Abstract
A mutant of human butyrylcholinesterase (BChE) with high activity against cocaine would be highly promising as a drug for therapeutic treatment of cocaine abuse and overdose. It is desirable to design a recombinant BChE mutant with a long half-life in human circulation. Studies showed that BChE subunits can be assembled by a peptide containing the proline-rich attachment domain (PRAD) to form a stable tetramer. The models of BChE tetramer complexed with PRAD with various sequences have been constructed, in the present study, based on homology modeling and molecular dynamics (MD) simulation of explicit water-solvated systems. The 3D models enable us to understand how the BChE subunits are arranged in the tetramer and how the tetramerization domain of BChE is associated with PRAD to form stable tetramer of human BChE. It has been shown that the six conserved hydrophobic residues located on the C-terminal of BChE are responsible for the key electrostatic and hydrophobic interactions between the tetramerization domain of BChE and PRAD. The simulated tetramer structures suggest that mutation of three residues, i.e. Phe547, Met554, and Phe561, to other hydrophobic residues may be beneficial for increasing the binding between the tetramerization domain of BChE and PRAD. Thus, the detailed structural insights obtained from this study may be valuable for rational design of a recombinant BChE tetramer with a longer residence time in circulation.
INTRODUCTION
Cocaine abuse causes serious medical and social consequences in our society. Studies show that cocaine causes its reinforcing and toxic effects by blocking neurotransmitter reuptake, i.e. by blocking the dopamine transporter which brings synaptic dopamine back to the presynaptic neuron located in the central nervous system (CNS).1–3 However, the classic pharmacodynamic approach has failed to yield small-molecule drugs due to the difficulties inherent in blocking a blocker. Human butyrylcholinesterase (BChE) is the principal plasma enzyme that catalyzes hydrolysis of cocaine into its biologically inactive metabolites, thus the BChE metabolic pathway is most suitable for amplification for an anti-cocaine medication.4–7 However, the catalytic efficiency of wild-type BChE is rather low (kcat = 4.1 ± 0.4 min−1, KM = 4.5 ± 0.3 µM).8 A few BChE mutants with significantly increased catalytic efficiency have been developed by our group7,9–13 based on our understanding of the mechanism of cocaine hydrolysis catalyzed by BChE and its mutants.6,14–16 In vitro and in vivo tests of BChE mutant A199S/F227A/S287G/A328W/Y332G showed that this mutant has greater than 2000-fold higher catalytic efficiency than the wild-type, and that a pretreatment with this mutant (using a dose of 0.01 mg per mouse) can fully protect mice from a lethal dose of cocaine (180 mg/kg).13
It has been known that BChE can exist in the forms of monomer, dimer, and tetramer and the half-life of BChE (wild-type or mutant) in circulation is dependent on the quaternary form of the protein. The residence time of the monomer is only on the order of minutes in the circulation of mice, whereas native human BChE has a half-life of ~46 hours in mice17–19 or ~12 days in human plasma.20 It has been shown that the predominant forms of recombinant BChE secreted from Chinese hamster ovary (CHO) cells were monomers and dimers, whereas native BChE consists more than 95% of tetramers that lead to the longer half-life of native BChE in plasma.19,21 However, recent studies indicated that association of recombinant BChE with a proline-rich peptide facilitated its formation into a tetramer. An N-terminal segment of the peptide, called proline-rich attachment domain (PRAD) is essential for the interaction.22 For example, a proline-rich peptide with PRAD sequence CCLLMPPPPPLFPPPFF (denoted as PRAD1 in this paper) was a modified form of the N-terminus of the rat collagen tail protein. Studies showed that coexpression of this peptide with recombinant BChE drives the formation of tetramers.19,21,23 A peptide with sequence PPPPPPPPPPPPPPPLP is denoted as PRAD2 in this paper. Addition of PRAD2 or poly-L-proline with mean molecular mass of 8 kDa to cultured medium was reported to increase the percentage of tetramer of recombinant BChE.18,23 Recently, Lockridge et al. found that the native plasma BChE tetramers were associated with various segments of a PRAD2-containing peptide which originated from a cytosolic protein called lamellipodin.18 This discovery indicated that association with a proline rich peptide leads to the stability of the BChE tetramer in blood.
So far, there has been no X-ray crystal structure showing how the BChE subunits are oriented in the tetramer and how they are associated with the proline-rich peptide. Previous efforts failed to get an X-ray structure of a full length BChE monomer with the tetramerization domain, let alone the BChE tetramer with or without the tetramerization domain.24,25 However, previous studies indicated that the tetramerization domain is located in the C-terminus of BChE, including the residues 534 to 574, while the catalytic domain comprises residues 1 to 533.21,23,24 Furthermore, based on experimental data, Lockridge et al. predicted that the four C-termini of the BChE subunits may form a four-helix bundle, with essential aromatic interaction between the conserved tryptophans, phenylalanines and tyrosines from the C-termini of the BChE subunits.21 According to their observation that the C-termini were readily accessible to biomedical and chemical agents, they proposed that the tetramerization domain is exposed to solvent and protrudes away from the catalytic domains.23
Other clues leading to the identification of the organization of the subunits of BChE tetramer can be found from the studies of the corresponding structure of acetylcholinesterase (AChE),26 a protein homologous to BChE. Studies have shown that four T-splice variants of AChE (AChET) are associated with a collagen-like subunit which makes the AChET tetramer stable. Similar to BChE, the tetramerization domain of AChET is also located at the C-terminus.22,27,28 Furthermore, studies indicated that the tryptophan amphiphilic tetramerization (WAT) sequence located at the C-terminus of AChET is responsible for the association with the collagen-like subunit, in which an N-terminal segment, called proline-rich attachment domain (PRAD) is essential for the interaction.22 Disulfide bonds are involved in the arrangement of the subunits, each of which has one cysteine close to the C-terminus. Two of the four subunits are disulfide-bonded with each other through the C-terminal cysteines, while the other two have disulfide-bond links with the collagen-like peptide.22,27,28 So far there have been a few X-ray structures of AChE dimers or monomers26,29–32 and three low-resolution X-ray structures of AChE tetramers,27,33 but all of them lack the structures of the tetramerization domain of AChET and the collagen-like subunit. However, a crystal structure released in 2004 shows the complex of four WAT sequences of human AChE together with a synthesized proline-rich peptide with sequence of PRAD, i.e. [WAT]4PRAD complex. In this structure, the four parallel alpha helices of WAT wrap around a single antiparallel PRAD helix to form a left-handed superhelix conformation. The middle PRAD helix resembles a left-handed polyproline II (PPII) helix, with 3.4 residues per turn.22 Based on the [WAT]4PRAD complex and a low-resolution structure of the catalytic domains of the tetramer of EeAChE,27 Sussman et al. built a preliminary structure of an AChET tetramer together with collagen-like protein (ColQ), i.e. [AChET]4-ColQ complex, by first manually docking the [WAT]4PRAD onto the structure of Electrophorus electricus AChE (EeAChE) and then adjusting the structure using CORELS.22,34,35 In this structure, the AChET tetramer is a dimer of dimers. The two dimers are oriented in an antiparallel fashion so that the subunits are diagonally equivalent, which accounts for the two-fold axis of the tetramer structure. In addition, the C-termini or the tetramerization domains of the four subunits wrap around the middle ColQ and project out of the body of the tetramer.22 The WAT chains stagger onto each other to form the super α-helix structure with offset N-termini of the WAT chains. Thus, there was difficulty docking the superhelix perpendicularly onto the two-fold symmetric body of AChET tetramer, which led to the 30° angle of the superhelical axis relative to the two-fold symmetry axis of the tetramer.22
Here we report three 3D structural models of BChE tetramer complexed with proline-rich peptide, forming the [BChE]4-PRAD complexes, for the first time, based on the homology modeling with the template of the structure of [AChET]4-ColQ.22 Next, we carried out the molecular dynamics (MD) simulation of the complexes solvated in explicit water molecules to get stable structures of [BChE]4-PRAD complexes. The obtained structures provide insights into the way the BChE subunits are oriented together with the proline-rich peptide, and the detailed information on the binding mode of the PRAD with the tetramerization domain of BChE, especially WAT. The simulated 3D structures of the BChE tetramers provide valuable insights about how to increase the interaction between WAT and PRAD by mutating the residues of PRAD or WAT, thus to possibly increase the half-life of recombinant BChE in blood so that a high-activity mutant of this enzyme could become a realistic drug for anti-cocaine medication.
METHODS
Construction of initial 3D model of BChE tetramer complexed with PRAD
The amino acid sequence and the X-ray structure of human BChE (hBChE) monomer25 were from Protein Data Bank (PDB ID code 1P0P).36 The construction of [BChE]4-PRAD complex was based on the homology modeling with the template of [AChET]4-ColQ complex.22 The involved structures for construction of the template were those of the EeAChE tetramer27 and [WAT]4PRAD22 from PDB, with ID numbers of 1C2O and 1VZJ, respectively. The sequence alignment between hBChE and EeAChE were generated with SIM37 from ExPASy.38 There are three proline-rich peptides with PRAD sequences of CCLLMPPPPPLFPPPFF,19 PPPPPPPPPPPPPPPLP,18 and PPPPPPPPPPPPPPPPP,23 denoted as PRAD1, PRAD2, and PRAD3, respectively. The initial structures of [BChE]4-PRAD complexes were built with InsightII software (Accelrys, Inc). First, four catalytic domains of BChE were superimposed onto the subunits of [AChET]4-ColQ complex. The missing residues of BChE catalytic domain were added based on their counterpart residues in AChET. The structures of the C-terminus, i.e. tetramerization domain of BChE and PRAD, were built by mutating the corresponding structures of [AChET]4-ColQ complex. Finally, the tetramerization domains of BChE subunits and PRAD were cut from the tetramer body and redocked onto it so that the axis of the supercoil is almost overlapping with the two-fold axis of the BChE tetramer. Due to the adjustment, there were some small gaps and steric clashes between the N-termini of the tetramerization domain and the C-termini of the catalytic domain of BChE subunits. Structural adjustments were also made to the C-termini of the catalytic domain of the BChE subunits where the α-helix structures were distorted for them to be connected to the N-termini of the tetramerization domain in the [AChET]4-ColQ template. The structure of [WAT]4PRAD22 could be a good reference in evaluating the interaction between the PRAD and the tetramerization domain of AChE. Therefore, the structure of this complex was constructed based on that of 1VZJ. All the structures were then minimized in gas phase using the Sander module of Amber version 8 package39 to be prepared for MD simulations of explicitly solvated systems.
Molecular dynamics simulation on explicitly solvated systems
To carry out the MD simulations, the topologic and coordinate files of the initial structures were built with LEaP module of the Amber, version 8 package. The Amber ff02 force field40,41 was used for the complexes. Each complex was neutralized by adding sodium counterions. The aqueous condition of blood could affect the features of our biological system. The main electrolytes in plasma are sodium and chloride ions with the concentrations of around 140 mM and 100 mM, respectively.42,43 Considering plasma takes up around 55% of blood volume,44 the ionic strength in blood is around 66 mM. Since the size of the periodic box used to simulate the model system is 71.2 × 93.5 × 67.6 Å, the ionic strength of ~66 mM accounts for about 20 pairs of sodium and chloride ions in the box. Therefore, 20 pairs of additional sodium and chloride ions were added to the reference structure of [WAT]4PRAD to imitate the affects of free ions in blood. For comparison, MD simulation on the other [WAT]4PRAD system without additional ions were also performed. All the systems were solvated in a rectangular box of TIP3P water molecules45 with a minimum solute-wall distance of 10 Å. As a result, around eighty-thousand explicit water molecules were added to each system, except [WAT]4PRAD. The energy minimization and MD simulation were performed by using the Sander module of the Amber, version 8 package. To resolve any possible bad contacts and improper bond lengths on the turn between the tetramerization domain and the catalytic domain of BChE in the initial structures, additional steps were added to the MD procedure used in our previous computational studies.6,7,9–11,46–53 First, the solvent molecules were minimized for 5000 cycles and equilibrated for 10 ps of MD simulation to make sure that they were in an equilibrated condition. Second, the structure of the [BChE]4-PRAD complex was energy-minimized in three steps. First, the side chains of the structure and water molecules were allowed to move during a 1000-cycle energy minimization to relieve possible unexpected side chain clashes. Next, besides the side chains and water molecules, the backbone of the last seven residues of the C-termini of the catalytic domain of BChE were allowed to move so as to adjust themselves to be connected with the N-termini of the tetramerization domain. Finally, the whole system was energy-minimized for 1000 cycles. The system was then gradually heated from T = 10 K to T = 298.15 K for 40 ps, followed by the MD simulation at 298.15 K for 20 ps with the backbone of the whole complex restrained with harmonic potential,54 while water molecules and the side chains were allowed to move. Then the backbone of the last seven residues of the C-termini of the catalytic domain of BChE was relaxed for 40 ps in order to get a smooth connection with the N-termini of the tetramerization domains of BChE. Since one hydrogen bond between WAT and PRAD was lost in the initial structure derived from [AChET]4-ColQ compared with the X-ray structure of [WAT]4PRAD,22 the distance relating to this hydrogen bond was restrained within 1.6 to 2.0 Å with NMR refinement55 during the heating and the first 20 ps of the MD simulation at 298.15 K. Then the whole complex was relaxed for 1 ns or longer to obtain a stable structure of [BChE]4-PRAD complex. The disulfide bonds between WAT and PRAD or between the WATs were defined either by LEaP or by restraining the bonding distance with NMR refinement. The time step used in the MD simulation was 2 fs. Periodic boundary condition was used in the NPT ensemble with Berendsen temperature coupling and P = 1 atm with isotropic molecular-based scaling.56 The SHAKE algorithm57 was used to fix all covalent bonds containing hydrogen atoms. The non-bonded pair list was updated every 25 steps. The particle mesh Ewald (PME) method was used to treat long-range electrostatic interactions.58 The non-bonded cutoff of 10 Å was used. During the MD simulation, the coordinates of the simulated complex were saved every 1 ps. The MD simulations of the [WAT]4PRAD systems were similar, except the steps involving the seven turn residues in [BChE]4-PRAD complex were eliminated. The other difference is that the MD simulations of the [WAT]4PRAD systems are much longer, with 10 ns for each of them, so that the interaction between WAT and PRAD can be investigated in a longer time scale.
Validation of the models
Procheck59,60 was used to verify/validate the initial models as well as the models derived from the MD simulation. The stereochemical quality of the structures was evaluated to determine whether the bond lengths and bond angles are within their normal ranges, and whether there are unfavorable dihedral angles in the models by using the G-factor generated by Procheck. This program was also used to identify bad contacts between non-bonded atoms. A Ramachandran plot was made to locate residues with disfavored phi-psi combinations. Whatif61–63 is a program capable of doing atomic contact analysis by comparing the atom contacts observed in the checked protein and the average distribution of atoms derived from the PDB.36 By examining the normality indices (z-score) generated by Whatif, the packing quality of a given structure can be evaluated.
RESULTS AND DISUSSION
Overall 3D structures of [BChE]4-PRAD complexes
The amino acid sequence alignment between EeAChE and hBChE revealed high homology, with 52.2% identity in 580 overlapping residues (Figure 1). The residues with stars below them refer to identical residues. The residues on the C-terminus of hBChE are indicated with bold font. By performing the MD simulation on a protein system solvated in explicit water, we for the first time constructed [BChE]4-PRAD complex models with initial structures constructed based on homology modeling of the [AChET]4-ColQ complex.22 There have been some studies of using molecular modeling for construction of AChE tetramers.21,29,64,65 One of them was the application of MD simulation of the explicit water solvated AChE tetramer with a long simulation time of 20 ns, so that the dynamic assembly of the subunits could be studied.65 In our study, the interaction between the tetramerization domain and the proline-rich peptide is the focus, and it is not supposed to change significantly during long time of MD simulation. Thus, we needed to simulate the BChE tetramer structure for a much shorter simulation time in order to obtain a stable MD trajectory for each structure. In our study, our initial structures were refined in a stepwise manner involving a series of consecutive energy minimizations, in order to relieve any unexpected steric clashes or improper contacts in the structures, and to allow smooth structural transition between the catalytic and tetramerization domains of the subunits. Simulations of 1 ns or longer at T = 298.15 K were performed to make sure that we obtained stable MD trajectories for each simulated [BChE]4-PRAD complex with different PRAD sequences.
Figure 1.
Sequence alignment of human BChE (hBChE) (pdb structure: 1P0P)25 with Electrophorus electricus AChE (EeAChE) (pdb structure: 1C2O) (Ref.27). Stars refer to identical residues. The C-terminus of hBChE is indicated in bold font. Resides in blue color indicate the six conserved residues on the C-terminal of hBChE.
The quality of the structures of [BChE]4-PRAD and [AChET]4-ColQ complexes were evaluated with Procheck. Since the structures of the three [BChE]4-PRAD complexes are similar except the various sequences of PRAD, there are small differences between the results of the validation of the structures. So in this paper only the results from the [BChE]4-PRAD1 complex are discussed. The stereochemical parameters calculated by Procheck can be found in the Supporting Information. According to Procheck, there are 79 bad contacts between non-bonded atoms in the [AChET]4-ColQ complex, whereas the number in the initial structure of [BChE]4-PRAD1 complex is reduced to 7. The reduced number of bad contacts is attributed to the energy minimization in gas phase to remove disfavored interactions before the structure was solvated in explicit water. The Ramachandran plot reveals that 85.9% of the residues of the initial structure of [BChE]4-PRAD1 complex are in the most favored regions, with 12.6% in allowed regions, giving a total of 98.5%. The corresponding plot of the [AChE]4-ColQ complex gives similar numbers of 84.4% and 14.3% for the percentages of the residues in the most favored regions and the allowed regions, respectively, with total of 98.7%. Other stereochemical parameters such as dihedral angles, covalent geometry, main-chain hydrogen bonding energy, and planarity were also examined. The G-factor was −0.21 for the initial structure of [BChE]4-PRAD1 complex, compared to the larger G-factor of −0.28 for [AChE]4-ColQ complex, revealing a better quality for our models of the [BChE]4-PRAD complexes. In fact, the most reasonable values for the G-factor in Procheck are between 0 and −0.5, with the best models displaying values closest to zero. Thus, the parameters obtained from the structural validation showed similar or better quality values for the initial structures of [BChE]4-PRAD1 complexes compared to those of [AChE]4-ColQ complex. We also obtained validation results with Procheck for the models of [BChE]4-PRAD1 complexes derived from MD simulation. These structures were minimized before the validation to remove bad contacts which stemmed from MD simulation. By comparing with the initial structure, the bad contacts between non-bonded atoms are reduced from 7 to 0, while the G-factors decrease from −0.21 to −0.29. The change indicates that MD simulation removed bad non-bonded contacts but increased the number of bad dihedral angles of the structures. The result of the phi-psi combination displayed by the Ramachandran plot of the MD structure of [BChE]4-PRAD1 complex is consistent with the change of G-factor, with a lower percentage of the residues in the most favored regions (80.6%) and higher percent of residue in additionally allowed regions (18.5%), giving a total of 99.1%. The lower number of favorable dihedral angles in the structure derived from the MD simulation is understandable, because MD simulation may cause unfavorable dihedral angle due to the temperature and velocity which enable the protein to overcome some high-energy barriers.
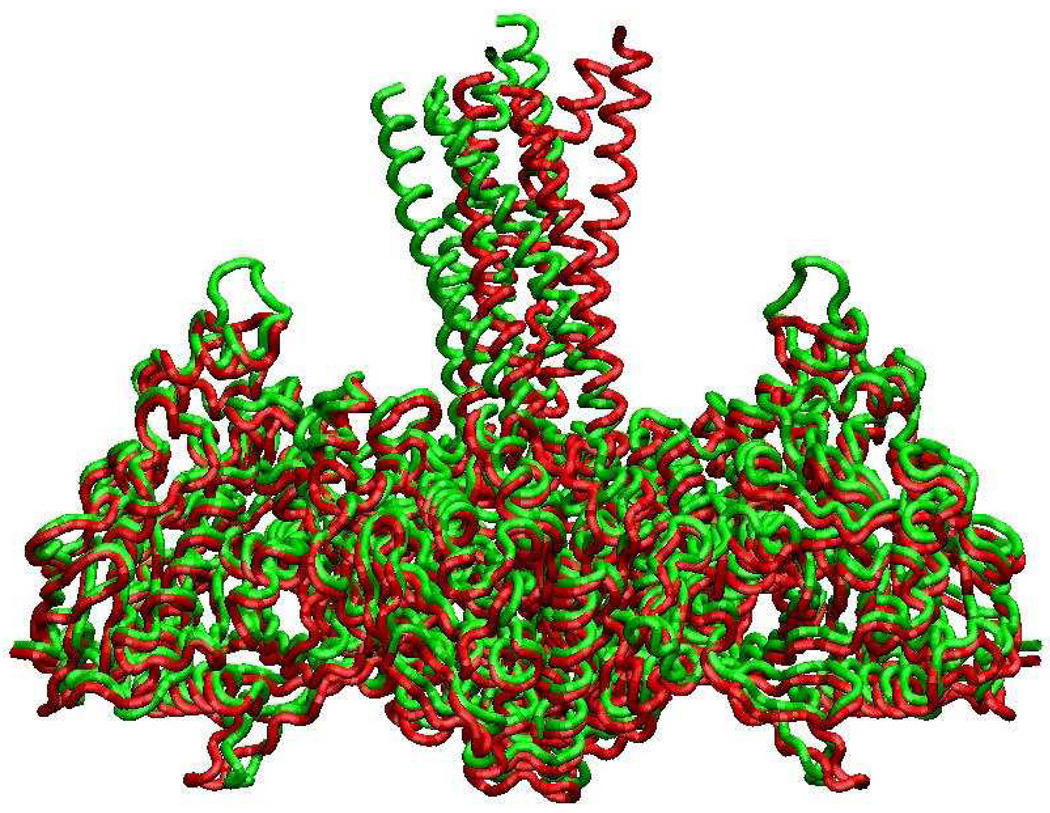
The packing quality of the model was also assessed by the atomic contact analysis of Whatif. The report of the packing quality of the initial structure of [BChE]4-PRAD1 complex showed an average z-score of −0.85 of individual residues, comparable with the z-score of −0.91 for the model of [AChE4]T-ColQ (see Supporting Information). These values fall in the acceptable range for a valid structure (z-score > −5.0). The z-score of the structure derived from the MD simulation of [BChE]4-PRAD1 complex is −1.32. The more negative number is possibly due to the dynamic movement of the structure that caused bad atom-atom interactions. Superimposing the initial structure of [BChE]4-PRAD1 with the [AChET]4-ColQ structure (Figure 2) shows that the overall initial structure of [BChE]4-PRAD1 overlaps well with the structure of [AChET]4-ColQ, except the axis of the superhelix formed by the C-termini of BChE is almost perpendicular to the plane of the body of the BChE tetramer while that of the [AChET]4-ColQ is tilted from the plane. This difference was due to the manual adjustment of the position of the protruding structure of [BChE]4-PRAD1 so that the axis of the superhelix could overlap with the two-fold axis of the BChE tetramer.
Figure 2.
Tube diagram of the superposition of the initial structure of [BChE]4-PRAD1 with template structure [AChET]4-ColQ (only the ColQ segment with PRAD sequence is shown here). [BChE]4-PRAD1 is shown in red color, [AChET]4-ColQ is in green color. This picture is viewed from the side of the two-fold symmetry axis of the tetramer body.
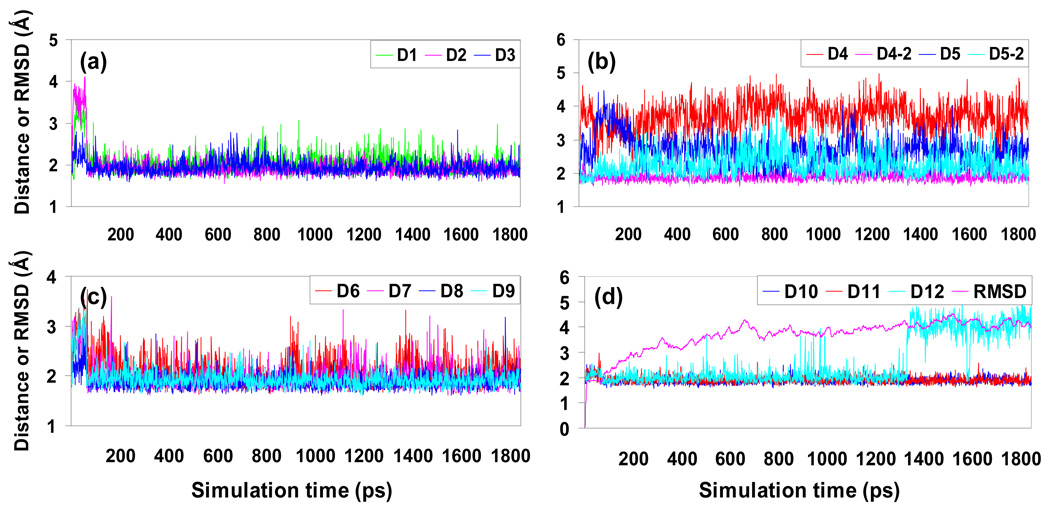
For all three complexes, our MD simulations became stable after ~600 ps of simulation time, which is indicated by the small fluctuation of the root-mean-square (rms) deviation of the simulated positions of the backbone atoms from those in the initial structure along simulation time. The figures of the [BChE]4-PRAD1 complexes are shown in Figure 3. The counterpart figures for the [BChE]4-PRAD2 and [BChE]4-PRAD3 complexes are shown in Supporting Information.
Figure 3.
Plots of the key distances versus time in the simulated structure of [BChE]4-PRAD1 complex. Traces D1, D2, D3 (a), D4, D4-2, D5, D5-2 (b), D6, D7, D8, D9 (c), D10, D11, and D12 (d) indicate the distances between the epsilon hydrogen atoms of tryptophan residues on WAT and the backbone carbonyl oxygen atoms on PRAD with the following residue numbers: A543-E13, B543-E12, C543-E11, D543-E11, D543-E10, A550-E10, A550-E9, B550-E9, C550-E8, D550-E7, A557-E6, B557-E5, C557-E4, and D557-E3. Here the letters “A”, “B”, “C”, and “D” represent the four BChE subunits whereas “E” represents PRAD chain. RMSD indicate the root-mean-square deviation of the coordinates of the backbone atoms of the [BChE]4-PRAD1 complex from MD simulation compared with those of the initial structure.
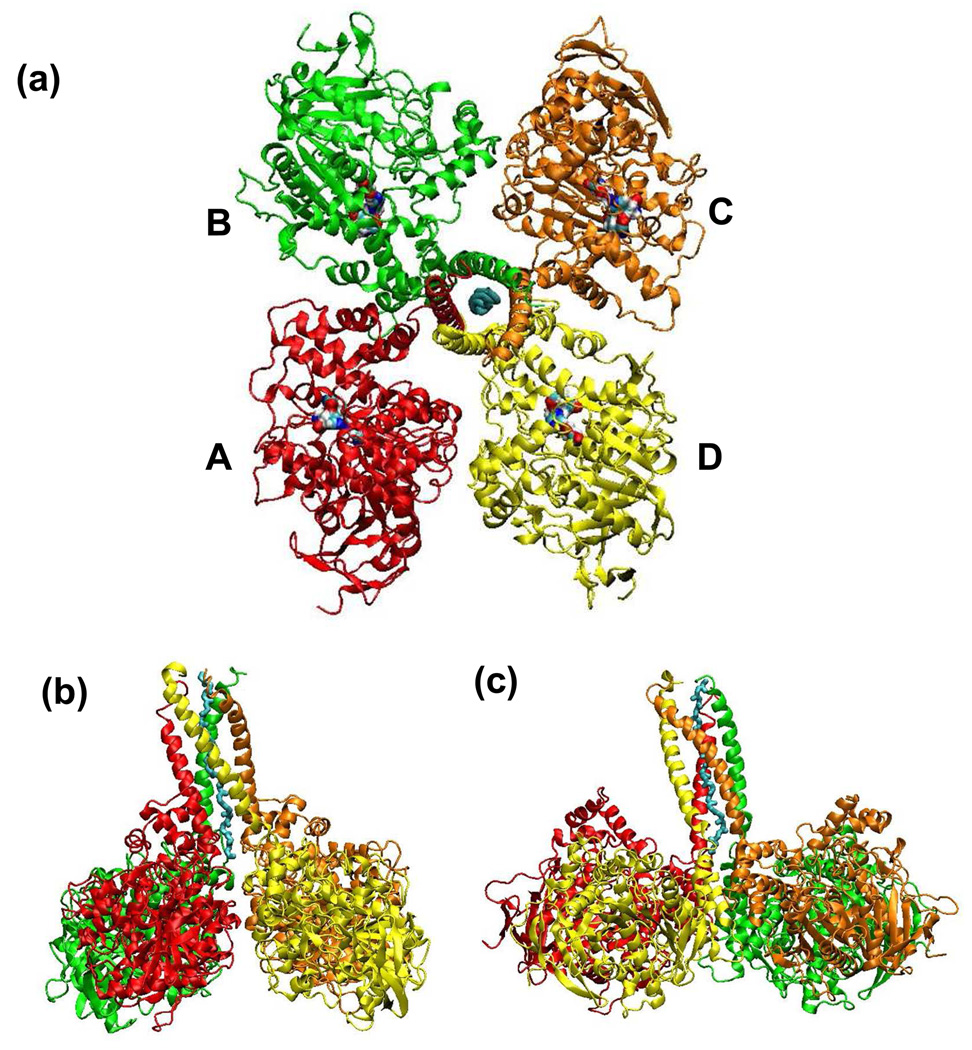
The three [BChE]4-PRAD complexes derived from MD simulations have similar quaternary structures. Figure 4 shows the arrangement of the BChE subunits associated with PRAD1. Similar to the structures of the [AChET]4-ColQ complex,22 the BChE tetramer is a dimer of dimers arranged as a compact, two-fold symmetric structure. The two dimers are oriented in an antiparallel fashion so that the subunits are diagonally equivalent, which accounts for the two-fold axis of the tetramer. Subunits of each dimer are opposite to each other, and interact with each other via a four-helix bundle, with two of the helixes located at the C-termini of the catalytic domain of the subunits. The tetramer is arranged in such way that the axes of the four-helix bundles are at angle of 30° to the two-fold axis of the tetramer, with C-termini of the catalytic domain of the subunits pointing towards the two-fold axis of the tetramer.
Figure 4.
Overall structural of [BChE]4-PRAD1 complex derived from MD simulation. The ribbons in color red, green, orange and yellow represent chains A, B, C and D of the BChE subunits, and the cyan tube represents chain E, or the PRAD chain. (a) Viewed from the top of tetramerization domain of BChE. The multi-colored regions refer to the active sites of the tetramer. Two of active sites in subunits indicated by chains A and C are exposed to outside solvent, while the other two face the central cavity of the BChE tetramer. (b). Viewed from the side of the 2 fold axis of BChE tetramer. This picture has 90° rotation from Figure (a). (c). This picture has 90° clockwise rotation from Figure (b) around the two- fold symmetry axis.
In this paper, four BChE subunits are indicated by chains A, B, C and D with red, green, orange and yellow, respectively, in Figure 4. The middle poly peptide or PRAD is denoted as chain E. Among the chains, A and B form a dimer, and chain C and chain D form a dimer. The catalytic triad in each subunit for this enzyme is shown in Figure 4(a) with a surface area colored by atom type. In our models the active sites of two diagonally opposed subunits, A and C, are exposed to the outside solvent, while the other two face the central cavity of the BChE tetramer, similar to the arrangement in the AChE tetramer models.22,27 The axis of the superhelix comprising the WAT chains of the BChE]4-PRAD complexes was overlapping with the two fold axis of BChE tetramer before the MD simulation. However, possibly due to the left-hand characteristic of the superhelix, the superhelix becomes tilted from the two-fold axis of the tetramer after MD simulation, for all three complexes, with a tilt of around 20° for the complex with PRAD1 indicated in Figure 4(b) and (c) and around 45° for the other two complexes. The root mean square deviation (RMSD) of the positions of the backbone atoms of the BChE tetramerization domain (shown in the Supporting Information) indicates that the tilt of [BChE]4-PRAD1 complex begins to develop during the energy minimization and then becomes stable after around 600 ps of MD equilibration, indicating a possible stabilized trajectory after 600ps of simulation. Figure 5 shows part of the tetramerization domain of BChE subunits (residues 543 to 561) together with PRAD1. In the staggering structure, as indicated in Figure 5(b), the ranking of the four chains is D, C, B, and A, according to the “height” of the C-terminal of each WAT chain.
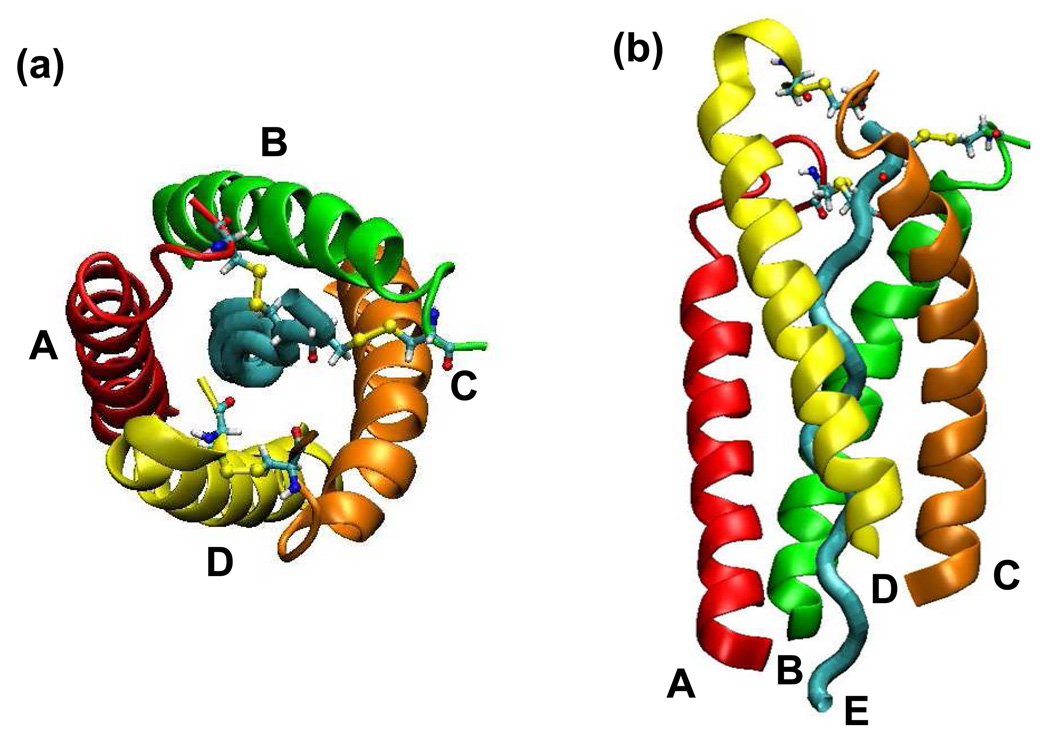
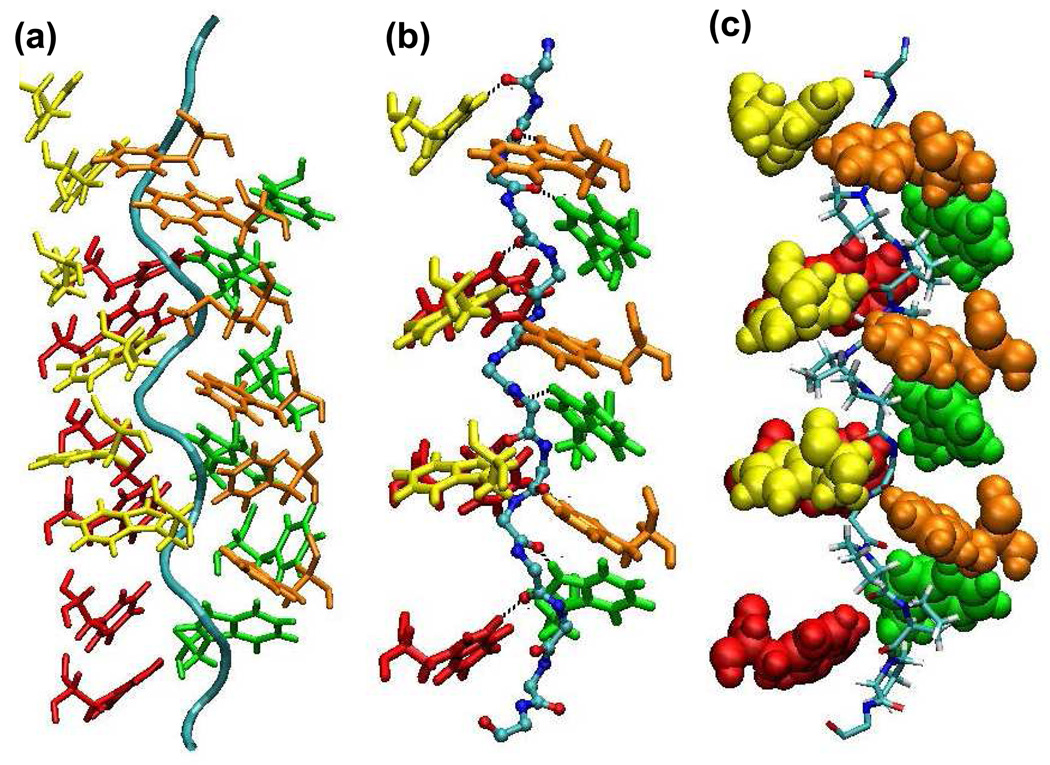
Figure 5.
Interchain disulfide bonds of [BChE]4-PRAD1 complex. These pictures show part of the C-terminus of BChE (residues 543 to 561) together with PRAD. The ribbons in red, green, orange, and yellow represent chains A, B, C, and D of the BChE subunits, respectively, and the cyan tube represents chain E, or the PRAD chain. The colorful ball and stick structures indicate the cysteine residues involved in the interchain disulfide bonds. Yellow sticks refer to the disulfide bonds between WAT and PRAD and between two WAT chains. The disulfide bonds are between Cys571 of chain A and Cys2 of chain E, and Cys571 of chain B and Cys1 of chain E; another disulfide bond is formed through the Cys571 of chains C and D. (a) Viewed from the top of the C-terminus of BChE. The C-terminus of BChE points to the reader. (b) Viewed from the side of the PRAD chain. The C-terminus of BChE points upside of the picture.
Organization of interchain disulfide bonds
Previous studies of AChET tetramer21–23 indicated there are interchain disulfide bonds between the C-termini of two AChET subunits and the ColQ and between two of the C-termini of the subunits. Although it has been shown that disulfide bonds are not essential for the association of AChET and BChE tetramer,21–23 our study does show that these bonds influence the conformation of the tetramerization domain of BChE together with PRAD, thus possibly influencing the binding between PRAD and the WATs. The colorful ball and stick structures in Figure 5 indicate the cysteine residues involved in the interchain disulfide bonds in [BChE]4-PRAD1. The Cys1 and Cys2 on the N-terminus of PRAD are able to form disulfide bonds with the Cys571 from the two lowest chains, B and A, due to the short distance between the PRAD cysteine residues and those from the two BChE subunits. For the two “highest” chains (C and D), since their Cys571 is far from the N-terminal cysteine of PRAD, these two cysteine residues are disulfide bonded with each other. For [BChE]4-PRAD2 and [BChE]4-PRAD3 complexes whose PRADs do not bear any cysteine, the interchain disulfide bonds are formed within the dimers. Due to the different organization of the interchain disulfide bonds among the three complexes, our models show that the axes of the superhelix of [BChE]4-PRAD2 and [BChE]4-PRAD3 are more tilted from the two-fold axis of BChE tetramer than that of the complex [BChE]4-PRAD1. The smaller tilt in the [BChE]4-PRAD1 complex could be due to the interchain disulfide bond between WAT and PRAD (absent in both PRAD2 and PRAD3) which reduces the deviation of the superhelix structure from the two-fold axis of BChE tetramer.
Interactions between PRAD and C-termini of BChE subunits
The key interaction between the WAT and PRAD involves residues 543 to 561 of each BChE subunit. Among these residues there are six hydrophobic residues conserved among cholinesterases,23 including three equally-spaced tryptophan residues, Trp543, Trp550, and Trp557, and three other hydrophobic residues Phe547, Met554, and Phe561. As indicated in Figure 6(a), the six residues are on the same side of the amphiphilic WAT helix, all pointing to the middle PRAD chain. This feature is consistent with the fact that the WAT helixes have 3.5 residues per turn,22 so that there are 2 residues pointing to the same side every 7 residues. Other than these six residues, according to our models, all the other residues numbered 543 to 561 are exposed to water, which is consistent with the hydrophilic property of most of the residues. The exceptions are residues Ala545, Gly546, and Met555, which are hydrophobic.
Figure 6.
(a) Arrangement of the six conserved hydrophobic residues (Trp543, Phe547, Trp550, Met554, Trp557, and Phe561) located on the C-terminus of BChE. The C-terminus is on the top of the pictures in Figure 6. Structures in red, green, orange, and yellow represent the side chains of the residues from chains A, B, C, and D, respectively. Chain E is represented by a cyan tube. The pictures show that the six residues reside on the same side of the chain pointing to the center of the superhelix comprising the four WAT chains. (b) Hydrogen bonding interactions between the tryptophans from WAT chains and the carbonyl oxygens on PRAD. The tryptophan residues Trp543, Trp550, and Trp557 from chains A, B, C, and D are represented as stick structures. The colors representing different side chains are the same as those in (a). The stick-and-ball structure with cyan, blue, and red backbone atoms represents chain E. The black dashed lines indicate the hydrogen bonds between the epsilon hydrogen of tryptophan from the four WAT chains and the carbonyl oxygen of backbone of consecutive residues of PRAD. (c) Hydrophobic interaction between the tryptophans from WAT chains and the PRAD. The tryptophan residues are represented as cloud-like structures indicating the van der Waals surfaces of the side chains of the residues. The indole ring of a tryptophan forming a hydrogen bond with residue numbered i on PRAD has a stacking interaction with the proline ring of residue numbered i+2 on PRAD.
The key distances involving the potential hydrogen bonds between WAT and PRAD1 along simulation time of the complex [BChE]4-PRAD1 are depicted in Figure 3. In the crystal structure of [WAT]4PRAD,22 there are 12 hydrogen bonds between the epsilon hydrogen atoms of tryptophans on the WAT chains and the backbone of PRAD. However, one hydrogen bond involving the conserved tryptophan equivalent to Trp557 in chain D of [BChE]4-PRAD is lost in the model of [AChET]4-ColQ complex,22 due to the rotation of the backbone of PRAD to form disulfide bond with two WAT chains. To examine if this hydrogen bond exists in the [BChE]4-PRAD complex, during our energy minimization of the initial structure the distance between the epsilon hydrogen of Trp557 of chain D and the carbonyl oxygen of the third residue of chain E was first restrained to hydrogen bond distance and then relaxed in the later MD simulation. The results show that the distance can be kept for a long simulation time for the three complexes. Therefore, for our three models of [BChE]4-PRAD complexes, from the N-terminus of PRAD down to its C-terminus, twelve hydrogen bonds have been formed between the eleven successive backbone carbonyls from PRAD and the conserved Trp543, Trp550, and Trp557 from the four WAT chains. The one fewer number of oxygen atoms from PRAD is attributed to the fact that the poly-proline peptide PRAD chain has 3.4 residues per turn,22 while the superhelix formed with the four WAT chains is four-fold screw symmetric. Thus one backbone oxygen has to make bifurcated hydrogen bonds with two tryptophan residues to “make up” the phase. In this paper, the H•••O distances involved in the potential hydrogen bonds between the PRAD and WAT are denoted as D1, D2, D3, D4, D5, D6, D7, D8, D9, D10, D11, and D12 indicating the distances between A543, B543, C543, D543, A550, B550, C550, D550, A557, B557, C557, and D557 from BChE subunits and E13, E12, E11, E11, E10, E9, E8, E7, E6, E5, E4, and E3 from PRAD (here letters A, B, C, D, and E indicate chain names, instead of residue names), respectively. Please notice that the backbone oxygen of residue 11 (E11) from PRAD make bifurcated hydrogen bonds with Trp543 from chains C and D. The distance associated with the lost hydrogen bond in the initial structure is denoted as D12, indicating the distance between Trp557 of chain D and the third residue of chain E.
For the complexes [BChE]4-PRAD2 and [BChE]4-PRAD3, where cysteine is not available on the N-terminus of PRAD, the hydrogen bond relating to distance D12 was well maintained during the MD simulations. However, as indicated by Figure 3(d), for the complex [BChE]4-PRAD1 with cysteine on the N-terminus of PRAD, this hydrogen bond was kept stable for ~1 ns before it was lost. To see if the loss of this hydrogen bond was due to an accidental fall into a local energy minimum, we energy-minimized the structure generated before the bond was lost and then performed MD simulation on the minimized structure. Our simulation shows that the hydrogen bond persists along all the equilibrium time. The availability of the disulfide bond between PRAD and WAT in [BChE]4-PRAD1 complex may be responsible for the less consistency of the hydrogen bond associated with D12, thus providing a negative influence on the binding between WAT and PRAD. On the other hand, the interchain disulfide bonds between PRAD and WAT may strengthen association between the tetramerization domain of BChE and the PRAD. The two opposite effects of interchain disulfide bond between PRAD and WAT chains indicated by our models are consistent with the experimental studies showing that disulfide bond involving residue C571 is not essential for the formation of BChE tetramer together with a poly-proline peptide.21,23
One interesting thing concerning the hydrogen bonds between the WAT chains and PRAD is, during the MD simulations of the three complexes, although there are always twelve hydrogen bonds, there have been a few hydrogen bond “shifts” between the tryptophan residues from adjacent WAT chains and the backbone carbonyls of PRAD chain. In other words, one tryptophan side chain, which originally formed a hydrogen-bond with a carbonyl oxygen, may turn to a neighbor carbonyl oxygen to form another hydrogen bond. Thus, we should evaluate the distances that are related to the “shifted” hydrogen bonds formed during the MD simulation. The distances associated with the “shifted” hydrogen bonds are denoted as D1-2, D4-2, D5-2, D6-2, D7-2, and D8-2, indicating the distances between the epsilon hydrogen of residues A543, D543, A550, B550, C550, and D550 and the backbone oxygen of E14, E10, E9, E8, E7, and E6, respectively. Here “−2” is added to an old distance name to represent a new one, suggesting that the tryptophan involved in the old distance name turns to another backbone oxygen to form a hydrogen bond. As indicated in Figure 3(b), the MD simulations show that in [BChE]4-PRAD1 complex, hydrogen bond related to distance D4 is replaced by the one with distance D4-2, indicating that Trp543 from chain D turns to residue E10 from PRAD to have hydrogen bond in the stable structure, instead of residue E11 from PRAD in the initial structure. In addition, for the [BChE]4-PRAD2 complex, after MD simulation the hydrogen bonds associated with distances D5 and D6 are replaced with those associated with D5-2 and D6-2 when Trp550 from chains A and B turn to Pro9 and Pro8 instead of Pro10 and Pro9 of PRAD to form hydrogen bonds. In addition, for all the complexes, there are cases when tryptophan residues alternately form hydrogen bonds with two carbonyl oxygen atoms during MD simulations. For example, in the [BChE]4-PRAD2 complex, the distances D7 and D7-2 indicate that residue Trp550 from chain C alternately hydrogen-bonds with Pro8 and Pro7 from PRAD. The same case happens to W550 from chain D, which takes turns forming hydrogen bonds with Pro7 and Pro6, as indicated by distances D8 and D8-2, respectively. The dynamic hydrogen bond network between the WAT chains and PRAD of [BChE]4-PRAD complex is not surprising, considering the slight mismatch in the construction of the superhelix wrapping around the middle peptide, in which PRAD with 3.4 residue per turn has to interact with a four-fold superhelix comprising four WAT chains.22
The other interaction between the WAT chains and the PRAD chain is hydrophobic interaction. Figure 5(c) shows the van der Waals surface of the side chains of the conserved tryptophan residues from the WAT chains, together with the prolines and the backbone of PRAD in [BChE]4-PRAD1 complex. As the other two models of [BChE]4-PRAD complexes, there is hydrophobic stacking interaction between the indole rings of the tryptophans and the proline rings on PRAD1. If a given tryptophan forms a hydrogen bond with a carbonyl oxygen from the PRAD at position i, the indole ring of the tryptophan will stack against a proline ring at position i+2 on PRAD (see Figure 5(c)). Besides the three conserved tryptophans, our models show that the side chain of Met554, Phe547, and Phe561 are involved in the hydrophobic packing interaction with the side chains of the residues of PRAD, and in the interaction between adjacent WAT chains.
The structures of [WAT]4-PRAD derived from the condition with or without additional sodium and chloride ions were obtained after 10 ns of MD simulations. Superimposing of the two structures does not reveal significant structural change, indicating that the influence of free ions on our simulated systems can be neglected, in terms of structure. The interactions between PRAD and WAT chains of the [WAT]4-PRAD systems are alike, and bear similar features as those in [BChE]4-PRAD complexes. There are both hydrophobic and hydrogen bond interactions between WAT and PRAD. There are 12 hydrogen bonds between the epsilon hydrogen atoms of conservative tryptophans on WAT and the backbone hydrogen atoms of PRAD, although the one closest to the C-terminus of PRAD is not as stable as that in the [BChE]4-PRAD complexes, possibly due to the omission of the catalytic domain of the enzyme in [WAT]4-PRAD. The hydrogen bond “shift” observed in [BChE]4-PRAD complexes are also available in the [WAT]4-PRAD systems. The figures regarding the simulations of the [WAT]4-PRAD systems are shown in the Supporting Information.
Insights on increasing the association between PRAD and the tetramerization domain of BChE
The insights into strengthening the association between the BChE tetramer and the polyproline peptide are based on our understanding of the binding mode between the C-terminus of BChE and the PRAD in the [BChE]4-PRAD complexes. Since residues 543 to 561 supply the key interactions between WAT and PRAD, our investigation focuses on these residues. Among these residues our models show that only six of the residues, Trp543, Phe547, Trp550, Met554, Trp557, and Phe561, point to the center of the superhelix and interact directly with the poly-proline peptide in the middle. Previous studies have shown that with addition of poly-L-proline, a BChE mutant with residues Trp543, Phe547, Trp550, Tyr553, Trp557, Phe561, and Tyr564 all mutated to alanine yielded no tetramer,23 but did not show what would happen if just one individual residue was mutated. Since the three tryptophans are shown to have hydrogen bonds with the backbone of PRAD, the remaining three residues, Phe547, Met554, and Phe561, are possible substitution candidates to increase the binding affinity between WAT and PRAD. Although these three residues are highly conserved among cholinesterases, our previous experience in discovering substitutions for design of high-activity mutants of BChE6,14–16 supplies us with understanding that mutation of a conserved residue can also lead to a mutant with desirable properties. Since the side chains of these three residues are involved in the packing interaction with the side chain of the residues from PRAD, and that between adjacent WAT chains, mutating these residues to other hydrophobic residues may be favorable for increasing the binding affinity. In addition, since our models indicate three hydrophobic residues, i.e. Ala545, Gly546, and Met555, are exposed to the outside water, mutating these residues to hydrophilic residues may benefit the stability of the BChE tetramer.
Concerning the idea of mutating the proline-rich peptide to increase the binding between WAT and PRAD, our study did not show significant difference among the binding between WAT and PRAD of the three complexes. Although there is some conformational difference due to the different arrangement of interchain disulfide bonds depending on the availability of the N-terminal cysteines on PRAD, the influence of interchain disulfide bonds is not significant. This conclusion is supported by experimental data, showing that usage of the three peptides with or without N-terminal cysteine for the association of the BChE subunits did not yield a significantly different percentage of tetramers.18,19,23 By examining the residues of the three PRAD sequences, it can be seen that those residues all have hydrophobic side chains, leading to their ability to form hydrophobic stacking interaction with side chains from the tetramerization domain of BChE. For this reason, when proline is mutated with leucine or phenylalanine, the formation of the tetramers was not affected significantly.18,19,23 These insights suggest that mutating the PRAD residues with other hydrophobic residues could enhance binding between PRAD and WAT.
CONCLUSION
Three comprehensive 3D structural models of human butyrylcholinesterase (BChE) tetramer complexed with proline-rich attachment domain (PRAD) have been developed by carrying out homology modeling followed by molecular dynamics simulation of the explicit water-solvated protein systems. The 3D structural models enable us to clearly identify the orientation of the BChE subunits in the tetramer and to study how the tetramerization domain of BChE binds with the proline-rich peptide to form stable tetramers of human BChE. The four helixes of tryptophan amphiphilic tetramerization (WAT) domain of BChE form a left hand superhelix wrapping around the proline-rich peptide in the middle. The six conserved hydrophobic residues located on the C-terminus of BChE are responsible for the key interaction between the tetramerization domain and PRAD. The three equally spaced tryptophan residues on adjacent WAT chains have a dynamic hydrogen-bond network with consecutive residues of PRAD to form 12 hydrogen bonds; there are hydrophobic interactions between the side chain of tryptophans and the proline rings located on the PRAD. The simulated models suggest that mutation of three residues, i.e. Phe547, Met554, and Phe561, to other hydrophobic residues may be beneficial for increasing the binding between the tetramerization domain of BChE and the proline-rich peptide. The structural insights obtained from the present study may be valuable for future rational design of recombinant wild-type or mutant BChE with a longer residence time in blood.
Supplementary Material
ACKNOWLEDGMENTS
The research was supported by NIH (grant R01 DA013930 to CGZ). The authors also acknowledge the Center for Computational Sciences (CCS) at University of Kentucky for supercomputing time on IBM X-series Cluster with 340 nodes and 1,360 processors.
Footnotes
Supporting Information Available One Table lists the phrases defined in this paper; three figures for the structural validation of the model of [AChET]4-ColQ, the initial structure of [BChE]4-PRAD1, and the [BChE]4-PRAD1 structure derived from MD simulation; three figures for the MD trajectories for BChE tetramer binding with the PRAD1, PRAD2, and PRAD3; one figure showing the position change of BChE tetramerization domain along simulation time; three figures regarding the simulations of [WAT]4-PRAD complexes with or without additional ions. This information is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Johanson CE, Fischman MW. Pharmacol. Rev. 1989;41:3. [PubMed] [Google Scholar]
- 2.Carroll FI, Howell LL, Kuhar MJ. J. Med. Chem. 1999;42:2721. doi: 10.1021/jm9706729. [DOI] [PubMed] [Google Scholar]
- 3.Gorelick DA, Gardner EL, Xi ZX. Drugs. 2004;64:1547. doi: 10.2165/00003495-200464140-00004. [DOI] [PubMed] [Google Scholar]
- 4.Gorelick DA. Drug. Alcohol. Depend. 1997;48:159. doi: 10.1016/s0376-8716(97)00119-1. [DOI] [PubMed] [Google Scholar]
- 5.Lockridge O, Blong RM, Masson P, Froment MT, Millard CB, Broomfield CA. Biochemistry. 1997;36:786. doi: 10.1021/bi961412g. [DOI] [PubMed] [Google Scholar]
- 6.Zhan CG, Zheng F, Landry DW. J. Am. Chem. Soc. 2003;125:2462. doi: 10.1021/ja020850+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pan Y, Gao D, Yang W, Cho H, Yang G, Tai HH, Zhan CG. Proc. Natl. Acad. Sci. U. S. A. 2005;102:16656. doi: 10.1073/pnas.0507332102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sun H, Pang YP, Lockridge O, Brimijoin S. Mol. Pharmacol. 2002;62:220. doi: 10.1124/mol.62.2.220. [DOI] [PubMed] [Google Scholar]
- 9.Gao D, Cho H, Yang W, Pan Y, Yang G, Tai HH, Zhan CG. Angew. Chem. Int. Ed. Engl. 2006;45:653. doi: 10.1002/anie.200503025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pan Y, Gao D, Yang W, Cho H, Zhan CG. J. Am. Chem. Soc. 2007;129:13537. doi: 10.1021/ja073724k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hamza A, Cho H, Tai HH, Zhan CG. J. Phys. Chem. B. 2005;109:4776. doi: 10.1021/jp0447136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yang W, Pan Y, Zheng F, Cho H, Zhan CG. Biophys. J. 2008 in press. [Google Scholar]
- 13.Zheng F, Yang W, Ko MC, Liu J, Cho H, Gao D, Tong M, Tai HH, Woods JH, Zhan CG. J. Am. Chem. Soc. 2008;130:12148. doi: 10.1021/ja803646t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gao D, Zhan CG. Proteins. 2006;62:99. doi: 10.1002/prot.20713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gao DQ, Zhan CG. J. Phys. Chem. B. 2005;109:23070. doi: 10.1021/jp053736x. [DOI] [PubMed] [Google Scholar]
- 16.Zhan CG, Gao D. Biophys. J. 2005;89:3863. doi: 10.1529/biophysj.105.070276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Saxena A, Ashani Y, Raveh L, Stevenson D, Patel T, Doctor BP. Mol. Pharmacol. 1998;53:112. doi: 10.1124/mol.53.1.112. [DOI] [PubMed] [Google Scholar]
- 18.Li H, Schopfer LM, Masson P, Lockridge O. Biochem. J. 2008;411:425. doi: 10.1042/BJ20071551. [DOI] [PubMed] [Google Scholar]
- 19.Duysen EG, Bartels CF, Lockridge O. J. Pharmacol. Exp. Ther. 2002;302:751. doi: 10.1124/jpet.102.033746. [DOI] [PubMed] [Google Scholar]
- 20.Huang YJ, Huang Y, Baldassarre H, Wang B, Lazaris A, Leduc M, Bilodeau AS, Bellemare A, Cote M, Herskovits P, Touati M, Turcotte C, Valeanu L, Lemee N, Wilgus H, Begin I, Bhatia B, Rao K, Neveu N, Brochu E, Pierson J, Hockley DK, Cerasoli DM, Lenz DE, Karatzas CN, Langermann S. Proc. Natl. Acad. Sci. U. S. A. 2007;104:13603. doi: 10.1073/pnas.0702756104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Blong RM, Bedows E, Lockridge O. Biochem. J. 1997;327(Pt 3):747. doi: 10.1042/bj3270747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dvir H, Harel M, Bon S, Liu WQ, Vidal M, Garbay C, Sussman JL, Massoulie J, Silman I. Embo J. 2004;23:4394. doi: 10.1038/sj.emboj.7600425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Altamirano CV, Lockridge O. Biochemistry. 1999;38:13414. doi: 10.1021/bi991475+. [DOI] [PubMed] [Google Scholar]
- 24.Ngamelue MN, Homma K, Lockridge O, Asojo OA. Acta. Crystallogr. Sect. F. Struct. Biol. Cryst. Commun. 2007;63:723. doi: 10.1107/S1744309107037335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nicolet Y, Lockridge O, Masson P, Fontecilla-Camps JC, Nachon F. J. Biol. Chem. 2003;278:41141. doi: 10.1074/jbc.M210241200. [DOI] [PubMed] [Google Scholar]
- 26.Sussman JL, Harel M, Frolow F, Oefner C, Goldman A, Toker L, Silman I. Science. 1991;253:872. doi: 10.1126/science.1678899. [DOI] [PubMed] [Google Scholar]
- 27.Bourne Y, Grassi J, Bougis PE, Marchot P. J. Biol. Chem. 1999;274:30370. doi: 10.1074/jbc.274.43.30370. [DOI] [PubMed] [Google Scholar]
- 28.Bourne Y, Taylor P, Bougis PE, Marchot P. J. Biol. Chem. 1999;274:2963. doi: 10.1074/jbc.274.5.2963. [DOI] [PubMed] [Google Scholar]
- 29.Dvir H, Wong DM, Harel M, Barril X, Orozco M, Luque FJ, Munoz-Torrero D, Camps P, Rosenberry TL, Silman I, Sussman JL. Biochemistry. 2002;41:2970. doi: 10.1021/bi011652i. [DOI] [PubMed] [Google Scholar]
- 30.Harel M, Hyatt JL, Brumshtein B, Morton CL, Yoon KJ, Wadkins RM, Silman I, Sussman JL, Potter PM. Mol. Pharmacol. 2005;67:1874. doi: 10.1124/mol.104.009944. [DOI] [PubMed] [Google Scholar]
- 31.Harel M, Kleywegt GJ, Ravelli RB, Silman I, Sussman JL. Structure. 1995;3:1355. doi: 10.1016/s0969-2126(01)00273-8. [DOI] [PubMed] [Google Scholar]
- 32.Hornberg A, Tunemalm AK, Ekstrom F. Biochemistry. 2007;46:4815. doi: 10.1021/bi0621361. [DOI] [PubMed] [Google Scholar]
- 33.Raves MLGK, Schrag JD, Schmide MF, Phillips GN, Chiu W, Howard AJ, Silman I, Sussman JL. Quanternary structure of tetrameric acetylcholinesterase. New York: Plenum Publishing; 1998. [Google Scholar]
- 34.Herzberg O, Sussman JL. J. Appl. Crystallogr. 1983;16:144. [Google Scholar]
- 35.Sussman JL, Holbrook SR, Church GM, Kim SH. Acta. Crystallographica. Section A. 1977;33:800. [Google Scholar]
- 36.Bernstein FC, Koetzle TF, Williams GJB, Meyer EF, Brice MD, Rodgers JR, Kennard O, Shimanouchi T, Tasumi M. J. Mol. Biol. 1977;112:535. doi: 10.1016/s0022-2836(77)80200-3. [DOI] [PubMed] [Google Scholar]
- 37.Huang XQ, Hardison RC, Miller W. Comp. Appl. Biosci. 1990;6:373. doi: 10.1093/bioinformatics/6.4.373. [DOI] [PubMed] [Google Scholar]
- 38. http://expasy.org/.
- 39.Case DA, Darden TA, Cheatham TEI, Simmerling CL, Wang J, Duck R, Luo KM, Merz KM, Wang B, Pearlman DA, Crowley M, Brozell S, Tsui V, Gohlke H, Mongan J, Hornak V, Cui G, Beroza P, Schafmeister C, Caldwell JW, Ross WS, Kollman PA. AMBER 8. San Francisco: University of California; 2004. [Google Scholar]
- 40.Cieplak P, Caldwell J, Kollman P. J. Comput. Chem. 2001;22:1048. [Google Scholar]
- 41.Wang JM, Cieplak P, Kollman PA. J. Comput. Chem. 2000;21:1049. [Google Scholar]
- 42.Normal Reference Range Table; The University of Texas Southwestern Medical Center at Dallas; pp Used in Interactive Case Study Companion to PATHOLOGIC BASIS of DISEASE.
- 43.Le TT, Bhushan V, Rao D. First Aid for the USMLE Step 1 2008 (First Aid for the Usmle Step 1) 18 ed. McGraw-Hill Medical; 2007. [Google Scholar]
- 44.Maton A, Hopkins J, McLaughlin CW, Johnson S, Warner MQ, LaHart D, Wright JD. Human Biology and Health. Englewood Cliffs, NJ, USA: Prentice Hall; 1993. [Google Scholar]
- 45.Jorgensen WL, Chandrasekhar J, MadurAa J, Klein ML. J. Chem. Phys. 1983;79:926. [Google Scholar]
- 46.Pan YM, Gao DQ, Zhan CG. J. Am. Chem. Soc. 2008;130:5140. doi: 10.1021/ja077972s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gao DQ, Zhan C-G. J. Phys. Chem. B. 2005;109:23070. doi: 10.1021/jp053736x. [DOI] [PubMed] [Google Scholar]
- 48.Pan YM, Gao DQ, Yang WC, Cho H, Yang GF, Tai HH, Zhan C-G. Proc. Natl. Acad. Sci. U.S.A. 2005;102:16656. doi: 10.1073/pnas.0507332102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gao D, Cho H, Yang W, Pan Y, Yang G, Tai HH, Zhan CG. Angew. Chem. Int. Ed. 2006;45:653. doi: 10.1002/anie.200503025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gao DQ, Zhan C-G. Proteins. 2006;62:99. doi: 10.1002/prot.20713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Koca J, Zhan C-G, Rittenhouse RC, Ornstein RL. J. Am. Chem. Soc. 2001;123:817. doi: 10.1021/ja000439r. [DOI] [PubMed] [Google Scholar]
- 52.Koca J, Zhan C-G, Rittenhouse RC, Ornstein RL. J. Comput. Chem. 2003;24:368. doi: 10.1002/jcc.10217. [DOI] [PubMed] [Google Scholar]
- 53.Zhan C-G, de Souza ON, Rittenhouse R, Ornstein RL. J. Am. Chem. Soc. 1999;121:7279. [Google Scholar]
- 54.Leach AR. Molecular modelling: principles and applications. second ed. Essex, England: Pearson Education Limited; 2001. [Google Scholar]
- 55.Case DA. http://www.wiley.com/legacy/wileychi/ecc/samples/sample09.pdf.
- 56.Berendsen HJC, Postma JPM, van Gunsteren WF, DiNola A, Haak JR. J. Chem. Phys. 1984;81:3684. [Google Scholar]
- 57.Ryckaert JP, Ciccotti G, Berendsen HJC. J. Comput. Phys. 1977;23:327. [Google Scholar]
- 58.Essmann U, Perera L, Berkowitz ML, Darden T, Lee H, Pedersen LG. J. Chem. Phys. 1995;103:8577. [Google Scholar]
- 59.Laskowski RA, MacArthur MW, Moss DS, Thornton JM. J. Appl. Crystallogr. 1993;26:283. [Google Scholar]
- 60.Morris AL, Macarthur MW, Hutchinson EG, Thornton JM. Proteins-Structure Function and Genetics. 1992;12:345. doi: 10.1002/prot.340120407. [DOI] [PubMed] [Google Scholar]
- 61.Vriend G. J. Mol. Graphics. 1990;8:52. doi: 10.1016/0263-7855(90)80070-v. [DOI] [PubMed] [Google Scholar]
- 62.Vriend G. J. Mol. Graph. 1990;8:52. doi: 10.1016/0263-7855(90)80070-v. [DOI] [PubMed] [Google Scholar]
- 63.Vriend G, Sander C. J. Appl. Crystallogr. 1993;26:47. [Google Scholar]
- 64.Zhang D, McCammon JA. PLoS. Comput. Biol. 2005;1:e62. doi: 10.1371/journal.pcbi.0010062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gorfe AA, Chang CE, Ivanov I, McCammon JA. Biophys. J. 2008;94:1144. doi: 10.1529/biophysj.107.117879. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.