Abstract
The molecular docking tools Autodock and Surflex accurately reproduce the crystallographic structures of a collection of small molecule ligands that have been shown to bind nucleic acids. Docking studies were performed with the intercalators daunorubicin and ellipticine and the minor groove binders distamycin and pentamidine. Autodock and Surflex dock daunorubicin and distamycin to their nucleic acid targets within a resolution of approximately 2 Å, which is similar to the limit of the crystal structure resolution. However, for the top ranked poses, Autodock and Surflex both dock ellipticine into the correct site but in a different orientation compared to the crystal structure. This appears not only to be partly related to the symmetry of the target nucleic acid, as ellipticine is able to dock from either side of the intercalation site, but also due to the shape of the ligand and docking accuracy. Surflex docks pentamidine in a symmetrically equivalent orientation relative to the crystal structure, while Autodock was able to dock this molecule in the original orientation. In the case of the Surflex docking of pentamidine, the initial rmsd is misleading, given the symmetrical structure of pentamidine. Importantly, the ranking functions of both of these programs are able to return a top pose within approximately 2 Å rmsd for daunorubicin, distamycin, and pentamidine and approximately 3 Å rmsd for ellipticine compared to their respective crystal structures. Some docking challenges and potential pitfalls are explored, such as the importance of hydrogen treatment on ligands as well as the scoring functions of Autodock and Surflex. Overall for this set of complexes, Surflex is preferred over Autodock for virtual screening, as although the results are comparable, Surflex has significantly faster performance and ease of use under the optimal software conditions tested. These experiments show that molecular docking techniques can be successfully extended to include nucleic acid targets, a finding which has important implications for virtual screening applications and in the design of new small molecules to target therapeutically relevant morphologies of nucleic acids.
INTRODUCTION
Molecular docking techniques have shown great promise as a new tool in the discovery of novel small molecule drugs for targeting proteins.1–4 Fewer molecular docking studies have been performed targeting nucleic acids structures, despite advances in the understanding of the functional importance and the unique structural features of duplex, triplex, and G-quadruplex morphologies.5–9 This is unfortunate since not only are there clinically used drugs that target nucleic acids but also many forms of nucleic acids are becoming an increasingly attractive target for antineoplastic and antimicrobial agents.8,10–18 The few docking studies in which nucleic acids are targeted have focused on such sites as the minor groove of DNA, a tetraloop structure of RNA, and the major groove of an RNA duplex, while rarely targeting intercalation sites which also hold therapeutic potential.19–24 The use of molecular docking has important implications for the synthesis and development of small molecule drugs that selectively target nucleic acids since these techniques have the potential to shed light on the interaction and mechanism of action of these ligands with targets that may have medicinal value.
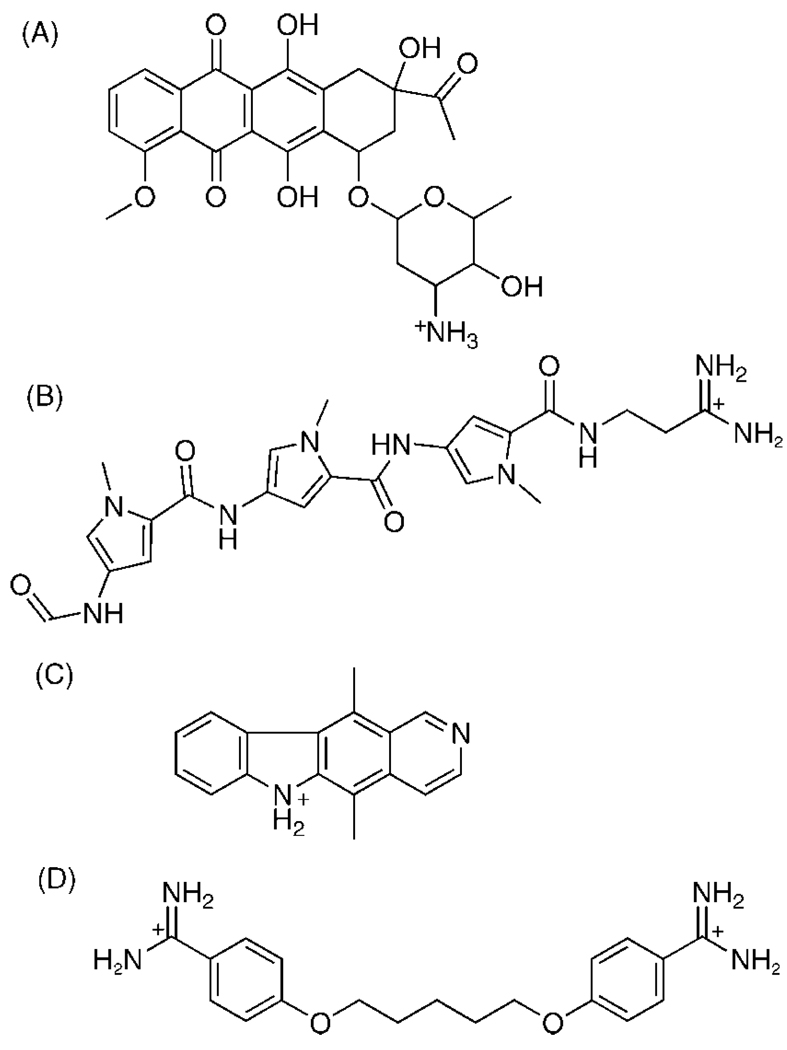
Small molecules can interact with various morphologies of nucleic acids at multiple sites to alter nucleic acid function.21,25,26 In the case of duplex DNA, one drug class binds within the minor groove, and a second class intercalates between existing base pairs of the nucleic acid structure.11,12,27 Intercalators and groove binders have distinctive thermodynamic signatures that indicate different driving forces for binding.28 The minor groove is a particularly attractive target for small molecules since this site has less competition from proteins and polymerases, which typically interact with the major groove.13 The closer proximity of the strands in the minor groove compared to the major groove allows more contact surface area for a small molecule to bind tightly.29 The unfavorable geometry of the major groove is another reason why few drugs target this groove.21 Two well-known and well-characterized minor groove binders are the anti-malarial drug pentamidine and the antiviral drug distamycin, which we selected for our studies.13,14,30,31 While only limited docking studies have been performed with minor groove binders, even fewer studies have tested whether drugs that act through intercalation can be modeled successfully using docking methods.2,12 We selected two prototypical intercalators, daunorubicin, a drug commonly used to treat certain forms of leukemia, and ellipticine, another antine-oplastic drug, for docking experiments using Autodock (4)32 and Surflex (2.11)33 (Figure 1).11,34
Figure 1.
Chemical structures of the four test ligands used in the Autodock and Surflex docking studies: (A) daunorubicin, (B) distamycin, (C) ellipticine, and (D) pentamidine.
Autodock 4 and Surflex 2.11 have been used previously for protein–ligand docking, but very few studies have been performed using nucleic acids as targets.8 Autodock is a logical selection for further exploration as it has been shown in some cases to be superior to DOCK, FlexX, and GOLD at reproducing the crystallographic pose of ligand–protein complexes.35 Surflex was chosen because it has rapid computational speed with protein–ligand docking which could prove useful for virtual screening.3 Autodock and Surflex have important differences in search algorithms and scoring functions. A search algorithm is initially used for conformationally sampling the ligand and target interactions, and scoring functions are used for evaluating and ranking the final poses of the ligand to determine the “correct” pose.36
Autodock performs molecular dockings by precalculating energy grids around a site of interest on the target.37 A stochastic search algorithm utilizing the Lamarkian Genetic Algorithm (LGA) for exploring the grid space is used to perform energy evaluations of the position of the ligand with respect to the target energy grids.37 This algorithm explores the various orientations and conformations of the whole ligand relative to the energy grids for the defined number of energy evaluations and returns the lowest energy conformation in the target site.37 The LGA has found particular utility in modeling systems with large numbers of rotatable bonds and possible numbers of conformations.37 Surflex uses a socalled “whole” molecule alignment algorithm based on morphological similarity between the ligand and target.3 This docking approach aligns the ligand to a “protomol” or idealized ligand in the active site of the target.3 The protomol is composed of a collection of fragments or probe molecules that characterize the surface morphology of the binding site.38 These probe molecules consist of CH4, C=O, and N–H fragments that model steric effects in the binding pocket, hydrogen bond acceptor groups, and hydrogen bond donor groups, respectively.3,38 The docking ligand fragments are checked for alignment and similarity against the protomol probes.3 This is referred to as a “whole” molecule approach because after the initial ligand fragmentation, both the small fragment and the rest of the “whole” ligand are carried into the protomol binding site.3 However, only the small fragment is checked for similarity and alignment against the protomol, while the rest of the “whole” ligand is assessed for steric interactions in the target site after optimal alignment of the fragment.3 This “whole” molecule approach is powerful because it considers the subsequent position of the rest of the “whole” molecule with respect to the target after the small fragment is optimally aligned with the protomol.3 This is an important difference between Autodock and Surflex, since Autodock involves evaluation of the conformations of the whole ligand without ligand fragmentation.37
The scoring functions for Autodock and Surflex are partially empirically based, with Autodock incorporating an Amber type force field and Surflex calculating atom to atom pairwise interactions between the ligand and target.3,37,39 Autodock evaluates pairwise interactions based on van der Waals radii of the atoms to determine the free energy of binding and returns the optimal lowest energy docked conformation as the best docked pose.36 The Surflex scoring function is parametrized by calculating van der Waals distances between protein and ligand, and parametrization of the scoring function was based on 34 protein–ligand complexes.40 Surflex assigns the atoms as either polar or nonpolar and then calculates a score based on hydrophobic and polar contacts between the two atoms.4 The docked poses are then ranked according to the maximal Surflex Overall score.
Aside from the algorithmic differences in Surflex and Autodock, there are several other aspects of molecular docking in general and these programs specifically that present challenges to successful docking of ligands to nucleic acids. First, because proteins have attracted the most interest as drug targets, proteins have also been the focus of most docking efforts compared to nucleic acids.36 This leads to the question of whether these protein-configured docking programs will work for nucleic acids because of the unique structural features of nucleic acids including their high charge density, exposed binding sites, and distinct geometrical symmetry.36,41 Another challenge is the dependence on crystal structures for visualizing how ligands interact with their targets and for assessing the accuracy of docking software. This approach relies on both the availability and resolution of the crystal structure. For nucleic acids, there are few crystal structures of ligand–nucleic acid complexes available, and even small variations in the resolution of the atomic positions of the crystals can significantly affect the modeling of important forces between the ligand and target such as hydrogen bonding.42 Differences in scoring functions also present a challenge for docking, as ranking of the poses is typically the most difficult aspect of docking.43,44 The coefficients and weighting for the scoring function terms are calibrated based on ligand–protein complexes, and it is unknown how well Autodock and Surflex would perform with ligand–nucleic acid complexes.39 Autodock and Surflex include entropic contributions by accounting for conformational and tortional changes as well as a term for solvation.3,39 However, the entropic contribution of solvation terms for most docking programs has been difficult to incorporate accurately in scoring functions and could contribute to erroneous pose ranking.36 Another traditionally challenging area for docking programs is accounting for target flexibility, since even small conformational changes of the ligand in the binding pocket can cause dramatic changes in the scoring function.4 While Autodock has the option to explore side chain flexibility for protein receptors, this function has not been extensively explored in the published literature for nucleic acids. Moreover, Surflex does not take target flexibility into account during molecular docking.3 To fairly compare the performance of these two programs, target flexibility was not considered in these experiments. These are important considerations when performing docking of ligands to nucleic acids using Autodock and Surflex and could significantly impact docking performance.
In spite of these challenges, however, we demonstrate that Autodock and Surflex can accurately dock small molecules with different binding modes to nucleic acid targets. More importantly, the ranking of the poses is also evaluated, which has been the more challenging aspect for many docking programs.43,44 The minor groove binders, distamycin and pentamidine, and the intercalators, daunorubicin and ellipticine, were selected for docking studies since these small molecules have crystal structures that are available in the Protein Data Bank (PDB). Autodock and Surflex software operating parameters were evaluated to determine which parameters increase docking accuracy and the successful ranking of the poses. Given the challenge of docking to nucleic acids, some reasons for suboptimal docking are detailed, including the importance of hydrogens on ligands, the scoring functions of the programs, and the quality of the crystal structure. This collection of experiments demonstrates the utility of these programs for molecular docking of ligands to target nucleic acids.
EXPERIMENTAL AND COMPUTATIONAL METHODS
Virtual Library Preparation
Ligand–nucleic acid complex crystal structures for daunorubicin, distamycin, ellipticine, and pentamidine were obtained from the Protein Data Bank with identification numbers of 152d, 2dnd, 1z3f, and 1d64, respectively. The resolutions of these structures are 1.4 Å, 2.2 Å, 1.5 Å, and 2.1 Å, respectively. Distamycin and pentamidine are bound to the minor groove of DNA duplex dodecamers d(CGCAAATTTGCG)2 and d(CGCGAATTCGCG)2, respectively. Daunorubicin and ellipticine intercalate between the cytosine and guanine nucleotides in the sequence d(CGATCG)2. For the ellipticine intercalation PDB structure, Maestro (8.0)45 was used to construct the symmetrical strand to form a complete, complementary, double stranded DNA. For the intercalator nucleic acid targets, there were two intercalation sites on the target. Thus, the 3′ terminal guanine residue was removed from the 6 base pair sequence so that there would only be a single intercalation site in the target nucleic acid structure. The ligand and nucleic acid targets were saved as separate files for docking purposes.
The PDB files were visually inspected using Macromodel (7.0)46 and all water molecules were removed. Amber ligand atom types were assigned using Sybyl (7.3),47 and hydrogen atoms were added as appropriate. The program Antechamber in the software suite Amber (8)48 was used to assign AM1-BCC charges to the atoms in each of the ligands and to also convert the files from PDB format to MOL2. Python scripts were used to prepare the nucleic acid structures in PDBQT format with Gasteiger charges for use in Autodock experiments, while MOL2 files were used for Surflex experiments.
Autodock 4 Methods
Autodock 4 and the graphical user interface AutoDockTools (1.4.6)49 were compiled for a Macintosh OS X PowerMac G5 and Linux workstations. AutoDockTools 1.4.6 was used for establishing the Autogrid points as well as visualization of docked ligand–nucleic acid structures. The target site on the nucleic acid was specified to encompass either the entire minor groove or the intercalation target site. The grid center was also established by centering the grid box on either the minor groove or the intercalation site. The grid maps had a spacing of 0.375 Å.
Several available docking parameter options in Autodock 4 were systematically varied to determine the optimal conditions for ligand–nucleic acid docking. These factors include the number of total energy evaluations per docking run and also the total number of docking runs performed. The total number of energy evaluations is the total number of ligand–target energy interaction evaluations before the lowest energy conformation is selected. These factors are suggested as logical starting areas of optimization as they have previously been shown to impact ligand–protein docking studies.50 The number of energy evaluations per docking run was varied as 200,000 (2E5), 2,000,000 (2E6), or 20,000,000 (2E7). Docking runs were varied as 5, 10, or 20 runs. Thus, a total of nine experiments were performed with varying numbers of energy evaluations and dockings to determine if these factors would impact docking accuracy and ranking. All other docking parameters were left at the default values. For the Autodock parametrization testing experiment with 50 docks and 5E7 energy evaluations, the “ga_num_generations” was set at 100,000. Normally, the docking run will terminate when either the “ga_num_generations” or the number of energy evaluations is reached,51 so the “ga_num_generations” was increased from 27,000 to 100,000 to ensure that 5E7 energy evaluations was reached for these docking experiments.
Surflex 2.11 Methods
Surflex 2.11 was compiled for a Macintosh OS X PowerMac G5 and Linux workstations. The protomol was generated using a ligand-based approach, where a small molecule is selected that fits into the site of interest. The structure of the molecule in the site is then used for protomol generation. The protomol represents a set of molecular fragments that characterizes the active site and to which the ligand of interest is fragmented and checked for both similarity and alignment.4 Furamidine was chosen as the ligand for protomol generation, as it has been previously shown to be a minor groove binder and is small enough to fit into the intercalation site to ensure adequate protomol generation.12,14,52 Importantly, this also reduces the bias of the evaluation by not using the actual ligands to be docked and is a more realistic, generalized docking approach. Two important factors that can significantly effect the size and extent of the protomol generated are “proto_thresh” and “proto_bloat” options. “Proto_thresh” determines how far the protomol extends into the concavity of the target site, while “proto_bloat” impacts how far the protomol extends outside of the concavity.53 For the purposes of these experiments, “proto_thresh” was set to 0.2 and “proto_bloat” was left at the default (0) for all protomols generated except for daunorubicin, where a “proto_bloat” of 0.5 was used. Protomols were visualized with Sybyl 7.3 to ensure proper coverage of the desired target area.
Surflex 2.11 offers many parameters that can be customized to help optimize ligand targeted docking. An investigation of all of the combinations of these factors is beyond the scope of this paper. Instead, two factors, the “Multistart 5” and “Random 5” options, were selected as these are thought to have the potential to most significantly impact the accuracy of the docked poses. The “Multistart 5” designation enables docking to begin from 5 different initial starting positions around the designated target. Previously, Jain et al. had observed little increase in successful docks with protein targets beginning at a value of 5 (“Multistart 5”), relative to the additional computational resources required for docking these extra conformations.53 A “Random 5” option ensures that the ligand adopts 5 random X,Y,Z coordinate conformations prior to initiating docking calculations. These options are both thought to be important since it minimizes the chance that the ligand may be randomly assigned to an energetically or conformationally unfavorable position from which it cannot recover during the docking. A total of three experiments were subsequently performed, with the first having default Surflex 2.11 options (“No Multistart”, “No Random”), the second with implementation of “Multistart 5”, and the last experiment with implementation of both “Multistart 5” and “Random 5” to test for a potential synergistic effect between these two options. All other parameters were left at the default values.
rmsd Calculations
One metric for evaluation of the quality of docking results is the difference in the X,Y,Z coordinates between the docked pose and the known crystal structure which can be used to calculate the Root Mean Square Deviation (rmsd) between the two poses. For consistency in evaluation of docked poses, the Surflex 2.11 software rmsd method was used for calculation of the rmsd differences for both Autodock and Surflex results based on only the heavy atoms. This method determines the rmsd between the docked pose and the crystallographic structure using a direct atom to atom comparison of the two structures. An additional Surflex rmsd function (Actual rmsd ISO) was used to account for internal ligand symmetry. This function is independent of atom numbering and computes isomorphisms between the crystal and docked poses, returning the lowest symmetrical rmsd value.54 The practice of accounting for ligand symmetry is fairly universal and has been documented in previous papers.54 To address nucleic acid target symmetry, Macromodel 7.0 was used to flip and superimpose the docked pose on the crystallographic pose. This involves copying the complex consisting of the ligand docked to the target nucleic acid and then selecting to superimpose DNA bases from the copied structure onto the opposite DNA base of the original structure. Molecular superposition was performed using the “Superimpose Atoms” (SuprA) function followed by the “Rigid Superposition” (RigSA) function. In all cases, the resolution of the superposition was less than 0.15 Å. The superimposed structures were saved, and the coordinates were used for rmsd calculations. Surflex docked poses are in a MOL2 file format which can be used directly by the Surflex program for rmsd calculations. Autodock docked poses are in a PDB file format and were converted to a MOL2 file format using OpenBabel (2.1.1)55 or iBabel (2.0)56 prior to rmsd calculations. Docked poses of Autodock and Surflex in the target binding site were visualized using AutoDockTools.
Autodock and Surflex Scoring Function Methods
Rescoring of all top ranked Autodock and Surflex poses and the crystal structure poses was performed using the Autodock and Surflex scoring functions. To rescore all of the poses using the Autodock scoring function, the files were converted to Autodock PDBQT file format by merging all of the nonpolar hydrogens. The Autodock epdb command was used to calculate a free energy of binding (kcal/mol) for each of the poses. The Surflex “score_list” command was used to rescore the top ranked poses using the Surflex scoring function. Macromodel was used to add hydrogens to the crystal structures and to the top ranked Autodock poses which normally only has polar hydrogens added for docking purposes. The Surflex scoring function ranks poses by an affinity score, pKd.53 To fairly compare the docking poses for these two programs, the Surflex pKd results were converted to free energy of binding (kcal/mol), as previously described, where RT = 0.59 kcal/mol:57
| (1) |
Macromodel Energy of Binding Methods
Macromodel was used as a third, independent software to calculate the energy of binding of the poses using different force fields and solvation. All hydrogens were added, as previously described. The energy of binding was determined in structures with and without energy minimization of the hydrogens as follows
| (2) |
where Ecomplex is the energy of the docked ligand in the target, and the Eligand and Enucleic acid represent the individually calculated energies. Energy minimization was performed by the Polak-Ribier Conjugate Gradient (PRCG) method for 1000 iterations with a convergence threshold of 0.05. The force fields were set at either Amber* or OPLS2005, with and without implicit water solvation to show the effects of these factors on the energy of binding. The experiments with no implicit water solvation were performed with distant dependent electrostatic treatment with a dielectric constant of 4.0 and an extended cutoff. The experiments with water solvation were performed with a constant dielectric electrostatic treatment with a dielectric constant of 1.0 and a normal cutoff.
RESULTS AND DISCUSSION
Few studies have been performed to determine if molecular docking techniques such as Autodock and Surflex can dock ligands accurately to nucleic acids. We compare two poses derived from the docking calculations, the lowest rmsd pose for accuracy comparisons, and the top ranked pose for ranking comparison. A common metric for evaluation of accurate dockings is to calculate the rmsd between the crystallographic pose and the docked conformation. A level of significance of 2 Å will be evaluated to facilitate a comparison of these data to docking data in other reports.8,40,44,58,59 When evaluating the ranking functions of the programs under different software conditions, only the single top ranking pose was used for comparing software conditions, as this is typically the mostly likely and facilitating pose that would be evaluated across large libraries of ligands that are used for virtual screening. The top pose was also inspected visually to determine the goodness of the ligand fit within the expected target site. Using these metrics, the optimal software conditions to maximize docking accuracy and ranking were “5 docks” and “2E7 energy evaluations” for Autodock and either the “Multistart 5” and “No Random” or the “Multistart 5” and “Random 5” for Surflex.
Autodock 4 Docking Accuracy
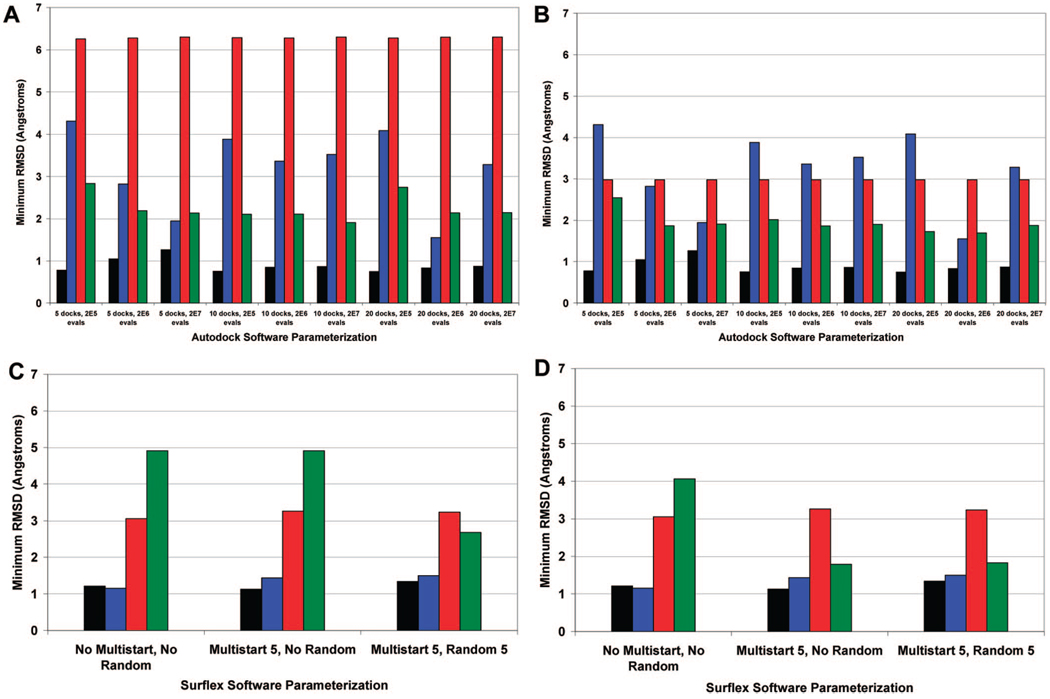
Close examination of the dock with the lowest rmsd for each software parametrization shows that Autodock is able to accurately reproduce the crystal structure of several ligand–nucleic acid complexes to a resolution of less than 2 Å (Figure 2A). Taking ligand and nucleic acid target symmetry into account results in even lower rmsd poses for pentamidine (ligand symmetry) and ellipticine (nucleic acid target symmetry). Of the four ligands tested, pentamidine is the only chemically symmetrical ligand. Accounting for this symmetry results in lower rmsd results since several of the poses that are docked in a flipped orientation relative to the crystal structure can be recalculated (Figure 2B). At first glance, the higher overall rmsd results for the optimal ellipticine pose can be misleading as this appears to be a relatively poor docking. Visualization of the dockings reveals that the ligand is actually docked successfully into the intercalation site but lies in a flipped orientation rotated 180 degrees relative to the crystal pose. This flipped orientation of ellipticine occurs for all of the lowest rmsd poses (Figure 2A) as well as the top ranked poses (Figure 3A). The orientation and quality of the docked ellipticine pose is partially explained by the symmetrical nature of the nucleic acid target, since ellipticine can dock into the intercalation site not only from the orientation observed in the crystal structure but also from a flipped orientation with intercalation from the opposite side of the nucleic acid. Given that the Surflex rmsd calculator is based solely on the ligand poses and is irrespective of the nucleic acid target structure symmetry, the rmsd for ellipticine is unusually high, even though ellipticine is positioned well inside the intercalation site compared to the crystal structure. Thus, flipping and superposition of the docked pose on the crystallographic pose using Macromodel were necessary for an accurate comparison to the crystal structure. The fact that ellipticine is docked in the intercalation site is encouraging, especially given the steric hindrance and tight fit typically associated with intercalation sites. Note that the Autodock grid is also large enough to allow for potential docking into the groove sites located near the intercalation site, so the intercalation dock is the preferred site. This emphasizes that rmsd values are only one metric for evaluating quality of docking poses and that the top poses should be visually inspected to check for ligand–target symmetry.
Figure 2.
Autodock and Surflex accuracy: The dock with the lowest rmsd is presented, regardless of ranking. Parts A and C present the rmsd calculated without taking into account ligand or nucleic acid symmetry, for Autodock and Surflex, respectively. Parts B and D include ligand and nucleic acid symmetry, for Autodock and Surflex, respectively. Black = daunorubicin, blue = distamycin, red = ellipticine, green = pentamidine.
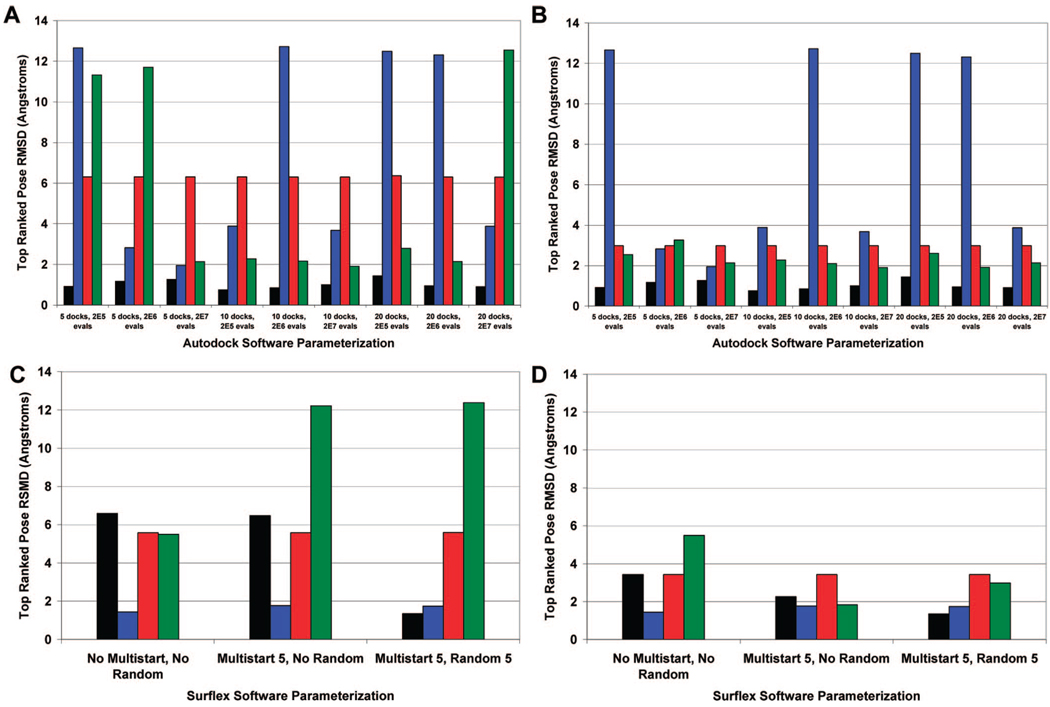
Figure 3.
The top ranked pose by Autodock and Surflex. Parts A and C present the rmsd calculated without taking into account ligand or nucleic acid symmetry, for Autodock and Surflex, respectively. Parts B and D include ligand and nucleic acid symmetry, for Autodock and Surflex, respectively. Black = daunorubicin, blue = distamycin, red = ellipticine, green = pentamidine.
The lowest rmsd docking pose for daunorubicin and pentamidine are close to the resolution of the crystal structures, especially at the software conditions of “5 docks” and “2E7 energy evaluations”. In particular, the rmsd for daunorubicin is almost always lower than 1 Å. The rmsd values for distamycin appear to be the most variable over the different software conditions, which is not surprising given that distamycin has the highest number of rotatable bonds (14) compared to daunorubicin (9), pentamidine (12), and ellipticine (0). The number of rotatable bonds for each molecule was defined by AutoDockTools using a united-atom representation that merges nonpolar hydrogens.49 AutoDockTools is used to automatically select the rigid “root” section of the ligand, and the “branches” off of the “root” are subsequently defined as rotatable bonds.51 Molecules with larger numbers of rotatable bonds are expected to take a larger number of energy evaluations to converge to an energy minimum due to a larger number of degrees of freedom and conformational states.58 The docking results for distamycin are especially encouraging considering that most small molecules that are tested for therapeutic utility typically have less than 12 rotatable bonds.59 The number of energy evaluations appears to be most important when the fewest number of docks (5) is used, and the accuracy of the distamycin docking increases significantly with an increasing number of energy evaluations. Moreover, once the number of energy evaluations used reaches 2E7, there appears to be no increase in docking accuracy when the number of docks is increased from 5 to 10 or 20. This finding is consistent with previous observations from ligand–protein studies that tested the effects of varying energy evaluations and the number of dockings on docking accuracy.50 Visualization of the distamycin docking poses that have a resolution of greater than 2 Å show that even though the rmsd is higher than the cutoff, the ligand still occupies a similar space in the minor groove relative to the crystal structure. These results suggest that a software parametrization of “5 docks” combined with “2E7 energy evaluations” is acceptable, as the resolution of all of these docks with the exception of ellipticine is less than 2 Å.
Autodock 4 Pose Ranking
The ability of Autodock to correctly rank the lowest rmsd docks must also be assessed as only one aspect of screening is to dock the pose correctly (low rmsd) but another aspect is to score the docked poses correctly. Autodock ranks the docked conformation by calculating a binding energy and sorting the results from lowest to highest energy. Ideally, the docked pose with the lowest binding energy would correspond to the docked pose with the lowest rmsd. In all software conditions, the top ranked dock for daunorubicin achieves the rmsd cutoff of 2 Å (Figure 3).
A number of poses with rmsd values less than 2 Å are produced for distamycin and pentamidine using several different software conditions. However, there are a number of top ranked poses for distamycin, ellipticine, and pentamidine in several software conditions that merit further discussion as these had higher rmsd values. It is critical to ascertain whether the high rmsd values associated with these poses are due to lack of consideration of either ligand or target symmetry or if the pose itself is of marginal quality. Visual inspection of the four top ranked poses for distamycin with a resolution of greater than 12 Å rmsd suggests that the flipped orientation of the ligand relative to the crystal structure is the main cause of the high rmsd. However, the high rmsd cannot be ascribed solely to nucleic acid target symmetry, as the crystal structure shows that distamycin is not centered around the minor groove and superposition of the docked pose results in poor visual overlap with the crystal structure. Instead there appears to be poor docking that is localized to the multiple terminal nitrogen groups, which float freely outside of the minor groove instead of the expected tight binding within the minor groove that is observed with the crystallographic structure. The marginal accuracy of these dockings may be influenced by the large number of rotatable bonds observed with distamycin. This significantly increases the degrees of freedom and number of possible conformations of the ligand, making it challenging to dock to the target.51,58 With respect to ellipticine, the high rmsd values appear to be due to a combination of the flipped orientation of the ligand which can be reassessed by accounting for nucleic acid target symmetry and also by marginal overall alignment of the docked pose relative to the crystal structure. Pentamidine is a unique case where consideration of ligand symmetry into the rmsd calculations dramatically reduces the rmsd values for several top ranking poses (Figures 3B). This shows that the high rmsd is ascribed to ligand symmetry rather than to marginal docking quality and atom overlap.
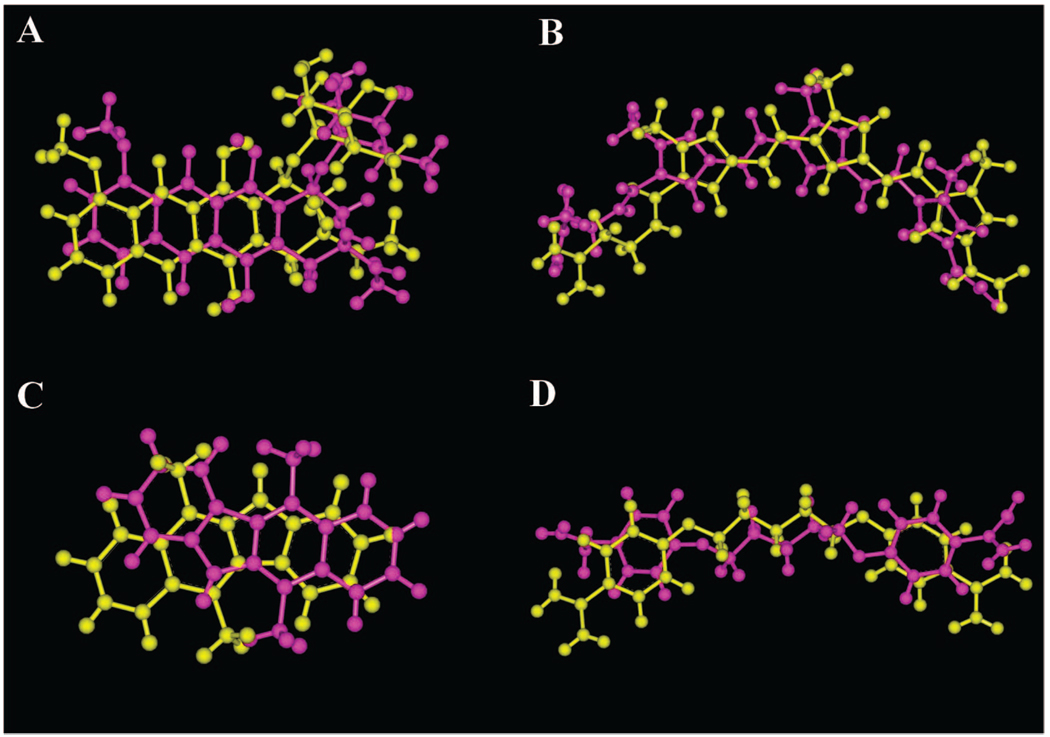
In summary, there are several software conditions that appear promising with respect to ranking of the poses including “5 docks” with “2E7 energy evaluations” and “10 docks” with “2E5 energy evaluations”. However, the real value in assessing Autodock performance lies in combining both docking accuracy and ranking of the docked results. A software parametrization of “5 docks” and “2E7 energy evaluations” appears best able to balance docking accuracy and ranking. By using this parametrization, docking of daunorubicin, distamycin, and pentamidine was achieved to a resolution of approximately 2 Å, while the intercalator ellipticine was the most challenging dock, with a top pose resolution of approximately 3 Å. These docked conformations are also visually in close agreement with the observed crystal structure (Figure 4).
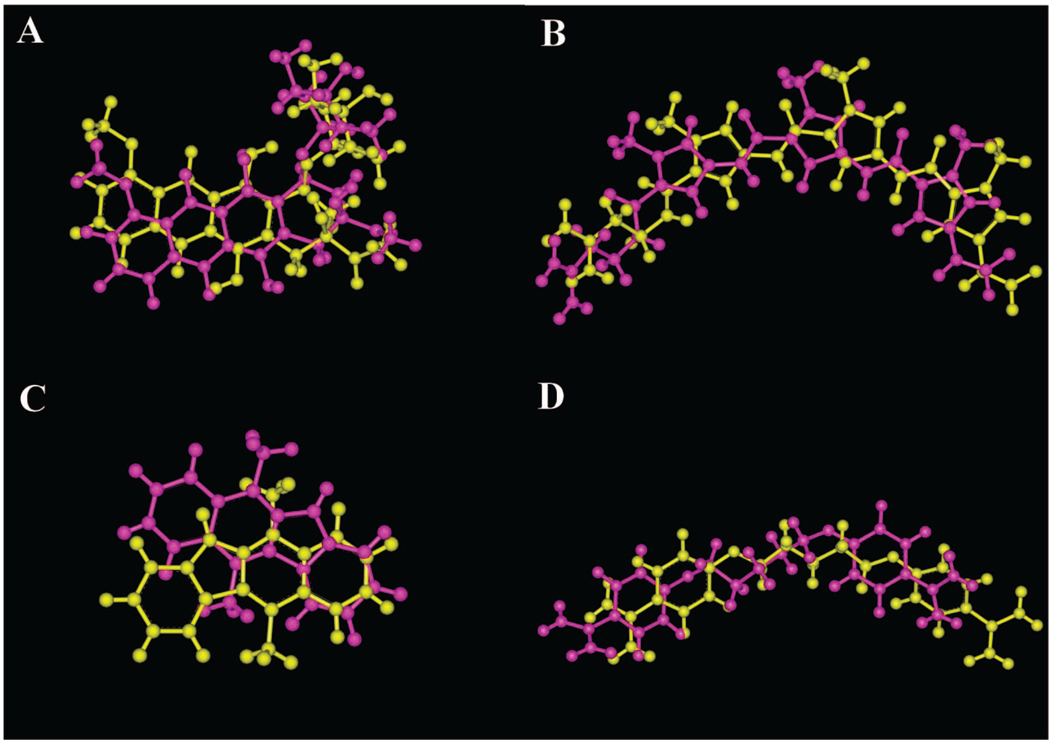
Figure 4.
Comparison of the top ranked Autodock pose (magenta) to the PDB crystallographic pose (yellow) for the experiment with a software parameterization of “5 docks” and “2E7 energy evaluations”: (A) daunorubicin, (B) distamycin, (C) ellipticine, and (D) pentamidine.
Surflex 2.11 Docking Accuracy
The Surflex docking results generally show that crystal structures are accurately reproduced (Figure 2). In all experiments, daunorubicin and distamycin are docked accurately to a resolution of less than 2 Å. Visualization of the lowest rmsd ellipticine pose demonstrates that ellipticine is docked in the correct orientation relative to the crystal structure. The higher rmsd for the top ellipticine pose relative to the other compounds appears to be due to the marginal alignment of the ligand structure with the crystal structure. A similar marginal overlap was observed for the Autodock ellipticine poses, as described previously. Importantly, ellipticine is located well inside the intercalation site. For pentamidine, incorporation of ligand symmetry into the rmsd calculation results in significant increases in the docking accuracy for all software conditions, with the lowest rmsd structures occurring with the “Multistart 5” only experiment and the “Multistart 5” and “Random 5” combination experiment. This is attributed to inclusion of poses that were docked in a flipped orientation that initially had rmsd values greater than 12 Å but subsequently have significantly lower rmsd values after taking into account ligand symmetry.
With respect to docking accuracy, addition of the “Multistart 5” option produces a better docked pose for pentamidine. This supports the hypothesis that initiating the docking of the ligand from multiple points surrounding the nucleic acid target increases the accuracy of the dockings. Interestingly, the addition of the “Random 5” option in combination with the “Multistart 5” option did not significantly impact the lowest rmsd dock produced for these test ligands. The “Random 5” option generates 5 randomized X,Y,Z coordinate positions of the atoms at the initial starting position of the ligand.53 Most importantly, Surflex is able to dock the ligands to the nucleic acid targets and produce docking results with rmsd values close to the resolution of the observed crystal structure.
Surflex 2.11 Pose Ranking
Ranking of Surflex results is performed by maximizing the Surflex Overall Score, which consists of an affinity score of the ligand for the target. Ideally, a maximal Surflex Overall Score would correspond with the lowest rmsd pose. Inspection of the rmsd values for the top Surflex docks ranked by maximal Surflex Overall Score are at first glance misleading (Figure 3). In particular, the experiment that included the “Multistart 5” and “No Random” options and the experiment with the “Multistart 5” and “Random 5” options initially appear to have a poor docking pose for pentamidine. However, closer visual inspection of the docked conformation relative to the crystal structure pose again emphasizes the use of symmetry for rmsd calculations where appropriate (Figure 5), which reduces the rmsd to under 2 Å.
Figure 5.
Comparison of the top ranked Surflex pose (magenta) to the PDB crystallographic pose (yellow) for the experiment with a software parameterization of “Multistart 5” and “Random 5”: (A) daunorubicin, (B) distamycin, (C) ellipticine, and (D) pentamidine.
For all of the experiments, ellipticine is docked in a flipped orientation in the intercalation site, which was initially thought to be the major factor influencing the high calculated rmsd value. However, even after accounting for nucleic acid target symmetry, ellipticine has still only minimal overlap with the crystal structure. Inspection of the top ranked dock for daunorubicin for the software parametrization with “No Multistart” and “No Random” and the software parametrization with “Multistart 5” and “No Random” options appears to show daunorubicin in a flipped orientation relative to the crystal structure. The daunosamine ring occupies the minor groove, which is similar to the ring location in the crystal structure. After taking into account the nucleic acid target symmetry, the docking pose rmsd values for both the “Multistart 5” and “No Random” experiment and the “Multistart 5” and “Random 5” experiment are dramatically improved, to a resolution of 3.4 Å and 2.3 Å, respectively. Finally, all Surflex software conditions docked distamycin to the target at a resolution of less than 2 Å. These results emphasize the importance of not only calculating rmsd values for docked poses but also visualizing results to check for reasonable docking conformations.
The software parametrization with “Multistart 5” alone and the software parametrization with both “Multistart 5” and “Random 5” appear to produce the top ranked results with the lowest rmsd structures, compared to the software parametrization of “No Multistart” and “No Random” options. The top ranked pose for daunorubicin using the “Multistart 5” and “Random 5” software parametrization has an rmsd of 1.3 Å and is superior to the top ranked dock for the other Surflex experiments. Both software conditions dock distamycin and ellipticine comparably with respect to the rmsd of the top ranked Surflex pose. For pentamidine, the top ranked pose for the “Multistart 5” and “No Random” option experiment has a marginally better rmsd for the top pose compared to the top pose from the “Multistart 5” and “Random 5” experiment. Overall, the performances of the “Multistart 5” and “No Random” experiment and the “Multistart 5” and “Random 5” experiments are comparable.
Extended Parameter Optimization for Autodock and Surflex
While the overall docking results for Surflex and Autodock generally show the ability to accurately reproduce the crystal structure and rank the results, it is important to determine the reason for some of the more challenging dockings such as ellipticine and distamycin. One possibility for the marginal docking accuracy could be an inadequate number of iterations (number of docks and energy evaluations for Autodock and multistart number and random parameters for Surflex) of the software. If this is the case, it would be expected that increased docking accuracy and ranking could be obtained by increasing the exploration of the Autodock and Surflex parameters.
To investigate this possibility for Autodock, the docking experiments with the four ligands were repeated after increasing the number of dockings from 5 to 50 and the number of energy evaluations from 2E7 to 5E7. The number of dockings were selected based on previous applications of the software.51 The number of energy evaluations was increased to 5E7, which is consistent with the number of energy evaluations used in previous protein docking experiments. 50 A similar approach was taken with Surflex by increasing the Multistart parameter from 5 to 10 and the Random parameter from 5 to 10. However, Jain et al. had previously seen only marginal improvement in increasing the Multistart parameter greater than 5 with protein docking.53
Evaluation of the docking accuracy (Figure SI7) and ranking (Figure SI8) results show that there is no benefit in docking accuracy or ranking for either Autodock or Surflex by extending dockings and evaluations of software parameters. Moreover, the Autodock experiments took approximately 25-fold longer under conditions of “50 docks” and “5E7 energy evaluations” compared to conditions of “5 docks” and “2E7 energy evaluations”. Surflex took approximately 5 times longer under conditions of “Multistart 10” and “Random 10” compared to “Multistart 5” and “Random 5”. Even if the extended experiments showed improved docking accuracy and ranking, the increase in computational time could be a limiting factor for use in virtual screening applications. In summary, the results suggest that the originally optimized Autodock conditions of “5 docks” and “2E7 energy evaluations” and Surflex conditions of “Multistart 5” and “Random 5” are optimized for molecular docking to nucleic acids.
Evaluation of the Autodock and Surflex Scoring Functions
As the docking accuracy and ranking does not appear to be related to suboptimal software parametrization, another possible contribution to marginal docking may be from the scoring functions of these programs. This is possible given that scoring functions are one of the major challenges of current docking programs.60 To investigate this possibility, the crystal structure and the top ranked poses for each method were rescored using both the Surflex and Autodock scoring functions. The poses were scored and ranked according to the lowest free energy of binding. An additional molecular mechanics method was selected to calculate the energy of binding of the crystal pose, Autodock, and Surflex poses. This was useful as the added hydrogens could also be selectively energetically minimized, which highlighted the hydrogen atom treatment as a potential pitfall. Macromodel 9.5 was used to determine the effects on the energy of binding of using either the OPLS2005 or Amber* force field with and without water as an implicit solvent. These experiments investigated if the limitations in the ranking of the software were related to the scoring functions for these programs.
Scoring of Poses by Autodock and Surflex
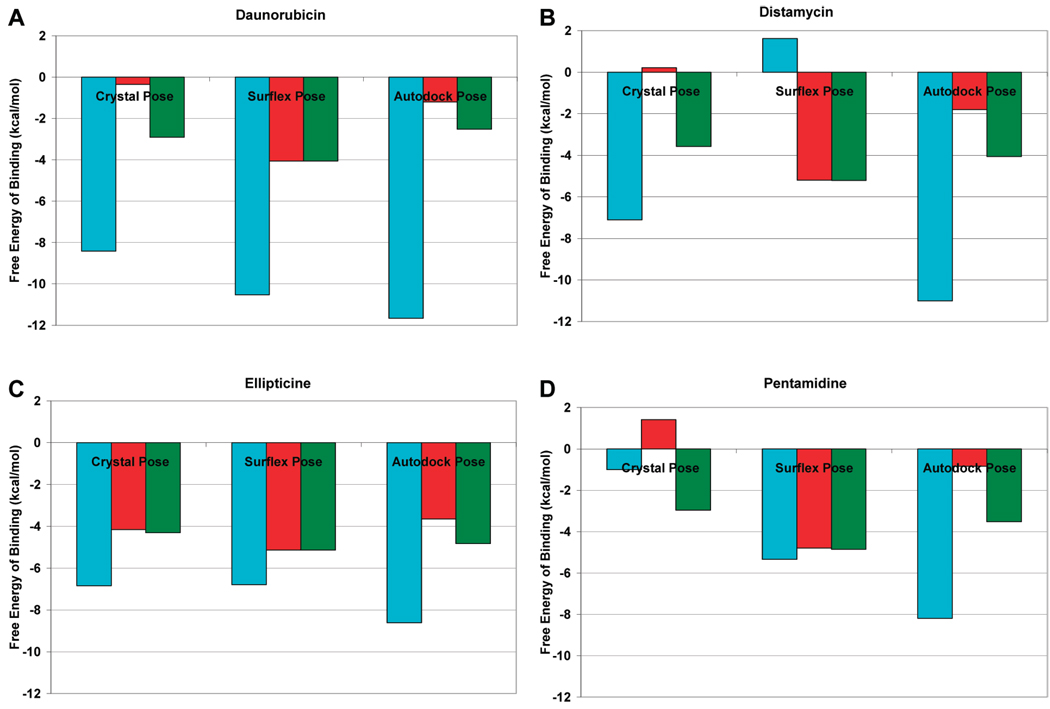
The direct comparison of the Surflex and Autodock Scoring Functions is shown in Figure 6. Unsurprisingly, the Surflex scoring function tends to score the Surflex poses the best, while the Autodock scoring function tends to score the Autodock poses the best. The Surflex scoring function scores the Autodock poses reasonably well, with a low free energy of binding. In general, the Autodock scoring function produces results with the lowest free energy of binding. Both Autodock and Surflex appear to typically score either the Autodock or Surflex poses as having lower free energy of binding compared to the crystal pose. The Surflex scoring function produces a “Static” score (red, Figure 6) and an “Optimized” score (green Figure 6) when scoring an individual pose. The “Static” score applies the Surflex scoring function directly to the input pose, with no energy minimization. The “Optimized” score performs a gradient energy minimization and subsequently scores the pose. Scoring of the Surflex poses using the Surflex scoring function reveals little difference between the Static score and Optimized score. On the other hand, Autodock and the crystal structure scores are significantly improved when comparing the “Static” score to the “Optimized” score. One possible explanation for this difference is how the hydrogens are accounted for by these docking programs.
Figure 6.
Comparison of the free energy of binding for the crystal pose, Autodock top ranked pose, and Surflex top ranked pose for various ligands using the Autodock and Surflex scoring functions: blue = Autodock scoring function, red = Surflex static scoring function, green = Surflex optimized scoring function. (A) daunorubicin, (B) distamycin, (C) ellipticine, and (D) pentamidine.
Hydrogen Atom Treatment of Poses Can Significantly Effect Free Energy of Binding
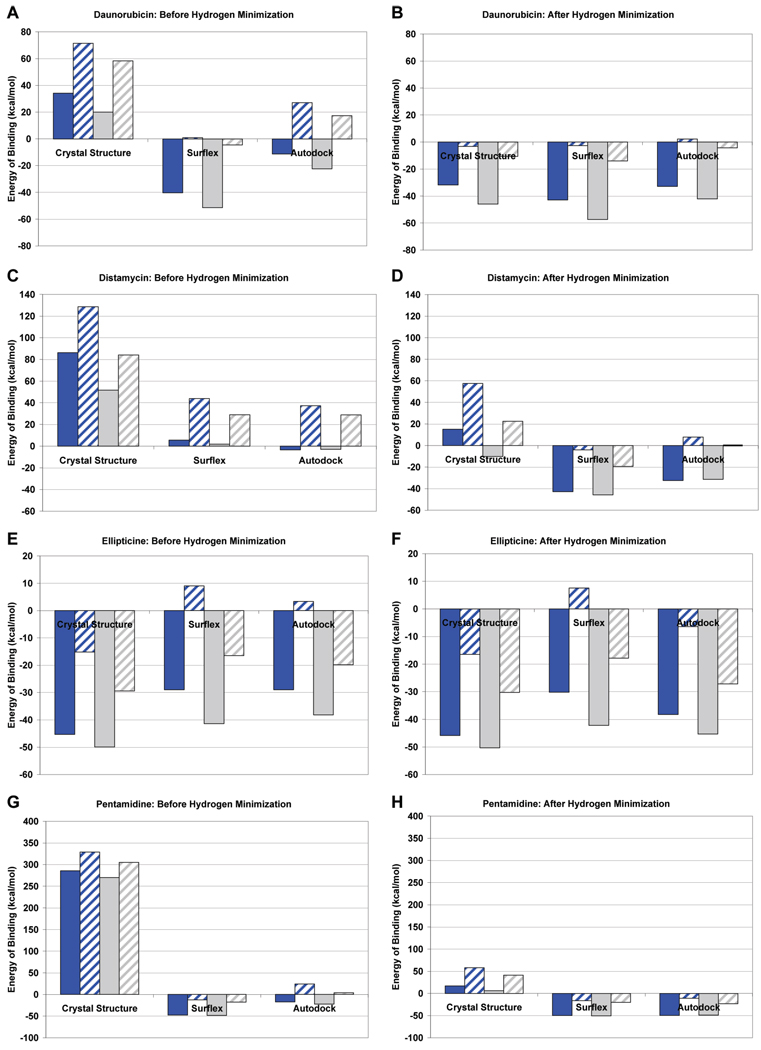
It appears from Figure 6 that the significant difference in the “Static” and “Optimized” Surflex scoring function scores for Autodock and the crystal poses could be influenced by the way hydrogen atoms are added to these structures. In order to determine if this is the case, it is important to first address the way hydrogens are normally accounted for by these programs. Surflex adds all hydrogens on the ligand prior to docking so all of the hydrogens are present during scoring. The crystal structure does not have any hydrogens added. Autodock uses a United Atom force field which takes into account “polar” hydrogens that are attached to electronegative atoms.51 “Nonpolar” hydrogens attached to carbon atoms are merged, and the charge is added to the nearby carbon atom.51 To evaluate whether the trends in Figure 6 could be influenced by the way hydrogens are handled by the docking programs, Macromodel was used to add all hydrogens to the ligands, and their binding energies were recalculated both before and after energy minimization of the hydrogens (Figure 7). Comparing the binding energy of the poses before and after minimization of the hydrogens shows that the most significant decrease in energy after minimization is seen with the crystal structure. However, there is also a substantial reduction in the energy of binding for Autodock. The Surflex binding energies appear to be the least effected presumably because all hydrogens were accounted for during docking and scoring. The results in Figure 7 are important because a molecular mechanics approach was used to assess each of the poses for the docking programs with two force fields and two solvation approaches. These results show that Surflex appears to consistently produce the docked poses with the lowest energy of binding. This suggests that the hydrogen atom treatment is an important consideration when scoring docked poses and can substantially influence scoring and energy calculations. It is interesting to note that ellipticine, which has the fewest number of rotatable bonds and hydrogen atoms, is least effected by hydrogen atom treatment.
Figure 7.
Calculated energy of binding by Macromodel for the crystal pose, Autodock top ranked pose, and Surflex top ranked pose for various ligands using the Amber* and OPLS2005 force fields with and without implicit water solvation: solid blue = OPLS2005, no implicit water solvation; blue with hatches = OPLS2005, with implicit water solvation; solid gray = Amber*, no implicit water solvation; gray with hatches = Amber*, with implicit water solvation. (A) and (B) daunorubicin, before and after hydrogen minimization, respectively; (C) and (D) distamycin, before and after hydrogen minimization, respectively; (E) and (F) ellipticine, before and after hydrogen minimization, respectively; and (G) and (H) pentamidine, before and after hydrogen minimization, respectively.
Effects of Force Field Choice and Solvation on Energy of Binding
A series of experiments was performed to test the effects of using either the Amber* and OPLS2005 force fields, with and without implicit water solvation, on the energy minimization and the calculated energy of binding of the ligands to the targets. The Amber* force field was selected because the Autodock force field is parametrized based on the Amber force field.39 OPLS2005 was chosen because it is an updated general force field from the original OPLSAA force field that has demonstrated utility in evaluating protein structures.61 The calculated energy of binding of the top ranked ligand poses, before and after energy minimization of the added hydrogens, is shown in Figure 7. The force field choice and solvation effects can significantly influence the calculated energy of binding. For the structures where the hydrogens were energetically minimized, the use of the Amber* force field with inclusion of water salvation appears to produce energy of binding results that are most consistent with the results in Figure 6 that were obtained using the Autodock and Surflex scoring functions. The energy of binding of the Autodock and Surflex poses appears significantly lower than the crystal structures, with the exception of ellipticine. It appears in these cases that for the Autodock and Surflex poses the addition of implicit salvation in just the energy minimization is not advantageous and not indicative of a favorable binding event. In total, this shows that force field selection and solvation factors can contribute substantially to scoring and ranking docked poses, and this could be one of the main challenges of docking ligands to nucleic acids.
Crystal Structure Energies Are Not Necessarily the “Minima”
The free energy of binding for the crystal structure and top ranked Autodock and Surflex poses, determined by either the Autodock or Surflex scoring function, is shown in Figure 6. The top ranked Autodock or Surflex pose almost universally has a comparable or lower free energy of binding compared to the reference crystal structure. This is true irrespective of the scoring function. These results are supported by the molecular mechanics results in Figure 7, where the calculated energy of binding for all ligands, apart from ellipticine, is comparable to or lower than the crystal structure. These results are important for several reasons. First, the crystal structures should not be assumed to be the energetically minimized conformation in the nucleic acid target as the structure is a product of experimental data and the original force field it is fitted to. Interestingly, the energy of the lower resolution crystals, distamycin (2.2 Å) (Figure 7D) and pentamidine (2.1 Å) (Figure 7H), appear to have more variability between the energy of the top ranked poses and the crystal structure compared to the higher resolution crystal structures daunorubicin (1.4 Å) (Figure 7B) and ellipticine (1.5 Å) (Figure 7F). This suggests that the quality and resolution of the crystal structure may be a consideration when performing docking studies and evaluating poses. However, it is also a function of flexibility of the ligand as distamycin and pentamidine are the most flexible. Another reason these results are important is that the docked poses such as distamycin that initially appeared to be of only marginal accuracy by rmsd compared to the crystal structure are better than initially thought with respect to the energy of binding, which implies that the crystal structure ligand pose may not be optimal to start with.
Overall Comparison of Autodock and Surflex Performance
In assessing the overall performance of Autodock and Surflex, several facets of docking must be compared including docking accuracy, docking ranking, computational speed, and even ease of use. Both Autodock and Surflex have comparable performance in accurately reproducing the crystal structure and ranking the poses, particularly with software conditions of “5 docks” with “2E7 energy evaluations” and “Multistart 5” and “Random 5”, respectively. However, one important factor where performance differs substantially is the computational resources required for docking. Using 2.0 GHz AMD Opteron 246 processors, Surflex performed the dockings significantly faster than Autodock for all ligands tested. The average time to complete each Surflex docking with a software parametrization of “Multistart 5” and “Random 5” was just under 8 min, while Autodock with a software parametrization of “5 docks” with “2E7 energy evaluations” took approximately 76 min. Given that the docking accuracy and ranking results were comparable, the significantly faster docking speed of Surflex makes it particularly well suited for virtual screening applications where large numbers of ligands are screened. Surflex is also superior with ease of use, as it is a single executable application with direct input from a MOL2 file format. Autodock requires file conversion from a MOL2 into a PDBQT file format prior to performing molecular dockings. For these reasons, under the tested software conditions, we show Surflex is a superior software package for virtual screening of nucleic acids in the system reported here.
Comparison of Results to Previous Studies
Relatively few molecular docking studies have been performed with nucleic acids. In comparing the data presented in this paper to other docking papers, we placed particular emphasis on the evaluation of the accuracy of the top ranked pose returned by either Surflex or Autodock. This is a logical approach for assessing docking software performance for virtual screening applications, since when screening a large ligand database, only the evaluation of the top ranked pose may be computationally feasible. Several previous studies have focused on utilization of the DOCK program for molecular docking of ligands to nucleic acids. Grootenhuis et al. used DOCK to target the minor groove, major groove, and an intercalation site on duplex DNA, while more recently Chen et al. successfully targeted the major groove of RNA.19,20,22 Yan et al. targeted an RNA tetraloop structure and demonstrated docking at a similar resolution to what was observed in our study of docking ligands to DNA targets.23 Rohs et al. recently developed a molecular docking approach utilizing a Monte Carlo algorithm that successfully demonstrated the binding of methylene blue to DNA by minor groove and intercalation binding modes.62 However, methylene blue has only four rotatable methyl groups with fewer degrees of freedom than several of the more conformationally complex ligands tested in this study.62
One report of docking studies to nucleic acids using Autodock was performed by Evans et al., who demonstrated the ability of a previous version of Autodock to accurately predict binding of minor groove binders to their respective nucleic acid targets.24 A direct comparison of all of the results from the Evans paper and this study is difficult due to different operating conditions and software versions for Autodock; however, some differences are noteworthy. One limitation of the previous study is that while the number of energy evaluations was varied, the maximum number of evaluations performed was only 2.5E6. Based on our studies, we found that 2E7 energy evaluations were optimal for docking accuracy and pose ranking. Another consideration is that in this previous study the number of dockings was kept constant. We evaluated the parameters by varying both the number of docks and energy evaluations to determine which combination of software parameters is best for virtual screening applications. Similar to the results in this paper, Evans did find that, in general, increasing the number of energy evaluations increased the accuracy of the predicted pose, with respect to the crystal structure.24 However, we also found that using fewer numbers of dockings while concurrently increasing the number of energy evaluations increases both pose accuracy and ranking. This is presumably due to a more complete exploration of the energetic landscape surrounding the ligand–target interaction. This has important implications for virtual screening where of crucial importance is the accuracy of the top ranked pose. Generally, the results of Evans et al. are consistent with results in this paper and show that Autodock is able to successfully predict the binding of multiple minor groove binders to their targets at a resolution of approximately 2 Å.24 However, based on the data herein, we recommend using more energy evaluations and fewer numbers of docks for virtual screening applications to produce the best top ranked dock. While the results in this paper expand and add value to previous Autodock work targeting nucleic acids, importantly, we show that the results with Surflex in particular are very useful, applicable, and the first published study to demonstrate successful molecular docking of intercalators or minor groove binders to nucleic acid targets using this software.
CONCLUSIONS
The results reported here support the primary objective of this work, which is to test Autodock 4 and Surflex 2.11 for accurately reproducing ligand-bound nucleic acid structures. This is the critical first step in validating these software for future use in targeting specific nucleic acid structures. Even given the aforementioned limitations and uncertainties of using Autodock 4 and Surflex 2.11 with nucleic acids, these results show that these software can accurately reproduce the crystal structures of both groove binders and intercalators. Ours is one of only a few studies to date that have shown that nucleic acids can be successfully targeted using these docking methods. Our results show that an Autodock 4 software condition of “5 docks” and “2E7 energy evaluations” is the best for combined docking accuracy and ranking. The Surflex 2.11 software conditions of “Multistart 5” and “No Random” and “Multistart 5” and “Random 5” appear equally good at producing top ranked structures with low rmsd values relative to the crystal structure. Extended experiments testing further increases in Autodock and Surflex parametrization did not improve docking accuracy or ranking. The most challenging ligand to dock accurately was ellipticine, which was no surprise given the small pocket in the nucleic acid and tight fit associated with the binding of ligands into the intercalation site. Given that the Autodock and Surflex scoring functions for ranking the docked poses were parametrized based on protein–ligand structures, the ranking results are particularly encouraging.8,40 Both programs are able to return a top ranked pose with approximately 2 Å rmsd for daunorubicin, distamycin, and pentamidine and a pose with approximately 3 Å rmsd for ellipticine. It is important to consider that while the docking accuracy and pose ranking of these programs is comparable, Surflex performs docking much faster than Autodock under the optimized software conditions in this paper. Surflex also requires less manipulation of input files, suggesting that Surflex is preferred for virtual screening applications for systems similar to those presented here.
Based on these docking studies, several points should be strongly considered when performing molecular docking with nucleic acids and evaluating docked poses. Docking parameters should be explored in detail since suboptimal software conditions can significantly impact the accuracy and ranking of the docked poses. When evaluating docked poses, visualization of the most promising docking poses should be performed as well as calculation of rmsd values. It is crucial to also account for both ligand and target symmetry by either including ligand symmetry in rmsd calculations or performing molecular superposition to account for nucleic acid target symmetry. Given the conformation and structural heterogeneity of proteins, target symmetry is less likely with respect to docking. However, nucleic acid targets are much more likely to exhibit symmetry due to the simple base pair composition and the nature and geometry of the nucleic acid strand associations. Another consideration when performing docking is the hydrogen atom treatment of the software, as this can significantly impact the free energy of binding. These studies also demonstrated that force field and salvation selection can dramatically effect the binding energy. Finally, selection of high quality and high resolution crystal structures is especially important when using these structures as reference conformations to evaluate docking poses. Based on the results in this paper, it is important to consider that the crystal structure does not necessarily represent the energetically minimized pose with respect to the poses generated by docking software.
These findings have important implications not only in the field of chemistry and computational biology but also in the area of organic small molecule synthesis using structure-based drug design. Many previous efforts at rational drug design have focused on time-consuming and expensive small molecule synthesis methods. If reliable, molecular docking allows for the construction of virtual libraries of molecules that can be docked against any nucleic acid target of interest. One of the logical next steps in molecular docking to nucleic acids is the development of rules to select ligands that may bind nucleic acid targets with affinity and specificity. These experiments suggest that molecular docking techniques may have particular value as a virtual screening precursor step to full chemical synthesis of drug candidates.
Supplementary Material
ACKNOWLEDGMENT
This work was supported by NIH grant 1R01GM077422 and by the James Graham Brown Foundation.
Footnotes
Supporting Information Available: Additional information on the chemical structure of furamidine, Autodock grid maps, and Autodock Docking parameter files, ranking of the lowest rmsd poses, other docking poses for several of the ligands, and Autodock and Surflex parametrization accuracy and ranking experiments. This material is available free of charge via the Internet at http://pubs.acs.org.
REFERENCES AND NOTES
- 1.Wang J, Kollman PA, Kuntz ID. Flexible ligand docking: a multistep strategy approach. Proteins. 1999;36:1–19. [PubMed] [Google Scholar]
- 2.Spitzer GM, Wellenzohn B, Laggner C, Langer T, Liedl KR. DNA minor groove pharmacophores describing sequence specific properties. J. Chem. Inf. Model. 2007;47:1580–1589. doi: 10.1021/ci600500v. [DOI] [PubMed] [Google Scholar]
- 3.Jain AN. Surflex: fully automatic flexible molecular docking using a molecular similarity-based search engine. J. Med. Chem. 2003;46:499–511. doi: 10.1021/jm020406h. [DOI] [PubMed] [Google Scholar]
- 4.Jain AN. Surflex-Dock 2.1: robust performance from ligand energetic modeling, ring flexibility, and knowledge-based search. J. Comput.- Aided Mol. Des. 2007;21:281–306. doi: 10.1007/s10822-007-9114-2. [DOI] [PubMed] [Google Scholar]
- 5.Lane AN, Jenkins TC. Structures and Properties of Multi-stranded Nucleic Acids. Curr. Org. Chem. 2001;5:845–869. [Google Scholar]
- 6.Hurley LH. Secondary DNA structures as molecular targets for cancer therapeutics. Biochem. Soc. Trans. 2001;29:692–696. doi: 10.1042/0300-5127:0290692. [DOI] [PubMed] [Google Scholar]
- 7.Chaires JB. Competition dialysis: an assay to measure the structural selectivity of drug-nucleic acid interactions. Curr. Med. Chem. Anticancer Agents. 2005;5:339–352. doi: 10.2174/1568011054222292. [DOI] [PubMed] [Google Scholar]
- 8.Detering C, Varani G. Validation of automated docking programs for docking and database screening against RNA drug targets. J. Med. Chem. 2004;47:4188–4201. doi: 10.1021/jm030650o. [DOI] [PubMed] [Google Scholar]
- 9.Hurley LH, Wheelhouse RT, Sun D, Kerwin SM, Salazar M, Fedoroff OY, Han FX, Han H, Izbicka E, Von Hoff DD. G-quadruplexes as targets for drug design. Pharmacol Ther. 2000;85:141–158. doi: 10.1016/s0163-7258(99)00068-6. [DOI] [PubMed] [Google Scholar]
- 10.Hurley LH. DNA and its associated processes as targets for cancer therapy. Nat. Rev. Cancer. 2002;2:188–200. doi: 10.1038/nrc749. [DOI] [PubMed] [Google Scholar]
- 11.Ren J, Chaires JB. Sequence and structural selectivity of nucleic acid binding ligands. Biochemistry. 1999;38:16067–16075. doi: 10.1021/bi992070s. [DOI] [PubMed] [Google Scholar]
- 12.Lauria A, Montalbano A, Barraja P, Dattolo G, Almerico AM. DNA minor groove binders: an overview on molecular modeling and QSAR approaches. Curr. Med. Chem. 2007;14:2136–2160. doi: 10.2174/092986707781389673. [DOI] [PubMed] [Google Scholar]
- 13.Reddy BS, Sondhi SM, Lown JW. Synthetic DNA minor groove-binding drugs. Pharmacol Ther. 1999;84:1–111. doi: 10.1016/s0163-7258(99)00021-2. [DOI] [PubMed] [Google Scholar]
- 14.Baraldi PG, Bovero A, Fruttarolo F, Preti D, Tabrizi MA, Pavani MG, Romagnoli R. DNA minor groove binders as potential antitumor and antimicrobial agents. Med. Res. Rev. 2004;24:475–528. doi: 10.1002/med.20000. [DOI] [PubMed] [Google Scholar]
- 15.Chaires JB, Ren J, Henary M, Zegrocka O, Bishop GR, Strekowski L. Triplex selective 2-(2-naphthyl)quinoline compounds: origins of affinity and new design principles. J. Am. Chem. Soc. 2003;125:7272–7283. doi: 10.1021/ja034181r. [DOI] [PubMed] [Google Scholar]
- 16.Bissler JJ. Triplex DNA and human disease. Front Biosci. 2007;12:4536–4546. doi: 10.2741/2408. [DOI] [PubMed] [Google Scholar]
- 17.Han H, Hurley LH. G-quadruplex DNA: a potential target for anticancer drug design. Trends Pharmacol. Sci. 2000;21:136–142. doi: 10.1016/s0165-6147(00)01457-7. [DOI] [PubMed] [Google Scholar]
- 18.Pfeffer P, Gohlke H. DrugScoreRNA--knowledge-based scoring function to predict RNA-ligand interactions. J. Chem. Inf. Model. 2007;47:1868–1876. doi: 10.1021/ci700134p. [DOI] [PubMed] [Google Scholar]
- 19.Grootenhuis PD, Roe DC, Kollman PA, Kuntz ID. Finding potential DNA-binding compounds by using molecular shape. J. Comput.- Aided Mol. Des. 1994;8:731–750. doi: 10.1007/BF00124018. [DOI] [PubMed] [Google Scholar]
- 20.Grootenhuis PD, Kollman PA, Seibel GL, DesJarlais RL, Kuntz ID. Computerized selection of potential DNA binding compounds. Anti-Cancer Drug Des. 1990;5:237–242. [PubMed] [Google Scholar]
- 21.Tse WC, Boger DL. Sequence-selective DNA recognition: natural products and nature’s lessons. Chem. Biol. 2004;11:1607–1617. doi: 10.1016/j.chembiol.2003.08.012. [DOI] [PubMed] [Google Scholar]
- 22.Chen Q, Shafer RH, Kuntz ID. Structure-based discovery of ligands targeted to the RNA double helix. Biochemistry. 1997;36:11402–11407. doi: 10.1021/bi970756j. [DOI] [PubMed] [Google Scholar]
- 23.Yan Z, Sikri S, Beveridge DL, Baranger AM. Identification of an aminoacridine derivative that binds to RNA tetraloops. J. Med. Chem. 2007;50:4096–4104. doi: 10.1021/jm070305p. [DOI] [PubMed] [Google Scholar]
- 24.Evans DA, Neidle S. Virtual screening of DNA minor groove binders. J. Med. Chem. 2006;49:4232–4238. doi: 10.1021/jm0601957. [DOI] [PubMed] [Google Scholar]
- 25.Reha D, Kabelac M, Ryjacek F, Sponer J, Sponer JE, Elstner M, Suhai S, Hobza P. Intercalators. 1. Nature of stacking interactions between intercalators (ethidium, daunomycin, ellipticine, and 4′,6- diaminide-2-phenylindole) and DNA base pairs. Ab initio quantum chemical, density functional theory, and empirical potential study. J. Am. Chem. Soc. 2002;124:3366–3376. doi: 10.1021/ja011490d. [DOI] [PubMed] [Google Scholar]
- 26.Nelson SM, Ferguson LR, Denny WA. DNA and the chromosome - varied targets for chemotherapy. Cell Chromosome. 2004;3:2. doi: 10.1186/1475-9268-3-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Moore MJ, Cuenca F, Searcey M, Neidle S. Synthesis of distamycin A polyamides targeting G-quadruplex DNA. Org. Biomol. Chem. 2006;4:3479–3488. doi: 10.1039/b607707b. [DOI] [PubMed] [Google Scholar]
- 28.Chaires JB. A thermodynamic signature for drug-DNA binding mode. Arch. Biochem. Biophys. 2006;453:26–31. doi: 10.1016/j.abb.2006.03.027. [DOI] [PubMed] [Google Scholar]
- 29.Wemmer DE. Designed sequence-specific minor groove ligands. Annu. Rev. Biophys. Biomol. Struct. 2000;29:439–461. doi: 10.1146/annurev.biophys.29.1.439. [DOI] [PubMed] [Google Scholar]
- 30.Broyles SS, Kremer M, Knutson BA. Antiviral activity of distamycin A against vaccinia virus is the result of inhibition of postreplicative mRNA synthesis. J. Virol. 2004;78:2137–2141. doi: 10.1128/JVI.78.4.2137-2141.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nelson SM, Ferguson LR, Denny WA. Non-covalent ligand/ DNA interactions: minor groove binding agents. Mutat. Res. 2007;623:24–40. doi: 10.1016/j.mrfmmm.2007.03.012. [DOI] [PubMed] [Google Scholar]
- 32.Autodock. version 4 ed. La Jolla, CA: The Scripps Research Institute; 2007. [Google Scholar]
- 33.Surflex. version 2.11 ed. St. Louis, MO: Tripos, Inc.; 2007. [Google Scholar]
- 34.Stiborova M, Rupertova M, Schmeiser HH, Frei E. Molecular mechanisms of antineoplastic action of an anticancer drug ellipticine. Biomed. Pap. Med. Fac. Univ. Palacky Olomouc Czech Repub. 2006;150:13–23. doi: 10.5507/bp.2006.002. [DOI] [PubMed] [Google Scholar]
- 35.Park H, Lee J, Lee S. Critical assessment of the automated AutoDock as a new docking tool for virtual screening. Proteins. 2006;65:549–554. doi: 10.1002/prot.21183. [DOI] [PubMed] [Google Scholar]
- 36.Moitessier N, Englebienne P, Lee D, Lawandi J, Corbeil C. Towards the development of universal, fast and highly accurate docking/scoring methods: a long way to go. Br. J. Pharmacol. 2007;153:S7–S26. doi: 10.1038/sj.bjp.0707515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Morris GM, Goodsell DS, Halliday RS, Huey R, Hart WE, Belew RK, Olson AJ. Automated Docking Using a Lamarkian Genetic Algorithm and an Empirical Binding Free Energy Function. J. Comput. Chem. 1998;19:1639–1662. [Google Scholar]
- 38.Ruppert J, Welch W, Jain AN. Automatic identification and representation of protein binding sites for molecular docking. Protein Sci. 1997;6:524–533. doi: 10.1002/pro.5560060302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Huey R, Morris GM, Olson AJ, Goodsell DS. A semiempirical free energy force field with charge-based desolvation. J. Comput. Chem. 2007;28:1145–1152. doi: 10.1002/jcc.20634. [DOI] [PubMed] [Google Scholar]
- 40.Pham TA, Jain AN. Parameter estimation for scoring protein-ligand interactions using negative training data. J. Med. Chem. 2006;49:5856–5868. doi: 10.1021/jm050040j. [DOI] [PubMed] [Google Scholar]
- 41.Halperin I, Ma B, Wolfson H, Nussinov R. Principles of docking: An overview of search algorithms and a guide to scoring functions. Proteins. 2002;47:409–443. doi: 10.1002/prot.10115. [DOI] [PubMed] [Google Scholar]
- 42.Acharya KR, Lloyd MD. The advantages and limitations of protein crystal structures. Trends Pharmacol. Sci. 2005;26:10–14. doi: 10.1016/j.tips.2004.10.011. [DOI] [PubMed] [Google Scholar]
- 43.Kapetanovic IM. Computer-aided drug discovery and development (CADDD): In silico-chemico-biological approach. Chem. Biol. Interact. 2008;171:165–176. doi: 10.1016/j.cbi.2006.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Warren GL, Andrews CW, Capelli AM, Clarke B, LaLonde J, Lambert MH, Lindvall M, Nevins N, Semus SF, Senger S, Tedesco G, Wall ID, Woolven JM, Peishoff CE, Head MS. A critical assessment of docking programs and scoring functions. J. Med. Chem. 2006;49:5912–5931. doi: 10.1021/jm050362n. [DOI] [PubMed] [Google Scholar]
- 45.Maestro. version 8.0 ed. San Diego, CA: Schrodinger, LLC; 2007. [Google Scholar]
- 46.Macromodel. version 7.0 ed. San Diego, CA: Schrodinger, LLC; 1999. [Google Scholar]
- 47.SYBYL. version 7.3 ed. St. Louis, MO: Tripos, Inc.; 2006. [Google Scholar]
- 48.AMBER. version 8 ed. San Francisco, CA: University of California, San Francisco; 2004. [Google Scholar]
- 49.AutoDockTools. version 1.4.6 ed. La Jolla, CA: The Scripps Research Institute; 2007. [Google Scholar]
- 50.Hetenyi C, van der Spoel D. Efficient docking of peptides to proteins without prior knowledge of the binding site. Protein Sci. 2002;11:1729–1737. doi: 10.1110/ps.0202302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. [accessed October 25, 2007];Autodock. http://autodock.scripps.edu/faqs-help/tutorial.
- 52.Bailly C, Arafa RK, Tanious FA, Laine W, Tardy C, Lansiaux A, Colson P, Boykin DW, Wilson WD. Molecular determinants for DNA minor groove recognition: design of a bis-guanidinium derivative of ethidium that is highly selective for AT-rich DNA sequences. Biochemistry. 2005;44:1941–1952. doi: 10.1021/bi047983n. [DOI] [PubMed] [Google Scholar]
- 53. [accessed November 1, 2007];Tripos. http://www.tripos.com/surflex/
- 54.Jain A. San Francisco, CA: University of California, San Francisco; 2008. personal communication. [Google Scholar]
- 55.OpenBabel. version 2.1.1 ed. Boston, MA: Free Software Foundation, Inc.; 2007. [Google Scholar]
- 56.iBabel. version 2.0 ed. Cambridgeshire, U.K: Cambridge MedChem Consulting; 2007. [Google Scholar]
- 57.Kim R, Skolnick J. Assessment of programs for ligand binding affinity prediction. J. Comput. Chem. 2008;29:1316–1331. doi: 10.1002/jcc.20893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kellenberger E, Rodrigo J, Muller P, Rognan D. Comparative evaluation of eight docking tools for docking and virtual screening accuracy. Proteins. 2004;57:225–242. doi: 10.1002/prot.20149. [DOI] [PubMed] [Google Scholar]
- 59.Gani OA. Signposts of docking and scoring in drug design. Chem. Biol. Drug Des. 2007;70:360–365. doi: 10.1111/j.1747-0285.2007.00571.x. [DOI] [PubMed] [Google Scholar]
- 60.Teramoto R, Fukunishi H. Supervised consensus scoring for docking and virtual screening. J. Chem. Inf. Model. 2007;47:536–534. doi: 10.1021/ci6004993. [DOI] [PubMed] [Google Scholar]
- 61. [accessed on April 15, 2008];Macromodel. http://www.schrodinger.com/
- 62.Rohs R, Bloch I, Sklenar H, Shakked Z. Molecular flexibility in ab initio drug docking to DNA: binding-site and binding-mode transitions in all-atom Monte Carlo simulations. Nucleic Acids Res. 2005;33:7048–7057. doi: 10.1093/nar/gki1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.