Abstract
Background
Newly infected subjects acquire a limited number of human immunodeficiency virus type 1 (HIV-1) variants with specific genotypic and phenotypic features from the array of viruses present in a chronically infected transmitting partner.
Methods
We examined HIV-1 envelope sequences from the earliest available serum sample after HIV-1 acquisition in 13 newly infected subjects and from their epidemiologically linked HIV-1–infected heterosexual partner. Samples from both members were collected on the same day in the Rakai Community Cohort Study.
Results
Ten couples were infected with subtype D HIV-1, and 3 pairs had subtype A HIV-1. Newly infected subjects acquired a subset of the viruses that were circulating in the transmitting partner; transmitted variants had less diversity and divergence and were more closely related to the ancestral sequences. The majority of signature amino acid differences among donor and recipient sequences were in and immediately following the V3 loop. Envelopes from recipients were significantly shorter and had a lower V3 charge than envelopes from donors, but there was no significant difference in the number of potential N-linked glycosylation sites.
Conclusion
A minority subset of HIV-1 variants with signature genotypes is favored for transmission in this population.
Regardless of the mode of HIV-1 acquisition and the subtype of the virus, newly infected subjects have only a limited number of variants [1–10]. Furthermore, early in infection, most viruses use the CCR5 receptor [5, 6, 11, 12], and this coreceptor restriction limits HIV-1 transmission to subjects with abnormal CCR5 receptor expression [13–15]. Over the course of infection, the virus evolves, leading to the presence of variants with diverse envelope genotypes and an ability to potentially use other coreceptors, such as CXCR4, for host-cell entry [10, 16–18]. Collectively, these observations suggest that certain variants are selected during transmission, presumably on the basis of specific viral properties and host factors. It is essential to elucidate the viral characteristics that confer fitness for transmission in order to develop vaccines or microbicides that specifically target the HIV-1 variants preferentially acquired by a newly infected subject.
Recent studies suggest that viruses with shorter and less glycosylated envelope variable loops were preferentially acquired by heterosexually infected subjects with subtype A and C HIV-1 [1, 19]. Similar signature envelope characteristics, however, were not observed in subjects newly infected with subtype B HIV-1 who primarily acquired their infection through injection drug use and/or homosexual contact [19, 20]. Because selection during transmission occurs during all modes of transmission and with every HIV-1 subtype, a signature viral phenotype may confer fitness for transmission; different subtypes, however, may have unique genotypic determinants for influencing this viral property. To provide further insight into the selection of specific HIV-1 envelope genotypes and phenotypes during transmission, we examined envelope sequences from 13 newly infected subjects and their epidemiologically linked heterosexual partners in the Rakai Community Cohort Study. The majority of the couples in this cohort are infected with HIV-1 subtype D, for which envelope characteristics early after transmission have not been elucidated.
SUBJECTS, MATERIALS, AND METHODS
Study subjects
Subjects were retrospectively identified from the previously described Rakai Community Cohort Study in the Rakai district of southwestern Uganda between 1994 and 1999 [21, 22]. In brief, consenting participants were offered free voluntary counseling and testing as individuals and as couples, were provided with health education on HIV prevention and free condoms, and had serum collected for HIV-1 serological analysis every 10 months. We investigated 13 newly infected monogamous subjects with their epidemiologically linked partner on the basis of the availability of serum samples and prior confirmation of molecular linkage of sequence data from the Gag and gp41 regions [23]. In the partner with incident infection, the HIV-1 seroconversion date was estimated as the midpoint between the last visit with an HIV-1–seronegative test result and the first visit during which HIV-1 antibody was detected. The study was approved by human subjects–research review boards at the Uganda Virus Research Institute, the AIDS Research Subcommittee of the Ugandan National Council for Science and Technology, Columbia University, and Johns Hopkins University.
Envelope analysis
HIV-1 RNA was isolated from 200−500 μL of the serum samples, and RT-PCR was used to amplify a library of full-length envelope genes as previously described [24]. Eight individual full-length envelope clones were isolated and sequenced from this library of envelope genes from each serum sample [25]. All unique sequences reported in this publication have been submitted to Genbank (accession numbers EU852934 – EU853141).
All sequences were aligned using Clustal X and further manually codon aligned using MacClade, version 4.01. Sequences were gap stripped, and neighbor joining phylogenetic analysis with a general time-reversible model and a gamma distribution rate parameter of 0.5 was used to assess relationships among sequences from newly infected subjects and their epidemiologically linked partners. Sequences were examined for intersubtype recombination, using RIP 3.0 (available at: http://www.hiv.lanl.gov/content/sequence/RIP/RIP.html). Genetic diversity in each subject was estimated using a Kimura 2-parameter model with a gamma rate distribution of 0.5, using MEGA, version 3.1 [26]. Heuristically searched maximum likelihood trees were generated for each transmission pair with outgroup sequences of the same subtype from the Los Alamos database, using the best-fit evolutionary model parameters selected by use of Modeltest, version 3.06 [27], and the software package PAUP, version 4.02b2a [28]. Each tree was also examined for segregation of recipient and donor sequences, using the Maddison-Slatkin test [29] as implemented in MacClade, version 4.01. The minimum number of mixing events between the recipient and donor sequences from the tree was compared with the distribution of migration events for 1000 randomly generated trees to estimate statistical significance.
For each sequence, V1-V2 (131−196), V3 (296−331), V1–V4 (131−418), and V1–V5 (131−463) segments corresponding to HXB2 envelope amino acids were defined. The number of amino acids and predicted N-linked glycosylation sites in each segment were determined through manual counting and by using the tool available at http://www.hiv.lanl.gov/content/sequence/GLYCOSITE/glycosite.html, respectively. Recipient and donor sequences in each partnership were examined for signature amino acid differences, using VESPA (available at: http://www.hiv.lanl.gov/content/sequence/VESPA/vespa.html) [30].
Coreceptor use was examined using the Monogram Biosciences Trofile assay as previously described [24]. Briefly, each subject's library of full-length envelopes was incorporated into HIV-1 pseudoviruses that express the firefly luciferase gene. Tropism of these pseudoviruses was designated by their ability to infect U87/CD4+/CCR5 and/or U87/CD4+/CXCR4 cells, as assessed by luciferase expression.
Statistical analysis
Viral diversity and divergence for the sequences from the newly infected subject and the transmitting partner were compared using the Wilcoxon matched-pairs signed rank test. The median value for other sequence characteristics from the 8 recipient and the 8 donor sequences from each partnership were compared using the Wilcoxon matched-pairs signed rank test. To account for the multiple sequences from each subject, we also compared the sequence characteristics in each pair, using the Wilcoxon rank sum test. The Wilcoxon rank sum test, stratified by pair, was further used for aggregate comparisons among all 13 couples. All statistical analyses were done with Intercooled Stata, version 8.0 (Stata), and SAS, version 8.2 (SAS Institute). All P values are for 2-sided tests.
RESULTS
Subjects and partnerships
HIV-1 envelope sequences from the 13 newly infected subjects were analyzed a median of 189 days (range, 142−359 days) after estimated HIV-1 seroconversion (table 1). All 13 transmitting partners had prevalent HIV-1 infection at the time of enrollment and were followed for a minimum of 2 years before the transmission event. Thus, transmission did not occur during the acute phase of the index partner's infection. Eight full-length envelope sequences were examined from each subject. Neighbor joining phylogenetic analysis showed that sequences from the newly infected subject and the transmitting partner clustered together with high bootstrap support, confirming the epidemiological linkage (figure 1). Ten couples had subtype D HIV-1, and subjects in the remaining 3 partnerships (120, 368, and 601) were infected with subtype A HIV-1. No sequences from any subject showed evidence of intersubtype recombination.
Table 1.
Demographic, viral, and coreceptor characteristics.
| Recipients |
Donors |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Plasma HIV-1 load, copies/mL |
Coreceptor level, RLUb |
Coreceptor level, RLUb |
|||||||||
| Couple | Transmission type | Time from seroconversion to HIV-1 detection, recipients, daysa | Subtype | Recipients | Donors | CCR5 | CXCR4 | Tropismc | CCR5 | CXCR4 | Tropismc |
| 9 | FTM | 359 | D | 146,000 | 16,800 | 334,690 | 55 | R5 | 338,413 | 73 | R5 |
| 32 | MTF | 142 | D | 487,000 | 362,000 | 602,717 | 58 | R5 | 1,058,435 | 152 | R5 |
| 108 | FTM | 161 | D | 483,000 | 258,000 | 223,954 | 56 | R5 | 1,354,111 | 59 | R5 |
| 120 | MTF | 189 | A | 389,000 | 141,000 | 18,727 | 54 | R5 | 188,528 | 85 | R5 |
| 183 | MTF | 202 | D | 1,020,000 | 421,000 | 271,742 | 61 | R5 | 1,542,808 | 116 | R5 |
| 295 | FTM | 180 | D | 1,630,000 | 56,000 | 211,202 | 29,007 | DM | 1,185,684 | 113 | R5 |
| 326 | MTF | 359 | D | 128,000 | 78,000 | 55,369 | 56 | R5 | 1,598,769 | 74 | R5 |
| 338 | MTF | 159 | D | 96,000 | 37,000 | 3,213,222 | 93 | R5 | 13,028 | 53 | R5 |
| 368 | MTF | 329 | A | 32,000 | 23,000 | 41,826 | 56 | R5 | 144,192 | 75 | R5 |
| 372 | FTM | 189 | D | 505,000 | 221,000 | 471,609 | 69 | R5 | 335,968 | 63 | R5 |
| 601 | MTF | 179 | A | 22,405 | 120,240 | 848,488 | 73 | R5 | 43,212 | 66 | R5 |
| 602 | MTF | 191 | D | 760,455 | 96,000 | 532,666 | 73 | R5 | 858,769 | 150 | R5 |
| 605 | MTF | 218 | D | 109,865 | 90,543 | 1,986,568 | 76 | R5 | 854,224 | 92 | R5 |
NOTE. DM, dual or mixed virus; FTM, female to male; MTF, male to female; RLU, relative light units; R5, R5 virus.
Seroconversion date was estimated as the midpoint between the last visit with an HIV-1–seronegative test result and the first visit during which HIV-1 antibody was detected.
Produced from envelope pools on U87/CD4+/CCR5 cells (for CCR5) or U87/CD4+/CXCR4 cells (for CXCR4).
See the text for a description of the methods and materials used for detection.
Figure 1.

Findings of neighbor joining phylogenetic analysis, showing that recipient sequences (closed circles) and donor sequences (open circles) cluster together. Bootstrap values from 1000 replicates are noted on each node of interest, and reference subtype sequences are specified. Subtype C reference sequence C.Br.92.BRO25-d was used as an outgroup. See the text for a description of methods and materials. FTM, female-to-male transmission; MTF, male-to-female transmission.
Sequence diversity and divergence
To better examine genetic relationships among the sequences from the partner with incident infection and the transmitting partner, maximum likelihood trees were constructed for each couple (figure 2). Newly infected subjects had a median genetic diversity of 0.7% (range, 0.3%−2.44%), which was significantly lower than the genetic diversity present in the transmitting partner (median, 1.4% [range, 0.4%−4.6%]; P < .001, by the Wilcoxon matched-pairs signed rank test). Recipient sequences (median distance to the recipient ancestor, 12 [range, 6.5−94]) also demonstrated significantly less divergence, compared with donor sequences (median distance to the donor ancestor, 102 [range, 20−165]; P < .001, by the Wilcoxon matched-pairs signed rank test). In each pair, recipient sequences demonstrated significantly separate clustering relative to the donor sequences, as examined by the Maddison-Slatkin test. In each couple, recipient and donor sequences could be separated by a maximum of 2 steps. Significantly less than 5% of the 1000 randomly generated trees from each partnership's sequences could be separated by ≤2 steps, which suggests that the recipient and donor sequences in each pair showed significantly different clustering (P < .005). The distinct clustering and decreased diversity and divergence suggest that a limited number of minority variants in the donors were preferentially favored for transmission during heterosexual transmission in Rakai, Uganda.
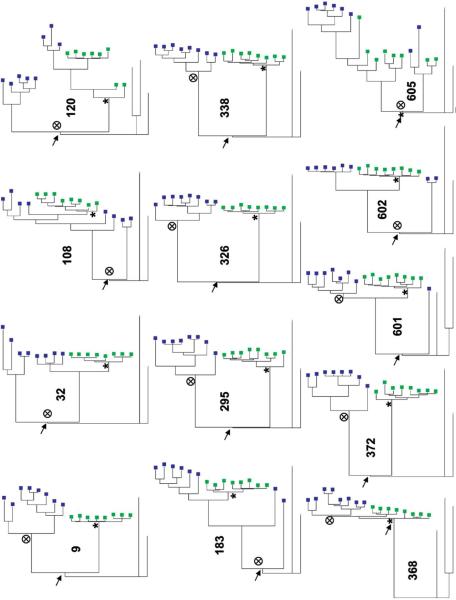
Figure 2.
Findings of maximum likelihood tree analysis of donor sequences (blue boxes) and recipient sequences (green boxes), showing that transmitted viruses are minority ancestral variants. See the text for a description of methods and materials. Arrows, most recent common ancestors; targets, donor ancestor nodes; asterisks, recipient ancestor nodes.
Sequence characteristics
To evaluate some of the characteristics of these minority variants, the maximum likelihood trees were used to compute branch lengths to the one common ancestor that linked all the donor and recipient sequences in each couple (figure 2). The median length of the newly infected subjects’ sequence branch to the common ancestor (median of the 13 recipient medians, 116.5 [range, 8−167.5]) was significantly shorter than that for the index partners’ sequence branch (median of the 13 donor medians, 133 [range, 25−175.5]; P = .04, by the Wilcoxon matched-pairs signed rank test). Because we isolated multiple sequences from every subject, we assessed the difference in the distribution of distance to the common ancestor between recipients and donors in each couple. In every pair except couple 108, the newly infected subject showed distributions with smaller distances to the common ancestor, compared with the donor. Recipients had significantly smaller branch lengths to the common ancestor than the donors in aggregate comparisons involving either 13 pairs or 10 pairs with subtype D HIV-1 (P < .001 for both comparisons, by the Wilcoxon rank sum test stratified by couple). Smaller branch lengths to the common ancestor suggest that recipient sequences were more closely related to ancestral sequences than the majority of viruses circulating in the donor.
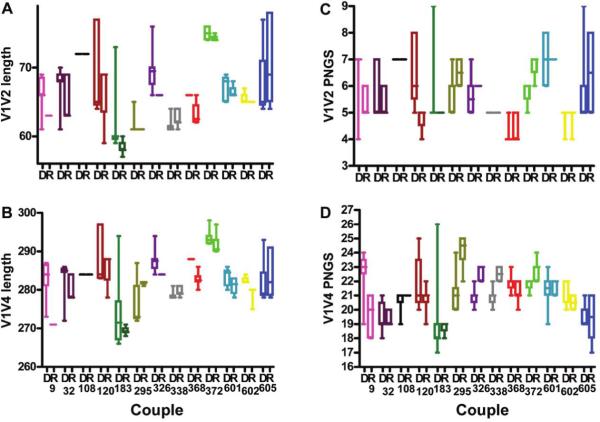
Sequences from each partnership were evaluated for envelope length and glycosylation. The median length and the number of predicted N-linked glycosylation sites of the donor and recipient sequences were not significantly different in any segment of the envelope (i.e., V1-V2, V3, V1–V4, and V1–V5) (P > .05, by the Wilcoxon matched-pairs signed rank test). To account for the multiple sequences isolated from each subject, we again compared the distribution of sequence characteristics among the newly infected individual and the transmitting partner within each pair. In general, envelope V1-V2, V1–V4, and V1–V5 lengths in recipient variants were shorter than those in donor variants in a majority of the partnerships. For instance, in only 3 of 13 couples (295, 338, and 605), the transmitting partner had shorter envelope V1–V4 regions than the newly infected subject (figure 3). In aggregate, recipient envelope V1-V2, V1–V4, and V1–V5 sequences were significantly shorter than donor sequences (P = .002, P < .001, and P = .001, respectively, by the Wilcoxon rank sum test stratified by couple). V1-V2, V1–V4, and V1–V5 sequences in newly infected subjects remained significantly shorter than those in their transmitting partners (P = .009, P = .004, and P = .006, respectively, by the Wilcoxon rank sum test stratified by couple), even when the analysis was limited to the 10 partnerships with subtype D HIV-1. Among all couples, however, there was no significant difference in the number of predicted N-linked glycosylation sites between recipient and donor sequences in the V1-V2, V1–V4, or V1–V5 segments (figure 3).
Figure 3.
V1-V2 length (A), V1–V4 length (B), V1V2 potential N-linked glycosylation sites (C), and V1-V4 potential N-linked glycosylation sites (D) for donors (D) and recipients (R), showing that transmitted viruses in Uganda have shorter envelopes. Boxes, interquartile ranges; horizontal lines, median values.
To identify other differences among envelope sequences from newly infected subjects and the viruses circulating in the transmitting partner, we examined sequences for signature amino acid differences, using VESPA [30]. VESPA calculates the frequency of each amino acid at each position in an alignment for the query and the reference set and selects the positions for which the most common character in the query set differs from that in the background set. A median of 18 signature amino acids (range, 8−50 amino acids) differentiated the consensus donor sequence from the consensus recipient sequence in different partnerships (table 2, which is available only in the electronic edition of the Journal). When all sequences from newly infected subjects were compared with all sequences from the transmitting partners, however, there was no consistent amino acid difference that differentiated recipient and donor envelopes. Thus, signature amino acid differences were specific to each partnership. We hypothesized that, although there were no consistent amino acid differences, signature amino acids may be concentrated in specific regions of the envelope gene. Among all couples, signature amino acid differences were recorded and tallied relative to the HXB2 envelope sequence [31] to identify a region of the envelope gene with a concentration of signature amino acid differences (figure 4). By visual inspection alone, the majority of predicated amino acid differences among the recipient and the donor sequences were in and immediately following the envelope V3 loop and in the heptad region 2 domain of gp41.
Figure 4.

Signature amino acid differences cluster around the V3 loop. Variable loops 1, 2, 4, and 5 were not included in the analysis because subject sequences could not be reliably aligned. The V3 loop and heptad region 2 (HR2) borders are delineated with red lines. Positions of the envelope gp120, gp41, C1-C2, C3–C5, and heptad region 1 (HR1) regions are also shown. See the text for a description of methods and materials.
Because signature amino acid differences among donor and recipient viruses were concentrated in and around the envelope V3 loop, we examined the V3 segment for specific characteristics known to influence its function. For subtype B HIV-1, the V3 loop charge [32] and the presence of positively charged amino acids at position 11 and 25 [33–35] have been associated with specific coreceptor use and cellular tropism. In all pairs except 108, 295, and 601, recipient sequences had a lower V3 charge, compared with donor envelopes (figure 5). In aggregate, recipient V3 loops had a significantly lower charge, compared with donor V3 sequences (P = .009, by the Wilcoxon rank sum test stratified by couple), and this difference remained significant when the analysis was restricted to the 10 couples with subtype D HIV-1 (P = .004, by the Wilcoxon rank sum test stratified by couple). The consensus donor sequence had a higher charge than the consensus recipient sequence at positions 11 and/or 25 of the V3 loop in only 3 couples (32, 326, and 605), suggesting that the charge differences were not primarily because of amino acid differences at these positions of the V3 loop. Because the ability to use the CXCR4 receptor as opposed to the CCR5 receptor has been associated with a higher V3 charge in subtype B HIV-1, we examined coreceptor tropism, using the Trofile assay [24]. Nearly all subjects had viruses that strictly used the CCR5 receptor; those from the newly infected individual from couple 295 showed dual or mixed tropism (table 1). Interestingly, V3 sequences of clones from this recipient had a positively charged amino acid at position 25 and a higher V3 charge, compared with V3 loops in the index partner (figure 5). CXCR4-using virus (X4 or dual) was detected in the virus population of the recipient but not the index partner of couple 295, suggesting that CXCR4-using variants may have arisen de novo or may have been transmitted from a minority of the virus population that was present in the partner. Taken together, our data suggest that the higher V3 charge differences observed in donors did not reflect that they had a greater number of CXCR4-using viruses than newly infected subjects. Indeed, previous studies have suggested that, within subtype D HIV-1, coreceptor use differences are not exclusively associated with envelope V3 loop sequence changes [25].
Figure 5.

V3 charges for viruses from donors (D) and recipients (R), showing that transmitted viruses have a lower V3 charge. Boxes, interquartile ranges; horizontal lines, median values.
DISCUSSION
In this study, we examined a minimum of 8 envelope sequences from 13 newly infected subjects and their chronically infected heterosexual partners from retrospectively identified discordant couples in a population-based cohort in the Rakai district of Uganda. Other publications have also investigated sequences from newly infected subjects and their sexual partners [1, 5, 20], but our study examined the largest number of discordant couples to date. Furthermore, subjects in this cohort were primarily infected with subtype D HIV-1, which has not been examined in detail previously. Our analyses showed that a limited number of minority HIV-1 variants closely related to ancestral sequences with condensed envelopes and less overall V3 charge are preferentially acquired by the naive host from the transmitting partner. In aggregate, our findings from the Rakai population, in which HIV-1 subtype D is the predominant variant transmitted, suggest that a small number of viruses with specific envelope genotypic characteristics are selected during heterosexual transmission, which accords with findings from other studies [1, 19].
We showed that the transmitted viruses are closely related to the ancestor sequences; this finding has important implications for the relationship between HIV-1 envelope evolution and pathogenesis. The HIV-1 envelope evolves in a host presumably to escape host immune responses and to increase host-cell range [36]. Because newly acquired viruses are more closely related to the ancestor sequences than the majority of viruses that have evolved in a chronically infected host, envelope modifications that occur in a host over the course of infection may decrease a virus’ fitness for transmission. Similar observations in heterosexual transmission pairs with subtype C HIV-1 [1, 37] and from an analysis of longitudinal samples from subjects infected with subtype B HIV-1 [37] suggest that HIV-1 of different subtypes are similarly constrained from productively infecting and disseminating in a naive host by changes that occur in the envelope glycoprotein over the course of infection. Further studies are needed to determine the biological mechanism for the disadvantage that these modifications confer during transmission.
An increase in the number or a shift in the position of the glycosylated residues in the envelope glycoprotein are changes that appear to occur in various HIV-1 subtypes in a host over the course of infection [38, 39]. Less glycosylated HIV-1 variants, however, are favored during heterosexual transmission of HIV-1 subtypes A and C [1, 19]. In our study, transmitted viruses had significantly smaller envelopes but no significant difference in predicted glycosylation sites, compared with variants circulating in the chronically infected partner. This suggests that, among the viruses circulating in Rakai, most of which are subtype D HIV-1, there is preferential selection for variants with compact but not necessarily less glycosylated envelopes. Samples from the newly infected subjects in our study, however, were obtained an average of ∼5 months after estimated seroconversion, which is longer than the sampling interval of ∼2−4 months after the last visit with a seronegative test result for individuals infected with subtypes A and C [1, 19]. Given that envelope characteristics, especially the level of glycosylation, can evolve in the early months after HIV-1 infection [10, 38, 40], it is possible that we failed to observe a significant difference in predicted glycosylation between recipient and donor viruses because the sampling interval for our newly infected subjects was longer than that for subjects in other studies. If modifications in the number and type of glycosylation sites are fixed in the virus population before changes in envelope length occur, the longer sampling interval may have precluded us from observing a significant difference in the number of predicted glycosylated sites between recipient and donor viruses.
We also showed that signature amino acid differences among recipient and donor sequences were primarily located in and immediately following the V3 loop. Interestingly, there was high variability in sequences immediately following the V3 loop in HIV-1 subtypes B and C between persons with early infection and those with chronic infection [41]. We further observed that variants in newly infected subjects had less overall V3 charge than HIV variants in the transmitting partner. Previously published subtype B and C envelope sequences from discordant couples did not show similar V3 charge differences [1, 20]. It is possible that HIV-1 subtypes B and C have a limited range of V3 charges among different variants, which may preclude the ability to document significant differences among viruses in the recipient and the donor. Although the proportion of variants that use the CXCR4 coreceptor is greater among HIV-1 subtype D than among other HIV-1 subtypes [25], V3 charge differences among donor and recipients was not associated with differences in coreceptor use.
We identified common sequence characteristics, such as smaller envelopes and less overall V3 charge, by examining aggregate differences among a cohort of discordant partners. These signature characteristics, however, were not observed in every couple in our study. For instance, in couple 108, HIV-1 variants in newly infected subjects did not have any significant differences in envelope length or V3 charge from those of viruses in their corresponding transmitting partners. Furthermore, signature genotypes, such as condensed and less glycosylated envelopes, have not been defined among all newly acquired viruses, particularly subtype B HIV-1 [19, 20]. Selection of a limited number of HIV-1 variants, however, occurs during all routes of HIV-1 acquisition and among all HIV-1 subtypes examined to date [1–7, 9, 10]. Taken together, these observations suggest that, regardless of the subtype, mode of HIV-1 acquisition, or index partners’ HIV-1 variants, only a limited number of viruses are successfully transmitted to a naive host. This selection during transmission may be based on a viral property that confers an advantage during transmission, although different HIV-1 subtypes and/or variants in an individual subject may have unique genotypic means for imparting this phenotypic characteristic.
It remains unclear how the differences in envelope length and V3 charge confer an advantage during transmission. Previous studies have suggested that envelope length may influence viral replicative capacity [39, 42]. In addition, V3 charge differences could affect the affinity of the envelope glycoprotein for the host cell coreceptor, although our studies suggest that it did not influence coreceptor use. Thus, differences in envelope length and V3 charge may augment the ability of envelopes to bind receptors and initiate host cell fusion, which, if true, may increase the capacity to productively infect early target cells and disseminate from the site of invasion. Further studies are needed to elucidate the mechanism of how the signature envelope genotypes confer an advantage during transmission. Detailed insight into the biological mechanisms of selection during transmission will greatly aid the ability to target the variants favored during transmission with a vaccine or a microbicide.
Supplementary Material
Acknowledgments
We thank the subjects who contributed samples for these studies as part of the Rakai Health Sciences Project and Simon Frost for providing aligned envelope sequences from transmission pairs with subtype B HIV-1 infection [20].
Financial support: Doris Duke Charitable Foundation (funds for use by M.S.); National Institutes of Health (NIH; grant AI077473 for use by M.S.); Division of Intramural Research, National Institute of Allergy and Infectious Diseases, NIH (funds for use by T.Q.; grants R01 A134826 and R01 A1342650 for data collection); National Institute of Child and Health Development, NIH (grant 5P30HD06826 for data collection); World Bank STI Project, Uganda; Fogarty Foundation (grant 5D43TW00010).
Footnotes
Potential conflicts of interest: none reported.
Presented in part: 12th Conference on Retroviruses and Opportunistic Infections, Boston, MA, 22−25 February 2005 (abstract 339); 15th Conference on Retroviruses and Opportunistic Infections, Boston, MA, 3−6 February 2008 (abstract 682).
References
- 1.Derdeyn CA, Decker JM, Bibollet-Ruche F, et al. Envelope-constrained neutralization-sensitive HIV-1 after heterosexual transmission. Science. 2004;303:2019–22. doi: 10.1126/science.1093137. [DOI] [PubMed] [Google Scholar]
- 2.Long EM, Martin HL, Jr, Kreiss JK, et al. Gender differences in HIV-1 diversity at time of infection. Nat Med. 2000;6:71–5. doi: 10.1038/71563. [DOI] [PubMed] [Google Scholar]
- 3.McNearney T, Hornickova Z, Markham R, et al. Relationship of human immunodeficiency virus type 1 sequence heterogeneity to stage of disease. Proc Natl Acad Sci U S A. 1992;89:10247–51. doi: 10.1073/pnas.89.21.10247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wolinsky SM, Wike CM, Korber BT, et al. Selective transmission of human immunodeficiency virus type-1 variants from mothers to infants. Science. 1992;255:1134–7. doi: 10.1126/science.1546316. [DOI] [PubMed] [Google Scholar]
- 5.Zhu T, Mo H, Wang N, et al. Genotypic and phenotypic characterization of HIV-1 patients with primary infection. Science. 1993;261:1179–81. doi: 10.1126/science.8356453. [DOI] [PubMed] [Google Scholar]
- 6.Zhang LQ, MacKenzie P, Cleland A, Holmes EC, Brown AJ, Simmonds P. Selection for specific sequences in the external envelope protein of human immunodeficiency virus type 1 upon primary infection. J Virol. 1993;67:3345–56. doi: 10.1128/jvi.67.6.3345-3356.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wolfs TF, Zwart G, Bakker M, Goudsmit J. HIV-1 genomic RNA diver-sification following sexual and parenteral virus transmission. Virology. 1992;189:103–10. doi: 10.1016/0042-6822(92)90685-i. [DOI] [PubMed] [Google Scholar]
- 8.Poss M, Martin HL, Kreiss JK, et al. Diversity in virus populations from genital secretions and peripheral blood from women recently infected with human immunodeficiency virus type 1. J Virol. 1995;69:8118–22. doi: 10.1128/jvi.69.12.8118-8122.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sagar M, Kirkegaard E, Long EM, et al. Human immunodeficiency virus type 1 (HIV-1) diversity at time of infection is not restricted to certain risk groups or specific HIV-1 subtypes. J Virol. 2004;78:7279–83. doi: 10.1128/JVI.78.13.7279-7283.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Keele BF, Giorgi EE, Salazar-Gonzalez JF, et al. Identification and characterization of transmitted and early founder virus envelopes in primary HIV-1 infection. Proc Natl Acad Sci U S A. 2008;105:7552–7. doi: 10.1073/pnas.0802203105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cheng-Mayer C, Seto D, Tateno M, Levy JA. Biologic features of HIV-1 that correlate with virulence in the host. Science. 1988;240:80–2. doi: 10.1126/science.2832945. [DOI] [PubMed] [Google Scholar]
- 12.Fenyo EM, Morfeldt-Manson L, Chiodi F, et al. Distinct replicative and cytopathic characteristics of human immunodeficiency virus isolates. J Virol. 1988;62:4414–9. doi: 10.1128/jvi.62.11.4414-4419.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dean M, Carrington M, Winkler C, et al. Genetic restriction of HIV-1 infection and progression to AIDS by a deletion allele of the CKR5 structural gene. Hemophilia Growth and Development Study, Multicenter AIDS Cohort Study, Multicenter Hemophilia Cohort Study, San Francisco City Cohort, ALIVE Study. Science. 1996;273:1856–62. doi: 10.1126/science.273.5283.1856. [DOI] [PubMed] [Google Scholar]
- 14.Liu R, Paxton WA, Choe S, et al. Homozygous defect in HIV-1 coreceptor accounts for resistance of some multiply-exposed individuals to HIV-1 infection. Cell. 1996;86:367–77. doi: 10.1016/s0092-8674(00)80110-5. [DOI] [PubMed] [Google Scholar]
- 15.Samson M, Libert F, Doranz BJ, et al. Resistance to HIV-1 infection in Caucasian individuals bearing mutant alleles of the CCR-5 chemokine receptor gene. Nature. 1996;382:722–5. doi: 10.1038/382722a0. [DOI] [PubMed] [Google Scholar]
- 16.Shankarappa R, Margolick JB, Gange SJ, et al. Consistent viral evolutionary changes associated with the progression of human immunodeficiency virus type 1 infection. J Virol. 1999;73:10489–502. doi: 10.1128/jvi.73.12.10489-10502.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tersmette M, Lange JM, de Goede RE, et al. Association between biological properties of human immunodeficiency virus variants and risk for AIDS and AIDS mortality. Lancet. 1989;1:983–5. doi: 10.1016/s0140-6736(89)92628-7. [DOI] [PubMed] [Google Scholar]
- 18.Tersmette M, Gruters RA, de Wolf F, et al. Evidence for a role of virulent human immunodeficiency virus (HIV) variants in the pathogenesis of acquired immunodeficiency syndrome: studies on sequential HIV isolates. J Virol. 1989;63:2118–25. doi: 10.1128/jvi.63.5.2118-2125.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chohan B, Lang D, Sagar M, et al. Selection for human immunodeficiency virus type 1 envelope glycosylation variants with shorter V1–V2 loop sequences occurs during transmission of certain genetic subtypes and may impact viral RNA levels. J Virol. 2005;79:6528–31. doi: 10.1128/JVI.79.10.6528-6531.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Frost SD, Liu Y, Pond SL, et al. Characterization of human immunodeficiency virus type 1 (HIV-1) envelope variation and neutralizing antibody responses during transmission of HIV-1 subtype B. J Virol. 2005;79:6523–7. doi: 10.1128/JVI.79.10.6523-6527.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wawer MJ, Sewankambo NK, Serwadda D, et al. Control of sexually transmitted diseases for AIDS prevention in Uganda: a randomised community trial. Rakai Project Study Group. Lancet. 1999;353:525–35. doi: 10.1016/s0140-6736(98)06439-3. [DOI] [PubMed] [Google Scholar]
- 22.Wawer MJ, Gray RH, Sewankambo NK, et al. A randomized, community trial of intensive sexually transmitted disease control for AIDS prevention, Rakai, Uganda. AIDS. 1998;12:1211–25. doi: 10.1097/00002030-199810000-00014. [DOI] [PubMed] [Google Scholar]
- 23.Wawer MJ, Gray RH, Sewankambo NK, et al. Rates of HIV-1 transmission per coital act, by stage of HIV-1 infection, in Rakai, Uganda. J Infect Dis. 2005;191:1403–9. doi: 10.1086/429411. [DOI] [PubMed] [Google Scholar]
- 24.Whitcomb JM, Huang W, Fransen S, et al. Development and characterization of a novel single-cycle recombinant-virus assay to determine human immunodeficiency virus type 1 coreceptor tropism. Antimicrob Agents Chemother. 2007;51:566–75. doi: 10.1128/AAC.00853-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huang W, Eshleman SH, Toma J, et al. Coreceptor tropism in human immunodeficiency virus type 1 subtype D: high prevalence of CXCR4 tropism and heterogeneous composition of viral populations. J Virol. 2007;81:7885–93. doi: 10.1128/JVI.00218-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kumar S, Tamura K, Nei M. MEGA3: integrated software for Molecular Evolutionary Genetics Analysis and sequence alignment. Briefings in Bioinformatics. 2004;5:150–63. doi: 10.1093/bib/5.2.150. [DOI] [PubMed] [Google Scholar]
- 27.Posada D, Crandall KA. MODELTEST: testing the model of DNA substitution. Bioinformatics. 1998;14:817–8. doi: 10.1093/bioinformatics/14.9.817. [DOI] [PubMed] [Google Scholar]
- 28.Swofford DL. Phylogenetic analysis using parsimony. 4th ed Sinauer Associates; Sunder-land, MA: 1998. [Google Scholar]
- 29.Slatkin M, Maddison WP. Detecting isolation by distance using phylogenies of genes. Genetics. 1990;126:249–60. doi: 10.1093/genetics/126.1.249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Korber B, Myers G. Signature pattern analysis: a method for assessing viral sequence relatedness. AIDS Res Hum Retroviruses. 1992;8:1549–60. doi: 10.1089/aid.1992.8.1549. [DOI] [PubMed] [Google Scholar]
- 31.Modrow S, Hahn BH, Shaw GM, Gallo RC, Wong-Staal F, Wolf H. Computer-assisted analysis of envelope protein sequences of seven human immunodeficiency virus isolates: prediction of antigenic epitopes in conserved and variable regions. J Virol. 1987;61:570–8. doi: 10.1128/jvi.61.2.570-578.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fouchier RA, Groenink M, Kootstra NA, et al. Phenotype-associated sequence variation in the third variable domain of the human immunodeficiency virus type 1 gp120 molecule. J Virol. 1992;66:3183–7. doi: 10.1128/jvi.66.5.3183-3187.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.De Jong JJ, De Ronde A, Keulen W, Tersmette M, Goudsmit J. Minimal requirements for the human immunodeficiency virus type 1 V3 domain to support the syncytium-inducing phenotype: analysis by single amino acid substitution. J Virol. 1992;66:6777–80. doi: 10.1128/jvi.66.11.6777-6780.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fouchier RA, Brouwer M, Broersen SM, Schuitemaker H. Simple determination of human immunodeficiency virus type 1 syncytium-inducing V3 genotype by PCR. J Clin Microbiol. 1995;33:906–11. doi: 10.1128/jcm.33.4.906-911.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Guay LA, Musoke P, Fleming T, et al. Intrapartum and neonatal single-dose nevirapine compared with zidovudine for prevention of mother-to-child transmission of HIV-1 in Kampala, Uganda: HIVNET 012 randomised trial. Lancet. 1999;354:795–802. doi: 10.1016/S0140-6736(99)80008-7. [DOI] [PubMed] [Google Scholar]
- 36.Overbaugh J, Bangham CR. Selection forces and constraints on retroviral sequence variation. Science. 2001;292:1106–9. doi: 10.1126/science.1059128. [DOI] [PubMed] [Google Scholar]
- 37.Herbeck JT, Nickle DC, Learn GH, et al. Human immunodeficiency virus type 1 env evolves toward ancestral states upon transmission to a new host. J Virol. 2006;80:1637–44. doi: 10.1128/JVI.80.4.1637-1644.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wei X, Decker JM, Wang S, et al. Antibody neutralization and escape by HIV-1. Nature. 2003;422:307–12. doi: 10.1038/nature01470. [DOI] [PubMed] [Google Scholar]
- 39.Sagar M, Wu X, Lee S, Overbaugh J. HIV-1 V1-V2 envelope loop sequences expand and add glycosylation sites over the course of infection and these modifications affect antibody neutralization sensitivity. J Virol. 2006;80:9586–98. doi: 10.1128/JVI.00141-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Frost SD, Wrin T, Smith DM, et al. Neutralizing antibody responses drive the evolution of human immunodeficiency virus type 1 envelope during recent HIV infection. Proc Natl Acad Sci U S A. 2005;102:18514–9. doi: 10.1073/pnas.0504658102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li M, Salazar-Gonzalez JF, Derdeyn CA, et al. Genetic and neutralization properties of subtype C human immunodeficiency virus type 1 molecular env clones from acute and early heterosexually acquired infections in Southern Africa. J Virol. 2006;80:11776–90. doi: 10.1128/JVI.01730-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shioda T, Oka S, Xin X, et al. In vivo sequence variability of human immunodeficiency virus type 1 envelope gp120: association of V2 extension with slow disease progression. J Virol. 1997;71:4871–81. doi: 10.1128/jvi.71.7.4871-4881.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.