Abstract
Common copy number variations (CNVs) represent a significant source of genetic diversity, yet their influence on phenotypic variability, including disease susceptibility, remains poorly understood. To address this problem in cancer, we performed a genome-wide association study (GWAS) of CNVs in the childhood cancer neuroblastoma, a disease where SNP variations are known to influence susceptibility1,2. We first genotyped 846 Caucasian neuroblastoma patients and 803 healthy Caucasian controls at 550,000 single nucleotide polymorphisms, and performed a CNV-based test for association. We then replicated significant observations in two independent sample sets comprised of a total of 595 cases and 3,357 controls. We identified a common CNV at 1q21.1 associated with neuroblastoma in the discovery set, which was confirmed in both replication sets (Pcombined = 2.97 × 10−17; OR = 2.49, 95% CI: 2.02 to 3.05). This CNV was validated by quantitative PCR, fluorescent in situ hybridization, and analysis of matched tumor specimens, and was shown to be heritable in an independent set of 713 cancer-free trios. We identified a novel transcript within the CNV which showed high sequence similarity to several “Neuroblastoma breakpoint family” (NBPF) genes3,4 and represents a new member of this gene family (NBPFX). This transcript was preferentially expressed in fetal brain and fetal sympathetic nervous tissues, and expression level was strictly correlated with CNV state in neuroblastoma cells. These data demonstrate that inherited copy number variation at 1q21.1 is associated with neuroblastoma and implicate a novel NBPF gene in early tumorigenesis of this childhood cancer.
Neuroblastoma is a pediatric cancer of the developing sympathetic nervous system that most commonly affects young children, and is often lethal5. Most neuroblastomas arise sporadically, with less than 1% of cases inherited in an autosomal dominant fashion5. We recently identified the anaplastic lymphoma kinase (ALK) gene as the major hereditary neuroblastoma predisposition gene1. For the vast majority of neuroblastomas that arise without a family history of the disease, we hypothesize that multiple common DNA variations cooperate to increase the risk for neuroblastic malignant transformation. By performing a genome-wide association study (GWAS) of single nucleotide polymorphism (SNP) genotypes, we recently identified common SNP alleles within the putative FLJ22536 and FLJ44180 genes at 6p22 and within the BARD1 gene at 2q35 associated with malignant neuroblastoma, providing the first evidence that childhood cancers also arise due to complex interactions of polymorphic variants1,2. Here, we investigate constitutional DNA copy number variations (CNVs) as another source of genetic diversity that may contribute to the development of this disease. CNVs have been shown to significantly influence mRNA expression levels6 and recent studies have described associations of CNVs with systemic autoimmunity7,8, autism9, schizophrenia10,11,12, and psoriasis13. In addition, Shlien and colleagues reported an increased number of CNVs in Li-Fraumeni families harboring TP53 mutations, but did not explore associations of individual CNVs with cancer susceptibility14.
To identify CNVs that are associated with neuroblastoma, we first genotyped a discovery set of 1,032 Caucasian neuroblastoma patients and 2,043 disease-free Caucasian control subjects, as previously described1,15. We next applied stringent quality control criteria necessary for accurate CNV detection and reliable association assessment (see Supplementary Methods). The final discovery set consisted of 846 Caucasian cases and 803 Caucasian controls (Supplementary Tables 1 and 2). These subjects showed tight clustering in a multi-dimensional scaling (MDS) analysis of SNPs not in linkage disequilibrium (LD; Supplementary Figure 1), demonstrating that population substructure was not likely to have a significant impact on association testing.
By comparing single marker binary copy number states at 531,689 SNPs mapped to autosomes, we observed a total of 131 SNPs showing significant association with neuroblastoma, defined as a two-sided Fisher’s exact test p-value below a genome-wide threshold of P = 1.0 × 10−7 (Supplementary Table 3). Associations with deletion polymorphisms were seen at chromosomes 1, 7, and 14 (Figure 1a); no duplication polymorphisms reached genome-wide significance (Figure 1b). Review of significant SNPs revealed four distinct regions of deletion (Table 1). We next sought to validate significant association signals in two independent replication sets, the first consisting of 363 Caucasian cases and 1,139 Caucasian controls, the second of 232 Caucasian cases and 2,218 Caucasian controls (Supplementary Tables 1 and 2). All deletion associations showed robust replication in both independent case-series (Table 2 and Supplementary Tables 4 and 5).
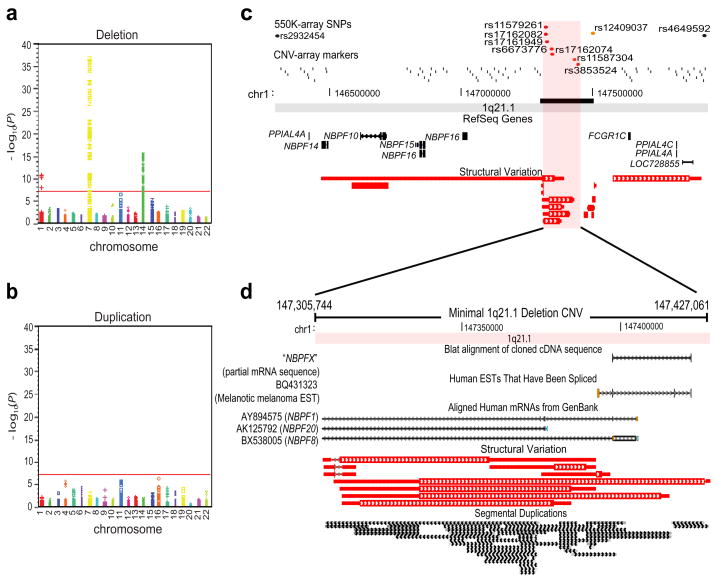
Figure 1. Discovery of 1q21.1 CNV associated with neuroblastoma.
(a) Summary of discovery results for deletions. Level of significance (log10 transformed P-values) for each SNP at each chromosome is plotted. Red line: genome-wide significance threshold. (b) Summary of discovery results for duplications. (c) Genomic map and refinement of 1q21.1 deletion boundaries. SNPs from the 550K array and markers from the CNV-12 array are plotted as dots or tick marks; those reaching genome-wide significance are colored red. Image borders represent maximal deletion boundaries based on 550K array data, and includes a cluster of NBPF genes and several other RefSeq transcripts. Refined maximal boundaries using CNV-12 data (black bar) is consistent with published structural variations. (d) Genomic map of 121-kb minimal 1q21.1 deletion covered by significant SNPs. The region contains a high level of segmental duplication which extends to flanking regions (not shown). The BQ431323 melanoma EST maps uniquely within this region, while several Genbank transcripts align to the region with > 95% identity, but map elsewhere on the genome. Blat alignment of PCR amplified and cloned sequence from fetal brain in this study is shown (NBPFX).
Table 1.
Significant copy number variable regions from the discovery phase.
| Chr | Start SNP End SNP |
Start Position End Position |
No. SNPs |
Size (bp) |
Gene(s) | Case % Loss |
Control % Loss |
Case % Gain |
Control % Gain |
P-value Range |
OR (95% CI) |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | rs11579261 rs3853524 |
147305744 147427061 |
7 | 121,317 | Putative NBPF | 15.1 (12.2–15.1) | 5.2 (3.6–5.2) | 3.2 (1.8–3.2) | 5.1 (1.6–5.1) | 2.38 × 10−11 1.31 × 10−8 |
3.23 (2.25–4.05) |
| 7 | rs7782269 rs733905 |
38285115 38346971 |
47 | 61,856 | TCRG | 24.5 (8.6–36.4) | 7.3 (0.5–16.2) | 0 | 0.1 (0.1–0.1) | 2.20 × 10−37 5.31 × 10−15 |
9.96 (6.40–15.48) |
| 7 | rs2213212 rs2367486 |
142086318 142192134 |
11 | 105,816 | TCRVB | 4.6 (3.9–5.2) | 0.2 (0.1–0.3) | 0.1 (0.0–0.1) | 0 | 5.80 × 10−11 2.85 × 10−9 |
21.97 (5.85–82.42) |
| 14 | rs979027 rs2128997 |
21558349 22030942 |
66* | 472,593 | TCRA, TCRD | 29.2 (5.2–45.1) | 14.1 (0.6–30.3) | 0.0 (0.0–0.1) | 0.0 (0.0–0.1) | 2.99×10−16 7.86×10−8 |
2.71 (2.12–3.46) |
Genomic coordinates based on UCSC Build 36.1 of human genome. P-value based on two-tailed Fisher’s exact test comparing deletion frequency in cases vs. controls. Median percent deletion and duplication are listed with range in parentheses. OR: Odds Ratio. ORs listed are from the most significant SNP in Discovery set within deleted region: 1q21.1 (NBPF): rs17162082, TCRG: rs718880, TCRVB: rs6959895, TCRA/D: rs741711. 95% confidence interval (CI) is indicated in parentheses. N Cases = 846, N Controls = 803.
SNPs not consecutive.
Table 2.
Replication of significant copy number variable regions.
| Chr | Start SNP End SNP |
Start End |
No. SN P |
Size (bp) | Gene | Case % Loss |
Control % Loss |
Case % Gain |
Control % Gain |
Replication P-value Range |
Combined P-value Range |
Combined OR (95% CI) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Replication - 1 | ||||||||||||
| 1 | rs11579261 rs3853524 |
147305744 147427061 |
7 | 121,317 | Putative NBPF | 14.3 (10.7–14.3) | 8.7 (6.4–8.7) | 1.9 (1.7–1.9) | 3.1 (1.9–3.1) | 2.58 × 10−3 8.15 × 10−3 |
1.70 × 10−11 7.27 × 10−11 |
2.23 (1.77–2.82) |
| 7 | rs7782269 rs733905 |
38285115 38346971 |
47 | 61,856 | TCRG | 14.9 (5.8–30.3) | 3.8 (0.2–9.3) | 0 | 0 | 8.61 × 10−23 2.19 × 10−11 |
1.17 × 10−63 6.29 × 10−33 |
10.74 (7.73–14.93) |
| 7 | rs2213212 rs2367486 |
142086318 142192134 |
11 | 105,816 | TCRVB | 3.9 (1.9–4.4) | 0 | 0 | 0 | 1.05 × 10−10 4.61 × 10−5 |
8.59 × 10−23 8.18 × 10−16 |
46.23 (12.40–172.34) |
| 14 | rs979027 rs2128997 |
21558349 22030942 |
66* | 472,593 | TCRA, TCRD | 24.7 (2.2–30.9) | 11.6 (0.3–19.9) | 0 | 0 | 1.47 × 10−11 0.11 |
3.54 × 10−32 8.04 × 10−9 |
2.82 (2.35–3.38) |
| Replication - 2 | ||||||||||||
| 1 | rs11579261 rs3853524 |
147305744 147427061 |
7 | 121,317 | Putative NBPF | 19.0 (14.2–19.0) | 10.6 (5.4–10.7) | 3.5 (2.6–3.5) | 4.1 (3.4–4.1) | 2.59 × 10−6 4.93 × 10−4 |
2.97 × 10−17 6.17 × 10−11 |
2.49 (2.02–3.05) |
| 7 | rs7782269 rs733905 |
38285115 38346971 |
47 | 61,856 | TCRG | 19.0 (7.7–31.0) | 3.1(0.4–7.3) | 0 | 0 | 2.22 × 10−23 6.45 × 10−13 |
1.64 × 10−103 6.36 × 10−51 |
12.62 (9.74–16.36) |
| 7 | rs2213212 rs2367486 |
142086318 142192134 |
11 | 105,816 | TCRVB | 3.0 (3.0–4.3) | 0.1 (0.1–0.2) | 0 | 0.0 (0.0–0.1) | 1.07 × 10−8 1.91 × 10−6 |
2.40 × 10−35 6.60 × 10−25 |
41.79 (16.83–103.80) |
| 14 | rs979027 rs2128997 |
21558349 22030942 |
66* | 472,593 | TCRA, TCRD | 22.2 (4.3–34.1) | 7.2 (0.4–14.5) | 0 | 0.0 (0.0–0.1) | 6.25 × 10−15 3.25 × 10−5 |
3.19 × 10−63 9.08 × 10−16 |
3.75 (3.22–4.36) |
Genomic coordinates based on UCSC Build 36.1 of human genome. P-value based on two-tailed Fisher’s exact test comparing deletion frequency in cases vs. controls. Median percent deletion and duplication are listed with range in parentheses. OR: Odds Ratio. ORs listed are from the most significant SNP in Discovery set within deleted region: 1q21.1 (NBPF): rs17162082, TCRG: rs718880, TCRVB: rs6959895, TCRA/D: rs741711. 95% confidence interval (CI) is indicated in parentheses. Replication-1: N Cases = 363, N Controls = 1,139. Replication-2: N Cases = 232, N Controls = 2,218.
SNPs not consecutive.
We observed a seven-SNP deletion at 1q21.1 which occurred in 15.6% of cases but only 9.1% of controls overall (Pcombined = 2.97 × 10−17; OR = 2.49, 95% CI: 2.02 to 3.05). This association remained significant after additional adjustment for potential population substructure captured by SNPs not in LD (Pdiscovery < 0.0001), and was driven by a difference in hemizygous deletion frequency (Pcombined = 1.83 × 10−19). The observed frequency of homozygous deletion was identical in cases and controls (1.3% overall). The maximal deletion defined by neighboring 2-copy SNPs spanned 1.6 Mb and contained a cluster of “Neuroblastoma Breakpoint Family” (NBPF) genes (Figure 1c). To refine the maximal deletion boundaries, we first genotyped 48 representative samples on the Illumina CNV-12 array (36 cases and 12 controls equally divided between those with and without the deletion CNV). These data reduced the maximal size of the deletion to approximately 300-Kb, consistent with published reports of CNVs at this location (Figure 1c)16,17,18. Lastly, use of the Illumina HumanHap610 SNP platform reduced the maximal deletion size to only 143-Kb, from 147,292,384 – 147,435,422 (Supplementary Figure 2). The minimal deletion based on significant SNPs in the association study spanned 121-Kb from 147,305,744–147,427,061and did not contain any known genes (Figure 1d).
We next sought to confirm that this CNV is indeed a heritable genetic variation. First, we genotyped an independent set of 713 trios from variable phenotypes on the same 550K SNP array and generated CNV calls using a family-based approach16. Deletion at 1q21.1 was observed in 125 offspring and confirmed by parental analysis in 123 trios, estimating the inheritance rate at 98.4%. Next, we genotyped paired tumor DNA in 226 cases (Supplementary Table 2) using the same 550K SNP platform, and confirmed existence of the CNV (deletion and duplication) in every tumor sample studied (Supplementary Figure 3a). We did not observe progression from hemizygous deletion in constitutional DNA to homozygous deletion in the matched tumor DNA for any case in this study, nor did we observe any expansion of CNV boundaries in tumor compared to matched constitutional DNA.
To investigate whether 1q21.1 deletions are associated with specific neuroblastoma phenotypes, we tested each for association with clinical covariates using the combined set of 1,441 cases (Supplementary Table 6). While deletions at 1q21.1 were observed more frequently in patients with aggressive disease; this trend did not reach statistical significance in this study. An additive effect on the odds ratio was observed for those harboring both the 1q21.1 deletion and the 6p22 risk alleles (Supplementary Figure 4), but no significant interaction effect was detected to suggest epistasis (Supplementary Table 7).
In addition to the 1q21.1 CNV, we observed highly significant associations of deletion within all four T-cell receptor (TCR) loci clustered on chromosomes 7 and 14. Contrary to the 1q21.1 CNV, TCR deletions were not observed in paired tumor DNA samples (Supplementary Figure 3b). Heterogeneity was observed in the areas of apparent deletion consistent with some cells harboring homozygous deletion, and others exhibiting 2-copy heterozygosity. Deletions within the T-cell receptors tended to co-occur in patients (P < 0.0001, Supplementary Table 7), and were significantly more common in blood-than bone marrow-derived DNA samples (PTCRG = 9.2 × 10−31, PTCRVB = 7.3 × 10−6, PTCRA/D = 8.0 × 10−50). Taken together, these findings suggest that we are detecting an oligoclonal expansion of T-cell lymphocytes in a subset of neuroblastoma patients, and this signal is diluted within the bone marrow compartment. Interestingly, TCR deletions showed a striking over-representation in the less aggressive subset of neuroblastoma (Supplementary Table 6, Supplementary Figure 5). It is possible that these events herald an immunologic response to neuroblastoma, however this hypothesis requires further investigation.
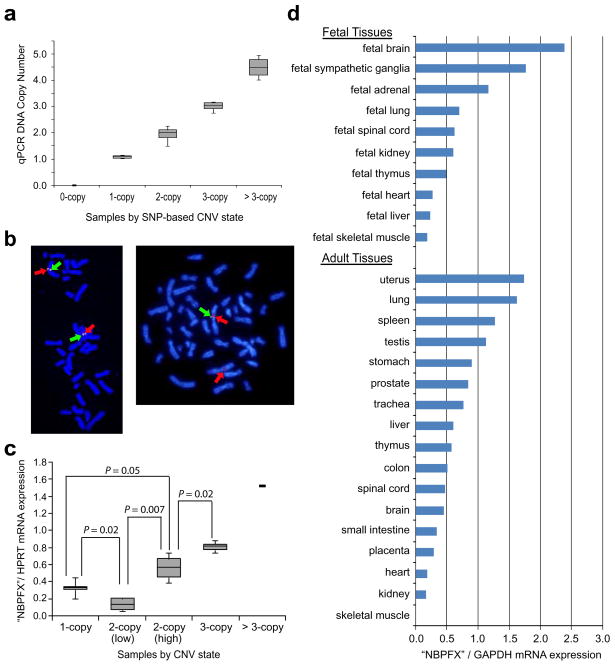
To validate the 1q21.1 CNV, we first performed quantitative PCR (qPCR) on 46 neuroblastoma cases (ten 0-copy, sixteen 1-copy, twelve 2-copy, and eight duplications as predicted by SNP analysis). We observed 100% concordant results when comparing copy number estimated by qPCR with copy number based on SNP genotyping (Figure 2a). To confirm that the detected CNV is not an artifact caused by segmental duplication of the 1p36 region, and that it indeed maps to 1q21.1, we validated the existence of the deletion in a sample harboring a single copy loss using fluorescent in situ hybridization (Figure 2b).
Figure 2. Validation and biological relevance of 1q21.1 CNV.
(a) Quantitative PCR validation of 1q21.1 CNV states shows 100% agreement with SNP genotyping data. Box plot is shown for 0-copy (n=10), 1-copy (n=16), 2-copy (n=12), 3-copy (n=6) and > 3 copies (n=2). (b) FISH validation of 1q21.1 deletion. Shown are metaphase spreads from lymphoblastoid cell lines established from an individual with normal 2-copy (left) and an individual with hemizygous deletion (right). Red arrows: BAC RP4-790G17 which maps just outside of the deletion CNV from 148,424,830–148,557,655 (two copies in both individuals). Green arrows: fosmid W12-1967b11 which maps within the deleted region from 147,382,742–147,420,968, encompassing the first two exons of the putative NBPF transcript (one copy in individual with hemizygous deletion). (c) Box plot showing correlation between CNV state and NBPFX transcript levels demonstrated using real-time quantitative PCR in eighteen neuroblastoma samples: 1-copy (n=5), 2-copy (low, n=4), 2-copy (high, n=3), 3-copy (n=5), and > 3 copies (n=1). 2-copy samples clustered into two distinct groups consistent with expression difference based on whether copies are in “cis” (2:0 CNV genotype) or “trans” (1:1 CNV genotype); see text for details. (d) Expression of NBPFX in normal fetal and adult tissues using real-time quantitative reverse transcriptase PCR showed highest expression in fetal brain (13–22 wks) and fetal sympathetic ganglia samples (18–22 wks) which were collected at the Children’s Hospital of Philadelphia.
Although no known genes mapped to the refined 1q21.1 CNV, we identified a spliced EST (BQ431323) from a melanoma library that mapped within the CNV with 100% identity across the entire sequence (Figure 1d). Using primers designed against exon-1 and exon-3 of BQ431323, we PCR amplified cDNA from fetal brain and a neuroblastoma cell line. These PCR products were cloned and sequenced; the resulting sequence mapped uniquely within the CNV with 100% identity across the entire sequence and showed splicing out of the predicted second exon of BQ431323 (Supplementary Figure 6). The top scoring hit from a Blastn19 search of this sequence against available human RefSeq transcripts was NBPF3 (E = 5.0 × 10−89), followed by NBPF1 (E = 1.0 × 10−75) and NBPF15 (E = 2.0 × 10−24). These data provide strong evidence for a novel NBPF transcript, termed “NBPFX” here, mapping within the 1q21.1 CNV associated with neuroblastoma.
We investigated the expression of NBPFX in both neuroblastoma cells and normal human fetal and adult tissues using realtime quantitative reverse transcriptase PCR. Analysis of eighteen neuroblastomas (tumors and cell lines) of known copy number at the 1q21.1 CNV showed a clear correlation between CNV state and transcript expression (Figure 2c). Notably, 2-copy samples clustered into two distinct expression classes (P = 0.007), the first with low expression and the second with high expression, and these likely represent different 1q21.1 CNV genotypes. There are two possible CNV genotypes for 2-copy samples, which we refer to as 2:0 (“cis”) and 1:1 (“trans”) based on the number of copies present on each chromosome in a diploid genome (Supplementary Figure 7a). Two neuroblastoma samples in the low-expression group can be demonstrated to be from the 2:0 constitutional CNV genotype because they have somatically acquired gain of chromosome 1q (3-copies) yet are 2-copies with heterozygous SNPs at the 1q21.1 CNV (see Supplementary Figure 7b–c for details). These findings are consistent with the hypothesis that two copies in “cis” (same chromosome) behave differently than two copies in “trans” (different chromosomes). Therefore, we propose a model whereby NBPFX expression is decreased when copies are in the “cis” configuration as opposed to the “trans” configuration. Importantly however, even when the 2-copy samples are not clustered in this manner, a statistically significant difference in transcript levels is observed between the 2- and 3-copy samples (P = 0.001). Finally, we analyzed expression of NBPFX in a panel of twenty-eight normal fetal and adult tissues (Figure 2d). We observed the highest transcript levels in fetal brain and fetal sympathetic ganglia from early gestation (13–22 wks), consistent with NBPFX being expressed in early sympathicoadrenal neurodevelopment.
NBPF genes were identified after the founding member, NBPF1, was determined to be disrupted via a constitutional chromosomal translocation in a neuroblastoma patient3,4. Subsequent scans of the genome identified three clusters of NBPF genes on chromosome 1 within areas of segmental duplication. The encoded proteins are recently evolved and primate-specific, share significant homology, and contain highly conserved domains of unknown function (DUF1220) that are thought to be neuronal-specific3,20. Constitutional deletions disrupting NBPF genes have been implicated in schizophrenia10,11,12 and autism10, and rare recurrent structural variants just upstream of the CNV identified in this study have also been reported in a variety of phenotypes including mental retardation, autism, and congenital anomalies21. Although the specific functions of NBPF genes are not known, recently evolved genes may be involved in cancer predisposition due to a lack of selective pressure12. Expression of NBPF1 was recently shown to suppress anchorage independent growth4. Conversely, over-expression of NBPF transcripts has been reported in several cancers including sarcomas22 and non-small-cell lung cancer23. A challenge for the field is to design experiments that clearly distinguish these highly homologous transcripts and define specific disease-causal mechanisms.
Neuroblastoma likely develops from the malignant transformation of partially committed sympathicoadrenal neuroblasts during fetal or early childhood development. We previously identified the ALK gene as the major familial neuroblastoma predisposition gene1 and showed that common variations at 6p22 and 2q35 are associated with sporadic neuroblastma1,2. In our current study, we show that a common CNV at 1q21.1 likewise contributes to neuroblastoma susceptibility, and that this CNV leads to altered expression of a novel NBPFX transcript. These data provide the first definitive evidence for a specific CNV predisposing to human cancer, and ongoing efforts will define remaining susceptibility variants in the human genome associated with neuroblastoma.
METHODS SUMMARY
A multi-phase GWAS of CNVs in neuroblastoma was performed. A discovery set of 846 Caucasian neuroblastoma patients and 803 Caucasian healthy controls were genotyped using a 550K SNP array and CNV profiles were generated24,16, providing a copy number state for each SNP on the array. Association with neuroblastoma was assessed at 531,689 individual SNPs on autosomes using Fisher’s exact test on the binary comparison of “deletion vs. no deletion” or “duplication vs. no duplication”. A genome-wide significance threshold was set at P = 1.0 × 10−7 and replication of significant findings was assessed using two independent replication sets, the first consisting of 363 Caucasian neuroblastoma cases and 1,139 Caucasian controls, the second of 232 Caucasian neuroblastoma cases and 2,218 Caucasian controls. Matched tumor DNA from 226 cases was genotyped to evaluate whether observed deletions were constitutional or somatically acquired rearrangements in the blood. An independent set of 713 trios with no reported incidence or history of cancer was genotyped and used to establish heritability of significant CNVs. Inherited CNVs were validated by quantitative realtime PCR and fluorescent in situ hybridization. Association with clinical and biological co-variates was assessed on the combined set of 1,441 neuroblastoma cases with available annotation. PCR amplification of cDNA from fetal brain and neuroblastoma cell lines was performed to identify a novel 1q21.1 transcript; products from these PCR reactions were sequenced and aligned to the human genome using BLAT25. Similarity to known NBPF transcripts was detected using Blastn19. Expression of the novel NBPF transcript was assessed using realtime quantitative reverse transcriptase PCR.
METHODS
Sample Selection
Cases were defined as a child diagnosed with neuroblastoma or ganglioneuroblastoma and registered through the Children’s Oncology Group (COG). All specimens were obtained at time of diagnosis and the majority were annotated with clinical and genomic information that included: age at diagnosis, site of origin, disease stage by the International Neuroblastoma Staging System26, INPC International Neuroblastoma Pathology Classification27, MYCN oncogene copy number28, DNA index (ploidy)29, registration on clinical trial(s), event-free and overall survival, second malignancies, and any associated conditions (e.g. congenital abnormalities). Additional eligibility criteria and quality control measures, including exclusion of samples with evidence of circulating tumor DNA, are detailed in Supplementary Methods.
Control subjects were recruited from the Philadelphia region through the CHOP Health Care Network, including four primary care clinics and several group practices and outpatient practices that included well child visits. Eligibility criteria for control subjects were: 1) self-reported as Caucasian; 2) availability of 1.5 μg of high quality DNA from peripheral blood mononuclear cells; and 3) no serious underlying medical disorder, including cancer.
Genome-wide SNP Genotyping
Genotyping for both discovery and replication phases was performed using the Illumina Infinium™ II HumanHap550 BeadChip according to methods detailed elsewhere15,30 and summarized in Supplementary Methods.
CNV Detection
Log R Ratio signal intensities were adjusted for “wave-like” artifacts using a regression-based approach described by Diskin and colleagues24 and PennCNV16 was utilized to call CNVs in each sample. Copy number at each SNP was then inferred based on projection of CNVs onto genomic sequence.
Statistical Tests
The single marker statistical analysis for the genome-wide data was carried out using Fisher’s exact test on binary copy number differences (“deletion vs. no deletion” or “duplication vs. no duplication”) between cases and controls. Two-sided p-values were reported and the Odds Ratio (OR) and corresponding 95% confidence intervals (CI) were calculated for genome-wide significant copy number association results. A threshold of 1.0 × 10−7 was set for genome-wide significance based on the fact that approximately 500,000 SNPs were tested (0.05/500,000 = 1.0 × 10−7).
CNVs reaching genome-wide significance were tested for association with clinical and biological co-variates using Fisher’s exact test. All comparisons were made based on binary comparison of “deletion vs. no deletion” or “duplication vs. no duplication, and two-sided p-values were reported. For deletions within the T-cell receptors, only cases from peripheral blood (not bone marrow) were considered in the clinical correlative analyses because we observed a significant association of T-cell receptor deletions with blood vs. bone marrow (PTCRG = 9.2 × 10−31, PTCRVB = 7.3 × 10−6, PTCRA/D = 8.0 × 10−50).
CNV heritability
We utilized Illumina HumanHap550 SNP genotyping data on 713 trios in order to establish heritability of the 1q21.1 CNV. Families consisted of individuals with no reported incidence or history of cancer; however other conditions were reported. The condition reported most frequently was autism (>50% of probands). Data from 565 trios was generated from lymphoblastoid cell lines and the remaining 148 were from peripheral blood. CNV profiles were generated for each offspring and parent in the trios using a family-based method in PennCNV to increase sensitivity of CNV detection16. The concordance frequency between offspring and parent was computed and reported as an estimated inheritance rate for the CNV.
Fluorescent In Situ Hybridization (FISH)
Metaphase spreads were prepared either from peripheral blood lymphocytes or subject-derived, lymphoblastoid cell lines using standard methodology. Chromosomes were visualized by counter-staining with DAPI. BAC and Fosmid clones used for FISH analysis were obtained from the BACPAC resources (Oakland, California, USA). FISH was performed using fosmid W12-1967b11 (GenBank:G248P87625A6) and BAC RP4-790G17(GenBank:AL138795). FISH analysis was carried out as described previously31. BACs and fosmids were isolated using the UltraClean ™ plasmid kit (MoBio Labs Inc., Carlsbad, California, USA) and probes were labeled with Spectrum Red or Green (Abbott Molecular Inc., Illinois, USA) by nick translation. FISH Images were captured using MacProbe software (Applied Imaging, San Jose, California, USA).
Realtime quantitative PCR validation of DNA copy number
Primers and probes to were designed using Primer Express 3.0 (Applied Biosystems, Foster City, CA) with default parameters. Amplification primers were synthesized by IDT (Coralville, IA) and probes were made by Applied Biosystems. Reactions were set up in triplicate using 10 ng of genomic DNA in a 10 ul reaction which contained 200 nM concentration of probe, 900 nM of each amplification primer and 1 X of Real-time PCR Master Mix (Applied Biosystems). Samples were amplified on an Applied Biosystems 7900HT Sequence Detection System using standard cycling conditions and data collected and analyzed with SDS 2.3 software. Standard curves were constructed using serial 2-fold dilutions of genomic DNA from an individual without NBPF copy number variation and used to estimate amounts of DNA in the experimental samples from their cycle threshold (Ct) values. Ratios of amounts were calculated from the assay designed in the area of variation (NBPF_DEL) and the assay in a region known to be present in all samples from the genome-wide association data (CTRL_2C). This normalized amount was then compared to the value in a control calibrator sample to produce a fold change ratio (normal =1) and multiplied by 2 to generate a copy number (normal = 2).
NBPF_DEL_Probe: 5′-6FAM- CACCACTGTCGTCCCTA -NFQ-3′
NBPF_DEL_ForwardPrimer 5′-CCCTAAACATATGTGGGTGTACACA -3′
NBPF_DEL_ReversePrimer 5′-TGCAGACAGACCCTATAGTGAGGTA -3′
CTRL_2C_Probe 5′-6FAM-AAAAGGCACTGGTTAGGGA-MGB-NFQ-3′
CTRL_2C_FowardPrimer 5′-CAAGTGCCAACAGAGTTGCTAGA-3′
CTRL_2C_ReversePrimer 5′-TAATGAAGGAAGAGAATCAGTTCAGATT-3′
PCR of cDNA and sequencing of 1q21.1 transcript
Standard PCR primers were designed using Primer3 with default parameters and synthesized by IDT (Coralville, IA) as follows:
Left Primer: CGTGCATTCATTTCCTTTGA
Right Primer: GTGCACTGAATGGGGAAGTT
cDNA was prepared from 4 μg of total RNA according to the SuperScript First-Strand Synthesis System for RT-PCR standard protocol using random hexamer primers (Invitrogen Life Technologies, Carlsbad, CA). PCR amplification of target cDNA was performed using the GC-RICH PCR system (Roche Applied Science, Indianapolis, IN) in a 50 μl reaction containing 5 μl cDNA, 0.2 mM dNTP mixture, 0.5 μM forward and reverse primers, 0.5 M GC-RICH Resolution Solution, 10 μl GC-RICH Reaction Buffer and 4 U GC-RICH Enzyme Mix. Amplification of the 1q21.1 transcript was performed in a Bio-Rad DNAEngine Peltier Thermal Cycler (Bio-Rad Laboratories, Hercules, CA) according to the following protocol: initial denaturation and enzyme activation at 95°C for 8 min.; denaturation at 95°C for 1 min., primer annealing at 54°C for 1 min., extension at 72°C for 45 sec for 40 total cycles; final extension at 72°C for 10 min. PCR products were cloned into a pCR4-TOPO vector according to standard protocols (Invitrogen Life Technologies) and bi-directional sequencing of the cloned products was performed using M13 standard forward and reverse primers.
RNA samples for “normal” panel
Fetal spinal ganglia were obtained at the Children’s Hospital of Philadelphia from perinatal autopsies, in non-macerated fetuses ranging in gestational age from 18 weeks to 22 weeks who had underwent intrauterine demise. Following removal of the lungs, the thoracic paraspinal sympathetic chains were identified in-situ, and entirely removed. Gestational age was recorded, but the specimens were otherwise anonymous. The specimens were frozen in optimum cutting temperature compound (OCT), and stored at minus 70 degrees. Validation of the presence of sympathetic ganglia was performed through examination of hematoxylin-eosin stained frozen sections by a pathologist. Total RNA for all other normal adult and fetal tissues was obtained from Clontech (Mountain View, CA) or BioChain (Hayward, CA).
Realtime quantitative reverse transcriptase PCR
Primers and probes to assess mRNA expression in the samples were designed using Primer Express 3.0 (Applied Biosystems, Foster City, CA) with default parameters. Amplification primers were synthesized by IDT (Coralville, IA) and probes were made by Applied Biosystems. Total RNA was DNase treated and cDNA was prepared from 1μg of total RNA according to the SuperScript First-Strand Synthesis System for RT-PCR standard protocol using random hexamer primers (Invitrogen Life Technologies, Carlsbad, CA) and diluted 1:4 for the unknowns. Reactions were set up in triplicate and also run +RT and −RT to confirm cDNA samples were free of genomic DNA contamination. Amplification was performed on an Applied Biosystems 7900HT Sequence Detection System using standard cycling conditions and data collected and analyzed with SDS 2.3 software. Standard curves were constructed using serial 2-fold dilutions of cDNA from either fetal brain or the neuroblastoma cell line IMR5. Ratios of mean quantities were normalized by comparing “NBPFX” expression to that of an endogenous control gene (GAPDH or HPRT). Primer and probe information is as follows:
NBPFX_ Probe: 5′-6FAM- TCCTCCTGAGGGTGTCT -NFQ-3′
NBPFX_ ForwardPrimer: 5′-TCACCAGCTGATAGTCCCTTACC -3′
NBPFX_ ReversePrimer: 5′-CCAAGAGCACACAGCACTGAA -3′
Supplementary Material
Acknowledgments
The authors acknowledge the Children’s Oncology Group (U10-CA98543) for providing neuroblastoma specimens and thank the many children who participated in this study. This work was supported in part by NIH Grants T32-HG000046 (SJD), R01-CA87847 (JMM) and R01-CA124709, the Giulio D’Angio Endowed Chair (JMM), the Alex’s Lemonade Stand Foundation (JMM), the Evan Dunbar Foundation (JMM), the Rally Foundation (JMM), Andrew’s Army Foundation (JMM), the Abramson Family Cancer Research Institute (JMM), Howard Hughes Medical Institute Medical Research Training Fellowship (KB), and the Center for Applied Genomics (HH) at the Joseph Stokes Research Institute of the Children’s Hospital of Philadelphia.
Footnotes
Supplementary Information is linked to the online version of the paper at www.nature.com/nature
Author Contributions
S.J.D and J.M.M. designed the study and drafted the manuscript. C.H., C.K., and H.H. performed the genotyping. S.J.D. analyzed SNP data and performed CNV association study. J.B., S.F.A.G., H.H., and H.L. performed the corrections for population stratification. S.J.D., E.F.A. and Y.P.M. analyzed and interpreted SNP data for tumor specimens. K.W. and S.J.D. analyzed SNP data for second replication set. J.G. analyzed SNP data from trios for inheritance estimates. C.H., S.J.D., A.W. and E.R.R. performed and/or analyzed qPCR experiments. E.A.G., K.C., and T.H.S. performed FISH experiments. S.J.D., M.L., K.B., K.P., M.D, and E.R.R. designed and/or performed experiments to identify and sequence transcript within 1q21.1 CNV. J.E.L., C.W., S.J.D. and E.R.R. performed and/or analyzed expression experiments. P.M. and W.L. analyzed clinical covariates. A.I.B. provided detailed endpoints for 1q21.1 CNV in an independent analysis of healthy controls using a custom high-resolution Agilent array. M.D., H.L., and HH contributed to overall GWAS study design. All authors commented on or contributed to the current manuscript.
Author Information
The authors declare no competing interests. Correspondence and requests for materials should be addressed to J.M.M. (maris@chop.edu).
References
- 1.Maris JM, et al. Chromosome 6p22 locus associated with clinically aggressive neuroblastoma. N Engl J Med. 2008;358(24):2585–2593. doi: 10.1056/NEJMoa0708698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Capasso M, et al. A genome-wide association study identifies common variations in the BARD1 tumor suppressor gene predisposing to high-risk neuroblastoma. Nature Genetics. 2009 In Press. [Google Scholar]
- 3.Vandepoele K, Van Roy N, Staes K, Speleman F, Van Roy F. A novel gene family NBPF: intricate structure generated by gene duplications during primate evolution. Mol Biol Evol. 2005;22:2265–2274. doi: 10.1093/molbev/msi222. [DOI] [PubMed] [Google Scholar]
- 4.Vandepoele K, et al. A constitutional translocation t(1;17)(p36.2;q11.2) in a neuroblastoma patient disrupts the human NBPF1 and ACCN1 genes. PLoS ONE. 2008;3(5):e2207. doi: 10.1371/journal.pone.0002207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Maris JM, Hogarty MD, Bagatell R, Cohn SL. Neuroblastoma. Lancet. 2007;369(9579):2106–2120. doi: 10.1016/S0140-6736(07)60983-0. [DOI] [PubMed] [Google Scholar]
- 6.Stranger BE, et al. Relative Impact of Nucleotide and Copy Number Variation on Gene Expression Phenotypes. Science. 2007;315:848–853. doi: 10.1126/science.1136678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Aitman TJ, et al. Copy number polymorphism in Fcgr3 predisposes to glomerulonephritis in rats and humans. Nature. 2006;439(7078):851–855. doi: 10.1038/nature04489. [DOI] [PubMed] [Google Scholar]
- 8.Fanciulli M, et al. FCGR3B copy number variation is associated with susceptibility to systemic, but not organ-specific, autoimmunity. Nat Genet. 2007;39(6):721–723. doi: 10.1038/ng2046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sebat J, et al. Stong association of de novo copy number mutations with autism. Science. 2007;316:445–449. doi: 10.1126/science.1138659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Walsh T, et al. Rare structural variants disrupt multiple genes in neurodevelopmental pathways in schizophrenia. Science. 2008;320(5875):539–543. doi: 10.1126/science.1155174. [DOI] [PubMed] [Google Scholar]
- 11.Stone JL, et al. Rare chromosomal deletions and duplications increase risk of schizophrenia. Nature. 2008 doi: 10.1038/nature07239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stefansson H, et al. Large recurrent microdeletions associated with schizophrenia. Nature. 2008 doi: 10.1038/nature07229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hollox EJ, et al. Psoriasis is associated with increased b-defensin genomic copy number. Nat Genet. 2008;40:23–25. doi: 10.1038/ng.2007.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shlien A, et al. Excessive genomic DNA copy number variation in the Li-Fraumeni cancer predisposition syndrome. Proc Natl Acad Sci U S A. 2008;105(32):11264–11269. doi: 10.1073/pnas.0802970105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Steemers FJ, et al. Whole-genome genotyping with the single-base extension assay. Nat Methods. 2006;3(1):31–33. doi: 10.1038/nmeth842. [DOI] [PubMed] [Google Scholar]
- 16.Wang K, et al. PennCNV: an integrated hidden Markov model designed for high-resolution copy number variation detection in whole-genome SNP genotyping data. Genome Res. 2007;17:1665–1674. doi: 10.1101/gr.6861907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Conrad DF, Andrews TD, Carter N, Hurles M, Pritchard JK. A high resolution survey of deletion polymorphisms in the human genome. Nat Genet. 2006;38:75–81. doi: 10.1038/ng1697. [DOI] [PubMed] [Google Scholar]
- 18.Pinto D, Marshall C, Feuk L, Scherer SW. Copy-number variation in control population cohorts. Hum Mol Genet. 2007;2:R168–173. doi: 10.1093/hmg/ddm241. [DOI] [PubMed] [Google Scholar]
- 19.Zhang Z, Schwartz S, Wagner L, Miller W. A greedy algorithm for aligning DNA sequences. J Comput Biol. 2000;7(1–2):203–214. doi: 10.1089/10665270050081478. [DOI] [PubMed] [Google Scholar]
- 20.Popesco MC, et al. Human lineage-specific amplification, selection, and neuronal expression of DUF1220 domains. Science. 2006;313:1304–1307. doi: 10.1126/science.1127980. [DOI] [PubMed] [Google Scholar]
- 21.Mefford HC, et al. Recurrent Rearrangements of Chromosome 1q21.1 and Variable Pediatric Phenotypes. N Engl J Med. 2008 doi: 10.1056/NEJMoa0805384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Meza-Zepeda LA, et al. Positional cloning identifies a novel cyclophilin as a candidate amplified oncogene in 1q21. Oncogene. 2002;21:2261–2269. doi: 10.1038/sj.onc.1205339. [DOI] [PubMed] [Google Scholar]
- 23.Petroziello J, et al. Suppression subtractive hybridization and expression profiling identifies a unique set of genes overexpressed in non-small-cell lung cancer. Oncogene. 2004;23:7734–7745. doi: 10.1038/sj.onc.1207921. [DOI] [PubMed] [Google Scholar]
- 24.Diskin SJ, et al. Adjustment of genomic waves in signal intensities from whole-genome SNP genotyping platforms. Nucleic Acids Res. 2008;36(19):e126. doi: 10.1093/nar/gkn556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kent WJ. BLAT--the BLAST-like alignment tool. Genome Res. 2002;12(4):656–664. doi: 10.1101/gr.229202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Brodeur GM, et al. Revisions of the international criteria for neuroblastoma diagnosis, staging, and response to treatment. Journal of Clinical Oncology. 1993;11(8):1466–1477. doi: 10.1200/JCO.1993.11.8.1466. [DOI] [PubMed] [Google Scholar]
- 27.Shimada H, et al. The International Neuroblastoma Pathology Classification (Shimada) System. Cancer. 1999;86:364–372. [PubMed] [Google Scholar]
- 28.Mathew P, et al. Detection of MYCN gene amplification in neuroblastoma by fluorescence in situ hybridization: a pediatric oncology group study. Neoplasia. 2001;3(2):105–109. doi: 10.1038/sj.neo.7900146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Look AT, et al. Clinical relevance of tumor cell ploidy and N-myc gene amplification in childhood neuroblastoma. A Pediatric Oncology Group Study. Journal of Clinical Oncology. 1991;9:581–591. doi: 10.1200/JCO.1991.9.4.581. [DOI] [PubMed] [Google Scholar]
- 30.Gunderson KL, Steemers FJ, Lee G, Mendoza LG, Chee MS. A genome-wide scalable SNP genotyping assay using microarray technology. Nat Genet. 2005;37(5):549–554. doi: 10.1038/ng1547. [DOI] [PubMed] [Google Scholar]
- 31.Shaikh TH, et al. Chromosome 22-specific low copy repeats and the 22q11.2 deletion syndrome: genomic organization and deletion endpoint analysis. Hum Mol Genet. 2000;9(4):489–501. doi: 10.1093/hmg/9.4.489. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.