Abstract
EphA2 is a transmembrane receptor tyrosine kinase that functions in the regulation of cell growth, survival, angiogenesis, and migration and EphA2 targeting has been proposed as a novel therapeutic strategy for neoplasms that overexpress this protein. EphA2 overexpression has been correlated with increased invasive and metastatic ability in pancreatic cancer cell lines. However, the patterns of EphA2 expression in human pancreatic cancers and associated metastases is unknown, as are the genetics of EphA2 in this tumor type. We collected clinicopathologic data and paraffin-embedded materials from 98 patients with primary and/or metastatic pancreatic cancer and performed immunohistochemical labeling for EphA2 protein. EphA2 protein immunolabeling was found in 207 of 219 samples (95%). The expression was predominantly cytoplasmic, although predominant membranous staining was observed in a minority of cases. When evaluated specifically for labeling intensity, primary and metastatic carcinomas were more strongly positive compared to benign ducts and PanIN lesions (P < 0.00001 and P < 0.01, respectively) and poorly differentiated carcinomas were more strongly positive for EphA2 than well and moderately differentiated tumors (P < 0.005). When primary carcinomas without metastatic disease were specifically compared to carcinomas with associated metastatic disease, the advanced carcinomas showed relatively less strong positive labeling for EphA2 (P < 0.008). Moreover, decreased EphA2 labeling was more commonly found in liver (P < 0.002), lung (P < 0.004) or peritoneal metastases (P < 0.01) as compared to distant lymph node metastases (P < 0.01). Genetic sequencing of the tyrosine kinase domain of EPHA2 in 22 samples of xenograft enriched pancreatic cancer did not reveal any inactivating mutations. However, EPHA2 amplification was found in 1 of 33 pancreatic cancers corresponding to a lymph node metastasis, indicating EPHA2 genomic amplification may underlie EphA2 overexpression in a minority of patients. Our data confirms that EphA2 is overexpressed in pancreatic cancer, but suggests a relative loss of EphA2 in co-existent pancreatic cancer metastases as well as a role for EPHA2 in organ specific metastasis.
Keywords: 1p36, Amplification, EphA2, Metastasis, Pancreatic cancer
Introduction
The EphA family of transmembrane receptor tyrosine kinases (RTK) bind ligands of the ephrinA family of glycosylphosphatidylinositol (GPI) linked membrane proteins. Eph and ephrin expression occur at tissue boundaries primarily during development. Ephrin binding results in tyrosine phosphorylation of EphA receptors followed by signals leading to cell–cell repulsion that prevent aberrant cellular migration across boundaries [1, 2].
EphA2, unlike other RTKs of the EphA family, is expressed in adult epithelial tissue [3]. The normal function of EphA2 is not fully understood, but it is thought to function in the regulation of cell growth, migration, survival, and angiogenesis [4, 5]. Also in contrast to other Eph RTKs, ligand binding and tyrosine phosphorylation of EphA2 are not necessary for its function [6, 7]. Instead, ephrinA binding seems to regulate EphA2 subcellular localization and interaction with other downstream signaling proteins including FAK and the MAP/ERK signaling pathways [8, 9].
EphA2 is overexpressed in a variety of human cancers, and high levels of EphA2 are associated with aggressive disease and poor clinical outcomes [10–19]. In vitro studies have shown that EphA2 is a powerful oncoprotein, sufficient to confer malignant potential on non-transformed epithelial cells [7]. In pancreatic cancers, EphA2 protein is also relatively overexpressed in pancreatic adenocarcinoma cell lines with higher metastatic potential [8] and suppression of EphA2 expression appears to attenuate the invasive phenotype of these same cell lines [20], providing support for a role for EphA2 targeted therapies in the treatment of pancreatic cancer.
Despite this compelling evidence of a role for EphA2 in pancreatic neoplasia, the expression of EphA2 in human tissues representing the full spectrum of pancreatic carcinogenesis and metastatic progression is unclear, as are the genetics of EPHA2 in these same tissues. To address these issues, we used immunohistochemical labeling to evaluate EphA2 expression in a series of normal (benign) pancreas, pancreatic intraepithelial neoplasia (PanIN), primary infiltrating and metastatic pancreatic cancer tissues, as well as applied conventional gene sequencing and copy number analyses to determine the relationship of EPHA2 genetics to its protein expression.
Materials and methods
Tissue samples
Clinicopathologic data and paraffin-embedded materials from 98 patients with primary and/or metastatic pancreatic cancer were collected from the Surgical and Autopsy Pathology Files of the Johns Hopkins Hospital, Baltimore, MD, or the Gastrointestinal Cancer Rapid Medical Donation Program (GICRMDP) [21]. These specimens included 20 samples of benign pancreatic duct epithelium, 10 samples of pancreatic intraepithelial neoplasia (PanIN), 98 primary infiltrating carcinomas and 91 matched pancreatic cancer metastases from the liver, lung, lymph node or peritoneum. Twenty-two xenograft-enriched pancreatic cancers were also used and created as previously described [21]. The project was approved by the Institutional Review Board.
Cell lines
A549, AsPC1, BxPC3, Capan 2, CF-Panc1, Hs678, Hs766t, MiaPACA2, Panc1, and Su86.86 cell lines were obtained from the ATCC (Manassas, VA). The IMIM-PC2 pancreas cancer cell line was generously provided by Dr. Paco Real [22], the PK8 and PK9 cell lines were generously provided by Dr. Akira Horii [23] the PL5, PL8 and PL12 lines were generously provided by Dr. Elizabeth Jaffee, and the normal duct epithelium cell line HPDE was generously shared by Dr. Michael Goggins. The A2.1, A2.4, A6L, A10.7 and A10.4 low passage cell lines were created in our own lab from pancreatic cancer metastasis tissues obtained from autopsied patients [21]. The immortal human pancreatic duct epithelial cell line, HPDE, was originally provided by Dr. Ming-Sound Tsao (University of Toronto, Ont., Canada). All cell lines were grown at 37°C in DMEM containing 10% FBS, 1% L-glutamine and 1% penicillin-streptomycin in a humidified atmosphere containing 5% CO2.
Immunohistochemical labeling
Unstained 5-μm sections were cut from tissue microarrays or the paraffin-embedded sections of each case and deparaffinized by routine techniques followed by steaming in sodium citrate buffer pH 6.0 for 20 min at 80°C. The slides were cooled for 15 min then incubated with a 1:25 dilution of monoclonal antibody to EphA2 (Santa Cruz Biotechnology, clone H-77) for 1 h at room temperature. Primary antibody was detected using the DAKO LSAB + System (DAKO, Carpinteria, CA), and sections were counterstained with hematoxylin. A negative control was used in each run in which the antibody was replaced by an equal volume of phosphate-buffered saline. Immunohistochemical labeling of each tissue section was scored on an intensity scale of 0–3, with 0 corresponding to no positive cells were observed, 1 corresponding to weak positive labeling (labeling best seen at 10× power or greater), 2 corresponding to unequivocal positive labeling, and 3 to intense positive labeling. The percentage of cells with positive labeling was also scored on a 10 tiered scale ranging from 10% to 100%. Scoring was performed by two of the authors (S.V.M. and C.I.D.) at a two-headed microscope.
Western blotting
Total protein was extracted from cell line pellets with RIPA Protein Lysis Buffer with Proteinase Inhibitors (Santa Cruz Biotechnology Inc., Santa Cruz, CA, USA) freshly added. The lysates were incubated on ice for 30 minutes followed by lysing using a sonic homogenizer for 5 s (Model 100, Fisher Scientific International Inc., AL, USA), then spun at 16000 × g for 30 min at 4°C. Protein concentrations of the supernatants were performed by the Bio-Rad Protein Assay (Bio-Rad Laboratories Inc., CA, USA). About 40 μg total protein was separated by electrophoresis in NuPAGE™ 4–12% Bis–Tris Gel 1.5 mm × 10-well (Invitrogen Inc., CA, USA) and then transferred onto Nitrocellulose Membrane Filter Paper Sandwich 0.45 μm pore size (Invitrogen Inc., CA, USA). Membranes were blocked with 5% skim milk in PBST (PBS in 0.1% Tween 20) for 1 h followed by incubation with a 1:100 dilution of primary antibody (anti-EphA2, clone H-77, or anti-actin, clone C-2, Santa Cruz Biotechnology) overnight in 5% skim milk in PBST. The membrane was washed with PBST for three times and 30 min, followed by incubation with 1:5000 ECL™ Anti-mouse IgG (Amersham Biosciences, Sweden) at room temperature. Detection was effected with an enhanced chemiluminescence (ECL) kit (Lumigen PS-3 detection reagent, Amersham Biosciences, Sweden) and the membranes were developed using Molecular Imager Gel Doc XR System (Bio-Rad Laboratories, CA, USA).
Sequencing of exons 10 through 13 of EPHA2
Genomic DNA was extracted from 22 xenograft enriched pancreatic cancers using a DNeasy Kit (Qiagen) and the coding sequence of exons 10–13 of the human EphA2 gene were amplified using intronic primers flanking each exon. All primer sequences and reaction conditions are available from the corresponding author upon request. PCR-amplified products were purified using QIAquick columns (Qiagen) and automated sequencing performed using nested primers and an ABI Prism model 3700 (Applied Biosystems, Foster City, CA). Sequence analysis employed Sequencher™ version 4.0.5 software (Gene Codes, Ann Arbor, MI). Potential mutations were verified by a second PCR amplification and sequencing of the original genomic DNA template.
Screening for genomic amplification of EphA2 using real-time PCR
Three primer sets were designed to determine the amplification status of EPHA2 on chromosome 1p36.12 using real-time PCR. Primers for EphA2 amplification of genomic DNA used were: forward 5′-GTGTACCGAGGAGAGGCTGA-3′ and reverse-5′-CCGACTCGGCATAGTAGAGG-3′. A second primer pair located approximate 90 Mb downstream from EphA2 on chromosome 1 was: primer1-sense-5′-ACTGCCAGCATCTCTCGTTT-3′ and primer1-antisense-5′-ATGCTCAACATCACCGATCA-3. The third primer pair was human genomic DNA specific primer derived from G protein-coupled receptor 15 (GPR15) on chromosome 3: primer2-sense-5′-GGTCCCTGGTGGCCTTAATT-3′ and primer2- antisense-5′-TTGCTGGTAATGGGCACACA-3′. For each real-time PCR reaction, 20–200 ng genomic DNA was added and each sample was run in duplicate. Real-time PCR Ct values obtained from control primers 1 and/or 2 and EphA2 primers using genomic DNA were first normalized to that of a diploid control (HPDE), and EphA2 amplification was expressed as the relative fold difference of EphA2 as compared to primer 1 and/or 2.
Statistics
All summary values are expressed as a mean ± standard deviation unless otherwise indicated. For frequency distributions a chi-squared test was used with modification by the Fishers exact test to account for frequency values less than 5. P values ≤ 0.05 were considered statistically significant.
Results
Relationship of EphA2 immunolabeling to pancreatic carcinogenesis and progression
The clinicopathologic features of the 98 patients whose tissues were analyzed for EphA2 are summarized in Table 1. There was no difference in the age or gender distribution among pancreatic cancers with or without associated metastases. Four infiltrating carcinomas arose in association with an intraductal papillary mucinous neoplasm (IPMN), one carcinoma had adenosquamous features, and the remaining cancers were conventional infiltrating ductal adenocarcinoma. More poorly differentiated carcinomas were present within patients with advanced stage disease than those with no metastases at diagnosis (P < 0.01).
Table 1.
Clinicopathological features of patients
| pT1–pT3 | pT4 | |
|---|---|---|
| Number of patients represented | 53 | 45 |
| Age (years) | 65.3 ± 11.4 | 62.7 ± 11.9 |
| Gender (M:F) | 36:17 | 30:15 |
| Pathologic diagnosis | ||
| Infiltrating ductal adenocarcinoma | 52 | 41 |
| Infiltrating carcinoma arising in IPMN | 0 | 4 |
| Infiltrating adenosquamous carcinoma | 1 | 0 |
| Tumor differentiation | ||
| Well/moderate | 38 | 18 |
| Poora | 23 | 29 |
P < 0.01, χ2-test
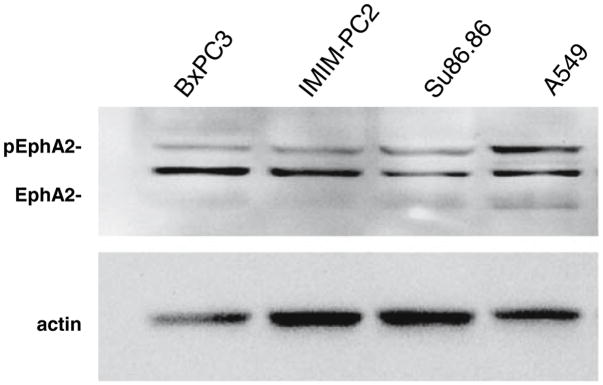
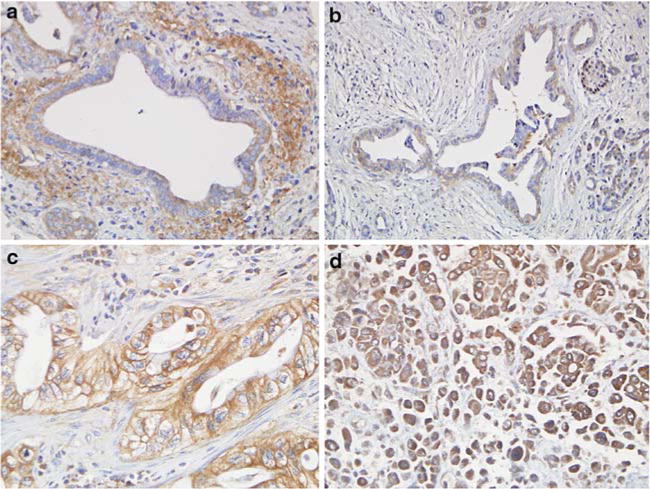
To determine the protein expression patterns of EphA2 in pancreatic cancer tissues, we used a rabbit polyclonal antibody raised against amino acids 422–498 of human EphA2. On Western blots, this antibody showed reactivity only for EphA2 protein (phosphorylated and unphosphorylated forms) in the EphA2 positive control lung cancer cell line A549 and in three different pancreatic cancer cell lines analyzed (Fig. 1). When this antibody was used to immunolabel a series of benign and malignant pancreatic cancer tissues, 207 of 219 samples (95%) showed positive immunolabeling for EphA2 (Table 2) corresponding to 16 of 20 (80%) benign ducts, 9 of 10 (90%) of PanINs, 98 of 98 (100%) primary infiltrating pancreatic cancers and 84 of 91 (92%) matched metastatic pancreatic cancers. The most frequent pattern of labeling was cytoplasmic with or without membranous accentuation, although predominant membranous staining was observed in a minority of carcinomas (Fig. 2). In the 207 positive labeling benign and neoplastic tissues the percentage of labeled cells was ≥90%. In addition, no significant differences in the sole presence or absence of EphA2 labeling among benign, PanIN, primary infiltrating or metastatic carcinomas was found, nor was a difference found in the pattern of labeling (cytoplasmic versus membranous) in these four groups. However, although EphA2 immunolabeling was detectable in most samples and was each positive sample showed homogeneously labeling of the epithelium, the intensity of the positive labeling was widely variable among the benign ducts and PanINs, primary carcinomas or metastatic carcinomas. Specifically, 71 of 219 cases (32%) showed weak positive labeling (1+ intensity), 93 of 219 (42%) showed positive labeling (2+ intensity) and 43 of 219 (20%) showed intense positive labeling (3+ intensity).
Fig. 1.
EphA2 detection by antibody H-77 on Western blot. Shown are three pancreatic cancer cell lines BxPC3, IMIM-PC2 and Su86.86 and the positive control lung cancer cell line A549. This antibody detects both the unphosphorylated and phosphorylated forms of EphA2. Relatively more of the phosph-EphA2 is present in A549 than in the pancreatic cancer cell lines
Table 2.
Relationship of EphA2 immunolabeling score to pathologic features
| Total samples | EphA2 score |
P value | ||||
|---|---|---|---|---|---|---|
| 0 | 1 | 2 | 3 | |||
| Pathology | ||||||
| Benign/PanINa | 30 | 5 | 16 | 9 | 0 | – |
| All Infiltrating Carcinomas | 98 | 0 | 26 | 49 | 23 | <0.00001 |
| All Metastases | 91 | 7 | 29 | 35 | 20 | <0.01b, <0.005c |
| Pathologic stage of primary carcinoma | ||||||
| pT1–pT3 (no metastatic disease) | 53 | 0 | 10 | 31 | 12 | <0.008 |
| pT4 (metastatic disease present) | 45 | 1 | 15 | 15 | 14 | |
| Differentiation of primary carcinoma | ||||||
| Well/moderate | 49 | 1 | 15 | 23 | 10 | <0.005 |
| Poor | 49 | 0 | 10 | 23 | 16 | |
| Advanced stage (pT4) carcinomas | ||||||
| Primary | 45 | 1 | 15 | 15 | 14 | <0.002 |
| All metastases | 91 | 7 | 29 | 35 | 20 | |
| Liverd | 37 | 4 | 11 | 15 | 7 | <0.002 |
| Lungd | 27 | 0 | 12 | 12 | 3 | <0.004 |
| Lymph noded | 13 | 1 | 2 | 4 | 6 | <0.01 |
| Peritoneumd | 14 | 2 | 4 | 4 | 4 | <0.01 |
Benign ducts and PanINs were combined due to small sample sizes in each category
All metastases compared to benign ducts and PanINs
All metastases compared to all primary carcinomas
Compared to the 45 matched primary carcinomas
Fig. 2.
EphA2 immunolabeling patterns in benign and neoplastic pancreatic tissues. (a) Benign pancreatic duct showing weak positive cytoplasmic labeling for EphA2 (intensity 1+). (b) Pancreatic Intraepithelial Neoplasia grade 2–3 with positive labeling for EphA2, also in a cytoplasmic distribution (intensity 1+). (c) Moderately differentiated infiltrating pancreatic adenocarcinoma showing prominent membranous labeling for EphA2 (intensity 2+). (d) Poor differentiated carcinoma with anaplastic features carcinoma showing intense positive cytoplasmic labeling for EphA2 (intensity 3+). All images are shown at 200× magnification
EphA2 expression has been reported to correlate with invasive and/or metastatic ability [8, 19, 24, 25]. Thus, we also compared EphA2 labeling patterns specifically among the benign ducts, PanINs, primary and metastatic carcinomas (Table 2). Primary infiltrating pancreatic carcinomas and matched metastases both showed a higher frequency of strong positive EphA2 labeling than benign duct epithelium and PanIN lesions (P < 0.00001 and <0.01, respectively). In addition, poorly differentiated carcinomas showed a higher frequency of strong positive EphA2 labeling than well to moderate differentiated carcinomas (P < 0.005). Immunolabeling was next compared among the primary and metastatic carcinoma tissues only. Overall, strong positive EphA2 expression was more frequently observed in the 98 primary infiltrating pancreatic cancers than in the 91 pancreatic cancer metastases (P < 0.005). Moreover, when EphA2 immunolabeling was evaluated among the 53 primary carcinomas without metastases to the 45 primary carcinomas with associated metastases, primary carcinomas without metastases more frequently showed strong positive labeling for EphA2 compared to advanced stage primary carcinomas (P < 0.008). We next compared EphA2 immunolabeling patterns specifically among the 45 primary carcinomas and their 91 matched metastases to liver, lung, lymph node or peritoneum, the most common sites of pancreatic cancer metastasis [21]. Overall, this subset of primary carcinomas again showed more frequent EphA2 immunolabeling than their matched metastases (P < 0.002). When each metastatic site was individually compared to the matched primary carcinomas, metastases to liver, lung and peritoneum showed less frequent strong positive labeling (P < 0.004, P < 0.002 and P < 0.01, respectively) whereas distant lymph node metastases showed more frequent strong positive labeling (P < 0.01). Thus, as liver, lung and peritoneal metastases are the predominant site of metastasis from pancreatic cancer, and the majority of metastatic carcinomas in this study were liver, lung and peritoneal metastases, the differences found for immunolabeling patterns among primary and metastatic carcinomas is likely due to an overrepresentation of metastases to these sites in our sample set. Finally, we determined the relationship of EphA2 immunolabeling among each matched set of primary and metastatic carcinoma. In 13 of 45 patients (29%), immunolabeling was identical among the primary carcinoma and all matched metastases regardless of metastatic site. However, in nine patients (20%) immunolabeling was greater in intensity in one or more metastases than the matched primary carcinoma, and in 21 patients (47%) it was decreased in intensity in one or more metastases than in the matched primary carcinoma. Thus, in the majority of patients, EphA2 labeling patterns in the primary carcinoma are not predictive of labeling patterns in the matched metastases and are most often decreased.
EphA2 genetic status in pancreatic carcinomas
In recent years, mutations of EPHB2 in prostate cancer and EPHA3 in colon cancer have been reported with many of these inactivating mutations corresponding to the tyrosine kinase domain of those receptors [26, 27]. Therefore, we sequenced the tyrosine kinase domain of EphA2 corresponding to exons 10 through 13 in 22 pancreatic cancer xenografts. In six xenografts, a silent single nucleotide polymorphism was present at codon 671 (CTC → CTT), and in another two xenografts a silent single nucleotide polymorphism was present at codon 678 (CTA → CTG). However, no potentially inactivating mutations were found to account for differential expression of EphA2.
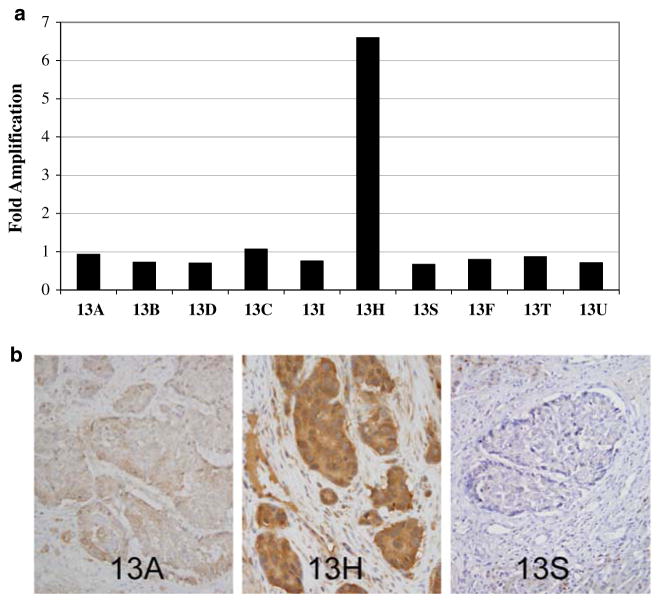
To determine if copy number alterations of EPHA2 play a role in its overexpression, we also performed quantitative PCR of genomic DNA at 1p36.12 in 33 pancreatic cancer cell lines, xenograft enriched primary or metastatic pancreatic cancers, and high quality microdissected pancreatic cancer tissues. One of 33 pancreatic cancers showed amplification of the region encompassing EPHA2 6.6-fold compared to a normal diploid control cell line, corresponding to a microdissected lymph node metastasis (sample A13H) (data not shown). To determine if EPHA2 amplification was also present in the matched primary carcinoma or seven other metastases available from this same patient, we also performed quantitative PCR for EphA2 in these tissues similarly microdissected (Fig. 3a). With exception of A13H, no other samples from this patient showed amplification for EPHA2. To determine if the EPHA2 amplification within this metastasis was associated with EphA2 protein overexpression, we performed immunolabeling for EphA2 in the paraffin-embedded tissues corresponding to these same samples whose genomic DNA was first analyzed by qPCR (Fig. 3b). Consistent with our findings at the genetic level, this lymph node metastasis showed intense positive (3+) labeling for EphA2 protein in contrast to the matched primary carcinoma or co-existent metastases to other organ sites. Interestingly, the liver metastases from this same patient showed decreased expression relative to the matched primary carcinoma, consistent with our findings of decreased expression in liver metastases in the larger sample set. Thus in a minority of pancreatic carcinomas, amplification of 1p36.12 may underlie EphA2 protein overexpression.
Fig. 3.
Relationship of genomic DNA copy number at 1p36.12, the locus encompassing EPHA2, to EphA2 protein expression. (a) A 6.6-fold amplification of this region is present in one lymph node metastasis (sample 13H), but not in the primary carcinoma (samples 13A and 13B), lung (sample 13D), additional lymph nodes (samples 13C and 13I) or liver metastases (samples 13S, 13F, 13T and 13U) of this same patient. (b) The primary carcinoma 13A shows positive labeling for EphA2, but intense positive labeling is seen in 13H, the lymph node metastasis with a 6.6 fold amplification of EphA2 shown in panel A. In contrast, virtual negative labeling is seen for sample 13S, a liver metastasis for this same patient (panel c)
Discussion
In 2006 in the United States, it is estimated that 33,730 individuals will be diagnosed with pancreatic cancer and 32,300 will die of pancreatic cancer [28]. The high mortality rate attributed to pancreatic cancer is largely due to the fact that most patients are diagnosed at an advanced stage of disease that is not amenable to surgical resection and pancreatic cancers are intrinsically drug-resistant [29]. The identification of molecular targets presents an opportunity for developing more effective treatment strategies and impacting upon patient survival. EphA2, a potent oncogene, has been suggested as a potential therapeutic target based on in vitro studies [30, 31]. However, the genetics of EphA2 and the patterns EphA2 overexpression in pancreatic carcinoma tissues have not been previously characterized, and thus those patients who may benefit most from EphA2 targeted therapies are unknown.
Unlike other members of the Eph receptor family that have been shown to be targeted by genetic inactivation such as EPHB2 in prostate cancer [26] and EPHA3 in colon cancer [27], we did not find mutations of the tyrosine kinase domain of EphA2. By contrast, we did find that in rare cases of pancreatic cancer, EphA2 protein overexpression may specifically result from amplification of 1p36.12. Chromosomal alterations involving 1p have been reported in a variety of human cancer types, including pancreatic carcinoma [32, 33], with most studies implicating the presence of a tumor suppressor gene(s) on this chromosome arm [34, 35]. However, the lack of inactivating mutations of EPHA2 reported in this study indicates that EPHA2 functions in a manner unlike that of the other Eph receptor family members targeted by inactivating mutations. Furthermore, genetic amplification of EPHA2, albeit an uncommon finding in this study, correlates with its role as a putative oncogene for this tumor type in which it may promote angiogenesis [36], loss of growth control and contact inhibition [6, 7]. Although the Eph family is among the largest receptor tyrosine kinase family in the human genome [37], the specific functions of each member remain incompletely understood. Thus, in addition to aiding our understanding of neoplasia, the patterns of activation or inactivation of Eph receptor tyrosine kinases may provide useful information regarding their role in development and growth regulation of normal tissues as well.
EphA2 overexpression has been reported to correlate with aggressive features in other human tumor types [10–19]. Consistent with these reports, we found a significant increase in EphA2 expression in pancreatic cancers as compared to benign ducts and PanIN lesions, as well as increased expression in poorly differentiated carcinomas compared to well or moderate differentiated carcinomas (P < 0.005). Due to the small number of PanINs available for evaluation a meaningful evaluation of EphA2 expression in precursor lesions was not possible, but nonetheless provides some insight into the upregulation of EphA2 in pancreatic cancer progression. EphA2 protein stability in cancer cells may arise as a result of decreased ligand-mediated degradation [38], or by decreased phosphotyrosine content [39], among others. Of interest, Western blotting for EphA2 in three pancreatic cancer cell lines (Fig. 1) indicated reduced phosphorylated EphA2 in all three lines as compared to A549, consistent with this possibility. EphA2 protein stability may also result from decreased cell adhesions that interfere with ligand binding [7]. We and others have shown that pancreatic cancers show features of aberrant cell–cell or cell–matrix interactions, including overexpression of claudin 4, a component of tight junctions [40–42]. Interestingly, a recent study by Tanaka et al. showed claudin 4 function is regulated in part by EphA2 [43] suggesting EphA2 may play a regulatory role in abnormal cell adhesion of pancreatic cancers as well.
EphA2 expression in metastases, while significantly greater than that of benign ducts and PanINs, is relatively less than for primary carcinomas indicating a possible increase in ligand binding by Ephrin A1 (the ligand for EphA2) within the metastatic site, or by decreased expression of phosphatases that regulate phosphotyrosine levels [2, 44]. Related to this finding, we have also made the novel observation of organ specific patterns of EphA2 expression by pancreatic cancer metastases, with relatively less expression of EphA2 seen in metastases to liver and relatively more expression seen in metastases to lymph nodes. This pattern was observed both among all metastases analyzed, but could also be found among analysis of an individual patients’ primary carcinoma and matched metastases (Fig. 3b). Organ specific patterns of EphA2 may indicate that, in addition to its known oncogenic properties, EphA2 may play a role in organ-specific metastasis. In a survey of Eph receptor/ephrin expression a large set of normal human tissues, Hafner et al. has shown that although all Eph/ephrin family members were expressed in all tissues, the levels of expression of individual Eph/ephrins was widely variable among different normal organs [45]. Thus, we cannot rule out the additional possibility that different levels of EphA2 expression in metastatic pancreatic cancers may indirectly indicate the degree of receptor ligand binding at each target organ site.
The finding of organ specific patterns of EphA2 expression has clinical implications. For example, Carles-Kinch et al. have proposed that antibody mediated targeting of EphA2 may mimic the actions of ligand binding, causing EphA2 internalization, degradation and loss of oncogenic signals [30]. Based on our observations of organ specific expression of EphA2, it is conceivable that antibody-mediated targeting may be more or less efficacious for different patients based on their metastatic burden and organ sites affected. At the very least, clinical trials in advanced stage patients designed to evaluate anti-EphA2 agents may best be evaluated with regard to these features of enrolled patients.
In conclusion, we provide evidence of a role for EphA2 in pancreatic carcinogenesis and metastasis, the lack of genetic alterations of EphA2 in pancreatic cancers, and further suggest a role of EphA2 in organ specific metastasis.
Acknowledgments
Supported by CA106610 (C.I.D.), a Career Development Award from the NIH SPORE (Specialized Programs of Research Excellence) in Gastrointestinal Cancer Grant CA62924 (C.I.D.), The George Rubis Family, The Jeff Zgonina Fund for Pancreatic Cancer Research, The Joseph C. Monastra Fund for Pancreatic Cancer Research, The Michael Rolfe Foundation and Sigma Beta Sorority.
References
- 1.Walker-Daniels J, Hess AR, Hendrix MJ, et al. Differential regulation of EphA2 in normal and malignant cells. Am J Pathol. 2003;162:1037–1042. doi: 10.1016/S0002-9440(10)63899-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Parri M, Buricchi F, Taddei ML, et al. EphrinA1 repulsive response is regulated by an EphA2 tyrosine phosphatase. J Biol Chem. 2005;280:34008–34018. doi: 10.1074/jbc.M502879200. [DOI] [PubMed] [Google Scholar]
- 3.Lindberg RA, Hunter T. cDNA cloning and characterization of eck, an epithelial cell receptor protein-tyrosine kinase in the eph/elk family of protein kinases. Mol Cell Biol. 1990;10:6316–6324. doi: 10.1128/mcb.10.12.6316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rosenberg IM, Goke M, Kanai M, et al. Epithelial cell kinase-B61: an autocrine loop modulating intestinal epithelial migration and barrier function. Am J Physiol. 1997;273:G824–G832. doi: 10.1152/ajpgi.1997.273.4.G824. [DOI] [PubMed] [Google Scholar]
- 5.Pandey A, Shao H, Marks RM, et al. Role of B61, the ligand for the Eck receptor tyrosine kinase, in TNF-alpha-induced angiogenesis. Science. 1995;268:567–569. doi: 10.1126/science.7536959. [DOI] [PubMed] [Google Scholar]
- 6.Zantek ND, Azimi M, Fedor-Chaiken M, et al. E-cadherin regulates the function of the EphA2 receptor tyrosine kinase. Cell Growth Differ. 1999;10:629–638. [PubMed] [Google Scholar]
- 7.Zelinski DP, Zantek ND, Stewart JC, et al. EphA2 overexpression causes tumorigenesis of mammary epithelial cells. Cancer Res. 2001;61:2301–2306. [PubMed] [Google Scholar]
- 8.Duxbury MS, Ito H, Zinner MJ, et al. EphA2: a determinant of malignant cellular behavior and a potential therapeutic target in pancreatic adenocarcinoma. Oncogene. 2004;23:1448–1456. doi: 10.1038/sj.onc.1207247. [DOI] [PubMed] [Google Scholar]
- 9.Pratt RL, Kinch MS. Activation of the EphA2 tyrosine kinase stimulates the MAP/ERK kinase signaling cascade. Oncogene. 2002;21:7690–7699. doi: 10.1038/sj.onc.1205758. [DOI] [PubMed] [Google Scholar]
- 10.Kinch MS, Moore MB, Harpole DH., Jr Predictive value of the EphA2 receptor tyrosine kinase in lung cancer recurrence and survival. Clin Cancer Res. 2003;9:613–618. [PubMed] [Google Scholar]
- 11.Hatano M, Eguchi J, Tatsumi T, et al. EphA2 as a glioma-associated antigen: a novel target for glioma vaccines. Neoplasia. 2005;7:717–722. doi: 10.1593/neo.05277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Abraham S, Knapp DW, Cheng L, et al. Expression of EphA2 and Ephrin A-1 in carcinoma of the urinary bladder. Clin Cancer Res. 2006;12:353–360. doi: 10.1158/1078-0432.CCR-05-1505. [DOI] [PubMed] [Google Scholar]
- 13.Wykosky J, Gibo DM, Stanton C, et al. EphA2 as a novel molecular marker and target in glioblastoma multi-forme. Mol Cancer Res. 2005;3:541–551. doi: 10.1158/1541-7786.MCR-05-0056. [DOI] [PubMed] [Google Scholar]
- 14.Xu F, Zhong W, Li J, et al. Predictive value of EphA2 and EphrinA-1 expression in oesophageal squamous cell carcinoma. Anticancer Res. 2005;25:2943–2950. [PubMed] [Google Scholar]
- 15.Han L, Dong Z, Qiao Y, et al. The clinical significance of EphA2 and Ephrin A-1 in epithelial ovarian carcinomas. Gynecol Oncol. 2005;99:278–286. doi: 10.1016/j.ygyno.2005.06.036. [DOI] [PubMed] [Google Scholar]
- 16.Herrem CJ, Tatsumi T, Olson KS, et al. Expression of EphA2 is prognostic of disease-free interval and overall survival in surgically treated patients with renal cell carcinoma. Clin Cancer Res. 2005;11:226–231. [PubMed] [Google Scholar]
- 17.Thaker PH, Deavers M, Celestino J, et al. EphA2 expression is associated with aggressive features in ovarian carcinoma. Clin Cancer Res. 2004;10:5145–5150. doi: 10.1158/1078-0432.CCR-03-0589. [DOI] [PubMed] [Google Scholar]
- 18.Wu D, Suo Z, Kristensen GB, et al. Prognostic value of EphA2 and EphrinA-1 in squamous cell cervical carcinoma. Gynecol Oncol. 2004;94:312–319. doi: 10.1016/j.ygyno.2004.05.019. [DOI] [PubMed] [Google Scholar]
- 19.Saito T, Masuda N, Miyazaki T, et al. Expression of EphA2 and E-cadherin in colorectal cancer: correlation with cancer metastasis. Oncol Rep. 2004;11:605–611. [PubMed] [Google Scholar]
- 20.Duxbury MS, Ito H, Zinner MJ, et al. Ligation of EphA2 by Ephrin A1-Fc inhibits pancreatic adenocarcinoma cellular invasiveness. Biochem Biophys Res Commun. 2004;320:1096–1102. doi: 10.1016/j.bbrc.2004.06.054. [DOI] [PubMed] [Google Scholar]
- 21.Embuscado EE, Laheru D, Ricci F, et al. Immortalizing the complexity of cancer metastasis: genetic features of lethal metastatic pancreatic cancer obtained from rapid autopsy. Cancer Biol Ther. 2005;4:548–554. doi: 10.4161/cbt.4.5.1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Moore PS, Sipos B, Orlandini S, et al. Genetic profile of 22 pancreatic carcinoma cell lines: Analysis of K-ras, p53, p16 and DPC4/Smad4. Virchows Arch. 2001;439:798–802. doi: 10.1007/s004280100474. [DOI] [PubMed] [Google Scholar]
- 23.Sun C, Yamato T, Furukawa T, et al. Characterization of the mutations of the K-ras, p53, p16, and SMAD4 genes in 15 human pancreatic cancer cell lines. Oncol Rep. 2001;8:89–92. doi: 10.3892/or.8.1.89. [DOI] [PubMed] [Google Scholar]
- 24.Brantley-Sieders DM, Fang WB, Hicks DJ, et al. Impaired tumor microenvironment in EphA2-deficient mice inhibits tumor angiogenesis and metastatic progression. FASEB J. 2005;19:1884–1886. doi: 10.1096/fj.05-4038fje. [DOI] [PubMed] [Google Scholar]
- 25.Fang WB, Brantley-Sieders DM, Parker MA, et al. A kinase-dependent role for EphA2 receptor in promoting tumor growth and metastasis. Oncogene. 2005;24:7859–7868. doi: 10.1038/sj.onc.1208937. [DOI] [PubMed] [Google Scholar]
- 26.Huusko P, Ponciano-Jackson D, Wolf M, et al. Nonsense-mediated decay microarray analysis identifies mutations of EPHB2 in human prostate cancer. Nat Genet. 2004;36:979–983. doi: 10.1038/ng1408. [DOI] [PubMed] [Google Scholar]
- 27.Bardelli A, Parsons DW, Silliman N, et al. Mutational analysis of the tyrosine kinome in colorectal cancers. Science. 2003;300:949. doi: 10.1126/science.1082596. [DOI] [PubMed] [Google Scholar]
- 28.Jemal A, Siegel R, Ward E, et al. Cancer statistics. CA Cancer J Clin. 2006;56:106–130. doi: 10.3322/canjclin.56.2.106. [DOI] [PubMed] [Google Scholar]
- 29.Laheru D, Yeo CJ. Role of adjuvant therapy in the management of pancreatic cancer. Adv Surg. 2005;39:223–244. doi: 10.1016/j.yasu.2005.05.003. [DOI] [PubMed] [Google Scholar]
- 30.Carles-Kinch K, Kilpatrick KE, Stewart JC, et al. Antibody targeting of the EphA2 tyrosine kinase inhibits malignant cell behavior. Cancer Res. 2002;62:2840–2847. [PubMed] [Google Scholar]
- 31.Noblitt LW, Bangari DS, Shukla S, et al. Decreased tumorigenic potential of EphA2-overexpressing breast cancer cells following treatment with adenoviral vectors that express EphrinA1. Cancer Gene Ther. 2004;11:757–766. doi: 10.1038/sj.cgt.7700761. [DOI] [PubMed] [Google Scholar]
- 32.Griffin CA, Hruban RH, Morsberger LA, et al. Consistent chromosome abnormalities in adenocarcinoma of the pancreas. Cancer Res. 1995;55:2394–2399. [PubMed] [Google Scholar]
- 33.Johansson B, Bardi G, Heim S, et al. Nonrandom chromosomal rearrangements in pancreatic carcinomas. Cancer. 1992;69:1674–1681. doi: 10.1002/1097-0142(19920401)69:7<1674::aid-cncr2820690706>3.0.co;2-l. [DOI] [PubMed] [Google Scholar]
- 34.Piaskowski S, Rieske P, Szybka M, et al. GADD45A and EPB41 as tumor suppressor genes in meningioma pathogenesis. Cancer Genet Cytogenet. 2005;162:63–67. doi: 10.1016/j.cancergencyto.2005.02.009. [DOI] [PubMed] [Google Scholar]
- 35.White PS, Thompson PM, Gotoh T, et al. Definition and characterization of a region of 1p36.3 consistently deleted in neuroblastoma. Oncogene. 2005;24:2684–2694. doi: 10.1038/sj.onc.1208306. [DOI] [PubMed] [Google Scholar]
- 36.Brantley-Sieders DM, Caughron J, Hicks D, et al. EphA2 receptor tyrosine kinase regulates endothelial cell migration and vascular assembly through phosphoinositide 3-kinase-mediated Rac1 GTPase activation. J Cell Sci. 2004;117:2037–2049. doi: 10.1242/jcs.01061. [DOI] [PubMed] [Google Scholar]
- 37.Dodelet VC, Pasquale EB. Eph receptors and ephrin ligands: embryogenesis to tumorigenesis. Oncogene. 2000;19:5614–5619. doi: 10.1038/sj.onc.1203856. [DOI] [PubMed] [Google Scholar]
- 38.Walker-Daniels J, Riese DJ, Kinch MS. c-Cbl-dependent EphA2 protein degradation is induced by ligand binding. Mol Cancer Res. 2002;1:79–87. [PubMed] [Google Scholar]
- 39.Davis S, Gale NW, Aldrich TH, et al. Ligands for EPH-related receptor tyrosine kinases that require membrane attachment or clustering for activity. Science. 1994;266:816–819. doi: 10.1126/science.7973638. [DOI] [PubMed] [Google Scholar]
- 40.Nichols LS, Ashfaq R, Iacobuzio-Donahue CA. Claudin 4 protein expression in primary and metastatic pancreatic cancer: support for use as a therapeutic target. Am J Clin Pathol. 2004;121:226–230. doi: 10.1309/K144-PHVD-DUPD-D401. [DOI] [PubMed] [Google Scholar]
- 41.Ryu B, Jones J, Hollingsworth MA, et al. Invasion-specific genes in malignancy: serial analysis of gene expression comparisons of primary and passaged cancers. Cancer Res. 2001;61:1833–1838. [PubMed] [Google Scholar]
- 42.Ryu B, Jones J, Blades NJ, et al. Relationships and differentially expressed genes among pancreatic cancers examined by large-scale serial analysis of gene expression. Cancer Res. 2002;62:819–826. [PubMed] [Google Scholar]
- 43.Tanaka M, Kamata R, Sakai R. EphA2 phosphorylates the cytoplasmic tail of Claudin-4 and mediates paracellular permeability. J Biol Chem. 2005;280:42375–42382. doi: 10.1074/jbc.M503786200. [DOI] [PubMed] [Google Scholar]
- 44.Kikawa KD, Vidale DR, Van Etten RL, et al. Regulation of the EphA2 kinase by the low molecular weight tyrosine phosphatase induces transformation. J Biol Chem. 2002;277:39274–39279. doi: 10.1074/jbc.M207127200. [DOI] [PubMed] [Google Scholar]
- 45.Hafner C, Schmitz G, Meyer S, et al. Differential gene expression of Eph receptors and ephrins in benign human tissues and cancers. Clin Chem. 2004;50:490–499. doi: 10.1373/clinchem.2003.026849. [DOI] [PubMed] [Google Scholar]