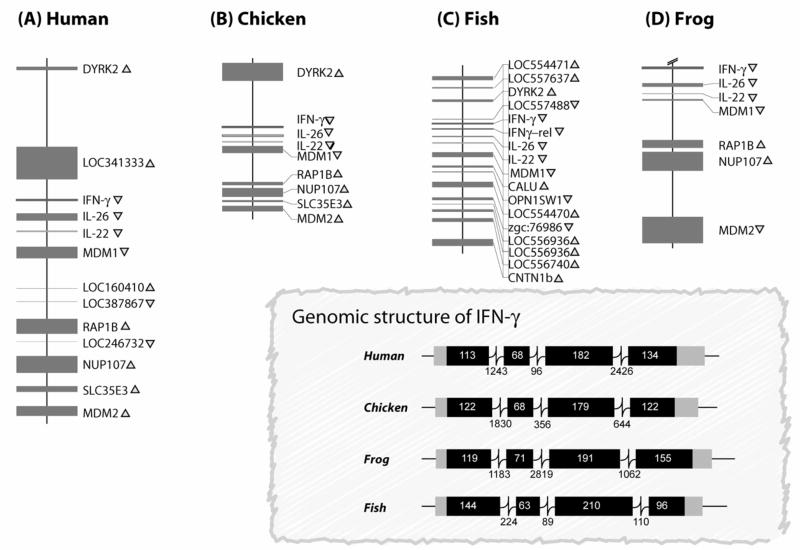
Fig. 1.
A schematic comparison showing syntenic conservation of the IFN-γ loci in human, chicken, fish and frog. A) Human IFNγ loci on chromosome 12 (NCBI map viewer; scale: 5 kb =1 cm), B) Chicken IFN-γ loci on chromosome1 build 1.1 (contig ID: NW_060207; NCBI map viewer; scale:10 kb =1 cm), C) Zebrafish IFN-γ loci on chromosome 4 Version Zv4 (contig ID: NW_634616.1; NCBI map viewer; scale; 10 kb =1 cm), and D) Frog (Xenopus tropicalis) IFN-γ loci on scaffold_380 (http://genome.jgi-psf.org/Xentr3/Xentr3.home.html; scale:10 kb =1 cm). The closed boxes and the arrowheads indicate the position and transcriptional orientations of the genes. The figure in the inset is a schematic diagram of the genomic structure of interferon gamma genes across species. The boxes represent exons and the lines connecting the boxes are introns, with numbers indicating length in base pairs. Abbreviations used: IFNγ, interferon gamma; IFNγ-rel, interferon gamma related; DYRK2, dual specificity tyrosine-(Y)-phosphorylation-regulated kinase, MDM, mouse double minute; RAP1B, Rab subfamily of small GTPases, NUP107, nucleoporin 107; SLC35E3, solute carrier family 35, member E3; CNTN1B, contactin 1b; OPN1SW1, opsin 1 short-wave-sensitive 1; CALU, calumin; IL, interleukin.
Fig. 2.
Origin of interferon gamma. A schematic depiction of the vertebrate lineages along with the occurrence of IFN-γ, nIFNγL, IFNγ1R1, IFNγR2, and IL-10 genes. The filled boxes indicate the presence of the gene and the empty boxes indicate the absence or lack of sufficient genomic/EST information to identify the gene.
3.0 Analysis of IFN-γ loci
3.1. The genomic loci of IFN-γ is conserved during evolution
A striking similarity of the gene order and orientation of both cytokine and non-cytokine genes was seen on the IFN-γ loci of human, mouse, chicken, frog and fish, when analyzed for their syntenic conservation. The IL-22 and IL-26 genes are also tightly linked to IFN-γ in these species. The non-immune genes surrounding the IFN-γ gene in mammals, fish, chicken and frog orthologs of MDM1, RAP1B, NUP107, SLC35E3 are present in the same gene order and orientation (Fig. 1). Thus, the general architecture of this region pre-existed 450 million years ago and has withstood evolution.
The promoter region of IFN-γ has been extensively studied and T-bet, GATA-3. NF-κB, NF-AT, STATs, AP-2, OCT-1 CREB/ATF-2 binding are present at the proximal promoter region of human and mouse IFN-γ. To analyze if there is a similar transcriptional mechanism in place for IFN-γ in other vertebrates, we screened the first 1000 bp from the transcriptional start site. As predicted due to the low sequence homology across species, we did not see a high level of conservation at nucleotide the level. However, when we scanned this region for transcription binding sites, we found that a striking conservation of sites for 15 families of transcription binding factors known to play a role in the immune system in vertebrates (Table 3). These include GATA, BRAC, NF-AT, STAT, OCT1, which have already been implicated to play a role in the regulation of IFN-γ gene expression in human and mouse. Furthermore, recent studies have shown that transcription factors like T-bet and GATA-3 are present in fish and may play important role in IFN-γ transcription. More importantly, there are other transcription factor members conserved in this region that have not been previously associated with IFN-γ (Table 3).
However, the genomic distances between the genes and intron lengths in fish are smaller than in tetrapod, which is attributed to the smaller genome size of fish (21). In addition, dinucleotide microsatellites (CA) have been described in the first intron of the human IFN- geneand the third intron in the mouse. Similar polymorphic regions are seen in the first intron of the IFN- gene in chicken and fish [21,6,22]. Polymorphisms in this region has been linked to diseases such as rheumatoid arthritis, lung transplant allograft fibrosis and graft-versus-host disease [23,24,25].
Post-transcriptional control of gene expression involves factors that determine the translation capacity of the generated mRNA transcript. Previous studies have indicated a role for the IFN-γ 3′-untranslated region (3′UTR) of the mRNA transcript in maintaining stability for protein translation [26]. The 3′UTR of the IFN-γ mRNA contains several cis-acting repetitive adenosine-uridine stretches (AUUUA), referred to as AU-rich elements (ARE), that destabilize the mRNA through ARE-mediated decay (AMD) process [27]. Along with IFN-γ, IL-3, G-CSF and TNF-α are additional cytokines that share a similar ARE region and are known to be regulated by AMD [28]. When we compared IFN-γ only from mammals, a high conservation of the 5′end of 3′UTR was observed and this region was AT-rich which harbored the ARE sites. A similar AT-rich region containing ARE sites is seen in the IFN-γ gene of bird, zebrafish and frogs, suggesting an evolutionary conservation of the post-transcriptional regulation.
3.2. Bony-fish show a lineage specific expansion of IFN-γ gene
After the initial identification of IFN-γ gene from Japanese pufferfish [10], the analysis of the complete genomic region was carried-out in order to identify fish orthologs of mammalian IL-22 and IL-26 genes [9]. The IFN-γ loci of zebrafish, Japanese pufferfish and spotted green pufferfish were highly conserved. Surprisingly, in zebrafish and pufferfish, a novel gene similar to fish IFN-γ was identified along this locus (see Fig. 6, which is published as supporting information)[9]. This gene has been putatively been named as IFNγ–rel (related) until further characterization. Furthermore, the IFNγ–rel shares a phase 0 acceptor and donor splice sites at the exon/intron boundaries. The predicted protein of IFNγ–rel does not share any of the motifs of the IL-10 family members and the genomic organization is different from the IL-26 and IL-22, so this may not be a new IL-10 family gene. The presence of the gene in tandem to IFN-γ and sharing the same genomic structure shows its common origin. A low comparative sequence identity of IFN-γ and IFNγ–rel suggests this duplication occurred very early in evolution [9]. This paralogous duplication might have taken place around 296 mya as zebrafish (Cypriniformes) and pufferfish (Tetrodontiformes) diverged during that time This tandem duplication event has not been observed in tetrapod lineages indicating this has occurred prior to the divergence in the rayfined fishes of the order Actinopterygii (before tetrapod-fish split). Furthermore, these paralogs might have duplicated at a very early stage of teleostean lineage because of the dis-similarity of the IFN-γ and IFNγ–rel genes among Cypriniformes and Tetrodontiformes. While the fish IFN-γ and IFNγ–rel genes themselves share low identities and form a separate cluster, they show an evolutionary link on a major IFN-γ clad in a phylogenetic tree. Although, during BLAST analysis, fish IFNγ–rel does not retrieve any IFN-γ genes of four legged vertebrate, they do retrieve fish IFN-γ which is considered to be the functional ortholog of mammalian IFN-γ. We hypothesize thatafter duplication, this novel IFNγ–rel protein might have diverged and acquired a specialized function and different receptor affinities. As there have been rapid changes of IFNγ–rel amino-acid sequence during a relatively short period of time, we speculate that this diversification might be subjected to positive selection. Recently, comprehensive expression analyses of these genes suggest that common-carp IFN-γ is expressed in T-cells when stimulated with phytohemagglutinin, while surprisingly IFNγ-rel is expressed in lipoly-saccharide stimulated IgM+ (B-cell enriched) cells [29]. Clearly, in future functional analysis of the IFNγ-rel gene is required to understand its significance in immune-system.
4.0 Comparative structural analyses of IFN-γ in evolutionarily-distant species
4.1 Evidence that the fold of IFN-γ protein is conserved in evolutionarily-distant species
The comparative sequence identities of IFN-γ among the mammals, bird, fish, and frog are very low (see Table 2). Structural information is imperative in understanding the protein function, but IFN-γ structures are currently available only for the mammals. Since the sequence identities and similarities among IFN-γ are so low, it is very challenging to use comparative modeling concepts to derive structures for these sequences[21,30,22]. It is, however, known that functionally similar genes and proteins often have higher sequence similarity in regions required for their function along with a common fold in their protein structures. In this study, we have adopted a novel approach of mapping the structural information extracted from the available mammalian protein structures onto the non-mammalian sequence data. This mapping along with the published experimental information relating to folding was used to understand the evolution of IFN-γ gene across these organisms. Secondary structures of the protein sequences for chicken, frog and fish were predicted using the PSIPRED [31,32] (see Figs 8 a–c, which is published as supporting information) server. The order and arrangement of the secondary structural elements in these sequences were similar to the available mammalian IFN-γ structures strongly suggesting the existence of a common fold across the species.
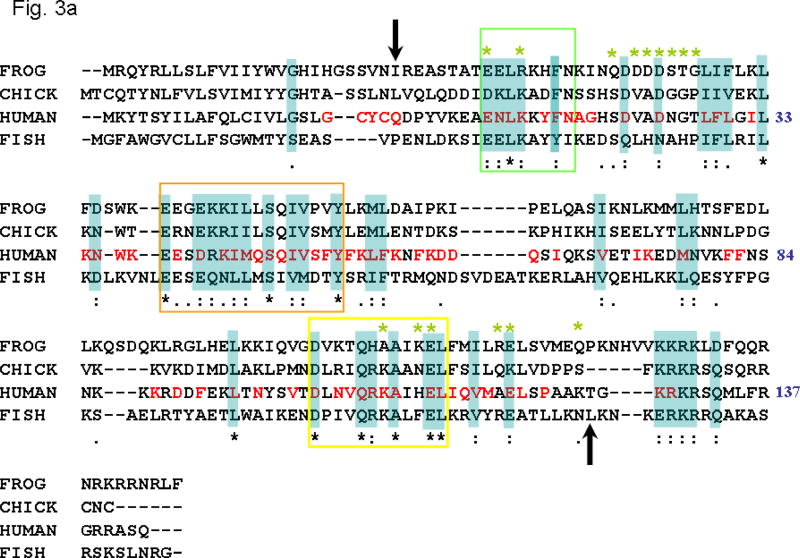
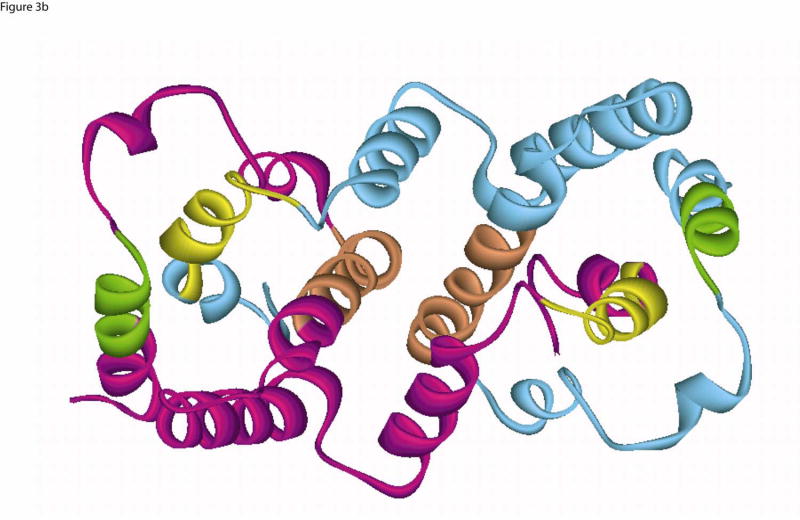
Experimental studies indicate that IFN-γ exists as an intercalating homo-dimer, and that dimer formation is crucial for the functional activity of the molecule. Structural alignment of interferon-γ structures of cow, rabbit (α-carbon only) and human (PDB: 1FG9) shows that the overall fold is conserved. From the mammalian IFN-γ structures, we have identified the functionally important regions such as dimer core, intercalating dimer interface, common dimer-receptor (IFNγ–Receptor complex, PDB:1fg9) interactions and the flexible ligand-receptor electrostatic interactions, and then extracted the relevant sequence motifs and compared them in the non-mammalian genes. We also aligned the mammalian IFN-γ protein sequences (see Fig. 7, which is published as supporting information) using Clustal W 1.8 to validate the sequence blocks extracted from the crystal structures and the non-mammalian sequences. The combined information from the mammalian sequence alignment, structural motifs, and the secondary structural information from the non-mammalian vertebrates were carried over to the sequence alignment of human, fish, chicken and frog (see Fig 3a). Despite the low overall sequence identity, the annotated sequence blocks (Fig 3a) showed high similarity (≥50%) in these functionally important regions (see Table 2). The areas of high similarity coincided with the helix, interface core, and intercalation regions of the protein (Fig. 3b). As mentioned above, these regions are functionally important for the core-structure of the dimer and, presumably, the function of IFN-. The helix and intercalation sites are important for the dimer stability, and the specific interactions leading to dimer formation are also conserved. This is consistent with the IFN-γ proteins sharing a common fold, interaction and function. This evidence lends support for the core structure and fold being conserved across species (see Figs 4a, b).
Fig. 3.
(a) Multiple sequence alignment of IFN-γ sequences from frog, chicken, human and fish. The human sequence numbering is based on the PDB entry, 1FG9, and the arrows show the location of the first and last residue of 1FG9. Cyan shaded residues identified to be conserved based on the multiple sequence alignment. The residues of human sequence colored red are identified to be conserved based on the mammalian (pig, cow, human, rat and mouse) IFN- multiple sequence alignment (see Fig. 7, which is published as supporting information). The green starred residues are located within a sphere of 3.5 Å from the receptor atoms (PDB: 1FG9). Multiple sequence alignment was carried out using Clustal W [version 1.8, GCG 10.3]. BLOSUM series was used for Protein weight matrix. “*” denote residues in that column that are identical in all sequences in the alignment. “:” denotes that conserved substitutions have been observed. “.” means that semi-conserved substitutions are observed.
(b) Ribbon diagram of IFN-γ dimer structure (PDB: 1FG9). Chain A of the dimer is shown in Cyan and the chain B in Magenta color. The core helical region [GLU9-ASN16] of the structure is colored in green, the residues forming the conserved core domain interface [GLU38-TYR53] in orange and the core helix [ASP102-LEU113] in yellow. The corresponding helical regions are marked by the same colored boxes in the multiple sequence alignment shown on Fig 3a.
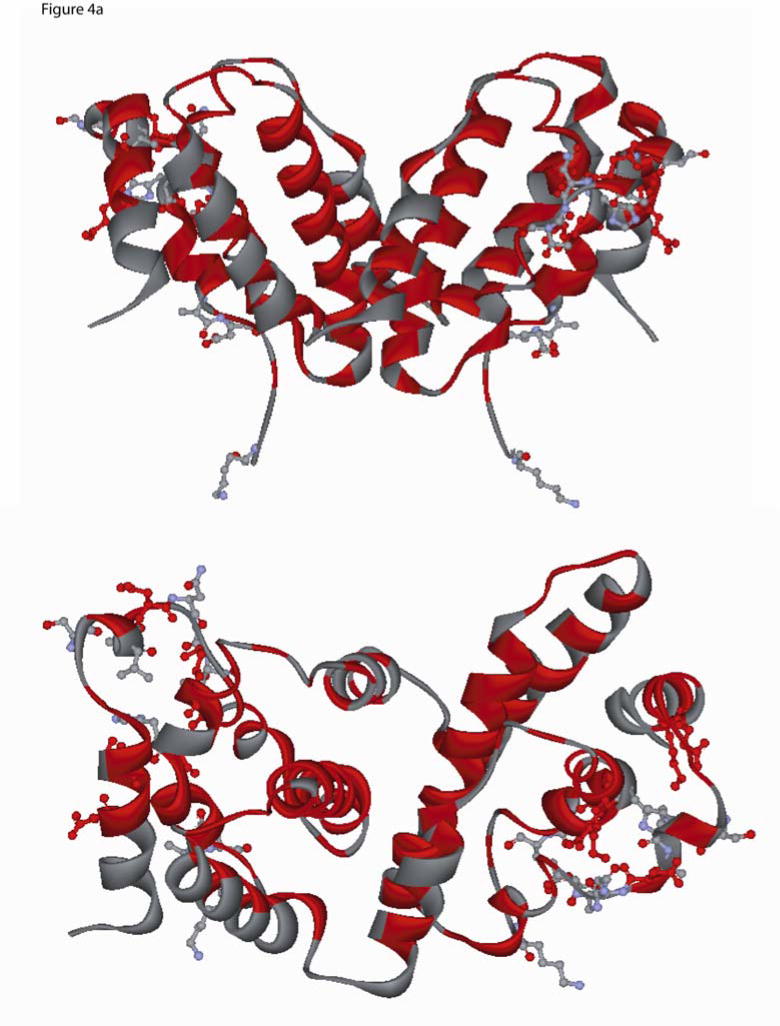
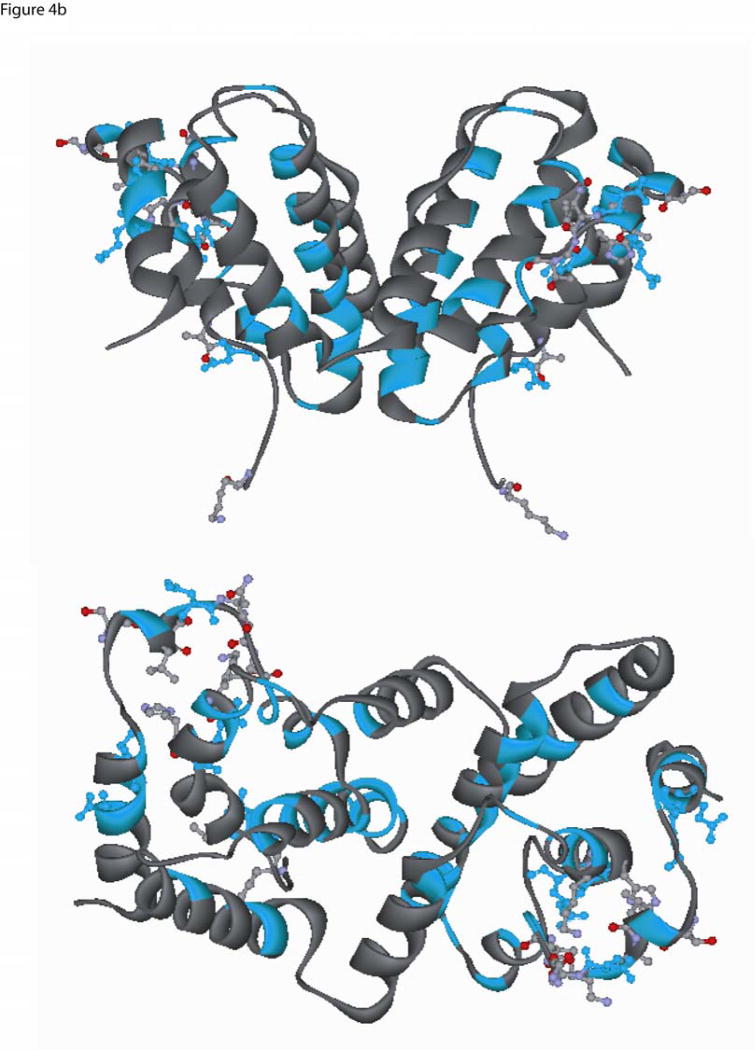
Fig. 4.
Two different ribbon view of the interferon-gamma dimer (PDB ID, 1FG9) (a) The location of the conserved residues identified from mammalian-only sequence alignment are shown in red (see Fig. 3a), and the residues that are within 3.5 Å from IFN-γ receptor atoms are shown in ball-and-stick and colored by element type (Carbon:light grey, Oxygen:red, Hydrogen:white, Nitrogen:light blue). Residues that belong to the mammalian conserved set and are present within 3.5 Å are shown in red color ball-and-stick (upper panel is top view and lower panel is side view (non stereo)). (b) Conserved residues based on the frog, chicken, human and fish sequence alignment were painted Cyan (Fig 3a), and the residues that are present within 3.5 Å from the IFN-γ receptor are shown in ball-and-stick colored by element type (Carbon:light grey, Oxygen:red, Hydrogen:white, Nitrogen:light blue). Residues which belong to the conserved set and are present within a 3.5 Å are shown in cyan color ball-and-stick (upper panel is top view and lower panel is side view (non stereo)).
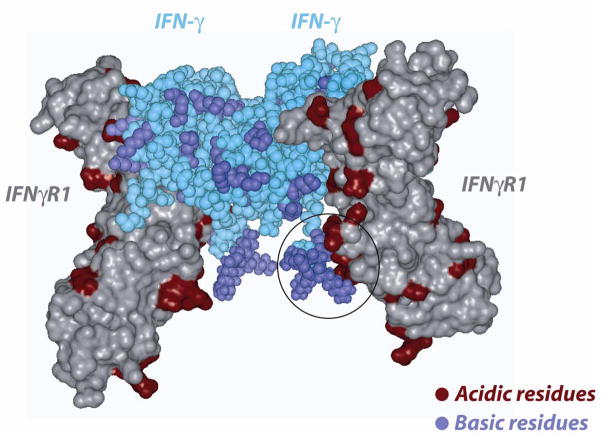
The location of the residues that are present within a 3.5 Å from the IFN-γ dimer and could play a role in the interaction and binding to IFNγ–IFNγR1 were identified from the PDB entry 1FG9 and are shown in Figs 4a,b. As can be seen from figures 3 and 4, most of these residues are non-conserved and may account for species specificity and lack of cross-reactivity (Figs 4a,b). The IFN-γ C-terminal tail harboring a basic stretch of alternating lysine and arginine residues is highly conserved in fish, frog, chicken and mammals (see Fig 3a). Previous studies have suggested that this region may bind to an acidic stretch on the receptor[33]. However, this region has not been observed in any of the NMR or X-ray structures, a fact that has been attributed to a highly flexible nature for this region [34]. In the current study, we added this highly conserved poly-cationic C-terminus tail [G127-R131] to the human IFN-γ crystal structure to assess its role in interacting with the IFN R1 receptor. This poly-cationic tail [KRKR] of IFN- occupied a region in close proximity to a highly acidic patch on the IFNγR1 receptor, as seen in figure 5. This electrostatic interaction is probably associated with a high affinity site specific for IFN- and its receptor since this sequence is not conserved across other cytokines[35]. The sequence conservation in this region of IFN-γ in mammals and birds, together with the recent findings in fish and frog, suggest that this region has been strictly conserved through evolution reinforcing the speculation that it might serve as an IFN-γ specific binding site.
Fig 5.
The figure shows the complementarity of the highly conserved basic segment of the C-terminal IFN-γ (space-filled, cyan color) with the receptor [PDB: 1FG9]. The receptor chains (C and D) are shown in solvent accessible surface representation. Receptor chain E is removed from the complex for clarity. The C-terminal basic segments of the IFN-γ chains have been modeled and added to the structure. Acidic and basic residues in the receptor/IFN-γ are colored red and blue, respectively.
4.2. Conservation of receptors is not seen at the IFN-γ ligand-receptor interaction sites
During this study, database mining for IFNγR1 and IFNγR2 homologs were conducted in divergent species. Among non-mammalian vertebrates, homologs of these receptors have been reported in chicken and recently in frog [34]. During our study, we could find receptor homologs for bony fish (IFNγR1 [CAD67765] and IFNγR2) and shark (IFNγR1; EB158140). Similar to the ligands, the receptors were not found in fly, tunicates, shrimp, lobster and oyster genomic sequences and/or EST databases. This suggests that that IFN-γ gene might have originated during early vertebrate evolution.
IFN-γ genes are highly species specific and the cross species reactivity is very low compared to other cytokines [36]. Previous studies using only mammalian sequences suggested that although the topology was conserved, differences in residues important for receptor interaction were responsible for the species specificity [37]. During this study, global approaches using the gene sequences of distantly related species were used to address this issue. As the core structure of the gene was fairly conserved in distant species, we felt that the receptor binding sites might be important for species specificity. The contact residues of IFN-γ homo-dimer with the receptor are shown in ball-and-stick representation (Fig. 4) and marked as green stars in the multiple sequence alignment (Fig. 3a). We found that several residues of the ligand contact sites were non-conserved among the representative species (see Fig. 3a). These non-synonymous changes in the contact sites can be attributed to the evolutionary pressure on immune genes. Dimer binding of IFN-γ to its receptor is necessary for its current function. It has been shown that the poly-cationic residues of the IFN-γ C-terminus are important for its biological activity and may be required for its binding to the extracellular domain of the receptors[38,39,40]. Recently, it was shown that the IFN-γ C-terminus is essential for its function in fish as well[22]. We speculate that there is an important interaction in this region and the flexibility might be required to facilitate the conformational changes of the cytoplasmic region of the receptor to accommodate the STAT 1 protein for subsequent phosphorylation. Although, this interaction has been postulated in previous studies [38,39,40], this is the first time evidence of this interaction has been demonstrated through modeling.
5.0 IFN-γ is an odd man in IL-10 family
The function, genomic structure and the position of interferon genes have led to a broad classification of these genes into type I (α, β, κ, ω IFNs), type II (IFN-γ) and type III (IFN-λ).
Interferons type I and III belong to the class II helical cytokines which are structurally similar with differences in that the type I are intronless while type III harbors introns. The intronless type I IFN’s are found as a cluster in tetrapod lineage and are thought to have independently expanded into several genes from an amniote specific-retrotransposition event after the tetrapod-fish split. Type I and III interferons might have a common evolutionary origin which underwent duplication and diversification during fish-tetrapod split. This theory can be supported by the fact that the fish type I interferon’s contain introns, in contrast to the mammalian genes.
Along with IFNs, the IL-10 family genes (IL-10, IL-19, IL-20, IL-24 and IL-26) also belong to class II helical cytokines. Lutfalla et. al. [39], proposed that the diversification of IL-10 family came up from a common ancestral gene. Genome duplications during evolution might have given rise to two separate gene clusters harboring IL-10 genes. This theory can be proved in fish, frog and chicken, where only two genes (IL-10 and IL-19/24 like genes) are present on the IL-10 loci compared to three genes as found in mammals. This shows that the lineage specific expansion may be in-complete in these organisms.
IFN-γ is a class II helical cytokine sharing some structural and functional properties of IFNs and IL-10 family. As IFN-γ is present in the IL-10 locus, we can speculate that this gene may be a product of a tandem duplication event during evolution. This reflects in its genomic organization as it shares similarities to IL-10 family except that the last two exons (EIII and EIV) are fused to form a single exon in the IL-10 family. The 3′-coding region of the IFN-γ gene must have undergone some evolutionary changes like recombination or deletion at the time of tandem duplication. This event might have lead to its diversification in its function and structural changes.
6.0 Concluding remarks
Analysis of the immune system of lower vertebrates, through comparative genomics, will not only help identifying human orthologs involved in regulating the immune system, but also such identifications at a molecular level will help in developing alternative disease and cancer models. Among lower vertebrates, zebrafish has been used as a model animal for toxicological studies and has been seen as a potential cancer model. The evolutionary and structural approaches in studying the immune genes will help us unravel the inter-species similarities and differences. During this study, an evolutionary history of IFN-γ, one of the most widely cloned cytokine genes, has been drawn and demonstrates a remarkable degree of conservation of its structure necessary for the function during evolution. Despite the low identities shared with mammalian counterparts, lower vertebrate IFN-γ shows a striking similarity in the regions of core structure. The function of IFN-γ in birds and fish are similar and comparable with mammals. The structural and functional conservation of IFN-γ suggests of the presence of an innate, NK like response or even an adaptive TH1 immune response in lower vertebrates. Conservation of the biological function of IFN-γ, during speciation, represents the resilience of the gene subjected to strong selective forces.
Supplementary Material
Supporting Fig. 6. A) Schematic diagram showing IFN-γ and IFNγ-rel in tandem in pufferfish. All the closed boxes indicate the exons of respective genes. The thin lines interrupting the exons are introns. Arrows indicate the transcriptional orientation of the genes. The numbers are the nucleotide lengths of exons, introns and the length between the genes. B) Phylogenetic tree showing the relationship between IFN-γ and IFNγ-rel genes. The nucleotide sequence of the coding regions were aligned using Clustal W 1.8 and the tree was constructed by NJ method supported by 1000 bootstrap replications using MEGA software[41].
Supporting Fig. 7. Alignment of deduced aminoacid sequence of mammalian IFNγ genes using clustal W 1.8 [GCG 10.3] software. BLOSUM series was used for protein. The core helical region, core domain and core helix are indicated in a square colored boxes as in Fig. 3b.
Supporting Fig. 8. Secondary structure predictions of IFNγ sequences of a) chicken, b) fish and c) frog using PSIPRED software.
Acknowledgments
R.S., S.R., J.R.C., M.S. and H.Y designed research; R.S., and S.R., performed research; R.S., and S.R., analyzed data; and R.S. wrote the paper. This work was funded by the intramural research program of the National Cancer Institute, NIH.
Biographies
 Ram Savan
Ram Savan
 Sarangan Ravichandran
Sarangan Ravichandran
 Jack Collins
Jack Collins
 Masahiro Sakai
Masahiro Sakai
 Howard Yound
Howard Yound
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Reference List
- 1.Lieberman LA, Hunter CA. Regulatory pathways involved in the infection-induced production of IFN-gamma by NK cells. Microbes Infect. 2002;4(15):1531–1538. doi: 10.1016/s1286-4579(02)00036-9. [DOI] [PubMed] [Google Scholar]
- 2.Shankaran V, Ikeda H, Bruce AT, White JM, Swanson PE, Old LJ, Schreiber RD. IFNgamma and lymphocytes prevent primary tumour development and shape tumour immunogenicity. Nature. 2001;410(6832):1107–1111. doi: 10.1038/35074122. [DOI] [PubMed] [Google Scholar]
- 3.Young HA, Hardy KJ. Role of interferon-gamma in immune cell regulation. J Leukoc Biol. 1995;58(4):373–381. [PubMed] [Google Scholar]
- 4.Young HA. Regulation of interferon-gamma gene expression. J Interferon Cytokine Res. 1996;16(8):563–568. doi: 10.1089/jir.1996.16.563. [DOI] [PubMed] [Google Scholar]
- 5.Kaiser P, Rothwell L, Avery S, Balu S. Evolution of the interleukins. Dev Comp Immunol. 2004;28(5):375–394. doi: 10.1016/j.dci.2003.09.004. [DOI] [PubMed] [Google Scholar]
- 6.Kaiser P, Poh TY, Rothwell L, Avery S, Balu S, Pathania US, Hughes S, et al. A genomic analysis of chicken cytokines and chemokines. J Interferon Cytokine Res. 2005;25(8):467–484. doi: 10.1089/jir.2005.25.467. [DOI] [PubMed] [Google Scholar]
- 7.Savan R, Sakai M. Genomics of fish cytokines. Comparative Biochemistry and Physiology - Part D: Genomics and Proteomics. 2005;1(1 SPEC ISS):89–101. doi: 10.1016/j.cbd.2005.08.005. [DOI] [PubMed] [Google Scholar]
- 8.Digby MR, Lowenthal JW. Cloning and expression of the chicken interferon-[gamma] gene. Journal of Interferon and Cytokine Research. 1995;15(11):939–945. doi: 10.1089/jir.1995.15.939. [DOI] [PubMed] [Google Scholar]
- 9.Igawa D, Sakai M, Savan R. An unexpected discovery of two interferon gamma-like genes along with interleukin (IL)-22 and -26 from teleost: IL-22 and -26 genes have been described for the first time outside mammals. Mol Immunol. 2006;43(7):999–1009. doi: 10.1016/j.molimm.2005.05.009. [DOI] [PubMed] [Google Scholar]
- 10.Zou J, Yoshiura Y, Dijkstra JM, Sakai M, Ototake M, Secombes C. Identification of an interferon gamma homologue in Fugu, Takifugu rubripes. Fish Shellfish Immunol. 2004;17(4):403–409. doi: 10.1016/j.fsi.2004.04.015. [DOI] [PubMed] [Google Scholar]
- 11.Randal M, Kossiakoff AA. The structure and activity of a monomeric interferon-gamma:alpha-chain receptor signaling complex. Structure. 2001;9(2):155–163. doi: 10.1016/s0969-2126(01)00567-6. [DOI] [PubMed] [Google Scholar]
- 12.Thiel DJ, le Du MH, Walter RL, D’Arcy A, Chene C, Fountoulakis M, Garotta G, et al. Observation of an unexpected third receptor molecule in the crystal structure of human interferon-gamma receptor complex. Structure. 2000;8(9):927–936. doi: 10.1016/s0969-2126(00)00184-2. [DOI] [PubMed] [Google Scholar]
- 13.Walter MR, Windsor WT, Nagabhushan TL, Lundell DJ, Lunn CA, Zauodny PJ, Narula SK. Crystal structure of a complex between interferon-gamma and its soluble high-affinity receptor. Nature. 1995;376(6537):230–235. doi: 10.1038/376230a0. [DOI] [PubMed] [Google Scholar]
- 14.Pfizenmaier K, Wiegmann K, Scheurich P, Kronke M, Merlin G, Aguet M, Knowles BB, et al. High affinity human IFN-gamma-binding capacity is encoded by a single receptor gene located in proximity to c-ros on human chromosome region 6q16 to 6q22. J Immunol. 1988;141(3):856–860. [PubMed] [Google Scholar]
- 15.Soh J, Donnelly RJ, Kotenko S, Mariano TM, Cook JR, Wang N, Emanuel S, et al. Identification and sequence of an accessory factor required for activation of the human interferon gamma receptor. Cell. 1994;76(5):793–802. doi: 10.1016/0092-8674(94)90354-9. [DOI] [PubMed] [Google Scholar]
- 16.Hibino Y, Mariano TM, Kumar CS, Kozak CA, Pestka S. Expression and reconstitution of a biologically active mouse interferon gamma receptor in hamster cells. Chromosomal location of an accessory factor. J Biol Chem. 1991;266(11):6948–6951. [PubMed] [Google Scholar]
- 17.Mariano TM, Kozak CA, Langer JA, Pestka S. The mouse immune interferon receptor gene is located on chromosome 10. J Biol Chem. 1987;262(12):5821–5814. [PubMed] [Google Scholar]
- 18.Krause CD, Mei E, Xie J, Jia Y, Bopp MA, Hochstrasser RM, Pestka S. Seeing the light: preassembly and ligand-induced changes of the interferon gamma receptor complex in cells. Mol Cell Proteomics. 2002;1(10):805–815. doi: 10.1074/mcp.m200065-mcp200. [DOI] [PubMed] [Google Scholar]
- 19.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215(3):403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 20.Venkatesh B, Gilligan P, Brenner S. Fugu: a compact vertebrate reference genome. FEBS Lett. 2000;476(1–2):3–7. doi: 10.1016/s0014-5793(00)01659-8. [DOI] [PubMed] [Google Scholar]
- 21.Greer J. Comparative modeling methods: application to the family of the mammalian serine proteases. Proteins. 1990;7(4):317–334. doi: 10.1002/prot.340070404. [DOI] [PubMed] [Google Scholar]
- 22.Zou J, Carrington A, Collet B, Dijkstra JM, Yoshiura Y, Bols N, Secombes C. Identification and bioactivities of IFN-gamma in rainbow trout Oncorhynchus mykiss: the first Th1-type cytokine characterized functionally in fish. J Immunol. 2005;175(4):2484–2494. doi: 10.4049/jimmunol.175.4.2484. [DOI] [PubMed] [Google Scholar]
- 23.Khani-Hanjani A, Lacaille D, Hoar D, Chalmers A, Horsman D, Anderson M, Balshaw R, et al. Association between dinucleotide repeat in non-coding region of interferon-gamma gene and susceptibility to, and severity of, rheumatoid arthritis. Lancet. 2000 Sep 2;356(9232):820–825. doi: 10.1016/s0140-6736(00)02657-x. [DOI] [PubMed] [Google Scholar]
- 24.Cavet J, Dickinson AM, Norden J, Taylor PR, Jackson GH, Middleton PG. Interferon-gamma and interleukin-6 gene polymorphisms associate with graft-versus-host disease in HLA-matched sibling bone marrow transplantation. Blood. 2001 Sep 1;98(5):1594–1600. doi: 10.1182/blood.v98.5.1594. [DOI] [PubMed] [Google Scholar]
- 25.Awad M, Pravica V, Perrey C, El GA, Yonan N, Sinnott PJ, Hutchinson IV. CA repeat allele polymorphism in the first intron of the human interferon-gamma gene is associated with lung allograft fibrosis. Hum Immunol. 1999;60(4):343–346. doi: 10.1016/s0198-8859(98)00133-5. [DOI] [PubMed] [Google Scholar]
- 26.Mavropoulos A, Sully G, Cope AP, Clark AR. Stabilization of IFN-gamma mRNA by MAPK p38 in IL-12- and IL-18-stimulated human NK cells. Blood. 2005;105(1):282. doi: 10.1182/blood-2004-07-2782. [DOI] [PubMed] [Google Scholar]
- 27.Khabar KS. The AU-rich transcriptome: more than interferons and cytokines, and its role in disease. J Interferon Cytokine Res. 2005;25(1):1. doi: 10.1089/jir.2005.25.1. [DOI] [PubMed] [Google Scholar]
- 28.Lai WS, Parker JS, Grissom SF, Stumpo DJ, Blackshear PJ. Novel mRNA targets for tristetraprolin (TTP) identified by global analysis of stabilized transcripts in TTP-deficient fibroblasts. Mol Cell Biol. 2006;26(24):9196. doi: 10.1128/MCB.00945-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stolte EH, Savelkoul HF, Wiegertjes G, Flik G, Lidy Verburg-van Kemenade BM. Differential expression of two interferon-gamma genes in common carp (Cyprinus carpio L) Dev Comp Immunol. 2008;32(12):1467–1481. doi: 10.1016/j.dci.2008.06.012. [DOI] [PubMed] [Google Scholar]
- 30.Martin AC, MacArthur MW, Thornton JM. Assessment of comparative modeling in CASP2. Proteins. 1997;(Suppl):114–28. doi: 10.1002/(sici)1097-0134(1997)1+<14::aid-prot4>3.3.co;2-f. [DOI] [PubMed] [Google Scholar]
- 31.Bryson K, McGuffin LJ, Marsden RL, Ward JJ, Sodhi JS, Jones DT. Protein structure prediction servers at University College London. Nucleic Acids Res. 2005;33(Web Server issue):W36–38. doi: 10.1093/nar/gki410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McGuffin LJ, Bryson K, Jones DT. The PSIPRED protein structure prediction server. Bioinformatics. 2000;16(4):404–405. doi: 10.1093/bioinformatics/16.4.404. [DOI] [PubMed] [Google Scholar]
- 33.Griggs ND, Jarpe MA, Pace JL, Russell SW, Johnson HM. The N-terminus and C-terminus of IFN-gamma are binding domains for cloned soluble IFN-gamma receptor. J Immunol. 1992;149(2):517–520. [PubMed] [Google Scholar]
- 34.Szente BE, Johnson HM. Binding of IFN gamma and its C-terminal peptide to a cytoplasmic domain of its receptor that is essential for function. Biochem Biophys Res Commun. 1994;201(1):215–221. doi: 10.1006/bbrc.1994.1691. [DOI] [PubMed] [Google Scholar]
- 35.Randal M, Kossiakoff AA. The 2.0 A structure of bovine interferon-gamma; assessment of the structural differences between species. Acta Crystallogr D Biol Crystallogr. 2000;56(Pt 1):14–24. doi: 10.1107/s0907444999014304. [DOI] [PubMed] [Google Scholar]
- 36.Szente BE, Soos JM, Johnson HW. The C-terminus of IFN gamma is sufficient for intracellular function. Biochem Biophys Res Commun. 1994;203(3):1645–1654. doi: 10.1006/bbrc.1994.2375. [DOI] [PubMed] [Google Scholar]
- 37.Szente BE, Weiner IJ, Jablonsky MJ, Krishna NR, Torres BA, Johnson HM. Structural requirements for agonist activity of a murine interferon-gamma peptide. J Interferon Cytokine Res. 1996;16(10):813–817. doi: 10.1089/jir.1996.16.813. [DOI] [PubMed] [Google Scholar]
- 38.Azumi K, De Santis R, De Tomaso A, Rigoutsos I, Yoshizaki F, Pinto MR, Marino R, et al. Genomic analysis of immunity in a Urochordate and the emergence of the vertebrate immune system: “waiting for Godot”. Immunogenetics. 2003;55(8):570–581. doi: 10.1007/s00251-003-0606-5. [DOI] [PubMed] [Google Scholar]
- 39.Lutfalla G, Crollius HR, Stange-Thomann N, Jaillon O, Mogensen K, Monneron D. Comparative genomic analysis reveals independent expansion of a lineage-specific gene family in vertebrates: the class II cytokine receptors and their ligands in mammals and fish. BMC Genomics. 2003;4(1):29. doi: 10.1186/1471-2164-4-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Milev-Milovanovic I, Long S, Wilson M, Bengten E, Miller NW, Chinchar VG. Identification and expression analysis of interferon gamma genes in channel catfish. Immunogenetics. 2006;58(1):70–80. doi: 10.1007/s00251-006-0081-x. [DOI] [PubMed] [Google Scholar]
- 41.Kumar S, Tamura K, Nei M. MEGA3: Integrated software for Molecular Evolutionary Genetics Analysis and sequence alignment. Brief Bioinform. 2004;5(2):150–163. doi: 10.1093/bib/5.2.150. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Fig. 6. A) Schematic diagram showing IFN-γ and IFNγ-rel in tandem in pufferfish. All the closed boxes indicate the exons of respective genes. The thin lines interrupting the exons are introns. Arrows indicate the transcriptional orientation of the genes. The numbers are the nucleotide lengths of exons, introns and the length between the genes. B) Phylogenetic tree showing the relationship between IFN-γ and IFNγ-rel genes. The nucleotide sequence of the coding regions were aligned using Clustal W 1.8 and the tree was constructed by NJ method supported by 1000 bootstrap replications using MEGA software[41].
Supporting Fig. 7. Alignment of deduced aminoacid sequence of mammalian IFNγ genes using clustal W 1.8 [GCG 10.3] software. BLOSUM series was used for protein. The core helical region, core domain and core helix are indicated in a square colored boxes as in Fig. 3b.
Supporting Fig. 8. Secondary structure predictions of IFNγ sequences of a) chicken, b) fish and c) frog using PSIPRED software.