Abstract
The X-ray structures of imidazolylcobalamin (ImCbl) and histidinylcobalamin (HisCbl) are reported. These structures are of interest given that the recent structures of human and bovine transcobalamin prepared in their holo forms from aquacobalamin show a histidine residue of the metalloprotein bound at the β-axial site of the cobalamin (Wuerges, J. et al. Proc. Natl. Acad. Sci. U.S.A. 2006, 103, 4386–4391). The β-axial Co–N bond distances for ImCbl and HisCbl are 1.94(1) and 1.951(7) Å, respectively. The α-axial Co–N bond distances to the 5,6-dimethylbenzimidazole are 2.01(1) and 1.979(8) Å for ImCbl and HisCbl, respectively, and are typical for cobalamins with weak σ-donor ligands at the β-axial site. The corrin fold angles of 11.8(3)° (ImCbl) and 12.0(3)° (HisCbl) are smaller than those typically observed for cobalamins.
Introduction
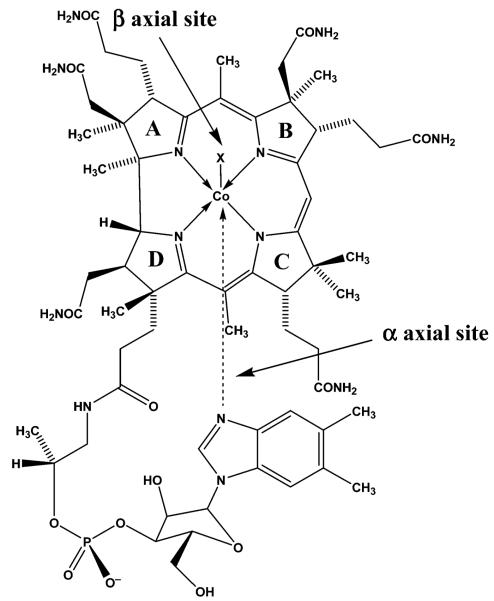
Imidazole-based ligands frequently directly coordinate, via the lone pair of electrons of the basic imidazole N atom, to metalloprotein metal centers, including Fe, Cu, and Zn.1 The Co-containing B12 cofactors known as cobalamins (Cbls; Figure 1) are comprised of a Co center coordinated at the equatorial sites by a macrocyclic tetrapyrrole ligand; the corrin ring. A nucleotide side chain extends from C17 of ring D of the corrin and terminates with a α-5,6-dimethylbenzimidazole moiety (α-DMB). α-DMB can either coordinate via the imidazole N atom to the lower (α)-axial site of the Cbl (“base-on” conformation; Figure 1) or remain uncoordinated (“base-off” conformation). In cob(III)alamins, various ligands can occupy the β-axial site, including methyl and 5′-deoxyadenosyl moieties. Methylcobalamin (MeCbl) and adenosylcobalamin (AdoCbl) are essential cofactors for the methionine synthase (MS) and methylmalonyl-CoA mutase (MCM) enzyme reactions in humans, respectively.2-4
Figure 1.
Structures of the major naturally occurring cob(III)alamins: X = 5′-deoxyadenosyl, adenosylcobalamin (coenzyme B12); X = CH3, methylcobalamin; X = H2O/OH, aquacobalamin/hydroxycobalamin.
In 1990, using 15N-labeling and EPR spectroscopy, it was discovered that during the MeCbl-dependent MS reaction, α-DMB is displaced by a histidine (His) imidazole group from a side chain of the metalloenzyme, the so-called “base-off/His-on” conformation.5 It is now known that this conformation occurs in several Cbl-dependent enzymes, including MCM and glutamate mutase,6-12 all of which have the conserved sequence DXHXXG.13 Mutagenesis studies show that substitution of His with another amino acid leads to enzyme inactivation for MS (His → Gly)14 and for MCM (His → Ala or Asn).15 For glutumate mutase, substituting His with Gly or Gln results in a significant decrease in enzyme turnover and a change in the rate-limiting step.14 Interestingly, several Cbl-dependent enzymes (class II AdoCbl-dependent enzymes) remain instead in the base-on conformation during catalysis (ribonucleotide reductase,16 diol dehydratase,17,18 and ethanolamine ammonia lyase19), and it is unclear what catalytic advantage(s) result from His displacement of α-DMB.14,15 Recently, it was shown that the rate of Co–C bond homolysis for 5′-deoxyadenosylimidazolylcobamide, in which the α-DMB of AdoCbl is replaced by imidazole, is only 4.3 times slower than that for AdoCbl (37 °C).20 Model studies and theoretical calculations also show that displacement of α-DMB trans to an alkyl ligand by imidazole-type ligands does not significantly increase the rate of homolytic cleavage or decrease the bond strength of the Co–C bond.21-24 Furthermore, although initial X-ray diffraction results suggested that the Co–N(Im) bond is abnormally long for some B12-dependent enzymes (2.5 Å for MCM7 and 2.3 Å for glutamate mutase10), the structures may actually represent Co in mixed-ligand and oxidation states (CoIII and CoII), due to X-ray-induced photoreduction.10,25-27
In protein-free Cbls, α-DMB is difficult to displace from the α-axial site but can be displaced by cyanide,28 by imidazole-type ligands,29,30 and by protonation of the basic N of the imidazole moiety.31 In comparison, substitution of the β-axial ligand of aquacobalamin (H2OCbl+, X = OH2; Figure 1) is rapid and thermodynamically favorable for many ligands,32 including imidazole-type ligands.29,30,33-38 These reactions drew little attention until recently, when kinetic studies using stopped-flow spectroscopy showed that binding of H2OCbl+ to the Cbl transporter protein transcobalamin (TC) occurs in two steps: a rapid step in which H2OCbl+ initially binds to TC (k ~ 6 × 107 M−1 s−1 at 37 °C) followed by a slower step (k ~ 0.3 s−1 at 37 °C) in which a His side chain of TC displaces the β-axial aqua ligand of H2OCbl+-bound TC.39,40 TC, one of three Cbl transporter proteins in mammals, transports Cbls from the circulation into tissues. It does this very efficiently via tight binding of Cbl to TC (K ~ 1010−1012 M−1 41) and by endocytosis of the TC–Cbl complex after binding to a specific extracellular surface protein; the TC receptor. Plasma and serum levels of holo TC are currently being explored as markers of B12 deficiency.42-47
The first crystal structures of a B12 transporter protein, histidinylcobalamin-bound TC from recombinant human and bovine TC, prepared in its holo form using H2OCbl+, were recently reported.48,49 In these structures, substitution of the β-axial H2O ligand by a His side chain of the metalloprotein is clearly observed. During the structural refinements, it was necessary to confine the β-axial Co–N(His) bond to 2.15 Å. Furthermore, a reliable estimate of the α-axial Co–N(DMB) bond length was not possible, due to the existence of mixed Co oxidation states (CoIII and CoII) in the crystal, probably as a consequence of CblIII reduction by the X-ray beam.27 In this paper we report the first structures of free Cbls bound at the β-axial site to an imidazole ligand: specifically, we report the crystal structures of imidazolylcobalamin and histidinylcobalamin.
Experimental Section
General Procedures and Chemicals
Hydroxycobalamin hydrochloride (HOCbl·HCl; stated purity by manufacturer is g96%; aromatic region of the 1H NMR spectrum shows that it contains at least ~6% impurities50) was purchased from Fluka BioChemica. Imidazole (99%) and l-His (98%) were purchased from Acros Organics. All chemicals were used without further purification. Water was purified using a Barnstead Nanopure Diamond water purification system, and/or HPLC-grade water was used.
UV–vis spectra were recorded on a Cary 5000 spectrophotometer equipped with a thermostatted cell holder, operating with WinUV Bio software (version 3.00). 1H NMR spectra were recorded on a Varian Inova 500 MHz spectrometer equipped with a 5 mm thermostatted (22.0 ± 0.5 °C) probe. Solutions were prepared in D2O, and TSP (3-(trimethylsilyl)propionic-2,2,3,3-d4 acid, Na+ salt), was used as an internal reference.
pH measurements were made at room temperature with a Corning model 445 pH meter equipped with a Mettler-Toledo Inlab 423 electrode. The electrode was filled with a 3 M KCl/saturated AgCl solution, pH 7.00, and standardized with standard BDH buffer solutions at pH 4.00 and 7.00.
Synthesis of Imidazolylcobalamin (ImCbl)
Imidazole (5.7 mg, 1.3 equiv) was added to HOCbl·HCl (100 mg) dissolved in TES buffer (1.0 mL, 0.10 M, pH 7.4), and the mixture reacted for 2 h at room temperature. The product was precipitated by the addition of cold (−20 °C) acetone, vacuum-filtered, and dried overnight under vacuum at 60 °C. 1H NMR in D2O, pD 8.80 (δ, ppm): 5.95, 6.20, 6.34 (d), 6.72, 6.80, 6.93, 7.08, and 7.27; ~95% purity (see Figure S1 in the Supporting Information).50 The percentage of non-Cbl impurities determined by converting the products to dicyanocobalamin, (CN)2Cbl− (0.10 M KCN, pH 10.0, ε368 = 3.04 × 104 M−1 cm−1),51 was 5 ± 1%. ES-MS (+ve), m/z: 1396.1 (calcd for [ImCbl]+, C65H92N15O14PCo = 1396.6); 698.6 (calcd for [ImCbl + H]2+, C65H93N15O14PCo = 698.3); 664.9 (calcd for [Cbl + 2H]2+, C62H90N13O14PCo = 665.3); 1329.5 (calcd for [Cbl + H]+, C62H89N13O14PCo = 1329.6).
Attempted Synthesis of Histidinylcobalamin (HisCbl)
The procedure was identical to that for ImCbl, except that 1.6 equiv of l-His was used. 1H NMR in D2O, pD 7.80 (δ, ppm): 7.28, 7.10, 6.81, 6.72, 6.32 (d), 6.20, and 5.75; ~70% purity (see Figure S2 in the Supporting Information). Free ligand (~25%) coprecipitated with [HisCbl]Cl; its chemical shifts appear at 7.91 and 7.12 ppm. (At pD 8.80, the chemical shifts of l-His and [HisCbl]Cl overlap.) ES-MS (+ve), m/z: 1484.3 (calcd for [HisCbl]+, C68H96N16O16PCo = 1484.6); 742.3 (calcd for [HisCbl + H]2+, C68H97N15O14PCo = 742.3); 664.9 (calcd for [Cbl + 2H]2+, C62H90N13O14PCo = 665.3); 1329.5 (calcd for [Cbl + H]+, C62H89N13O14PCo = 1329.6). ES-MS (−ve): 1482.5 (calcd for [HisCbl − 2H]−, C68H94N16O16PCo = 1482.6).
X-ray Diffraction Studies
Crystals of ImCbl were grown from water. Briefly, HOCbl·HCl (8.85 mg) and imidazole (4.00 mg) were dissolved in water (0.50 mL), NaClO4 (1.50 mg) was added, and the pH was adjusted to ~7.5 using concentrated HClO4. The sample was kept at room temperature, and crystals appeared after ~24 h. A crystal specimen was mounted in a glass capillary, and diffraction data were collected at room temperature on a modified Stoe four-circle diffractometer using Mo Kα radiation (λ = 0.71073 Å).
Crystals of HisCbl were grown by adding a solution of histidine (0.20 mL, ~100 mg/mL, pH = 7.5) to an aqueous solution of HOCbl·HCl (0.10 mL, 100 mg/mL, pH = 7.4). The sample was kept at room temperature, in a closed-cap vial, and crystals appeared after about 32 h. X-ray diffraction data for HisCbl were measured at 100 K (Oxford 700 series cryostream) on a Bruker AXS platform single-crystal X-ray diffractometer upgraded with an APEX II CCD detector. Graphite-monochromatized Mo Kα radiation was used. Crystals were mounted on a thin glass fiber from a pool of Fluorolube and placed under a stream of nitrogen.
Both structures were solved by direct methods to yield the Co atoms plus most remaining atoms of the structure. Missing atoms (mostly in the solvent region) were located in subsequent electron density maps. Full-matrix least-squares refinement on F2 was performed with the program SHELXL-97.52 No absorption correction was applied to the data. Scattering factors, including real and imaginary dispersion corrections, were taken from the International Tables of Crystallography. H-atom positions were calculated and refined as “riding” on their respective non-H atom. The isotropic atomic displacement parameter (adp) for each H atom was set to 1.5 times the equivalent isotropic adp of the adjacent non-H atom. Crystallographic residuals at the close of the refinement are also given in Table 1.
Table 1.
Crystallographic Data for ImCbl and HisCbl
| ImCbl | HisCbl | |
|---|---|---|
| empirical formula | C65H91N15O14-PCo·22H2O | C68H96N16O16P-Co·20H2O·0.5HCl |
| H2O sites | 22 | 27 |
| formula weight | 1792.8 | 1862.1 |
| cryst syst | orthorhombic | orthorhombic |
| space group | P212121 | P212121 |
| unit cell dimens | ||
| a [Å] | 15.834(10) | 15.697(1) |
| b [Å] | 22.469(10) | 22.422(1) |
| c [Å] | 25.766(10) | 25.417(1) |
| V [Å3] | 9167(8) | 8946(1) |
| Z | 4 | 4 |
| Dcalc [g cm−3] | 1.299 | 1.383 |
| μ [mm−1] | 0.293 | 0.318 |
| F(000) | 3840 | 3980 |
| cryst size [mm] | 0.3 × 0.1 × 0.1 | 0.35 × 0.20 × 0.15 |
| θ range for data collection [deg] | 2.83–22.51 | 1.21–18.80 |
| T [K] | 298 | 103 |
| wavelength [Å] | 0.71073 | 0.71073 |
| no. of unique reflns | 5976 | 6966 |
| no. of reflns with I > 2σI | 3952 | 6050 |
| R[int] | 0.089 | 0.079 |
| data/restraints/ params | 5976/1787/1076 | 6966/2611/1249 |
| final R indices: | ||
| R1 (I > 2σI, all data) | 0.0811 (0.1420) | 0.0734 (0.0869) |
| R2 (I > 2σI, all data) | 0.1862 (0.2424) | 0.1885 (0.2023) |
| largest diff peak/hole [e Å−3] | 0.61/−0.48 | 0.56/−0.45 |
In ImCbl, the B12 complex was very well defined except for the hydroxymethyl group of the ribose in the nucleotide loop, which was found to be disordered over two alternate conformations. The solvent electron density was modeled with 22 fully occupied and anisotropically refined H2O molecules.
In HisCbl, the Co-bound histidine is disordered over two alternate conformations. Discrete disorder was also observed for the amide side chains A and E and for the ribose hydroxymethyl group. The solvent comprises 20 (partially occupied) water molecules distributed over 27 sites and a half-occupied Cl ion.
The CCDC database (ImCbl, CCDC 627001; HisCbl, CCDC 627000) contains the supplementary crystallographic data for this paper. These data can be obtained free of charge via www.ccdc.cam.ac.uk/data_request/cif by sending an data_request@ccdc.cam.ac.uk or by contacting The Cambridge Crystallographic Data Centre, 12 Union Road, Cambridge CB2 1EZ, U.K.; fax, +44 1223 336033.
Results and Discussion
Synthesis and Characterization of ImCbl and HisCbl
Synthesis of [ImCbl]Cl from aquacobalamin and imidazole was achieved in good purity and yield using a general procedure developed for the synthesis of non-alkylcobalamins.53 Upon the addition of imidazole (or histidine) to aquacobalamin, a rapid color change from red to purple is observed. [ImCbl]Cl was synthesized in 96% purity as assessed by a 1H NMR spectrum of the product redissolved in buffer (see Figure S1, Supporting Information) and in 90% yield. The percentage of non-cobalamin impurities was assessed by converting the products to dicyanocobalamin, (CN)2Cbl− (0.10 M KCN, pH 10.0, ε368 = 3.04 × 104 M−1 cm−1),51 and was found to be 5 ± 1%. However, using a similar procedure to synthesize [HisCbl]Cl resulted in a product that was only ~70% pure, as assessed by 1H NMR spectroscopy, with the major impurity being l-His (Figure S2, Supporting Information). Using a smaller equivalent of l-His resulted in incomplete conversion of aquacobalamin to histidinylcobalamin. Attempts to remove L-His from the product by column chromatography (C8-SPE or Varian HF Bond Elut C18 cartridges) were unsuccessful, with a mixture of aquacobalamin, l-His, and HisCbl observed in the final product (1H NMR spectroscopy). Purification of the product using reverse-phase HPLC under acidic and neutral conditions was also unsuccessful, with aquacobalamin and HisCbl coeluting even at long (>20 min) retention times under isocratic conditions. This suggests that an exchange between the two species occurs on the column.
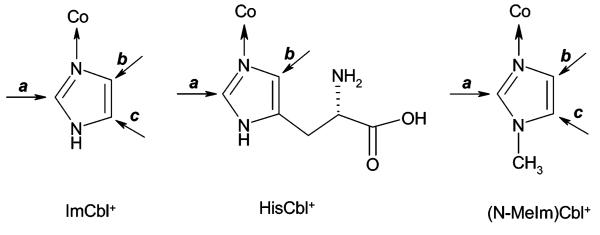
1H NMR chemical shifts and assignments for the signals observed in the aromatic region for ImCbl+ and HisCbl+ are given in Table 2 (see Scheme 1 for the labeling scheme of the β-axial ligand) and are similar to those previously observed for N-MeImCbl+.29 Signal assignments were possible with the additional information provided by heteronuclear single-quantum coherence, heteronuclear multiple-bond correlation, and heteronuclear multiple-quantum coherence total-correlation spectroscopy experiments. The products were also characterized by electrospray mass spectrometry (see Experimental Section) and UV–vis spectroscopy (Figure S3, Supporting Information). Wavelength maxima for ImCbl+ (357, 413, and 536 nm) and HisCbl+ (358, 414, and 538 nm) are in agreement with literature values.30
Table 2.
1H NMR Assignments of the Protons in the Aromatic Region for ImCbl+, HisCbl+, and (N-MeIm)Cbl+ d
Scheme 1.
Labeling Scheme for the β-Axial Ligand of ImCbl+, HisCbl+, and N-MeImCbl+
X-ray Structural Characterization of ImCbl and HisCbl
The three-dimensional structures of ImCbl and HisCbl were determined by X-ray crystallography. Crystal data and structural refinement parameters are given in Table 1. ImCbl and HisCbl crystallize in the orthorhombic space group P212121, the most common space group observed for Cbl structures.54,55 Cbl structures can be classified into three to four packing types, based on the unit cell ratios b/a and c/a.54-56 Both ImCbl and HisCbl fall into the range of cluster type II (for ImCbl, b/a and c/a = 1.419 and 1.627, respectively; for HisCbl, b/a and c/a = 1.428 and 1.619, respectively).56
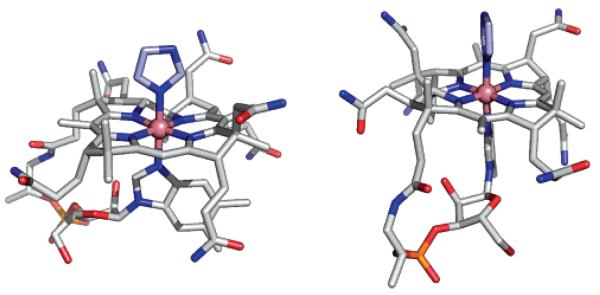
Both structures show the B12 complex in its standard base-on form with imidazole and histidine as the β-axial ligand (Figures 2 and 3). The distances between Co and the imino N of the β-ligands (see Scheme 1) are 1.94(1) Å (in ImCbl) and 1.951(7) Å (to N ε2 in HisCbl). The Co–N bond distances to the lower α-DMB ligand are 2.01(1) and 1.979-(8) Å, respectively. Since the α-DMB is a weaker nucleophile,57 in addition to being more sterically demanding, it is not surprising that the Co–N(Im) and Co–N(His) bond distances are shorter than those of the Co–N(α-DMB) in both structures. The Co–N(α-DMB) bond length of cobalamins can vary from 1.95 to 2.27 Å depending on the σ-donor strength of the ligand trans to Co–N(α-DMB); values of 2.01 and 1.98 Å fall within the range expected for weak σ-donor ligands such as imidazole and histidine.55
Figure 2.
Two approximately perpendicular views of the molecular structure of ImCbl in a stick representation. The central Co atom is depicted as a pink sphere. N, O, and P atoms are drawn in blue, red, and orange, respectively. C atoms are drawn in gray in the B12 trunk and in light blue in the β-ligand (imidazole conformation with higher occupancy). H atoms have been omitted for clarity. The figure was prepared using the program PyMol (http://www.pymol.org/).
Figure 3.
Molecular structure of HisCbl depicting the two alternate conformations observed for the upper histidine ligand. The central Co atom is depicted as a pink sphere. N, O, and P atoms are drawn in blue, red, and orange, respectively. C atoms are drawn in gray in the B12 trunk and in yellow or orange in the β-ligand. H atoms have been omitted for clarity. The figure was prepared using the program PyMol.
In both structures, the Co atom lies in the least-squares plane through the four corrin nitrogen atoms (maximum deviation, 0.01 Å in HisCbl). The fold angles of the corrin rings are very similar in the two structures and were measured as 11.8(3)° and 12.0(3)° for ImCbl and HisCbl, respectively. The fold angles are smaller than those typically observed for cobalamin structures with relatively short Co–N(α-DMB) bond distances.54 The imidazole rings of ImCbl and HisCbl are oriented along the line connecting atoms C5 and C15 of the corrin ring, bisecting the angles between N21 and N22 as well as those between N23 and N24. Both imidazole rings in each Cbl structure are aligned with the lower α-DMB ligand.
An ambiguity exists with respect to the orientation of the imidazole in ImCbl. Two different orientations related by a 180° rotation about the Co–N bond were tested in the refinement and yielded R1 values of 0.0811 and 0.0816, respectively. The rotation about the Co–N bond interchanges two pairs of atoms of the imidazole ring (C2 and C5; N3 and C4). Because of the interchange between a nitrogen and a carbon atom, the correct orientation of the imidazole ligand should be detectable from refined adp's. These values were 0.083(4) and 0.072(4) (conformation 1) vs 0.089(4) and 0.066(4) (conformation 2). Thus, conformation 1 fits the data better, but the difference in the R value and the dissymmetry of the adp's are not large enough to rule out the presence of two alternate conformations of the imidazole. Therefore, the β-ligand in ImCbl was finally modeled by two alternate conformations with refined occupancies of 0.64 and 0.36 (Figure 2).
In principle, the β-ligand in ImCbl can be either a neutral imidazole or an anionic imidazolate. The compound itself was crystallized at neutral pH. Since the pKa of Cbl bound imidazole is ~10,30,36,58,59 only a very minor fraction is present as imidazolate in solution under these conditions. The current version of the Cambridge Structural Database (version 5.27, Nov 2005) holds 46 entries with the term “cobalamin” in the compound name. In most of these entries, the Cbl complexes are overall neutral in charge. There are only five examples of nonneutral Cbls in the database: ammonium sulfitocobalamin (refcodes: DEMSIX and NOJKIG),55,60 sodium thiosulfatocobalamin (EJADIC),61 thioureacobalamin chloride (DEMTUK),55,60 thioureacobalamin hexafluorophosphate (NOJNOP),55,60 and aquocobalamin perchlorate (SUNYEF).62 In the present crystal structure of ImCbl, there is no indication of a counteranion, which would be expected to be Cl− or ClO4− (see method of synthesis). Of course, the occurrence of hydroxide at one of the modeled water sites cannot be ruled out, but it is also conceivable that the minor fraction of imidazolatocobalamin present in solution preferentially crystallized.
In HisCbl, the histidine ligand is clearly disordered over two approximately equally occupied (0.54 vs 0.46) alternate conformations related by a 180° rotation around the Co–N bond (Figure 3). With respect to the protonation state of the β-ligand, it is assumed that the amino group is protonated and the carboxyl group is deprotonated. The difference electron density close to the carboxylate of one of the alternate histidine conformers was modeled as a chloride ion with ~50% occupancy indicating the presence of a mixture of histidine ligands with neutral and anionic imidazole moieties.
For ImCbl, the Co-bound imidazole does not form any direct interactions with symmetry-equivalent B12 molecules. Two weak H bonds exist between N3 (of the major component) and two water molecules with heavy-atom distances of 3.3 and 3.5 Å. In HisCbl, most of the interactions of the β-ligand are formed with water molecules. Only one H bond exists between one of the carboxylate O atoms and the amide N of side chain d of a symmetry-equivalent B12 molecule.
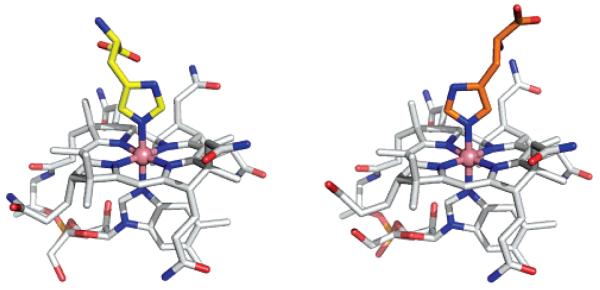
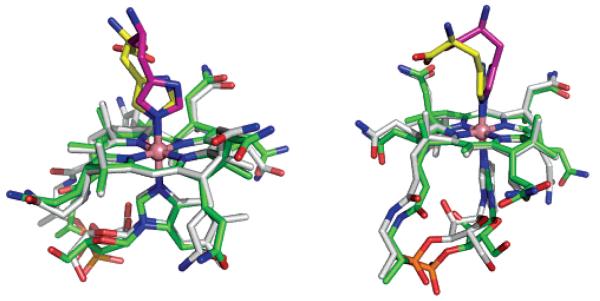
Recently, the structures of human and bovine TC were determined and revealed a histidine residue from the protein as the upper (β)-axial ligand of the cobalamin. Of the available TC structures, the monoclinic and triclinic forms of the bovine proteins (PDB entries, 2BBC and 2BB6) are the most accurate, and Figure 4 shows a superposition of a TC-bound Cbl (taken from PDB entry 2BB6) and our HisCbl structure. The orientation of the imidazole with respect to the corrin ring is the same in the bound protein and in the isolated cofactor. The Co–N distances to the β-axial His ligand were restrained to 2.1 Å (with an estimated standard deviation of 0.03 Å) during the refinement of the protein structures. The resulting mean distance (averaged over five crystallographically independent molecules of bovine TC) was 2.13(2) Å, significantly longer than the bond distances observed in ImCbl and HisCbl (1.94(1) and 1.951(7) Å, respectively). The distance to the α-DMB is also longer (2.08 Å) in the protein structure compared with our structures of the isolated Cbls (2.01(1) Å and 1.979(8) Å, respectively). The fold-angle of TC-bound B12 is 9(1)° (average value for five independent Cbl moieties present in the two structures of bovine TC), which is slightly smaller than that found for ImCbl and HisCbl (11.8(3)° and 12.0(3)° for ImCbl and HisCbl, respectively). Minor differences are also observed for the conformations of the nucleotide loop and of some amide side chains.
Figure 4.
Two approximately perpendicular views of the superposition of one conformer of HisCbl with one of the B12 moieties present in the monoclinic structure of bovine TC (PDB entry, 2BB6). The B12 trunks are shown in white (HisCbl) and green (TC). The histidine residues are shown in yellow and magenta (His-175 in TC).
Of the three known B12 transporter proteins (haptocorrin, intrinsic factor, and TC), displacement of the β-axial ligand of protein-bound H2OCbl+ by a side-chain residue from the protein only occurs for TC.40 Furthermore, there is no evidence for β-axial ligand substitution for other Cbl derivatives upon binding to TC.39,40 The biological significance of this reaction, if any, is currently unclear. Replacing the β-axial ligand of H2OCbl+ limits the possibility of otherwise rapid β-axial ligand substitution chemistry, which occurs for H2OCbl+ with a wide range of nucleophiles, regardless of whether H2OCbl+ is free or protein-bound.39,40 It has also been suggested that the resulting conformational change of the protein from an open form (with respect to the β-axial site of the Cbl) to a closed form, which occurs upon substitution of the β-axial H2O of TC-bound H2OCbl+ for a histidine side chain, may be important in B12 trafficking and/or provide a mechanism by which extracellular surface TC receptors can distinguish between the various Cbl forms of holo-TC.39
Supplementary Material
Acknowledgment
We wish to acknowledge funding of this research from Kent State University (N.E.B.), The Alexander von Humboldt Foundation (N.E.B.), and the National Heart Lung and Blood Institute of the National Institutes of Health (HL52234) (D.W.J.).
Footnotes
Supporting Information Available: 1H NMR spectra of [ImCbl]Cl (Figure S1) and [HisCbl]Cl (Figure S2); UV–vis spectra of HOCbl·HCl, [HisCbl]Cl, and [ImCbl]Cl (Figure S3). This information is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Bertini I, Sigel A, Sigel H, editors. Handbook on Metalloproteins. Marcel Dekker; New York: 2001. [Google Scholar]
- 2.Banerjee R. Chem. Biol. 1997;4:175–86. doi: 10.1016/s1074-5521(97)90286-6. [DOI] [PubMed] [Google Scholar]
- 3.Banerjee R, Ragsdale SW. Annu. Rev. Biochem. 2003;72:209–47. doi: 10.1146/annurev.biochem.72.121801.161828. [DOI] [PubMed] [Google Scholar]
- 4.Banerjee R, editor. Chemistry and Biochemistry of B12. John Wiley & Sons; New York: 1999. [Google Scholar]
- 5.Stupperich E, Eisinger HJ, Albracht SP. Eur. J. Biochem. 1990;193:105–9. doi: 10.1111/j.1432-1033.1990.tb19310.x. [DOI] [PubMed] [Google Scholar]
- 6.Drennan CL, Huang S, Drummond JT, Matthews RG, Ludwig ML. Science. 1994;266:1669–74. doi: 10.1126/science.7992050. [DOI] [PubMed] [Google Scholar]
- 7.Mancia F, Keep NH, Nakagawa A, Leadlay PF, McSweeney S, Rasmussen B, Bosecke P, Diat O, Evans PR. Structure. 1996;4:339–50. doi: 10.1016/s0969-2126(96)00037-8. [DOI] [PubMed] [Google Scholar]
- 8.Mancia F, Smith GA, Evans PR. Biochemistry. 1999;38:7999–8005. doi: 10.1021/bi9903852. [DOI] [PubMed] [Google Scholar]
- 9.Padmakumar R, Taoka S, Padmakumar R, Banerjee R. J. Am. Chem. Soc. 1995;117:7033–7034. [Google Scholar]
- 10.Reitzer R, Gruber K, Jogl G, Wagner UG, Bothe H, Buckel W, Kratky C. Structure. 1999;7:891–902. doi: 10.1016/s0969-2126(99)80116-6. [DOI] [PubMed] [Google Scholar]
- 11.Tollinger M, Eichmuller C, Konrat R, Huhta MS, Marsh EN, Krautler B. J. Mol. Biol. 2001;309:777–91. doi: 10.1006/jmbi.2001.4696. [DOI] [PubMed] [Google Scholar]
- 12.Zelder O, Beatrix B, Kroll F, Buckel W. FEBS Lett. 1995;369:252–4. doi: 10.1016/0014-5793(95)00762-x. [DOI] [PubMed] [Google Scholar]
- 13.Marsh EN, Holloway DE. FEBS Lett. 1992;310:167–70. doi: 10.1016/0014-5793(92)81321-c. [DOI] [PubMed] [Google Scholar]
- 14.Chen HP, Marsh EN. Biochemistry. 1997;36:7884–9. doi: 10.1021/bi970169y. [DOI] [PubMed] [Google Scholar]
- 15.Vlasie M, Chowdhury S, Banerjee R. J. Biol. Chem. 2002;277:18523–7. doi: 10.1074/jbc.M111809200. [DOI] [PubMed] [Google Scholar]
- 16.Lawrence CC, Gerfen GJ, Samano V, Nitsche R, Robins MJ, Retey J, Stubbe J. J. Biol. Chem. 1999;274:7039–42. doi: 10.1074/jbc.274.11.7039. [DOI] [PubMed] [Google Scholar]
- 17.Shibata N, Masuda J, Tobimatsu T, Toraya T, Suto K, Morimoto Y, Yasuoka N. Structure. 1999;7:997–1008. doi: 10.1016/s0969-2126(99)80126-9. [DOI] [PubMed] [Google Scholar]
- 18.Yamanishi M, Yamada S, Muguruma H, Murakami Y, Tobimatsu T, Ishida A, Yamauchi J, Toraya T. Biochemistry. 1998;37:4799–803. doi: 10.1021/bi972572a. [DOI] [PubMed] [Google Scholar]
- 19.Abend A, Bandarian V, Nitsche R, Stupperich E, Retey J, Reed GH. Arch. Biochem. Biophys. 1999;370:138–41. doi: 10.1006/abbi.1999.1382. [DOI] [PubMed] [Google Scholar]
- 20.Brown KL, Zou X, Banka RR, Perry CB, Marques HM. Inorg. Chem. 2004;43:8130–8142. doi: 10.1021/ic040079z. [DOI] [PubMed] [Google Scholar]
- 21.Puckett J, Mitchell M, Hirota S, Marzilli L. Inorg. Chem. 1996;35:4656–4662. doi: 10.1021/ic9602613. [DOI] [PubMed] [Google Scholar]
- 22.Dorweiler JS, Matthews RG, Finke RG. Inorg. Chem. 2002;41:6217–24. doi: 10.1021/ic010265u. [DOI] [PubMed] [Google Scholar]
- 23.Brown KL, Marques HM. J. Inorg. Biochem. 2001;83:121–32. doi: 10.1016/s0162-0134(00)00188-4. [DOI] [PubMed] [Google Scholar]
- 24.Dölker N, Maseras F, Lledós A. J. Phys. Chem. B. 2001;105:7564–7571. [Google Scholar]
- 25.Trommel JS, Warncke K, Marzilli LG. J. Am. Chem. Soc. 2001;123:3358–66. doi: 10.1021/ja004024h. [DOI] [PubMed] [Google Scholar]
- 26.Champloy F, Jogl G, Reitzer R, Buckel W, Bothe H, Beatrix B, Broeker G, Michalowicz A, Meyer-Klaucke W, Kratky C. J. Am. Chem. Soc. 1999;121:11780–11789. [Google Scholar]
- 27.Champloy F, Gruber K, Jogl G, Kratky C. J. Synchrotron Radiat. 2000;7:267–273. doi: 10.1107/S0909049500006336. [DOI] [PubMed] [Google Scholar]
- 28.Reenstra WW, Jencks WP. J. Am. Chem. Soc. 1979;101:5780–5791. [Google Scholar]
- 29.Cregan AG, Brasch NE, van Eldik R. Inorg. Chem. 2001;40:1430–8. doi: 10.1021/ic0009268. [DOI] [PubMed] [Google Scholar]
- 30.Marques HM, Marsh JN, Mellor JR, Munro OQ. Inorg. Chim. Acta. 1990;170:259–269. [Google Scholar]
- 31.Hamza MSA, Zou X, Brown KL, van Eldik R. Eur. J. Inorg. Chem. 2003:268–276. [Google Scholar]
- 32.Pratt JM. Inorganic Chemistry of Vitamin B12. Academic Press, Inc.; London: 1972. [Google Scholar]
- 33.Marques HM, Bradley JC, Campbell LA. Dalton Trans. 1992:2019–2027. [Google Scholar]
- 34.Hill HA, Pratt JM, Thorp RG, Ward B, Williams RJ. Biochem. J. 1970;120:263–9. doi: 10.1042/bj1200263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Marques HM, Egan TJ, Marsh JH, Mellor JR, Munro OQ. Inorg. Chim. Acta. 1989;166:249–255. [Google Scholar]
- 36.Hanania GIH, Irvine DH. J. Chem. Soc. Suppl. 1964;1:5694–5697. [Google Scholar]
- 37.Knapton L, Marques HM. Dalton Trans. 2005:889–95. doi: 10.1039/b416083e. [DOI] [PubMed] [Google Scholar]
- 38.Randall WC, Alberty RA. Biochemistry. 1967;6:1520–5. doi: 10.1021/bi00857a038. [DOI] [PubMed] [Google Scholar]
- 39.Fedosov SN, Fedosova NU, Nexo E, Petersen TE. J. Biol. Chem. 2000;275:11791–8. doi: 10.1074/jbc.275.16.11791. [DOI] [PubMed] [Google Scholar]
- 40.Fedosov SN, Berglund L, Fedosova NU, Nexo E, Petersen TE. J. Biol. Chem. 2002;277:9989–96. doi: 10.1074/jbc.M111399200. [DOI] [PubMed] [Google Scholar]
- 41.Rothenberg SP, Quadros EV, Regec A. In: Chemistry and Biochemistry of B12. Banerjee R, editor. John Wiley & Sons; New York: 1999. pp. 441–473. Chapter 17. [Google Scholar]
- 42.Baker H, Leevy CB, DeAngelis B, Frank O, Baker ER. J. Am Coll. Nutr. 1998;17:235–8. doi: 10.1080/07315724.1998.10718752. [DOI] [PubMed] [Google Scholar]
- 43.Tisman G, Vu T, Amin J, Luszko G, Brenner M, Ramos M, Flener V, Cordts V, Bateman R, Malkin S, Browder T. Am. J. Hematol. 1993;43:226–9. doi: 10.1002/ajh.2830430313. [DOI] [PubMed] [Google Scholar]
- 44.Vu T, Amin J, Ramos M, Flener V, Vanyo L, Tisman G. Am. J. Hematol. 1993;42:202–11. doi: 10.1002/ajh.2830420212. [DOI] [PubMed] [Google Scholar]
- 45.Herzlich B, Herbert V. Lab. Invest. 1988;58:332–7. [PubMed] [Google Scholar]
- 46.Herbert V, Fong W, Gulle V, Stopler T. Am. J. Hematol. 1990;34:132–9. doi: 10.1002/ajh.2830340210. [DOI] [PubMed] [Google Scholar]
- 47.Ulleland M, Eilertsen I, Quadros EV, Rothenberg SP, Fedosov SN, Sundrehagen E, Orning L. Clin. Chem. 2002;48:526–32. [PubMed] [Google Scholar]
- 48.Garau G, Fedosov SN, Petersen TE, Geremia S, Randaccio L. Acta Crystallogr., Sect. D: Biol. Crystallogr. 2001;57:1890–2. doi: 10.1107/s0907444901015360. [DOI] [PubMed] [Google Scholar]
- 49.Wuerges J, Garau G, Geremia S, Fedosov SN, Petersen TE, Randaccio L. Proc. Natl. Acad. Sci. U.S.A. 2006;103:4386–91. doi: 10.1073/pnas.0509099103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Brasch NE, Finke RG. J. Inorg. Biochem. 1999;73:215–9. doi: 10.1016/s0162-0134(99)00018-5. [DOI] [PubMed] [Google Scholar]
- 51.Barker HA, Smyth RD, Weissbach H, Toohey JI, Ladd JN, Volcani BE. J. Biol. Chem. 1960;235:480–8. [PubMed] [Google Scholar]
- 52.Sheldrick GM, Schneider TR. In: Methods Enzymology. Sweet RM, Carter CWJ, editors. Vol. 277. Academic Press; Orlando, FL: 1997. pp. 319–343. [PubMed] [Google Scholar]
- 53.Suarez-Moreira E, Hannibal L, Smith CA, Chavez RA, Jacobsen DW, Brasch NE. Dalton Trans. 2006:5269–77. doi: 10.1039/b610158e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gruber K, Jogl G, Klintschar G, Kratky C. In: Vitamin B12 and B12-Proteins: Lectures Presented at the 4th European Symposium on Vitamin B12 and B12-Proteins. Kräutler B, Arigoni D, Golding BT, editors. Wiley-VCH; Weinheim, Germany: 1998. pp. 335–347. [Google Scholar]
- 55.Randaccio L, Geremia S, Nardin G, Slouf M, Srnova I. Inorg. Chem. 1999;38:4087–4092. [Google Scholar]
- 56.Randaccio L, Geremia S, Nardin G, Wuerges J. Coord. Chem. Rev. 2006;250:1332–1350. [Google Scholar]
- 57.Fasching M, Schmidt W, Krautler B, Stupperich E, Schmidt A, Kratky C. Helv. Chim. Acta. 2000;83:2295–2316. [Google Scholar]
- 58.Eilbeck WJ, West MS. Dalton Trans. 1976:274–278. [Google Scholar]
- 59.Hamza MSA, Pratt JM. Dalton Trans. 1994:1367–1371. [Google Scholar]
- 60.Randaccio L, Geremia S, Stener M, Toffoli D, Zangrando E. Eur. J. Inorg. Chem. 2002:93–103. [Google Scholar]
- 61.Perry BP, Fernandes MA, Brown KL, Zou X, Valente EJ, Marques HM. Eur. J. Inorg. Biochem. 2003:2095–2107. [Google Scholar]
- 62.Kratky C, Farber G, Gruber K, Wilson K, Dauter Z, Nolting HF, Konrat R, Krautler B. J. Am. Chem. Soc. 1995;117:4656–4670. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.