Abstract
Bone metabolism results from a balance between osteoclast-driven bone resorption and osteoblast-mediated bone formation. Diseases such as periodontitis and rheumatoid arthritis are characterized by increased bone destruction due to enhanced osteoclastogenesis1,2. Here we report that interferon regulatory factor 8 (IRF8), a transcription factor expressed in immune cells, is a key regulatory molecule for osteoclastogenesis. IRF8 expression in osteoclast precursors was downregulated during the initial phase of osteoclast differentiation induced by receptor activator of nuclear factor κB ligand (RANKL, also called TRANCE, ODF, and OPGL), which is encoded by the Tnfsf11 gene. Mice deficient in IRF8 exhibited severe osteoporosis due to increased numbers of osteoclasts, and enhanced bone destruction following lipopolysaccharide (LPS) administration. Irf8–/– osteoclast precursors underwent increased osteoclastogenesis in response to RANKL and tumor necrosis factor α (TNFα). IRF8 suppressed osteoclastogenesis by inhibiting the function and expression of nuclear factor of activated T cells c1 (NFATc1). Our results show that IRF8 inhibits osteoclast formation under physiological and pathological conditions, and suggest a model where downregulation of inhibitory factors like IRF8 contributes to RANKL-mediated osteoclastogenesis.
Osteoclasts are multinucleated giant cells derived from the monocyte/macrophage lineage. Their differentiation is triggered by RANKL in the presence of macrophage colony-stimulating factor (M-CSF, encoded by Csf-1 gene), which is produced by osteoblasts1,2. RANKL induces intracellular signals via its receptor RANK, and upregulates the expression of various genes, such as Nfatc1,fos, Oscar, Ctsk, and Calcr that encode proteins for NFATc1, c-Fos, OSCAR, cathepsin K and calcitonin receptor, respectively3–5. Numerous studies have focused on these upregulated genes and their roles in osteoclastogenesis. On the other hand, the expression levels of various genes are simultaneously downregulated during osteoclastogenesis6. The biological significance of the downregulated expression of these genes following RANK activation, however, has not been fully elucidated.
To identify genes that show reduced expression levels in response to RANK signaling, we performed a genome-wide screening of mRNAs from osteoclast precursors and osteoclasts using a DNA microarray technique (data not shown). Among the identified genes, expression of the transcription factor Irf8 [also called interferon consensus sequence binding protein (ICSBP)] was found to be downregulated during the initial phase of osteoclastogenesis triggered by RANKL (data not shown). IRF8 is known to be specifically expressed in immune cells, including monocytes/macrophages, B lymphocytes, and activated T lymphocytes7–9. It is a member of the IRF family and has been shown to regulate myeloid cell development by interacting with the Ets family transcription factors10,11. Hence, Irf8–/– mice show an increased number of myeloid progenitors, defects in macrophage function including impaired IL-12 production, developmental defects in CD8α+ and plasmacytoid dendritic cells, and systemic expansion of the granulocyte population, which frequently leads to a fatal blast crisis12–14.
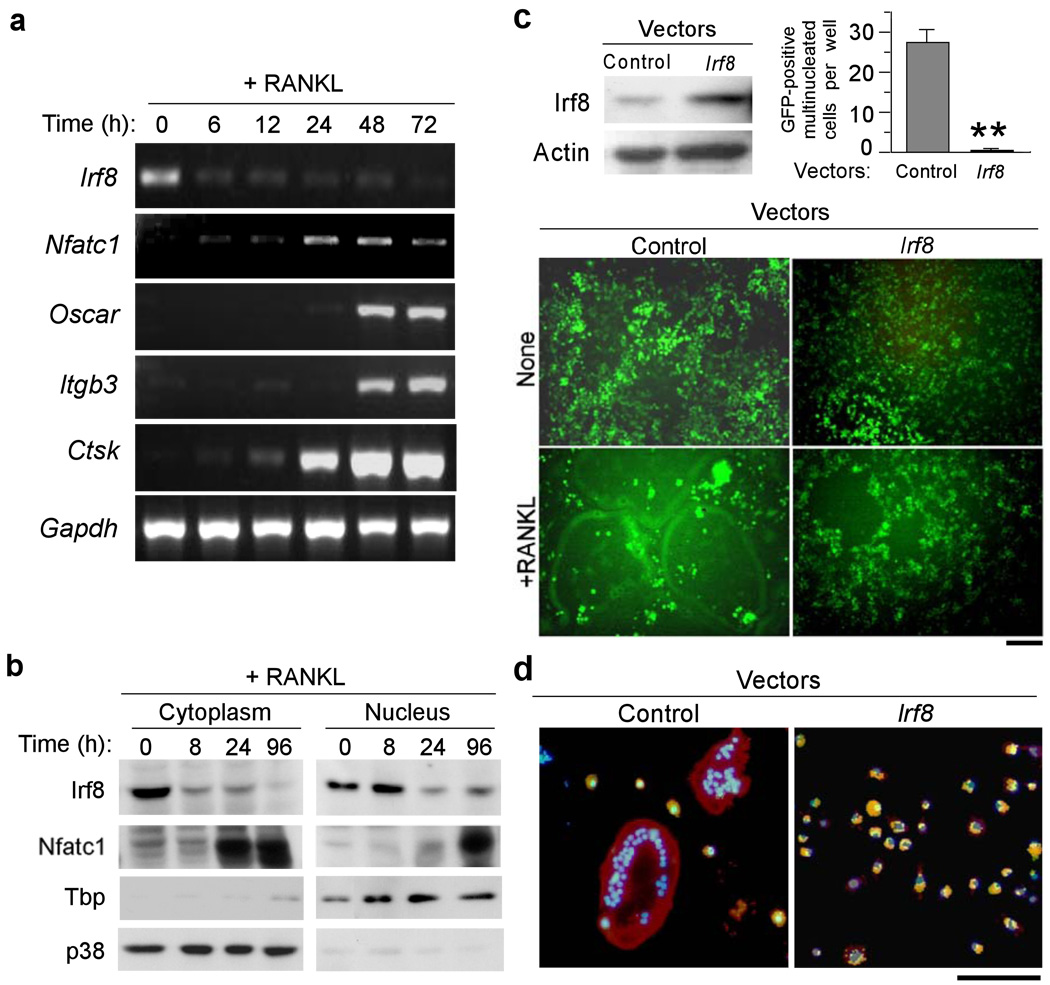
Using RT-PCR and immunoblot analysis, IRF8 expression was detected in bone marrow-derived macrophages (BMMs) and spleen-derived macrophages, which are capable of differentiating into osteoclasts (Fig. 1a,b). As previously shown3, NFATc1 was modestly expressed at a basal level in unstimulated BMMs and NFATc1 expression was strongly induced after RANKL stimulation (Fig. 1b); robust induction of NFATc1 by RANKL is a necessary and pivotal step for osteoclast differentiation characterized by enhanced expression of osteoclast marker genes such as Oscar, Itgb3 that encodes integrin β3 protein, and Ctsk3. In contrast, IRF8 expression decreased after RANKL stimulation (Fig. 1a,b), resulting in substantially lower nuclear IRF8 protein 24 h after RANKL addition (Fig. 1b). We hypothesized that IRF8 downregulation may be required for osteoclast differentiation and that IRF8 may inhibit early stages of osteoclastogenesis.
Figure 1.
IRF8 inhibits osteoclastogenesis in BMMs stimulated with M-CSF and RANKL. (a) mRNA expressions of Irf8, Nfatc1, Oscar, Itgb3, Ctsk and Gapdh in BMMs 0, 6, 12, 24, 48 and 72 h after M-CSF (50 ng/ml) and RANKL (150 ng/ml) stimulation (RT-PCR). (b) Immunoblot analysis of IRF8 and NFATc1 expressions in cytoplasmic and nuclear fractions of BMMs obtained 0, 8, 24 and 96 h after stimulation with M-CSF and RANKL. Expression levels of TBP and p38 were measured as loading controls for nuclear and cytoplasmic fractions, respectively. (c) Inhibition of GFP-positive multinucleated cell (osteoclast) formation due to retrovirus-mediated overexpression of IRF8 in BMMs. Overexpression of IRF8 protein in BMMs was confirmed by immunoblotting (top left). GFP-positive multinucleated cells were counted (top right). GFP-positive multinucleated cells appear green giant cells (bottom). Control, pMX-IRES-EGFP; Irf8, pMX-Irf8-IRES-EGFP. **P<0.01. Bar, 50 µm. (d) BMMs were transduced with pMX-puro (control) or pMX-Irf8-puro (Irf8) and stimulated with M-CSF and RANKL for 3 days. Phagocytic activity was examined using FITC-conjugated zymosan bioparticles (green). F-actin was labeled with rhodamine-conjugated phalloidin (red). Nuclei were stained with DAPI (blue). Colocalization of zymosan and actin is denoted by the yellow fluorescent signals. Bar, 100 µm.
To examine the role of IRF8 in osteoclastogenesis, we constructed a retroviral vector, pMX-Irf8-IRES-EGFP, which was engineered to express both IRF8 and enhanced green fluorescence protein (EGFP). Macrophage-like osteoclast precursors were transduced with retroviral particles encoding Irf8 or control viral particles that lacked the Irf8 sequence (pMX-IRES-EGFP). IRF8–overexpressing precursors failed to differentiate into osteoclasts in response to RANKL stimulation (Fig. 1c). Furthermore, these cells were able to phagocytize zymosan particles, whereas multinucleated osteoclasts failed to do so (Fig. 1d). These results suggest that IRF8 inhibits osteoclastogenesis from precursor cells, which instead retain the characteristics of phagocytic macrophages.
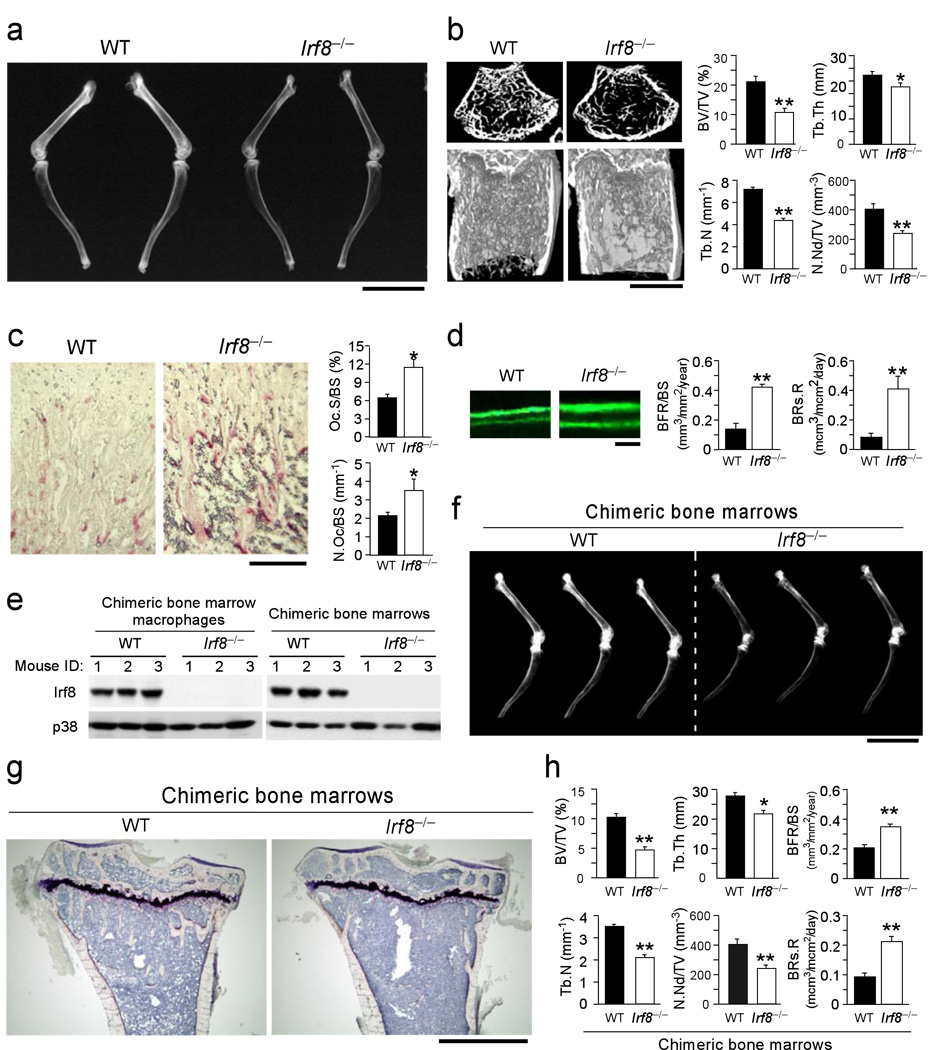
Next, we analyzed the Irf8–/– mice for abnormal bone phenotypes. Radiographic and microcomputed tomographic analyses showed that these mice had severe osteoporosis accompanied by dramatic decreases in trabecular bone volume, number, and thickness, as well as the number of bone nodules (Fig. 2a, b). Histomorphometric analysis also revealed reduced bone mass in the Irf8–/– mice (data not shown). Importantly, increases in the osteoclast number and surface area were observed in these mice (Fig. 2c). There were no significant differences between wild-type and Irf8–/– mice in the serum level of osteoprotegerin (OPG encoded by Tnfrsf11b gene), a decoy RANKL receptor that inhibits osteoclastogenesis, or in RANKL levels in primary cultured osteoblasts (Supplementary Fig. 1 online). Moreover, an increased rate of bone formation accompanying the accelerated bone resorption rate was observed in the Irf8–/– mice, suggesting that their osteoporosis was caused by enhanced bone turnover and remodeling (Fig. 2d). Together, these observations indicated that IRF8 plays a suppressive role in osteoclastogenesis during in vivo bone remodeling. To address whether the bone phenotype could be explained by Irf8 deficiency in osteoclast precursors, or whether a cell autonomous effect of Irf8 deficiency in osteoblasts or bone marrow stromal cells could play a role, we established chimeric mouse models in which either wild-type or Irf8–/– littermate bone marrow was transplanted into lethally irradiated wild-type recipients. Consistent with the global Irf8 deficient mice, Irf8–/– bone marrow chimeric mice showed severe high turnover osteoporosis (Fig. 2e–h). IRF8 is mainly expressed in hematopoietic cells7–9. Indeed, IRF8 expression was not detected in primary calvarial osteoblasts (data not shown). Furthermore, primary Irf8–/– osteoblast differentiation and matrix calcification were not affected compared to wild-type cells (Supplementary Fig. 2 online). Thus, the osteoporosis resulted from enhanced osteoclastogenesis, which is a cell autonomous consequence of Irf8 deficiency in osteoclast precursors but not osteoblasts.
Figure 2.
Irf8−/− mice exhibit severe osteoporosis due to enhanced osteoclast formation. (a) Radiographic analysis of the femur and tibia. Bar, 1 cm. (b) Microcomputed tomography of the femurs of 8-week-old wild-type and Irf8−/− mice (left), and bone morphometric analysis of femurs isolated from8-week-old mice (n = 6 in each group) (right). BV/TV, bone volume per tissue volume; Tb.Th, trabecular bone thickness; Tb.N, trabecular number; N.Nd/TV, number of nodules per tissue volume. **P<0.01; *P<0.05. Bar, 500 µm (c) Histology of femurs from 8-week-old mice in which osteoclasts were stained using TRAP, an enzymatic marker of osteoclasts (left), and histomorphometric analysis of tibias from 8-week-old mice (n=5 in each group) (right). Oc.S/BS, osteoclast surface per bone surface; N.Oc/BS, number of osteoclasts per bone surface. *P<0.05. Bar, 100 µm. (d) Histological photographs of bone formation that show tetracycline-calcein double labeling, which were administered with an interval of 72 h (left), and histomorphometric analysis of the bone formation rate and bone resorption rate in 8-week-old mice (right). BFR/BS, bone formation rate per bone surface; BRs.R, bone resorption rate. **P<0.01. (e) Analysis of IRF8 protein levels in BMs or M-CSF induced BMMs from recipient chimeric mice. (f) Soft-X ray photographs of long bones (tibias and femurs) isolated from chimeric mice. (g) Histological analysis of tibias isolated from chimeric mice (Villanueva bone staining). Bar, 500 µm. (h) Bone morphometric analysis of femurs isolated from chimeric mice. Representative data from one of two independent experiments is shown [n=3 (WT) and 3 (Irf8−/−) in each experiment]. Data are expressed as the mean+SD (n=3). **P<0.01; *P<0.05.
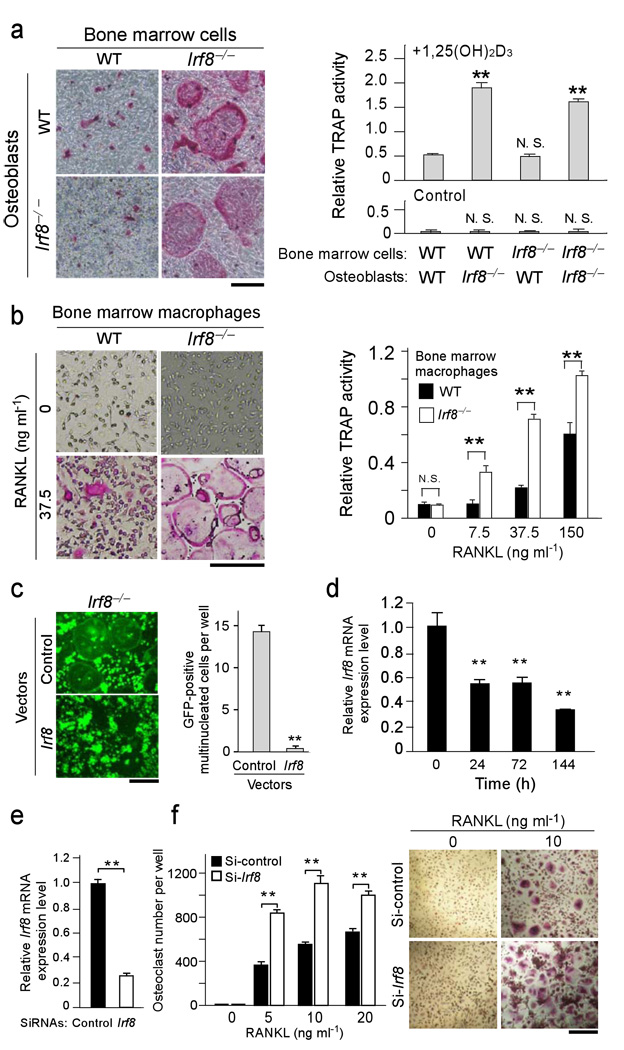
On the basis of these results, we examined the differentiation potential of osteoclast precursors obtained from Irf8–/– mice. When wild-type or Irf8–/– osteoclast precursors were cocultured with wild-type or Irf8–/– primary osteoblasts in the presence of active vitamin D3 (an inducer of RANKL expression in osteoblasts), a greater number of osteoclasts formed from the Irf8–/– precursors than from the wild-type cells, independent of whether the osteoblasts were from Irf8–/– or wild-type mice (Fig. 3a). These data further support the notion that there is no difference in osteoclastogenic activity between wild-type and Irf8–/– osteoblasts, and that Irf8 deficiency in osteoclast precursors plays a decisive role in promoting osteoclast differentiation. Indeed, macrophage cultures prepared from Irf8–/– mice also exhibited augmented osteoclastogenesis in the presence of RANKL and M-CSF (Fig. 3b), which led to an increase in resorption on dentin slices in vitro (data not shown). On the other hand, retrovirus-mediated reconstitution of IRF8 expression in Irf8–/– macrophages inhibited RANKL-induced osteoclastogenesis (Fig. 3c). These results demonstrate that IRF8 in osteoclast precursors is involved in the inhibition of osteoclastogenesis in vitro and in vivo. Furthermore, we found that IRF8 expression was also decreased during RANKL-induced human osteoclastogenesis, although with slower kinetics (Fig. 3d). Silencing of Irf8 mRNA in human osteoclast precursors by siRNAs resulted in enhanced osteoclast differentiation (Fig. 3e–f), indicating the function of IRF8 in osteoclastogenesis is well conserved in humans and mice.
Figure 3.
IRF8 deficiency or RNAi-mediated silencing in osteoclast precursors leads to enhanced osteoclast formation. (a) Primary calvarial osteoblasts and bone marrow cells obtained from wild-type and Irf8−/− mice were cocultured in the presence of 108 M 1,25(OH)2D3 and 10−6 M prostaglandin E2 (inducers of RANKL expression in osteoblasts) for 6 days. TRAP staining (left) and TRAP activity (right) of cultures are shown. Bar, 50 µm. (b) Osteoclast formation induced by 50 ng/ml M-CSF and the indicated doses of RANKL in BMM cultures. TRAP staining (left) and TRAP activity (right) of cultures are shown. Bar, 50 µm. (c) Irf8−/− macrophages were transduced with the vectors such as pMX-Irf8-IRES-EGFP (Irf8) or pMX-IRES-EGFP (Control) and stimulated with 150 ng/ml RANKL for 3 days (left). GFP-positive multinucleated cells were counted as osteoclasts (right). **P<0.01. Bar, 50 µm. (d) Kinetics of Irf8 mRNA expression during human osteoclastogenesis induced by 40 ng/ml RANKL at indicated time points. (e) Human CD14-positive monocytic cells were transfected with human Irf8-specific short interfering RNAs (si-hIrf8) or non-targeting control siRNAs (si-control), cultured for 2 days in the presence of 20 ng/ml M-CSF, and efficiency of silencing of Irf8 mRNA was examined by quantitative real time-PCR. (f) The cells were further stimulated with indicated concentrations of RANKL for 6 days. Number of TRAP-positive multinucleated cells was counted as osteoclasts (left). TRAP-positive cells appear red in the photograph (right). **P<0.01. Representative data from one of three donors is shown; similar results were obtained using a distinct Irf8-specific siRNA in an additional three experiments. Data are expressed as the mean+SD of quad-duplicate cultures. **P<0.01; n.s., no statistical difference. Bar, 50 µm.
Consistent with the results regarding osteoclastogenesis, the mRNA expression profiles of various osteoclast markers were more strongly upregulated by RANKL stimulation in macrophages prepared from Irf8–/– mice compared with those obtained from wild-type mice (Supplementary Fig. 3 online). However, Rank and c-fms mRNA expression and RANK cell surface protein expression were comparable in wild-type and Irf8–/– osteoclast precursors (Supplementary Fig. 4a online and data not shown). During osteoclast precursor generation from bone marrow cells, M-CSF treatment did not lead to a greater increase in Rank mRNA expression in Irf8–/– cells than in control cells (Supplementary Fig. 4b online). These results suggest that enhanced osteoclastogenesis in Irf8–/– macrophages is not due to changes in these receptor levels.
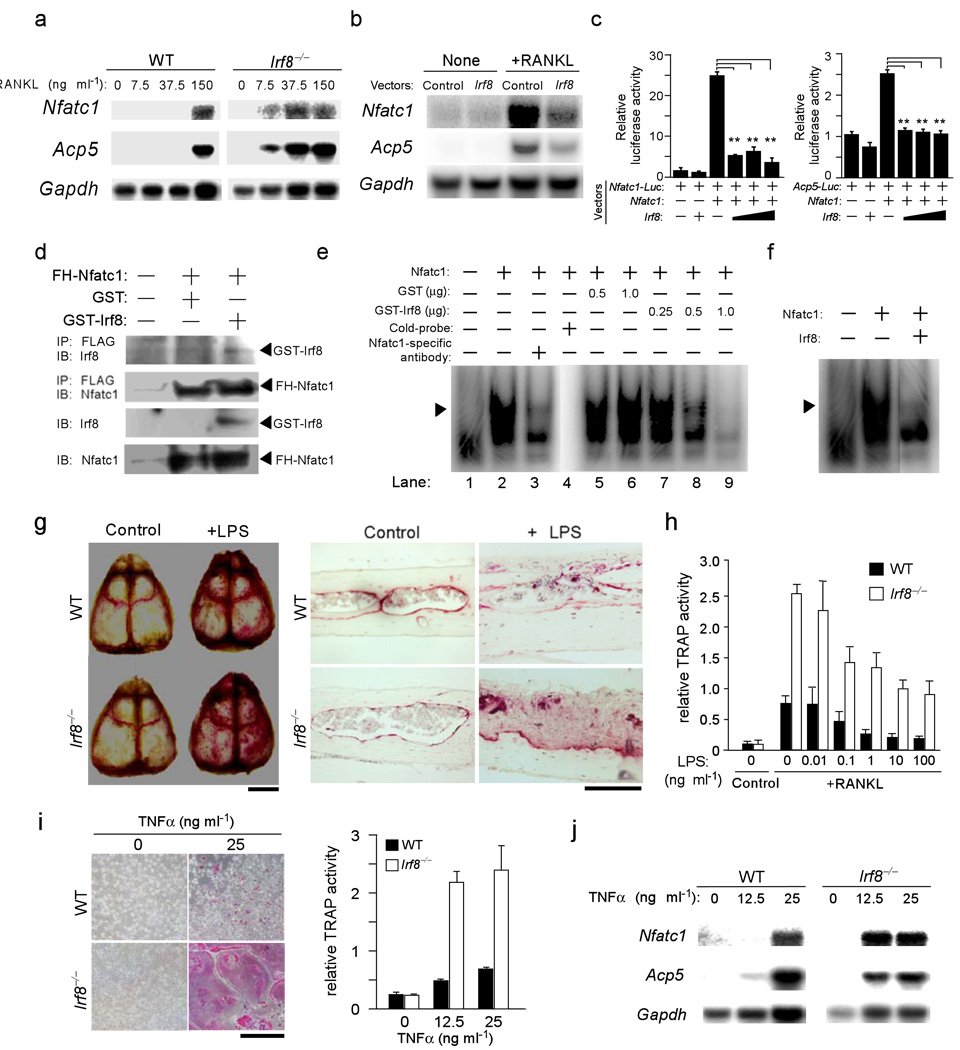
Expression of Nfatc1 gene and the osteoclast marker Acp5 gene that encodes tartrate-resistant acid phosphatase (TRAP) protein in Irf8–/– precursors was induced by concentrations of RANKL that were 5%–25% of those required for the expression of these genes in wild-type precursors (Fig. 4a). Furthermore, overexpression of IRF8 in the precursor cells repressed the RANKL-induced expression of Nfatc1 and Acp5 mRNAs (Fig. 4b).
Figure 4.
IRF8 inhibits NFATc1 transcriptional activity and expression, and reduced IRF8 expression may contribute to pathological bone destruction. (a) Expression of Nfatc1 and Acp5 mRNAs induced by 24 h stimulation of BMMs derived from wild-type and Irf8−/− mice with 50 ng/ml M-CSF and the indicated doses of RANKL (Northern blotting). (b) Expression of Nfatc1 and Acp5 mRNAs in retrovirus-infected BMMs (Control, pMX-puro; Irf8, pMX-Irf8-puro) in the absence or presence of 150 ng/ml RANKL with 50 ng/ml M-CSF for 24 h (Northern blotting). Gapdh was used as an internal control. (c) A luciferase activity assay to examine the effect of IRF8 on the transcriptional activity of NFATc1. **P<0.01. (d) The interaction between NFATc1 and IRF8. IP, immunoprecipitation; IB, immunoblotting. (e,f) Analysis of the inhibition of NFATc1 binding to its target DNA sequences using EMSAs (see supplementary methods on line). The arrow heads indicate a specific binding complex, which included the NFATc1–DNA complex; this was confirmed in a supershift assay (lane 1–3) and a cold competition experiment (lane 4). The intensity of NFATc1–DNA bands decreased with increasing amounts of GST-IRF8 (lanes 7–9), or when nuclear extracts from HEK293 cells cotransfected with pcDNA3-Nfatc1 and pcDNA3-Irf8 were used (f), but not with GST alone (lanes 5–6). (g) TRAP staining of mouse whole calvaria (left) and histological sections (right) obtained from wild-type and Irf8−/− mice with or without administration of LPS. (h) Osteoclast formation in BMMs stimulated with various doses of LPS in the presence of M-CSF (50 ng/ml) and RANKL (150 ng/ml) for 5 days. Data are expressed as the mean+SD of 4 cultures. (i) Left, BMM cultures in the presence of 50 ng/ml M-CSF and 0 or 25 ng/ml TNFα on day 3 shown by TRAP staining. Right, TNFα dose-dependent induction of osteoclast formation examined using TRAP activity assays. (j) Expression of Nfatc1 and Acp5 mRNAs in BMMs derived from wild-type and Irf8−/− mice stimulated with 50 ng/ml M-CSF and the indicated doses of TNFα for 24 h (Northern blotting).
These observations led us to examine the effect of IRF8 on the transcriptional activity of NFATc1, which was previously reported to interact with IRF815. For these experiments, we employed luciferase reporter plasmids driven by three copies of the NFATc1 binding site from the human IL-2 distal promoter (p3x Nfatc1-Luc) or by the mouse Acp5 promoter (pAcp5-Luc). Overexpression of Nfatc1 gene activated these promoters, whereas simultaneous expression of Irf8 gene reduced the activities of the promoters to control levels (Fig. 4c), indicating that IRF8 inhibits the transcriptional activity of NFATc1. When GST-IRF8 proteins were incubated with nuclear lysates containing FLAG-hemagglutinin-tagged NFATc1 (FH-NFATc1), anti-FLAG antibodies resulted in the coimmunoprecipitation of GST-IRF8 and FH-NFATc1, suggesting that IRF8 physically interacts with NFATc1 (Fig. 4d). Furthermore, association of endogenous IRF8 and NFATc1 was identified by co-immunoprecipitation from nuclear extracts of human monocytic cells (Supplementary Fig. 5 online). We further examined the effects of IRF8 on the binding of NFATc1 to its target DNA elements in an electrophoretic mobility shift assay (EMSA); NFATc1-DNA complexes were detected, which was confirmed by the addition of competitive probes or anti-NFATc1 antibodies. Increased levels of GST-IRF8 or nuclear lysates containing excess IRF8, however, significantly decreased the binding of NFATc1 to the probes (Fig. 4e, f), demonstrating the inhibitory effect of IRF8 on NFATc1 binding to its target DNA elements.
We then attempted to examine the roles of IRF8 in the processes underlying pathological bone destruction. Because several members of the IRF family of transcription factors, including IRF8, have been demonstrated to play crucial roles in toll-like receptor (TLR) signaling in response to such microbial components as LPS and unmethylated CpG DNA16,17, we examined the role of IRF8 in the bone destruction observed during TLR-mediated inflammation. Administration of LPS to the calvarial periosteum resulted in enhanced osteoclast formation in wild-type mice, whereas more extensive bone destruction was observed in Irf8–/– mice (Fig. 4g). These results suggest that IRF8 is a critical negative regulator of osteoclastogenesis and a mediator of the maintenance of bone integrity during inflammatory bone destruction. We also examined the effects of TLR ligands on osteoclastogenesis in wild-type and Irf8–/– osteoclast precursors. As previously reported18, LPS at high doses completely inhibited RANKL-induced osteoclastogenesis in wild-type cell cultures. In contrast, corresponding high doses of LPS, and also of the TLR3 ligand poly(I:C) and the TLR9 ligand CpG DNA, only partially inhibited osteoclast differentiation in Irf8–/– cell cultures (Fig. 4h and Supplementary Fig. 6 on line). Irf8-deficient cells were almost completely refractory to the inhibitory effects of the TLR2 ligand peptidoglycan (Supplementary Fig. 6a). These results suggest a potential inhibitory role of IRF8 in the regulation of osteoclastogenesis by TLRs, although TLRs can also activate IRF8-independent inhibitory mechanisms. We further found that Ifnα/β expression was not diminished in Irf8-deficient cells, and that IFN-γ completely inhibited osteoclastogenesis in Irf8–/– macrophages, similar to wild-type cells (data not shown), thus suggesting that IRF8 can inhibit osteoclastogenesis independently of IFN-γ.
Finally, we examined the effect of TNFα on osteoclastogenesis using Irf8–/– precursors, because TNFα is a critical mediator of inflammation induced by TLRs19–21 and has been suggested to be able to induce osteoclastogenesis22–24. Consistent with previous reports, TNFα induced the development of a small number of osteoclasts in cultures of wild-type precursor cells (Fig. 4i). Notably, osteoclastogenesis was enhanced in cultures of Irf8–/– precursor cells treated with TNFα (Fig. 4i). The mRNA expression levels of Nfatc1 and Acp5 in the Irf8–/– osteoclast precursors were also augmented by TNFα (Fig. 4j), indicating that IRF8 also plays a suppressive role in TNFα–induced osteoclastogenesis.
Our data provide a mechanism by which IRF8 suppresses osteoclastogenesis (depicted in Supplementary Fig. 7 on line). In osteoclast precursors, abundant IRF8 interacts with basally expressed NFATc1 to suppress its transcriptional activity and thus prevent its activation of target genes, including autoamplification of its own promoter. Stimulation of osteoclast precursors with RANKL results in the activation of NF-κB and AP-1 that bind to the Nfatc1 promoter to induce its activity. At the same time, RANKL induces the downregulation of IRF8, thereby releasing NFATc1 from IRF8-mediated suppression and augmenting NFATc1-mediated auto-amplification of its own expression. Together, these mechanisms result in robust NFATc1 expression and induction of downstream genes required for osteoclast differentiation.
Bone erosion that occurs in the setting of infection and chronic inflammation, termed inflammatory osteolysis, contributes to the pathogenesis of infectious and inflammatory diseases such as rheumatoid arthritis25. Inflammatory bone erosion is driven by microbial products such as LPS and inflammatory cytokines including TNFα that activate osteoclastogenesis directly or indirectly via activation of stromal cells and osteoblasts25. Our findings show that IRF8 plays an important role in attenuating LPS-induced inflammatory bone resorption and LPS- and TNFα-induced osteoclastogenesis. This homeostatic role of IRF8 may be important during acute infections and also in chronic inflammatory conditions such as rheumatoid arthritis. Identification of additional factors and mechanisms that augment IRF8 expression or function may represent a fruitful approach to therapeutic suppression of inflammatory bone erosion.
Overall, our findings support a model of RANKL-induced NFATc1 expression and osteoclast differentiation that involves cooperation between RANKL-mediated induction of positive regulators of osteoclastogenesis and suppression of negative regulators of osteoclast differentiation such as IRF8.
Methods
Mice and analysis of bone phenotypes
The Irf8–/– mice (C57BL/6) used in this study have been described previously12. Tetracycline hydrochloride (20 mg/kg; Sigma) and, 72 h later, calcein (10 mg/kg; Wako) were injected subcutaneously into 8-week-old wild-type and Irf8–/– mice. The mice were then euthanized 32 h after the second injection. The LPS-induced model of bone loss has been described previously26, except that 12.5 mg/kg LPS (Sigma) was used in the present study. The mice were subjected to histomorphometric and microradiographic examinations as described previously27. All mice were born and maintained under specific pathogen-free conditions. All animal experiments were approved by and conducted according to the guidelines of the Showa University Animal Care and Use Committee (approval number: 17079).
Generation of bone marrow chimeric mice
Donor bone marrow cells from wild-type or Irf8–/– littermate mice (on C57/BL6 background) were harvested and one-fourth of total bone marrow cells from each donor were injected intravenously via tail vein into each of the irradiated wild-type recipients. Recipient mice (3-week-old C57/BL6 mice) were obtained from Jackson Laboratory and were lethally irradiated with a single dose of 875 rads 1 day prior to transplantation. Chimeric mice were sacrificed 8 weeks after bone marrow transplantation. The experiments using chimeric mice were approved by the Hospital for Special Surgery Institutional Animal Care and Use Committee.
In vitro assays of osteoclast differentiation and macrophage function
The method used to analyze osteoclast differentiation in vitro has been described previously28. Briefly, mouse bone marrow or spleen cells were cultured with 50 ng/ml M-CSF for 3 days. The obtained BMMs or spleen-derived macrophages were further stimulated with 150 ng/ml RANKL in the presence of 50 ng/ml M-CSF for 3–4 days. We also cocultured bone marrow cells, BMMs, or spleen-derived macrophages with primary osteoblasts derived from mouse calvaria in the presence of 108 M 1,25 (OH)2D3 and 10−6 M prostaglandin E2 for 6 days. Media were changed every 2 days. Generation of osteoclasts was evaluated by TRAP staining and its activity assays28. Methods used for the phagocytosis assay and labeling of actin have been described previously28.
Human osteoclast culture system and RNAi
Human fresh peripheral blood mononuclear cells (PBMCs) were obtained from whole blood from disease-free volunteers. Human CD14+ monocytes were then purified from PBMCs with anti-CD14 magnetic beads (Miltenyi Biotec) and were cultured in α-MEM medium with 10% FBS (Hyclone) and 20 ng/ml of M-CSF (Peprotech) for 2 days to induce osteoclast precursors that were further stimulated with RANKL in the presence of 20 ng/ml of M-CSF for 3–6 days to generate human osteoclasts. The experiments using human cells were approved by the Hospital for Special Surgery Institutional Review Board. Short interfering RNAs (siRNAs) specifically targeting human Irf8 or control siRNAs (Dharmacon, Invitrogen) were transfected into primary human CD14+ monocytes with the Amaxa Nucleofector device set to program Y-001 using the Human Monocyte Nucleofector kit (Amaxa). Two different targeting siRNAs and control siRNAs were used with comparable results in a total of 6 independent experiments with different blood donors.
Plasmid constructs
Irf8 cDNA was prepared and amplified in RT-PCRs using RNA from BMMs. The coding region of Irf8 was PCR-amplified using the following primers: 5’-GCAGGATGTGTGACCGGAAC-3’ (sense) and 5’-ACTGAGGCTTAGACGGTGAT-3’ (antisense). The amplified PCR fragment was subcloned into a pCR-Blunt vector to produce pCR-Blunt-Irf8 using a Zero Blunt PCR Cloning kit (Invitrogen). The pMX-Irf8-IRES-EGFP retroviral vector was constructed by inserting a 1.3-kb EcoRI/EcoRI DNA fragment encoding Irf8 from pCR-Blunt-Irf8 into the same site in the pMX-IRES-EGFP vector. The 1.3-kb BamHI/XhoI DNA fragment encoding IRF8 from pMX-Irf8-IRES-EGFP was then inserted into the same site in the pMX-puro vector to construct the pMX-Irf8-puro retroviral vector. The pcDNA3-Irf8 expression vector was constructed by inserting a 1.3-kb BamHI/NotI DNA fragment encoding IRF8 from pMX-Irf8-IRES-EGFP into the same site in the pcDNA3 vector. pcDNA3-Nfatc1 was constructed by inserting an EcoRI/EcoRI DNA fragment encoding NFATc1 from pMX-Nfatc1-IRES-EGFP 3 into the same site in the pcDNA3 vector. The pFH-Nfatc1, p3×Nfatc-Luc, and pAcp5-Luc vectors were previously described3,29.
Statistical analysis
Statistical analysis was performed using Student t-tests (p < 0.05 was taken as statistically significant) and all data are presented as the mean ± s.e.m. Results are representative of more than four individual experiments.
Additional methods
Details of the methods including retroviral gene transduction, GeneChip, RT-PCR, Northern blot assay, Real-time PCR, Luciferase reporter assay, preparation of GST fusion proteins, immunoprecipitation and immunoblot analyses, and EMSA are described in Supplementary Methods on line.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to Dr. M. Asagiri (Tokyo Medical and Dental University) for great technical assistance. We thank Dr. T. Kitamura (University of Tokyo) for providing the retrovirus expression system. We also thank Dr. A. Mochizuki and all members of the Department of Biochemistry, School of Dentistry, Showa University for valuable discussion. This work is supported in part by High-Tech Research Center Project for Private Universities from the Ministry of Education, Culture, Sports, Science and Technology, Japan, 2005–2009, and by Grants-in-Aid for Scientific Research from the Japan Society for the Promotion of Science (20390474 to M.T.), and by grants from the NIH (AR053843 and DE19381 to Y.C., and DE019420 and AR46713 to L.B.I.).
Footnotes
COMPETING INTERESTS STATEMENT
The authors declare that they have no competing financial interests.
REFERENCES
- 1.Wada T, Nakashima T, Hiroshi N, Penninger JM. RANKL-RANK signaling in osteoclastogenesis and bone disease. Trends Mol Med. 2006;12:17–25. doi: 10.1016/j.molmed.2005.11.007. [DOI] [PubMed] [Google Scholar]
- 2.Lee SK, Lorenzo J. Cytokines regulating osteoclast formation and function. Curr Opin Rheumatol. 2006;18:411–418. doi: 10.1097/01.bor.0000231911.42666.78. [DOI] [PubMed] [Google Scholar]
- 3.Takayanagi H, et al. Induction and activation of the transcription factor NFATc1 (NFAT2) integrate RANKL signaling in terminal differentiation of osteoclasts. Dev Cell. 2002;3:889–901. doi: 10.1016/s1534-5807(02)00369-6. [DOI] [PubMed] [Google Scholar]
- 4.Grigoriadis AE, et al. c-Fos: a key regulator of osteoclast-macrophage lineage determination and bone remodeling. Science. 1994;266:443–448. doi: 10.1126/science.7939685. [DOI] [PubMed] [Google Scholar]
- 5.Kim N, Takami M, Rho J, Josien R, Choi Y. A novel member of the leukocyte receptor complex regulates osteoclast differentiation. J Exp Med. 2002;195:201–219. doi: 10.1084/jem.20011681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ishida N, et al. Large scale gene expression analysis of osteoclastogenesis in vitro and elucidation of NFAT2 as a key regulator. J Biol Chem. 2002;277:41147–41156. doi: 10.1074/jbc.M205063200. [DOI] [PubMed] [Google Scholar]
- 7.Kantakamalakul W, et al. Regulation of IFN consensus sequence binding protein expression in murine macrophages. J Immunol. 1999;162:7417–7425. [PubMed] [Google Scholar]
- 8.Nelson N, et al. Expression of IFN regulatory factor family proteins in lymphocytes. Induction of Stat-1 and IFN consensus sequence binding protein expression by T cell activation. J Immunol. 1996;156:3711–3720. [PubMed] [Google Scholar]
- 9.Driggers PH, et al. An interferon gamma-regulated protein that binds the interferon-inducible enhancer element of major histocompatibility complex class I genes. Proc Natl Acad Sci U S A. 1990;87:3743–3747. doi: 10.1073/pnas.87.10.3743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tamura T, Ozato K. ICSBP/IRF-8: its regulatory roles in the development of myeloid cells. J Interferon Cytokine Res. 2002;22:145–152. doi: 10.1089/107999002753452755. [DOI] [PubMed] [Google Scholar]
- 11.Marecki S, Fenton MJ. PU.1/Interferon Regulatory Factor interactions: mechanisms of transcriptional regulation. Cell Biochem Biophys. 2000;33:127–148. doi: 10.1385/CBB:33:2:127. [DOI] [PubMed] [Google Scholar]
- 12.Holtschke T, et al. Immunodeficiency and chronic myelogenous leukemia-like syndrome in mice with a targeted mutation of the ICSBP gene. Cell. 1996;87:307–317. doi: 10.1016/s0092-8674(00)81348-3. [DOI] [PubMed] [Google Scholar]
- 13.Tamura T, Nagamura-Inoue T, Shmeltzer Z, Kuwata T, Ozato K. ICSBP directs bipotential myeloid progenitor cells to differentiate into mature macrophages. Immunity. 2000;13:155–165. doi: 10.1016/s1074-7613(00)00016-9. [DOI] [PubMed] [Google Scholar]
- 14.Schiavoni G, et al. ICSBP is essential for the development of mouse type I interferon-producing cells and for the generation and activation of CD8alpha(+) dendritic cells. J Exp Med. 2002;196:1415–1425. doi: 10.1084/jem.20021263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhu C, et al. Activation of the murine interleukin-12 p40 promoter by functional interactions between NFAT and ICSBP. J Biol Chem. 2003;278:39372–39382. doi: 10.1074/jbc.M306441200. [DOI] [PubMed] [Google Scholar]
- 16.Tsujimura H, et al. Toll-like receptor 9 signaling activates NF-kappaB through IFN regulatory factor-8/IFN consensus sequence binding protein in dendritic cells. J Immunol. 2004;172:6820–6827. doi: 10.4049/jimmunol.172.11.6820. [DOI] [PubMed] [Google Scholar]
- 17.Zhao J, et al. IRF-8/interferon (IFN) consensus sequence-binding protein is involved in Toll-like receptor (TLR) signaling and contributes to the cross-talk between TLR and IFN-gamma signaling pathways. J Biol Chem. 2006;281:10073–10080. doi: 10.1074/jbc.M507788200. [DOI] [PubMed] [Google Scholar]
- 18.Takami M, Kim N, Rho J, Choi Y. Stimulation by toll-like receptors inhibits osteoclast differentiation. J Immunol. 2002;169:1516–1523. doi: 10.4049/jimmunol.169.3.1516. [DOI] [PubMed] [Google Scholar]
- 19.O'Neill LA. How Toll-like receptors signal: what we know and what we don't know. Curr Opin Immunol. 2006;18:3–9. doi: 10.1016/j.coi.2005.11.012. [DOI] [PubMed] [Google Scholar]
- 20.Akira S, Uematsu S, Takeuchi O. Pathogen recognition and innate immunity. Cell. 2006;124:783–801. doi: 10.1016/j.cell.2006.02.015. [DOI] [PubMed] [Google Scholar]
- 21.Takaoka A, et al. Integral role of IRF-5 in the gene induction programme activated by Toll-like receptors. Nature. 2005;434:243–249. doi: 10.1038/nature03308. [DOI] [PubMed] [Google Scholar]
- 22.Azuma Y, Kaji K, Katogi R, Takeshita S, Kudo A. Tumor necrosis factor-alpha induces differentiation of and bone resorption by osteoclasts. J Biol Chem. 2000;275:4858–4864. doi: 10.1074/jbc.275.7.4858. [DOI] [PubMed] [Google Scholar]
- 23.Kobayashi K, et al. Tumor necrosis factor alpha stimulates osteoclast differentiation by a mechanism independent of the ODF/RANKL-RANK interaction. J Exp Med. 2000;191:275–286. doi: 10.1084/jem.191.2.275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim N, et al. Osteoclast differentiation independent of the TRANCE-RANK-TRAF6 axis. J Exp Med. 2005;202:589–595. doi: 10.1084/jem.20050978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lorenzo J, Horowitz M, Choi Y. Osteoimmunology: interactions of the bone and immune system. Endocr Rev. 2008;29:403–440. doi: 10.1210/er.2007-0038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Takayanagi H, et al. T-cell-mediated regulation of osteoclastogenesis by signalling cross-talk between RANKL and IFN-gamma. Nature. 2000;408:600–605. doi: 10.1038/35046102. [DOI] [PubMed] [Google Scholar]
- 27.Takayanagi H, et al. RANKL maintains bone homeostasis through c-Fos-dependent induction of interferon-beta. Nature. 2002;416:744–749. doi: 10.1038/416744a. [DOI] [PubMed] [Google Scholar]
- 28.Mochizuki A, et al. Identification and characterization of the precursors committed to osteoclasts induced by TNF-related activation-induced cytokine/receptor activator of NF-kappa B ligand. J Immunol. 2006;177:4360–4368. doi: 10.4049/jimmunol.177.7.4360. [DOI] [PubMed] [Google Scholar]
- 29.Koga T, et al. NFAT and Osterix cooperatively regulate bone formation. Nat Med. 2005;11:880–885. doi: 10.1038/nm1270. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.