Abstract
Many tRNA molecules that recognize the purine-ending codons but not the pyrimidine-ending codons have a modified uridine at the wobble position, in which a methylene carbon is attached directly to position 5 of the uracil ring. Although several models have been proposed concerning the mechanism by which the 5-substituents regulate codon-reading properties of the tRNAs, none could explain recent results of the experiments utilizing well-characterized modification-deficient strains of Escherichia coli. Here, we first summarize previous studies on the codon-reading properties of tRNA molecules with a U derivative at the wobble position. Then, we propose a hypothetical mechanism of the reading of the G-ending codons by such tRNA molecules that could explain the experimental results. The hypothesis supposes unconventional base pairs between a protonated form of the modified uridines and the G at the third position of the codon stabilized by two direct hydrogen bonds between the bases. The hypothesis also addresses differences between the prokaryotic and eukaryotic decoding systems.
INTRODUCTION
During protein biosynthesis, the ribosomes select correct aminoacyl-tRNA molecules one-by-one by recognizing the anticodon triplet of the tRNA molecule that fits to the A-site codon triplet. According to the wobble hypothesis (1), when a tRNA molecule is recognized as a correct one, the third and second nucleosides of the anticodon (positions 36 and 35, respectively) form Watson–Crick base pairs with the first and second nucleosides of the codon, respectively, and the nucleoside at the first position of the anticodon (position 34) forms a Watson–Crick or a wobble base pair with the nucleoside at the third position of the codon (position III) (1). The base pairs allowed between position 34 and position III were assumed to be only those that could form two or more than two direct hydrogen bonds between them with a small displacement from the position for the Watson–Crick base pair. A U-G pair with the U at position 34 could be formed with a small displacement of the uracil base toward the major groove side, while it was known at that time that uridines at position 34 are quite often post-transcriptionally modified.
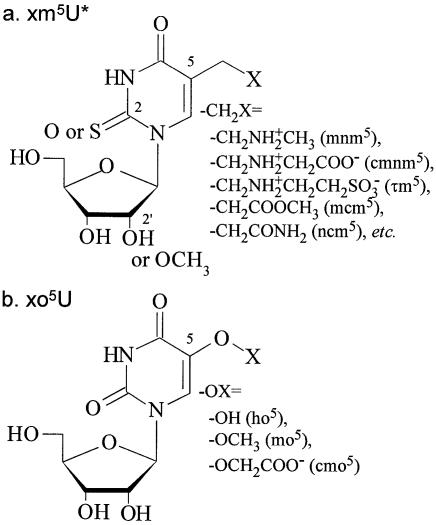
Modified uridines found at position 34 of naturally occurring tRNA species are classified into two groups (Fig. 1) (2). A modified uridine with a methylene carbon directly bonded to the C5 atom (xm5U) (Fig. 1a) is often found in tRNA species that recognize only the purine-ending codons. The xm5U nucleosides are often thiolated at position 2 (xm5s2U) or methylated at the ribose 2′-hydroxyl group (xm5Um). A modified uridine with an oxygen atom directly bonded to the C5 atom of the uracil ring (xo5U) (Fig. 1b) is often found in tRNA species that recognize the U-, A- and G-ending codons.
Figure 1.
Chemical structures of modified uridines found at position 34 of tRNA species. Symbols of the 5-substituents are shown in parentheses.
In the present paper, we use an asterisk to represent any substituent. For example, U* represents any of the naturally occurring modified and unmodified uridines. In the same way, xm5U* stands for any of the xm5U derivatives including xm5U, xm5s2U and xm5Um. Parts of the nucleoside symbols, such as xm5 and s2, may also be used to stand for the substituents, as in Figure 1. 5′-Nucleotides may be symbolized such as pxo5U. The position of a codon nucleoside may be shown in parentheses in Roman numerals, such as G(III), and the position of the anticodon nucleoside may be shown in the same way in Arabic numerals, such as U(34).
The puckering equilibrium of the ribose ring of the nucleosides in RNA molecules is generally biased to the C3′-endo form instead of the C2′-endo form, which is required for the formation of the typical A-form helices. This bias is also observed in mononucleotides and nucleosides. In xm5U*, the ribose puckering is biased to the C3′-endo conformation to a greater extent than in unmodified U (3–6). On the other hand, the puckering equilibrium of pxo5U is much shifted to the C2′-endo form (3). It was also shown that a U in the C2′-endo conformation could basepair with another U through two direct base–base hydrogen bonds by a model building study (3). Therefore, it was proposed that the modifications in xm5U* restricts, and the xo5 modification promotes, the formation of the U*(34)–U(III) pair (3). It has been shown that the substitution of U(34) of the unmodified form of Escherichia coli tRNA1Ser by mo5U(34) enhances the in vitro reading of the UCU codon (7).
It is noteworthy that this theory of the regulation of codon recognition at the level of the dynamic conformation of the nucleotides is based on the wobble hypothesis proposed by Crick (1): two direct hydrogen bonds are required between the bases at positions 34 and III. A mechanism that does not require the two direct base–base hydrogen bonds has also been proposed to contribute to the codon reading and is named as the ‘two out of three’ mechanism (8,9) (see below).
Recently, the physicochemical effects of the mnm5 and s2 modifications were elucidated in detail by NMR structural analyses of anticodon stem–loop (ASL) oligonucleotides from E.coli tRNALys (10). The comparison of different ASLs with different modifications showed that the s2 modification enhances the stacking of the bases in positions 35 and 36 onto the 3′ side of the anticodon to elevate the interaction of these bases with the first and second bases of the codon. The mnm5 modification also reduces the flexibility of the anticodon and contributes to ‘preorganize’ the anticodon into an A-form structure ready to interact with the codon in collaboration with the s2 modification. This clearly explained the effects of each modification on the misreading of the AAU/C codons by the tRNA observed in vivo under an Asn starvation condition (11), with the assumption that the misreading primarily depends on the ‘two out of three’ mechanism. Therefore, in this case, the ‘two out of three’ mechanism may dominate over the wobble mechanism. It is reasonable that, in such cases, the decoding properties of the modification-deficient tRNAs could not be predicted from the conformational properties of the nucleotide at position 34.
On the other hand, the in vivo effects of the lack of each modification on the reading of the GAA/G codons by E.coli tRNAGlu with an mnm5s2U at position 34 were also measured with the modification mutants (12). However, the data could not be explained completely even with the dynamic 3D structures of the ASLs (10), as described in detail below. This may mean that some unknown mechanism, different from the conformational regulation, has some contribution to the codon reading by the tRNAs with mnm5s2U(34).
In the present paper, we first summarize the known experimental results and theories on the effects of the xm5 modification and on other related subjects. Then, we propose a physicochemical model of xm5U(34)–G(III) pairing that could explain the in vivo effects of the mnm5 and s2 modifications on the reading of the purine-ending codons.
EXPERIMENTAL FACTS AND THEORIES
Views from tRNA composition
Distribution and properties of xm5U*(34). In E.coli, mnm5s2U(34) is found in tRNALys, tRNAGlu and one of the two tRNAsGln (13). tRNA4Leu and tRNA4Arg have cmnm5Um(34) and mnm5U(34), respectively (6,14). Many eubacteria also have the cmnm5 or mnm5 modification in Leu (UUA/G), Gln, Lys, Glu and Arg (AGA/G) tRNA species (13). At least E.coli tRNALys, tRNAGlu and tRNA4Leu could read the G-ending codons: tRNALys and tRNAGlu are the single tRNA species for the amino acids (15), and a su6 strain in which only tRNA4Leu could read the UUG codon grows very well (6,16). tRNA4Arg was suggested to read the AGG codon only weakly (17), although the experimental results are not very conclusive: overproduction of tRNA4Arg might have caused undermodification, which might have led to overproduction of tRNA molecules that do not recognize the AGG codon; and the tRNA species that compete with tRNA4Arg in their frameshifting assay were different between the assay of the AGA and AGG codons, which made direct comparison of the activities on the different codons difficult. Instead, the above in vivo experiment clearly showed that the tRNAGlu mutant with mnm5U(34) could read the GAG codon efficiently (12). Human mitochondrial tRNALeu (UUA/G) and tRNALys are also the single tRNAs for the codons and have τm5U(34) and τm5s2U(34), respectively (18).
Difference in prokaryotic and eukaryotic systems. The substituents in the xm5U derivatives in prokaryotes and eukaryotes are different. Prokaryotic tRNAs have derivatives of mnm5U, and eukaryotic tRNAs have those of mcm5U or ncm5U (13). Although many bacteria dispense with some tRNAs with C(34) that would read the CAG, AAG or GAG codon, all eukaryotes so far investigated have the C(34)-containing tRNAs for these codons (19). Therefore, it is possible that the eukaryotic xm5U derivatives do not pair with G(III). It has also been suggested that eukaryotes cannot decode G-ending codons with tRNAs having a U derivative at position 34, based on the fact that they have at least a copy of an Ile tRNA gene with a T at the first position of the anticodon (20), although the post-transcriptional modifications of the Us are unknown (the tRNA would insert Ile for the Met codon if it could read the G-ending codon). This difference between prokaryotes and eukaryotes could be ascribed to the difference in the ribosomes as well as the difference in the tRNA modifications. As for prokaryotes, we focus on the modifications in eubacteria, as information on archaebacterial modifications is limited.
Undiscriminating reading by mitochondria and mycoplasma tRNA species with U(34). In mitochondria and mycoplasmas, many family codon boxes (a codon box is a set of four different codons that have the first two bases in common, and if it specifies a single amino acid in the genetic code, it is a family box) are each translated by only one tRNA species with unmodified U(34) (21–24). Therefore, it was proposed that this kind of undiscriminating codon reading is based on the ‘two out of three’ mechanism. In some cases, this undiscriminating codon reading was shown to be less significant in the split codon boxes (a split codon box is a codon box that specifies more than one amino acid) than in family boxes (8,9,25).
Unmodified U
In vitro translation assay and properties of undiscriminating tRNA species with unmodified U(34). It is important to understand the codon-reading properties of tRNA species with an unmodified U(34) before discussing the properties of the modified species. Most studies on such tRNA molecules utilize an in vitro translation system. It is well known that the discrimination of the third bases of the codons will be ambiguous if only one aminoacyl-tRNA species is used in excess to introduce radiolabeled amino acid into proteins (26). This could be so even in a split codon box (27). It is also known that the accuracy of in vitro translation is affected by the reaction conditions. For example, pH and the concentrations of magnesium ions and polyamines could affect the fidelity of translation (28). Therefore, it is necessary to control the experimental conditions carefully. However, such assays have been used successfully to determine the relative efficiency of a tRNA species in reading a codon as compared with that of another competing tRNA species.
tRNAGly from Mycoplasma mycoides with U(34) is a single tRNA species for the four Gly codons (29). This tRNA reads all the four Gly codons even in an in vitro translation system from E.coli. It has also been demonstrated that this undiscriminating codon reading requires a C at the first position of the anticodon loop (position 32) of the tRNA (30–33). However, C(32) is very often found in tRNA molecules. Therefore, it also seems essential that the interaction of the ‘two out of three’ is strong enough to support such ambiguous reading (25). These results also suggested that tRNAs with U(34) behave differently in different situations concerning the discrimination of the third bases of the codons.
On the other hand, Mycoplasma capricolum has two Thr tRNAs with A(34) and U(34) (24). Although the tRNA with U(34) reads all the four Thr codons, the reading of the ACC codon is weak when the tRNA with A(34) is competing in an in vitro translation system from M.capricolum (34). The readings of the ACU and ACG codons by the tRNA with U(34) are also weaker than those by the tRNA with A(34). Thus, codon preferences could be observed in this case. Therefore, some interaction between U(34) and the third base of the codon contributes to the undiscriminating reading.
As it seems that there is some confusion in some of the literature concerning the use of the terms ‘wobble mechanism’ and ‘two out of three’ mechanism, we define the meanings within this paper as follows. The wobble mechanism is a class of those mechanisms by which two or more direct hydrogen bonds are formed between the bases at positions 34 and III while two Watson–Crick base pairs are formed between the last two positions of the anticodon and the first two positions of the codon. The ‘two out of three’ mechanism is a class of those mechanisms by which less than two direct base–base hydrogen bonds are formed between positions 34 and III while two Watson–Crick base pairs are formed at the other two positions. A ‘two out of three’ mechanism is one that satisfies the criteria for this particular mechanism. Therefore, a ‘two out of three’ mechanism may involve some interaction without two direct base–base hydrogen bonds between positions 34 and III. A ‘base pair’ in this paper means a pair of bases with two or more than two direct hydrogen bonds between them, unless mentioned otherwise. Some researchers use the term ‘four-way wobble’. However, we do not use this term because it is for the mechanism of the ambiguous recognition of the four different codons but is not for the mechanism of recognition of individual codons.
Unexpected inefficiency in the reading of G-ending codons by unmodified tRNAs with U(34). Artificial unmodified tRNAs with U(34) have also been investigated with an in vitro translation system. The in vitro transcript of E.coli tRNA1Ser reads the UCA codon half as efficiently as the fully modified molecules, but does not read the UCU and UCG codons (the UCU codon may be recognized weakly) (35). A sample of the same tRNA prepared by chemical synthesis followed by enzymatic ligation had the same codon-reading properties, and the substitution of the U(34) by an mo5U enhanced the reading of the UCU and UCG codons (7). The transcripts with base changes at the second and third positions of the anticodon into AA, UC and CU, respectively, also read the A-ending codons and did not read the G-ending codons (36). Thus, the U(34)–G(III) interaction is very weak at least with the structural context of the tRNA. As all of these unmodified tRNAs discriminate well the UCX codons, irrespective of C(32) that the tRNAs have, it is unlikely that any of the codon readings depends on a ‘two out of three’ mechanism. Therefore, U(34), which primarily assumes the C3′-endo form, cannot form a base pair with G(III). Something other than the ribose puckering equilibrium should be different between U(34) and xm5U*(34).
In vitro analyses of the effects of the xm5U* modifications
A-site binding of ASL. Oligonucleotide-dependent ribosome-binding experiments have been used most conveniently for the determination of codon-specificity of tRNAs. However, researchers should choose an experimental condition that gives reasonable results, as some tRNAs bind strongly and the others bind weakly. Nevertheless, many researchers have obtained reasonable results. Recently, 17mer ASL oligonucleotides were found to bind to the ribosomes. Although they mainly bind to the P site in the original techniques, the binding to the A site could also be measured as the tetracycline-sensitive binding (37). With this technique, the effects of the mnm5s2U modifications were investigated (37). The results clearly showed that the mnm5 and s2 modifications enhance the A-site binding to the AAA and AAG codons, and that the efficiency of the A-site binding is always in parallel with that of the P-site binding. The results from the P-site binding assay showed that the ASLs with U(34) bind only in the cases where the codon is from a family codon box. It is noteworthy that, although the GUG codon could bind the corresponding ASL with U(34) with a low affinity, the GCG, UCG and CCG codons did not bind the ASLs with a measurable affinity. This is consistent with the inefficiency observed for the reading of the G-ending codons by the unmodified tRNAs with U(34) during the in vitro translation assay mentioned above. Anyway, the modifications are required for the U*(34)-containing ASLs to bind to the G-ending codons.
xm5U* from eukaryotes. An early work has shown that a yeast tRNAGlu specifically translates the GAA codon in an in vitro translation system from rabbit reticulocytes (38). This tRNA has mcm5s2U(34), and this may fit to the above idea that eukaryotes do not use the U*(34)–G(III) wobbling (20). This tRNA was also investigated by the conventional ribosome-binding method (38). The results were consistent with those from the in vitro translation assay, while the source of the ribosomes was E.coli. Therefore, it may be the difference in tRNA but not that in the ribosomes that causes the difference in the codon specificity between eukaryotes and eubacteria at least in this case. It was also shown that yeast tRNA3Arg with mcm5U(34) does not recognize the G-ending codon either (39). Therefore, the inability to recognize G(III) in tRNAGlu may be independent of the s2 modification.
In vivo experiments using modification mutant strains
Rates of translation of the GAA/G codons. The effects of the mnm5 and s2 modifications on the rates for the translation of the GAA and GAG codons were measured with the use of well-characterized E.coli mutants of the mnm5s2U modifications (12). The rates at which the GAG codon was translated in the strains with mnm5s2U(34), s2U(34) and mnm5U(34) were 7.7, 1.9 and 6.2 codons/s, respectively, and the rates of the reading of the GAA codon were 18, 47 and 4.5 codons/s, respectively. Therefore, the s2 modification of mnm5U elevates the reading of the GAA codon and has only a small effect on the reading of the GAG codon, and the mnm5 modification of s2U restricts the reading of the GAA codon and enhances the reading of the GAG codon. Although the level of the available undermodified aminoacyl-tRNA species in each mutant was not clear, the elongation rates during the translation of the whole lacZ coding sequence were almost the same for these mutants. Therefore, we believe that the results are highly reliable. However, these results cannot be explained completely by the 3D structures of the ASL variants as described in detail below.
Results from frameshift assays. Brierley et al. (40) also utilized the modification mutants of E.coli to determine the in vivo correlation between the mnm5s2U modification and frameshifting efficiency at a coronavirus frameshift site. In their assay, the frameshift efficiencies at the AAA/G codons in the modification mutant strains were measured. They tried to interpret the results with the assumption that the frameshift efficiency should be negatively correlated with the stability of the codon binding by the tRNALys species with different modifications. However, the frameshift efficiency may also be affected by the efficiency of the tRNA molecules to shift to the AAA codon in the –1 frame, and, if this was the rate-limiting step of the whole process, the frameshift efficiency should have reflected the relative efficiency to bind to the –1 frame codon as compared to the 0 frame codon. Therefore, interpretation of these results as related to the efficiency of the A-site codon binding is quite difficult, as pointed out in other papers (10,12).
Misreading of pyrimidine-ending codons. The same E.coli strains were used to analyze the efficiency of the misreading of the AAU/C Asn codons by the tRNALys modification mutants under an Asn starvation condition (11). The results indicate that both mnm5 and s2 modifications enhance the misreading. These results were unexpected, because the modifications were thought to restrict wobbling at that time (3), but were rationalized when the NMR structures of the wild type and mutant ASLs were revealed in detail (10) (see below).
Physicochemical aspects
Conformational preferences of uridine derivatives and the expected basepairing pattern. As described above, the conformation of xm5s2U is biased to the C3′-endo form (3,4,41). This conformational preference is mainly due to the s2 modification, which would enhance steric repulsion between the O2′ and the atom at position 2 in the C2′-endo form. Therefore, 2′-O-methylation was also suggested to stabilize the C3′-endo form. The xm5 modification also contributes to the stabilization of the C3′-endo form (5,10). By contrast, pxo5U is much more in its C2′-endo form than pU. Although the mechanism of the preference is not clear, it was suggested to be due to the interaction between the 5′-phosphate and the oxygen atom of the xo5 substituent (3). With the C2′-endo form, U*(34) could basepair with U(III) if the codon and the second and third positions of the anticodon are in the A form, as shown by a model building study. Therefore, it was proposed that the conformational restriction into the C3′-endo form in xm5U*(34) should prevent mispairing with U(III) and stabilize the correct pair with A(III) (3).
As long as the A-type RNA is assumed in the other parts of the codon–anticodon duplex, U(34) could not form a base pair with C(III) because of steric hindrance (41,42). Therefore, the C2′-endo form could not explain the reading of the C-ending codons by mitochondrial and mycoplasma tRNAs with U(34). Although the anticodon loop of E.coli tRNALys was recently shown to have a remarkable flexibility (10), it is well known that the 2′-hydroxyl groups of the five nucleosides of the codon–anticodon duplex except the first one of the anticodon are hydrogen-bonded to the ribosome at the A site (43). Thus, it is reasonable to consider that these five nucleosides should be in the C3′-endo form on the A site, no matter how flexible the conformation of the unbound anticodon loop is.
In the original hypothesis (3), the U*(34)-G(III) was thought to be possible with both of the C2′-endo and C3′-endo forms. However, the above in vitro translation experiments (4,35,36) showed that the U(34)–G(III) pair with the C3′-endo form of the U should be weak. Thus, the xm5s2U(34)–G(III) pair should be very weak, because the C2′-endo form of xm5s2U should be less stable than that of U and the S…H-N hydrogen bond required for the base pair should be weaker than the O…H-N bond required for the U-G pair. Therefore, the reading of the AAG and GAG codons by E.coli tRNALys and tRNAGlu, respectively, could not be rationalized by this theory, as pointed out in an excellent review (44).
NMR structures of the modification variants of the tRNALys ASL. As described above, the physicochemical effects of the modifications in mnm5s2U in the tRNALys ASL have been elucidated by NMR analyses (10). The s2 modification enhances the stacking of the ‘two out of three’ onto the 3′-side of the anticodon, and the mnm5 modification decreases the flexibility of the loop.
As described above, the stabilization of the A-form structure of the anticodon resulted in the elevated misreading of the AAU/C Asn codons during the Asn starvation. This enhancement of the misreading could not be predicted from the conformational properties of the nucleotides at position 34 (11). The prediction implied that the misreading should depend primarily on the wobble mechanism with the U*(34)–U(III) base pair, instead of the ‘two out of three’ mechanism. The fact that the AAC codons were also misread under the starvation condition indicates that at least the misreading of the AAC codons depended on the ‘two out of three’ mechanism. It is likely that the conformational restriction into the C3′-endo form by the modifications reduced the efficiency of the misreading by the wobble mechanism to the extent that it was lower than that by the ‘two out of three’ mechanism.
The s2 modification could be predicted to enhance the reading of the GAA codon from its structural effects, and it did in the in vivo experiment. In the case of the GAG codon, the substitution of O2 with a sulfur atom would destabilize the wobble base pair because the O2…H-N hydrogen bond would be substituted by a weak S2…H-N hydrogen bond, if any, while the stacking enhancement should more or less compensate for the destabilization (though this point is not described explicitly in the published material). Therefore, the small effect on the reading of the GAG codon observed in the in vivo experiment could be rationalized. On the other hand, the mnm5 modification should stabilize the interaction with A(III), as it should stabilize the ‘preorganized’ conformation. This contradicts to the in vivo data. As for G(III), if the mnm5s2U(34)–G(III) base pair is formed with the C2′-endo form of the mnm5s2U, then the mnm5 modification should reduce the reading of the G-ending codon because it should destabilize the C2′-endo form, which is again contradictory to the in vivo result. This may mean that the mnm5s2U(34)–G(III) pair with the C3′-endo form of the mnm5s2U is stabilized by the mnm5 modification through some unknown mechanism.
Thermodynamic analyses. Stabilities of RNA duplexes could be estimated by measuring the melting profiles of the duplexes. Many oligonucleotides containing modified nucleosides have been studied by this method. It should be noted that the contribution of a single base pair to the stability of the duplex could not be defined because a base pair should affect the neighboring base pair interactions. However, a sum of free energy parameters for all the pairs of neighboring two base pairs in the duplex, plus the parameters for the terminal base pairs and other constants, could be a good estimation of the stability of the duplex (45,46). Substitution of an A-U pair in the middle of an RNA duplex by a G-U pair would usually destabilize the duplex, and the free energy difference could be estimated easily if the neighboring base pairs are known. In the same way, the effect of the substitution of a terminal A-U pair by a G-U pair could be estimated. However, this substitution turns out to be non-destabilizing or even stabilizing, in general, when the A and G are at the 5′-ends of the duplexes and the Us are at the 3′-ends (46). Codon–anticodon duplexes with U(III) are in a similar situation to the latter case, as position 34 is at the 5′-end of the codon–anticodon duplex. It has also been observed that the substitution of A(34) by a G in a series of unmodified tRNAs raises, or does not change, the efficiencies to read the U-ending codons in an in vitro translation system (36). Therefore, the effects of a modification of U(34) could not be estimated from the stabilities of RNA duplexes with the modified uridines in the middle or at the 3′-end of the duplexes. There have been no experiments in which the effects of the 5-substitution of uridines at the 5′-end of RNA duplexes are estimated, although even this type of experiment would not necessarily be promising in terms of the estimation of the effects of the modification on tRNA codon recognition.
Model-building study (Lim’s model). Lim and coworkers have proposed a model that explains the codon-reading patterns as related to the properties of the nucleosides at position 34 (42,47). In the model, U(34) could interact with U(III) and C(III) through water bridges, and a tRNA with U(34) reads all the four bases at position III. Therefore, the model may be useful to predict codon preferences when the family codons are not fully discriminated. The mnm5 substitution would break some bonds needed for the water-bridged pairs, and this loss of the stabilizing interaction would not be compensated. Therefore, the modification should restrict the formation of the water-bridged pairs. However, it is obvious that this model does not take into account that the strength of the interaction between the first two codon positions and the last two anticodon positions could be changed by the s2 and xm5 modifications (10). Therefore, the model could not predict the in vivo effects of the modification (11,12).
A MODEL OF THE xm5U(34)–G(III) BASEPAIRING
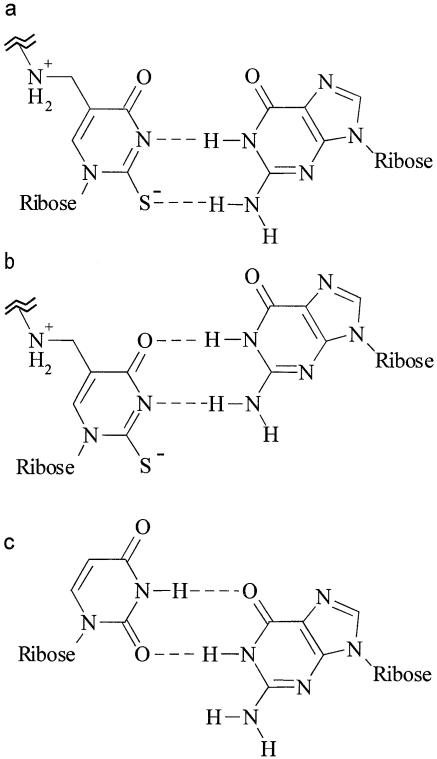
Here, we propose a model of the xm5U*(34)–G(III) pairing, by which the in vivo effects of each modification on the codon-reading rates and the prokaryote/eukaryote difference of the wobble rule concerning the U*(34)–G(III) pairing could be rationalized. The xm5U nucleosides from prokaryotes and mitochondria are derivatives of 5-aminomethyluridine (xnm5U*), while those from eukaryotes are not (13). As for xnm5U*, the 5-substituent is likely to lower pKa at the N3 position of the uracil ring, because the positively charged nitrogen atom of the substituent should withdraw electrons from the uracil ring. Thus, xnm5U* may partially ionize under the physiological condition. The ionized form of the nucleoside (xnm5U*-) could base pair with G(III) in two different configurations (Fig. 2a and b). In the case of the 2-thiolated uridines, the ionization could confer a negative charge on the sulfur atom and convert it to an efficient proton acceptor. The ionized form would be able to pair only with G(III), and the neutral form, which may be symbolized here as xnm5U*0, could pair only with A(III). Both pairs could be formed with the C3′-endo conformation. Thus, the stabilization of the C3′-endo form by the modifications would stabilize both pairs. We suppose that the ionization should be partial. The relative efficiency in the reading of the G-ending codon to that of the A-ending codon would not only depend on the degree of the ionization, but would also depend on the difference in the intrinsic stabilities between the xnm5U*-(34)–G(III) and xnm5U*0(34)–A(III) pairs. Therefore, an xnm5U* could pair with G(III) more efficiently than with A(III) even when the neutral form is the major species. In the case of mnn5s2U, the pairing in the neutral form with A(III) may be still more efficient, in total, than the pairing in the ionized form with G(III). The ionized modified uridine would not pair with U(III) or C(III). As the eukaryotic xm5 substituents do not withdraw electrons as xnm5 may do, the eukaryotic tRNAs do not recognize the G-ending codons.
Figure 2.
The proposed base pairs between a deprotonated modified U and a G proposed in the present study (a and b) and the conventional wobble U–G pair (c). (a) The proposed xnm5U*-–G pair with the Watson–Crick configuration; (b) the alternative xnm5U*-–G pair with the xnm5U*- displaced toward the minor groove side from the Watson–Crick configuration; (c) the conventional wobble U-G pair. The sulfur atom in (a) and (b) could be substituted with an oxygen atom, and the negative charge could be delocalized within the π-electron system.
HOW THE MODEL FITS TO THE KNOWN FACTS AND THEORIES
Hammett equation and the known pKa values for uridine and uracil derivatives. The pKa values of a series of substituted compounds could be well predicted using the Hammett equation (48), as described in many organic chemistry textbooks. pKa of a substituted molecule is predicted to be lower than the unsubstituted one by σ times ρ, where σ is a constant specific to the substituent and its position, and ρ is a constant specific to the core acid. As judged from the known pKa values for several uracil derivatives (U, 9.3; m5U, 9.7; and 1-methyl-5-bromouracil, 7.8) (49) and the σ values for the meta position (methyl, –0.07 and bromo, 0.39), ρ for uracil should be positive (and ∼5).
The effects of substitutions at aromatic rings are mainly ascribed to two factors: inductive electron withdrawal and the resonance electron donation by the substituent. In the case of the xnm5 substituent, the resonance effect may be small because of the methylene group directly attached to the uracil ring, and the inductive effect may be large because the nitrogen atom is protonated and is positively charged. Therefore, it is expected that the inductive effect dominates over the resonance effect, and the pKa of mnm5U should be significantly lower than that of U. In fact, the pKa values of cmnm5Um and cmnm5U have been measured to be 8.3 and 8.2, respectively (6). As the negative charge of the carboxyl group of the cmnm5 substituent may somewhat neutralize the electron withdrawing effect of the charged nitrogen, it is expected that the pKa value for mnm5U is not ≤8.2. Furthermore, as pKa for s2U (8.8) is lower than that for U by ∼0.5, it is expected that pKa for mnm5s2U should be <8.
pKa values for the uracil N3 position are lower in nucleosides than in the corresponding nucleotides, in general, because of the negative charge of the phosphate group. In fact, the measured values (pU, 9.7; pm5U, 10.1; 5-bromouridine 5′-monophosphate, 8.1) (50) are higher than those of the corresponding nucleosides by ∼0.4. We have to be careful when comparing pKa values measured under different conditions because the effect of the charge of the phosphate may depend on the ion strength and probably on the conformational preference of the nucleotide. However, it is still reasonable to assume that the pKa value for pmnm5s2U may be higher than that of the nucleoside by ∼0.4.
The environment around the uracil ring on the decoding site of the ribosome may also affect the pKa values. However, from the recent results of the X-ray crystallographic analyses of the ribosome (51), we could see that the charged group nearest to the first base of the anticodon of the A-site-bound tRNA is its 5′-phosphate, and that the distance of the phosphate from the uracil ring may be almost the same as in the free nucleotide. Therefore, there is no reason at present to consider that the pKa values on the ribosome are much higher than those in the corresponding nucleotides.
If the pKa value of a molecule is 8.4, for instance, a 9% fraction of the molecule is ionized under pH 7.4. Therefore, it is reasonable to assume that a considerable fraction of mnm5s2U in tRNA is ionized under the physiological condition.
Two possible configurations. Two different configurations might be possible for the pair between xnm5U*-(34) and G(III) (Fig. 2a and b). We expect that the one with the Watson–Crick configuration (a) should be more stable than the one with the wobble configuration in which the xnm5U*- is displaced toward the minor groove side (b). However, it is still possible that the latter configuration could contribute significantly. The pair may be possible with the same ribose–phosphate conformation as in the G(34)–U(III) wobble pair, which could be stabilized by the stacking onto the neighboring base pair (2), and was shown to be no less stable than the A(34)–U(III) pair in a cell-free translation assay (36), as stated above. Thus, the wobble form of the xm5U*-(34)–G(III) pair might be stabilized by the same mechanism. In addition, as the sulfur atom of the wobble xnm5s2U*-(34)–G(III) pair should not participate in the hydrogen bonding interactions, the pair could be more stable than the pair with the Watson–Crick configuration. The possibility of the dual-mode base pairing itself might also contribute to the enhancement of the reading of the G-ending codon.
Physicochemical and biological effects of the mnm5s2U(34) modifications. As mentioned above, it is expected that the s2 modification of xnm5U decreases the pKa value. Therefore, the modification should promote the deprotonation. In addition, the s2 modification should promote the stacking of the anticodon as in tRNALys (10) and stabilize the C3′-endo form of the nucleoside. If, for example, the conformational effect stabilizes the pair by 5-fold and the fraction of the ionized form increased by the thiolation is 20%, then the total effect should be stabilization by 4-fold (though this may be an oversimplification). Therefore, the xnm5U-to-xnm5s2U modification should enhance the reading of the A-ending codon. This is consistent with the case of E.coli tRNAGlu (12). As for the G-ending codon, the xnm5s2U-–G pair could be weaker in itself than the xmn5U-–G pair, because at least one of the two possible base pair configurations in the former (Fig. 2) needs a hydrogen bond that involves the sulfur atom. Therefore, it is possible that this effect cancels the conformational effects. This is not contradictory to the fact that the s2 modification of tRNAGlu had only a small effect on the reading of the GAG codon in vivo (12). The experimental results also indicate that the reading of the GAG codon by the tRNA with mnm5U(34) is more efficient than the reading of the GAA codon (12), and this could be reasoned if we assume that the mnm5U-–G pair might be intrinsically much more stable than the mnm5U0–A pair.
The mnm5 modification of s2U would cause the partial ionization, which would destabilize the pair with A(III) and stabilize the pair with G(III), and the restriction of the flexibility of the anticodon, which would stabilize both pairs through stabilizing the C3′-endo form. Therefore, the pair with G(III) should be stabilized, while the prediction of the effects on the pair with A(III) is more difficult. The conformational effect might be smaller for the s2U-to-mnm5s2U modification than for the U-to-mnm5U modification, as the s2U-containing anticodon is already much biased to the preorganized conformation (10). On the other hand, the decrease in the fraction of the neutral form required for the pairing with A(III) by the lowering of pKa should be larger in the presence of the s2 modification, as s2U has a lower pKa value than U. Thus, the mnm5 modification might be more destabilizing as for the pair with A(III) in thiolated uridines than in non-thiolated uridines. In the case of tRNAGlu (12), the effect through the deprotonation may have been larger than the conformational effect. As xnm5U-* could form no base pair with U(III) or C(III), the observed misreading of the AAU/C codons by tRNALys during Asn starvation (11) should not have been due to the ionized form.
2-Selenouridine derivatives. Bacteria modifies mnm5s2U(34) into 5-methylaminomethyl-2-selenouridine (mnm5Se2U), when selenium is available (52). The pKa value for this nucleoside is ∼7.1, and a glutamate tRNA with mnm5Se2U(34) binds efficiently to the GAG codon (53). Thus, it has been proposed that the ionized form is responsible for the base pairing with G(III) (53). It seems that the low pKa was considered to be mainly due to the 2-seleno substitution, but the mnm5 substitution might also contribute.
xo5U. xo5U(34) has been proposed to recognize G(III) with the C2′-endo conformation (3,7). If the pKa value for pxo5U is low enough, it would be also possible that xo5U(34) could also be partially ionized. In fact, the pKa value for pmo5U has been measured to be 8.96 (50). The pKa values for the xo5U nucleosides could also be predicted based on Hammett equation: pKa for mo5U and ho5U could be predicted to be ∼8.7, using the σ values for the methoxy and hydroxyl substituents at the meta position (0.12) and the above-estimated ρ value for uracil. Therefore the predicted value is consistent considering the effect of the 5′-phosphate. In cmo5U, the pKa could be higher than in mo5U because of the negative charge in the carboxyl group. Thus, only a small fraction could be ionized in these nucleotides under the physiological condition. In addition, the C3′-endo conformation is destabilized in these nucleotides. Therefore, it may be reasonable to consider that the primary mechanism for the G(III) recognition should be the formation of the neutral xo5U(34)–G(III) pair with the C2′-endo form of xo5U.
Eukaryotic xm5U* nucleosides. In mcm5U* and ncm5U* found in eukaryotic tRNAs (13,54), the substituents are unlikely to withdraw electrons as well as the xnm5 group may do. Therefore, it could be predicted that the tRNAs with the nucleoside could not pair efficiently with G(III). The experimental facts are as described above, and the G-ending codons in eukaryotes seem to be recognized by tRNAs with C(34) in general (19,54). This could be an explanation for the prokaryote/eukaryote difference of the wobble rule, if any, though it is still possible that the ribosomes are also different. It is possible that the tRNA modifications and ribosome functions have coevolved to optimize the translational function. If the present model is correct, it would mean that the eukaryotic ribosomes dispense with the C2′-endo conformation of U*(34) and a wobble pair with the anticodon base shifted toward the major groove, as eukaryotes do not have xo5U(34) (13). Thus, it is possible that the eukaryotic ribosomes have lost the ability to accept eubacterial tRNAs with xo5U(34) for the reading of the U- and G-ending codons.
TESTING THE MODEL
An obvious experimental test of the model is to measure the pKa values of the modified uridines. This could disprove the model if the pKa values for mnm5U* or τm5U* are not lower than ∼8.5, which is unlikely considering the values for cmnm5U and cmnm5Um.
Another possible experiment may be to measure the pH dependence of the relative efficiency of the U*(34)-containing tRNA in the reading of the G-ending codon as compared to the efficiency of the tRNA with C(34) (35). However, the experiment could be difficult because the overall fidelity in cell-free translation from E.coli depends on pH (28). Conventional ribosome-binding experiments and other advanced A-site binding methods may also be possible (37,55).
It is known that mnm5s2U(34) in tRNA molecules could be reversibly oxidized by iodine (56). Therefore, it may be possible to test the pH sensitivity of this reaction using prokaryotic and eukaryotic tRNA molecules, as the fraction of the ionized species should directly correlate with the reactivity. It may also be possible to estimate the pKa values of xnm5U* in tRNA molecules by measuring the pH-dependence of the aminoacylation reactions catalyzed by the corresponding aminoacyl-tRNA synthetases. Fortunately, bacterial Lys-, Glu- and Gln-tRNA synthetases are in direct contact with position 34 when complexed with the cognate tRNA molecules (57–59).
It may also be possible to measure the efficiency of the reading of a codon with an inosine at the third position. If the displacement of the uracil ring of the modified uridine to the major groove side is possible, it is expected that the I(III)-containing codon also could be recognized (see Fig. 2c). On the other hand, if the ionization is responsible for the pairing with G(III), the codon with I(III) could not be recognized. Some other methods may also be possible that could determine whether the 2-amino group of G(III) participates in the base pair hydrogen bonding.
The 3D structure of the decoding site is emerging in detail in these years (43,51,60,61). However, the position of the base of G(34) in the crystal was somewhat deviated from the normal wobble configuration. Therefore, it may still take some time to localize the uracil ring of some of the modified uridines on the A site precisely enough to tell the base pair configuration.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Dr Mitsuo Sekine (Tokyo Institute of Technology) and Dr Kensaku Sakamoto (Tokyo University) for discussion. This work was supported in part by the Sumitomo Foundation, Tokyo, Japan (no. 020764).
REFERENCES
- 1.Crick F.H.C. (1966) Codon-anticodon pairing: the wobble hypothesis. J. Mol. Biol., 19, 548–555. [DOI] [PubMed] [Google Scholar]
- 2.Mizuno H. and Sundaralingam,M. (1978) Stacking of Crick wobble pair and Watson–Crick pair: stability rules of G-U pairs at ends of helical stems in tRNAs and the relation to codon–anticodon wobble interaction. Nucleic Acids Res., 5, 4451–4461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yokoyama S., Watanabe,T., Murao,K., Ishikura,H., Yamaizumi,Z., Nishimura,S. and Miyazawa,T. (1985) Molecular mechanism of codon recognition by tRNA species with modified uridine in the first position of the anticodon. Proc. Natl Acad. Sci. USA, 82, 4905–4909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Agris P.F. (1991) Wobble position modified nucleosides evolved to select transfer RNA codon recognition: a modified wobble hypothesis. Biochimie, 73, 1345–1349. [DOI] [PubMed] [Google Scholar]
- 5.Sakamoto K., Kawai,G., Watanabe,S., Niimi,T., Hayashi,N., Muto,Y. Watanabe,K., Satoh,T., Sekine,M. and Yokoyama,S. (1996) NMR studies of the effects of the 5′-phosphate group on conformational properties of 5-methylaminomethyluridine found in the first position of the anticodon of Escherichia coli tRNA4Arg. Biochemistry, 35, 6533–6538. [DOI] [PubMed] [Google Scholar]
- 6.Horie N., Yamaizumi,Z., Kuchino,Y., Takai,K., Goldman,E., Miyazawa,T., Nishimura,S. and Yokoyama,S. (1999) Modified nucleoside in the first positions of the anticodons of tRNA4Leu and tRNA5Leu from Escherichia coli. Biochemistry, 38, 207–217. [DOI] [PubMed] [Google Scholar]
- 7.Takai K., Okumura,S., Hosono,K., Yokoyama,S. and Takaku,H. (1999) A single uridine modification at the wobble position of an artificial tRNA enhances wobbling in an Escherichia coli cell-free translation system. FEBS Lett., 447, 1–4. [DOI] [PubMed] [Google Scholar]
- 8.Lagerkvist U. (1978) ‘Two out of three’: an alternative method for codon reading. Proc. Natl Acad. Sci. USA, 75, 1759–1762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lagerkvist U. (1981) Unorthodox codon reading and the evolution of the genetic code. Cell, 23, 305–306. [DOI] [PubMed] [Google Scholar]
- 10.Sundaram M., Durant,P.C. and Davis,D.R. (2000) Hypermodified nucleosides in the anticodon of tRNALys stabilize a canonical U-turn structure. Biochemistry, 39, 12575–12584. [DOI] [PubMed] [Google Scholar]
- 11.Hagervall T.G., Pomerantz,S.C. and McCloskey,J.A. (1998) Reduced misreading of asparagine codons by Escherichia coli tRNALys with hypermodified derivatives of 5-methylaminomethyl-2-thiouridine in the wobble position. J. Mol. Biol., 284, 33–42. [DOI] [PubMed] [Google Scholar]
- 12.Krüger M.K., Pedersen,S., Hagervall,T.G. and Sørensen,M.A. (1998) The modification of the wobble base of tRNAGlu modulates the translation rate of glutamic acid codons in vivo. J. Mol. Biol., 284, 621–631. [DOI] [PubMed] [Google Scholar]
- 13.Rozenski J., Crain,P.F. and McCloskey,J.A. (1999) The RNA Modification Database: 1999 update. Nucleic Acids Res., 27, 196–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sakamoto K., Kawai,G., Niimi,T., Satoh,T., Sekine,M., Yamaizumi,Z., Nishimura,S., Miyazawa,T. and Yokoyama,S. (1993) A modified uridine in the first position of the anticodon of a minor species of arginine tRNA, the argU gene product, from Escherichia coli. Eur. J. Biochem., 216, 369–375. [DOI] [PubMed] [Google Scholar]
- 15.Komine Y., Adachi,T., Inokuchi,H. and Ozeki,H. (1990) Genomic organization and physical mapping of the transfer RNA genes in Escherichia coli K12. J. Mol. Biol., 212, 579–598. [DOI] [PubMed] [Google Scholar]
- 16.Yoshimura M., Inokuchi,H. and Ozeki,H. (1984) Identification of transfer RNA suppressors in E. coli. IV. Amber suppressor Su+6 a double mutant of a new species. J. Mol. Biol., 177, 627–644. [DOI] [PubMed] [Google Scholar]
- 17.Spanjaard R.A., Chen,K., Walker,J.R. and van Duin,J. (1990) Frameshift suppression at tandem AGA and AGG codons by cloned tRNA genes: assigning a codon to argU tRNA and T4 tRNAArg. Nucleic Acids Res., 18, 5031–5036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Suzuki T., Suzuki,T., Wada,T., Saigo,K. and Watanabe,K. (2002) Taurine as a constituent of mitochondrial tRNAs: new insights into the functions of taurine and human mitochondrial diseases. EMBO J., 21, 6581–6589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Marck C. and Grosjean,H. (2002) tRNomics: analysis of tRNA genes from 50 genomes of Eukarya, Archaea and Bacteria reveals anticodon-sparing strategies and domain-specific features. RNA, 8, 1189–1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Percudani R. (2001) Restricted wobble rules for eukaryotic genomes. Trends Genet., 17, 133–135. [DOI] [PubMed] [Google Scholar]
- 21.Barrell B.G., Anderson,S., Bankier,A.T., de Bruijn,M.H., Coulson,A.R., Drouin,J., Eperon,I.C., Nierlich,D.P., Roe,B.A., Sanger,F., Schreier,P.H., Smith,A.J., Staden,R. and Young,I.G. (1980) Different pattern of codon recognition by mammalian mitochondrial tRNAs. Proc. Natl Acad. Sci. USA, 77, 3164–3166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bonitz S.G., Berlani,R., Coruzzi,G., Li,M., Macino,G., Nobrega,F.G., Nobrega,M.P., Thalenfeld,B.E. and Tzagoloff,A. (1980) Codon recognition rules in yeast mitochondria. Proc. Natl Acad. Sci. USA, 77, 3167–3170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Heckman J.E., Sarnoff,J., Alzner-DeWeerd,B., Yin,S., RajBhandary,U.L. (1980) Novel features in the genetic code and codon reading patterns in Neurospora crassa mitochondria based on sequences of six mitochondrial tRNAs. Proc. Natl Acad. Sci. USA, 77, 3159–3163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Andachi Y., Yamao,F., Muto,A. and Osawa,S. (1989) Codon recognition pattern as deduced from sequences of the complete set of transfer RNA species in Mycoplasma capricolum. J. Mol. Biol., 209, 37–54. [DOI] [PubMed] [Google Scholar]
- 25.Lustig F., Elias,P., Axberg,T., Samuelsson,T., Tittawella,I. and Lagerkvist,U. (1981) Codon reading and translational error. Reading of the glutamine and lysine codons during protein synthesis in vitro. J. Biol. Chem., 256, 2635–2643. [PubMed] [Google Scholar]
- 26.Mitra S.K., Lustig,F., Åkesson,B., Axberg,T., Elias,P. and Lagerkvist,U. (1979) Relative efficiency of anticodons in reading the valine codons during protein synthesis in vitro. J. Biol. Chem., 254, 6397–6401. [PubMed] [Google Scholar]
- 27.Takai K., Horie,N., Yamaizumi,Z., Nishimura,S., Miyazawa,T. and Yokoyama,S. (1994) Recognition of UUN codons by two leucine tRNA species from Escherichia coli. FEBS Lett., 344, 31–34. [DOI] [PubMed] [Google Scholar]
- 28.Bartetzko A. and Nierhaus,K.H. (1988) Mg2+/NH4+/polyamine system for polyuridine-dependent polyphenylalanine synthesis with near in vivo characteristics. Methods Enzymol., 164, 650–658. [DOI] [PubMed] [Google Scholar]
- 29.Samuelsson T., Axberg,T., Borén,T. and Lagerkvist,U. (1983) Unconventional reading of the glycine codons. J. Biol. Chem., 258, 13178–13184. [PubMed] [Google Scholar]
- 30.Lustig F., Borén,T., Guindy,Y.S., Elias,P., Samuelsson,T., Gehrke,C.W., Kuo,K.C. and Lagerkvist,U. (1989) Codon discrimination and anticodon structural context. Proc. Natl Acad. Sci. USA, 86, 6873–6877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Claesson C., Samuelsson,T., Lustig,F. and Borén,T. (1990) Codon reading properties of an unmodified transfer RNA. FEBS Lett., 273, 173–176. [DOI] [PubMed] [Google Scholar]
- 32.Lustig F., Borén,T., Claesson,C., Simonsson,C., Barciszewska,M. and Lagerkvist,U. (1993) The nucleotide in position 32 of the tRNA anticodon loop determines ability of anticodon UCC to discriminate among glycine codons. Proc. Natl Acad. Sci. USA, 90, 3343–3347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Claesson C., Lustig,F., Borén,T., Simonsson,C., Barciszewska,M. and Lagerkvist,U. (1995) Glycine codon discrimination and the nucleotide in position 32 of the anticodon loop. J. Mol. Biol., 247, 191–196. [DOI] [PubMed] [Google Scholar]
- 34.Inagaki Y., Kojima,A., Bessho,Y., Hori,H., Ohama,T. and Osawa,S. (1995) Translation of synonymous codons in family boxes by Mycoplasma capricolum tRNAs with unmodified uridine or adenosine at the first anticodon position. J. Mol. Biol., 251, 486–492. [DOI] [PubMed] [Google Scholar]
- 35.Takai K., Takaku,H. and Yokoyama,S. (1996) Codon-reading specificity of an unmodified form of Escherichia coli tRNA1Ser in cell-free protein synthesis. Nucleic Acids Res., 24, 2894–2899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Takai K., Takaku,H. and Yokoyama,S. (1999) In vitro codon-reading specificities of unmodified tRNA molecules with different anticodons on the sequence background of Escherichia coli tRNA1Ser. Biochem. Biophys. Res. Commun., 257, 662–667. [DOI] [PubMed] [Google Scholar]
- 37.Yarian C., Townsend,H., Czestkowski,W., Sochacka,E., Malkiewicz,A.J., Guenther,R., Miskiewicz,A. and Agris,P.F. (2002) Accurate translation of the genetic code depends on tRNA modified nucleosides. J. Biol. Chem., 277, 16391–16395. [DOI] [PubMed] [Google Scholar]
- 38.Sekiya T., Takeishi,K. and Ukita,T. (1969) Specificity of yeast glutamic acid transfer RNA for codon recognition. Biochim. Biophys Acta, 182, 411–426. [DOI] [PubMed] [Google Scholar]
- 39.Weissenbach J. and Dirheimer,G. (1978) Pairing properties of the methylester of 5-carboxymethyl uridine in the wobble position of yeast tRNA3Arg. Biochim. Biophys Acta, 518, 530–534. [DOI] [PubMed] [Google Scholar]
- 40.Brierley I., Meredith,M.R., Bloys,A.J., Hagervall,T.G. (1997) Expression of a coronavirus ribosomal frameshift signal in Escherichia coli: influence of tRNA anticodon modification on frameshifting. J. Mol. Biol., 270, 360–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yokoyama S. and Nishimura,S. (1995) Modified nucleosides and codon recognition. In: Söll,D. and RajBhandary,U. (eds), tRNA: Structure, Biosynthesis and Function. ASM Press, Washington, DC, pp. 207–223. [Google Scholar]
- 42.Lim V.I. and Venclovas,C. (1992) Codon-anticodon pairing. A model for interacting codon-anticodon duplexes located at the ribosomal A- and P-sites. FEBS Lett., 313, 133–137. [DOI] [PubMed] [Google Scholar]
- 43.Ogle J.M., Carter,A.P. and Ramakrishnan,V. (2003) Insights into the decoding mechanism from recent ribosome structures. Trends Biochem. Sci., 28, 259–266. [DOI] [PubMed] [Google Scholar]
- 44.Curran J.F. (1998) Modified nucleosides in translation. In: Grosjean,H. and Benne,R. (eds), Modification and Editing of RNA. ASM Press, Washington, DC, pp. 493–516. [Google Scholar]
- 45.Tinoco I. Jr, Borer,P.N., Dengler,B., Levine,M.D., Uhlenbeck,O., Crothers,D.M. and Gralla,J. (1973) Improved estimation of secondary structure in ribonucleic acids. Nature New Biol., 246, 40–41. [DOI] [PubMed] [Google Scholar]
- 46.Turner D.H., Sugimoto,N. and Freier,S.M. (1988) RNA structure prediction. Ann. Rev. Biophys. Biophys. Chem., 17, 167–192. [DOI] [PubMed] [Google Scholar]
- 47.Lim V.I. and Curran,J.F. (2001) Analysis of codon:anticodon interactions within the ribosome provides new insights into codon reading and the genetic code structure. RNA, 7, 942–957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hammett L.P. (1970) Physical Organic Chemistry, 2nd Ed. McGraw Hill, New York, NY. [Google Scholar]
- 49.Saenger W. (1984) Principles of Nucleic Acid Structure, Springer-Verlag, New York, NY. Japanese translation by Nishimura,Y., Springer-Verlag Tokyo, Japan. [Google Scholar]
- 50.Shibaev V.N., Eliseeva,G.I. and Kochetkov,N.K. (1975) Interaction of uridine diphosphate glucose analogs with calf liver uridine diphosphate glucose dehydrogenase. Influence of substituents at C-5 of pyrimidine nucleus. Biochim. Biophys Acta, 403, 9–16. [DOI] [PubMed] [Google Scholar]
- 51.Ogle J.M., Brodersen,D.E., Clemons,W.M.,Jr, Tarry,M.J., Carter,A.P., Ramakrishnan,V. (2001) Recognition of cognate transfer RNA by the 30S ribosomal subunit. Science, 292, 897–902. [DOI] [PubMed] [Google Scholar]
- 52.Wittwer A.J., Tsai,L., Ching,W.M. and Stadtman,T.C. (1984) Identification and synthesis of a naturally occurring selenonucleoside in bacterial tRNAs: 5-[(methylamino)methyl]-2-selenouridine. Biochemistry, 23, 4650–4655. [DOI] [PubMed] [Google Scholar]
- 53.Ching W.M. (1986) Characterization of selenium-containing tRNAGlu from Clostridium sticklandii. Arch. Biochem. Biophys., 244, 137–146. [DOI] [PubMed] [Google Scholar]
- 54.Björk G. (1998) Appendix 6: Modified nucleosides at positions 34 and 37 of tRNAs and their predicted coding capacities. In: Grosjean,H. and Benne,R. (eds), Modification and Editing of RNA. ASM Press, Washington, DC, pp. 577–581. [Google Scholar]
- 55.Phelps S.R., Jerinic,O. and Joseph,S. (2002) Universally conserved interactions between the ribosome and the anticodon stem-loop of A site tRNA important for translocation. Mol. Cell, 10, 799–807. [DOI] [PubMed] [Google Scholar]
- 56.Carbon J.A., Hung,L. and Jones,D.S. (1965) A reversible oxidative inactivation of specific transfer RNA species. Proc. Natl Acad. Sci. USA, 53, 979–986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cusack S., Yaremchuk,A. and Tukalo,M. (1996) The crylstal structures of T.thermophilus lysyl-tRNA synthetase complexed with E.coli tRNALys and a T.thermophilus tRNALys transcript: anticodon recognition and conformational changes upon binding of a lysyl-adenylate analogue. EMBO J., 15, 6321–6334. [PMC free article] [PubMed] [Google Scholar]
- 58.Rould M.A., Perona,J.J., Söll,D. and Steitz,T.A. (1989) Structure of E. coli glutaminyl-tRNA synthetase complexed with tRNAGln and ATP at 2.8 Å resolution. Science, 246, 1135–1142. [DOI] [PubMed] [Google Scholar]
- 59.Sekine S., Nureki,O., Shimada,A., Vassylyev,D.G. and Yokoyama,S. (2001) Structural basis for anticodon recognition by discriminating glutamyl-tRNA synthetase. Nat. Struct. Biol., 8, 189–191. [DOI] [PubMed] [Google Scholar]
- 60.Ramakrishnan V. (2002) Ribosome structure and the mechanism of translation. Cell, 108, 557–572. [DOI] [PubMed] [Google Scholar]
- 61.Ogle J.M., Murphy,F.V., Tarry,M.J. and Ramakrishnan,V. (2002) Selection of tRNA by the ribosome requires a transition from an open to closed form. Cell, 111, 721–732. [DOI] [PubMed] [Google Scholar]