Abstract
Recently, artificial oriC plasmids containing the chromosomal dnaA gene and surrounding DnaA box sequences were obtained for the mollicutes Spiroplasma citri and Mycoplasma pulmonis. In order to study the specificity of these plasmids among mollicutes, a set of similar oriC plasmids was developed for three mycoplasmas belonging to the mycoides cluster, Mycoplasma mycoides subsp. mycoides LC (MmmLC), M.mycoides subsp. mycoides SC (MmmSC) and Mycoplasma capricolum subsp. capricolum. Mycoplasmas from the mycoides cluster, S.citri and M.pulmonis were used as recipients for transformation experiments by homologous and heterologous oriC plasmids. All five mollicutes were successfully transformed by homologous plasmids, suggesting that the dnaA gene region represents the functional replication origin of the mollicute chromosomes. However, the ability of mollicutes to replicate heterologous oriC plasmids was found to vary noticeably with the species. For example, the oriC plasmid from M.capricolum did not replicate in the closely related species MmmSC and MmmLC. In contrast, plasmids harbouring the oriC from MmmSC, MmmLC and the more distant species S.citri were all found to replicate in M.capricolum. Our results suggest that the cis-elements present in oriC sequences are not the only determinants of this host specificity.
INTRODUCTION
Chromosomal replication is a key event in the life cycle of all organisms. Among the trans-acting factors involved in DNA replication, the DnaA protein has a central role in the initiation of the process and is essential in eubacteria (1), with the possible exception of Wigglesworthia glossinidia (2). The specific interaction of DnaA proteins with 9 bp DnaA boxes (5′TTATCCACA3′) results in a local unwinding of the DNA double helix and creates the correct structural arrangement for the subsequent loading of proteins (DnaB, DnaC) required for the formation of the ‘pre-priming complex’. DnaA acts as a replisome organiser and is necessary for the priming mechanism that leads to the separation of the DNA strands, the entry of the replication machinery and the formation of the replication forks (3).
In Bacillus subtilis, localisation of the chromosomal oriC was first attempted by constructing a replication order map from measurements of the relative frequencies of various genetic markers, and by direct determination of their time of replication (4). Some years later, cloning of autonomous replicating sequence (ARS) into plasmid vectors showed that the ARS activity coincided with a region containing the DnaA boxes and, in many bacterial genomes, the dnaA gene. More recently, new methods have been developed such as GC skew analysis or two-dimensional replicon mapping which allows prediction of the location of the oriC from the whole genome sequence (5). The second approach not only leads to the unambiguous identification of the initiation site but also documents the progression of the replication fork (6,7).
With the development of oriC plasmids, the compatibility of replication origin sequences with heterologous replication machineries has been examined, showing that, in some cases, the oriC from one species functions in another species (8–10). However, examples of compatible oriC plasmids between different genera are rather unusual (11–13), and the molecular basis for the oriC plasmid specificity remains unclear.
In this study, we have constructed oriC plasmids for different species belonging to the Mycoplasma genus. Mycoplasmas belong to the mollicutes class which includes the simplest, wall-less, self-replicating organisms (14). Part of the knowledge on the replication origins of mollicute chromosomes comes from genome analyses; however, functional determination of the oriC was achieved for Mycoplasma capricolum oriC by studying the progression of the replication fork (15,16), and for both Spiroplasma citri (17) and Mycoplasma pulmonis (18) by obtaining replicative oriC plasmids. Although oriC-based plasmids are considered as the most promising tools for genetic studies of these bacteria known for the scarcity of suitable genetic vectors (19), the host specificity of these plasmids from one species to another was so far unknown. Therefore, we decided to investigate the specificity of oriC plasmids among mollicutes by reciprocal transformation of five species strategically chosen from their phylogenetic positions. Mycoplasma mycoides subsp. mycoides LC (MmmLC), M.mycoides subsp. mycoides SC (MmmSC) and M.capricolum subsp. capricolum are three highly related species belonging to the mycoides cluster (20). All three are aetiological agents of respiratory diseases of cattle. Spiroplasma citri is a plant pathogen belonging to the same Spiroplasma phylogenetic group as members of the mycoides cluster, and M.pulmonis is a rodent pathogen which belongs to the distinct Hominis phylogenetic group.
MATERIALS AND METHODS
Bacterial strains and culture conditions
The mycoplasma strains used in this study were M.mycoides subsp. mycoides LC (strain Y-goatT), M.mycoides subsp. mycoides SC (strain PG1T), M.capricolum subsp. capricolum (M.capricolum, strain California kidT) and M.pulmonis [UAB CTIP (21)]. Mycoplasmas were grown at 37°C in Hayflick medium (22) without thallium acetate and supplemented with BBL IsoVitalex Enrichment (Becton Dickinson). For growth in solid medium, mycoplasmas were incubated at 37°C under anaerobic conditions. Spiroplasma citri GII3 (23) was grown at 32°C in SP4 medium as described previously (24). Escherichia coli DH10B [(F′-mcrA Δ(mrr-hsdRMS-mcrBC) φ80dlacZΔM15 ΔlacX74 deoR recA1 endA1 araD139 Δ(ara, leu)7697 galU galK l– rpsL nupG] (Stratagene) served as the host strain for cloning procedures and plasmid propagation. Escherichia coli cells were grown in Luria–Bertani (LB) broth medium or in LB agar at 37°C. The E.coli cells transformed with plasmids were grown in LB medium supplemented with 50 µg/ml ampicillin and 5 µg/ml tetracycline.
Cloning procedures
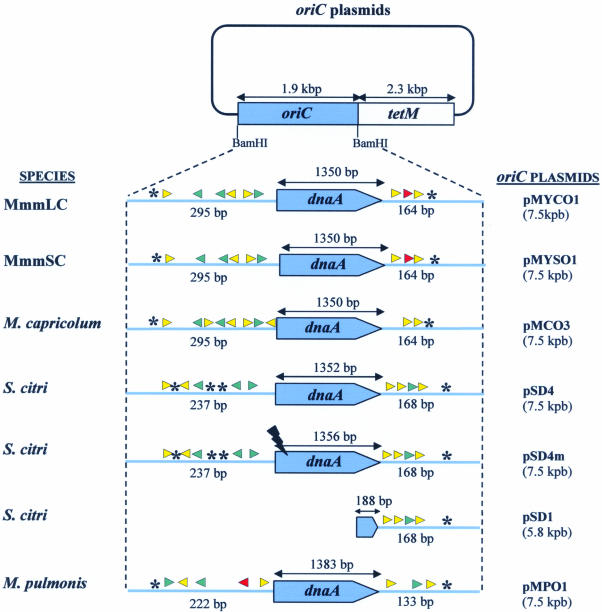
All the plasmids constructed in this study were derived from pSRT2 (24) which harbours the tetM gene from transposon Tn916 inserted into the pBS(+) plasmid (Stratagene). To construct the pMCO3 plasmid, the M.capricolum oriC region, which was previously identified and sequenced by others (25), was PCR amplified using the primers CAPO1 (5′-GAC GGGATCCTTTAGTAGCCATTCTTGCTC-3′) and CAPO2 (5′-AGGCGGATCCCAATTACTTTGGCAGC-3′) that include a BamHI site (underlined). The primers CAPO1 and CAPO2 were designed from the sequence of the genes rpmH and dnaN, located upstream and downstream from the oriC region, respectively. Consequently, the PCR product contained 76 bp from the rpmH gene at one end and 61 bp from the dnaN gene at the other. After BamHI digestion of the amplified DNA, the 1951 bp BamHI-restricted DNA fragment was inserted into the BamHI-linearised pSRT2 (Fig. 1). Similarly, the putative MmmSC oriC region was PCR amplified using the oligonucleotides MYCO1 (5′-GATCGGATCCTAGCCATTCTTGCTCTAAATC-3′) and MYCO2 (5′-GATCGGATCCTCAATTACTTTAGCTGCTTTTG-3′). These primers were chosen from the unpublished MmmSC genome sequence which overlaps the dnaA gene and which was kindly provided by Anja Persson (Royal Institute of Technology, Stockholm, Sweden). Their positions were similar to those of CAPO1 and CAPO2, respectively. Cloning the BamHI-restricted PCR fragment into pSRT2 yielded plasmid pMYSO1 (Fig. 1). Primer pair MYCO1 and MYCO2 was also used to amplify the oriC region of the closely related mycoplasma, MmmLC. The identity of this sequence was confirmed by sequencing. Plasmid pMYCO1 was obtained by cloning the BamHI-restricted MmmLC oriC into pSRT2 (Fig. 1). The oriC plasmids pMPO1 and pSD4 which replicate respectively in M.pulmonis and in S.citri have been previously described (18,24,26). The pSD1 plasmid is identical to the previously described pSD2, except that the S.citri oriC fragment was cloned in the opposite orientation (24). To generate the dnaA-disrupted pSD4m plasmid, the BglII site located at position 396 in the dnaA gene of pSD4 was cleaved and filled-in using the Klenow fragment before recircularising the plasmid (Fig. 1). Prior to being used for transformation, the purified plasmids were verified by restriction analyses and the integrity of the dnaA gene sequences was checked by DNA sequencing.
Figure 1.
Structure of oriC plasmids used in this study. Regions cloned from chromosomal oriC are indicated in blue. Putative DnaA boxes are represented by arrowheads, and AT-rich regions by asterisks. Match with the consensus (TTATCCACA) is symbolised by the following: red, 9/9; green, 8/9; yellow, 7/9. OriC regions are not drawn to scale.
Transformation of mollicutes
Mycoplasmas were transformed by the PEG-mediated method as previously described by others (19). For each transformation, ∼109 c.f.u. were transformed with 10 µg of plasmid DNA. Transformants were selected on Hayflick solid medium containing 5 µg/ml tetracycline. The cultures were incubated at 37°C and examined for colony development from the fifth day of incubation. Transformants were subcultured for 15 passages in 1 ml of Hayflick liquid medium, with the tetracycline concentration gradually increased from 5 to 20 µg/ml. Transformation of S.citri was achieved by electroporation, as described previously (27).
DNA isolation and Southern blot hybridisation
Mycoplasma genomic DNA was prepared from 10 ml cultures using the Wizard genomic DNA purification kit (Promega). For Southern blot hybridisation, 1.5 µg of genomic DNA or 15 ng of purified plasmid were digested by the appropriate restriction enzyme, and submitted to electrophoresis in a 0.8% agarose gel. After alkali transfer of the DNA fragments to a positively charged nylon membrane (Nytran Super Charge, Schleicher and Schuell), hybridisation was performed in the presence of 20 ng/ml digoxigenin-labelled DNA probes. Detection of hybridised probes was achieved using anti-digoxigenin antibodies coupled to alkaline phosphatase and the fluorescent substrate HNPP (2-hydroxy-3-naphthoic acid-2′-phenylanilide phosphate) (Roche Molecular Biochemicals). Chemi-fluorescence was detected by using a high resolution camera (Fluor-S, Biorad) and Quantity One, a dedicated software for image acquisition (Biorad).
Sequence analysis of the oriC regions of mollicutes
In silico comparisons of mollicute oriC regions were performed using either already published sequences (S.citri, accession no. Z19108; M.capricolum, D90426; M.pulmonis, NC_002771), unpublished sequences from MmmSC (kindly provided by A.Persson) or sequences obtained during this work (MmmLC, accession no. AY277700). DnaA boxes were searched within the oriC regions of mollicutes using the MEME/MAST software (28) as previously described (18). Sequence alignments were performed with the ClustalW program (29) and similarity between DnaA proteins was calculated with the Edtaln software (http://www.infobiogen.fr/services/analyseq/cgi-bin/edtaln/) using the PAM250 Dayhoff substitution matrix. Secondary structure predictions of DnaA proteins were obtained with the PHD software (30); boundaries of α-helices and β-sheets were specified using the SOPMA program (31).
RESULTS
Construction of oriC plasmids for the mycoplasmas belonging to the mycoides cluster
OriC plasmids have been previously obtained for the mollicutes S.citri (pSD4) (24) and M.pulmonis (pMPO1) (18). They have a similar architecture which includes the tetM selection marker and an oriC region comprising the dnaA gene and the flanking regions which contain the putative DnaA boxes (Fig. 1). In order to compare the host specificity of the oriC plasmids in mollicutes, we chose to extend these studies to three other mycoplasmas belonging to the mycoides cluster, within the same phylogenetic group as S.citri. Putative oriC regions of M.capricolum, MmmLC and MmmSC were analysed, and oriC plasmids were constructed and named pMCO3, pMYCO1 and pMYSO1, respectively (Fig. 1).
Conservation of the oriC regions containing the putative DnaA boxes in the genomes of mollicutes from the Spiroplasma group
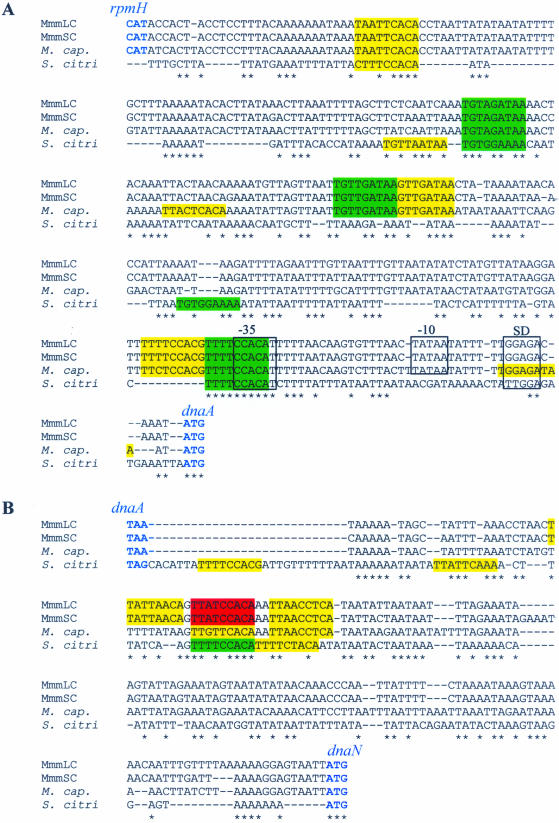
The oriC region of S.citri and the homologous region in M.capricolum have been shown to include seven (S.citri) and 10 (M.capricolum) putative DnaA boxes flanking the dnaA gene (17,25). In order to determine the variability in the organisation of putative oriC regions, the intergenic sequences flanking the dnaA gene from MmmSC, MmmLC, M.capricolum and S.citri were aligned and compared (Fig. 2). The sequences located upstream and downstream from the dnaA gene were highly conserved within the mycoides cluster. The percentage identities were ∼95% between MmmLC and MmmSC, and 75–85% between MmmLC (or MmmSC) and M.capricolum (Fig. 2). The homologous sequences from S.citri were much less conserved.
Figure 2.
Alignment of the non-coding sequences located upstream (A) and downstream (B) from the dnaA gene of MmmLC, MmmSC, M. capricolum (M.cap.) and S.citri. Putative DnaA boxes are coloured according to their matching score to the DnaA box consensus (TTATCCACA): red, 9/9; green, 8/9; yellow, 7/9. Putative promoter sequences (–35, –10), Shine–Dalgarno (SD) are positioned in reference to Seto et al. (36). Start and stop codons of the flanking genes (rpmH and dnaN) are in blue. Note that the genes located upstream from dnaA in the S.citri genome have not been characterised yet. Positions with identical nucleotide in the four sequences are noted by an asterisk.
In E.coli, the DnaA replication initiator protein recognises and binds DnaA boxes that match the consensus sequence TTATCCACA (1). Using this sequence as a reference, putative DnaA boxes were searched within the oriC regions of M.capricolum, MmmLC, MmmSC and S.citri. Of the 10 putative DnaA boxes predicted in the non-coding sequences flanking the dnaA gene of M.capricolum (25), eight were also found in the oriC region of MmmLC and MmmSC (Figs 1 and 2). Two additional DnaA boxes were predicted upstream from the dnaA gene in M.capricolum, and one additional box, specific to MmmLC and MmmSC, was found in the downstream region. For these two latter mycoplasmas, one DnaA box exactly matches the consensus sequence, three others match in eight positions out of nine and the last five match in seven positions out of nine. Putative DnaA boxes identified in the oriC of M.capricolum show a slightly more marked drift from the consensus; three with a match score of 8/9 and seven of 7/9. In S.citri, five and four putative DnaA boxes were identified in the sequences upstream and downstream from the dnaA gene, respectively; only seven of these nine boxes had been previously described (17). In S.citri, five DnaA boxes out of nine are located at similar positions in the mycoides cluster (Fig. 2). With the more phylogenetically distant M.pulmonis, five and three putative DnaA boxes have been found in the regions upstream and downstream from dnaA, respectively (Fig. 1) (18). However, sequences in these regions were too divergent to generate significant alignments with those of the Spiroplasma phylogenetic group mentioned above. Studying the replication of the E.coli chromosome has revealed that additional GATC and AGATCT sequences were involved in the initiation process (1). Similar sequences were not found in the dnaA-flanking regions of the studied mollicutes.
Transformation of mycoplasmas with homologous oriC plasmids
In order to evaluate whether predicted oriC derived from M.capricolum, MmmLC and MmmSC could promote plasmid replication, the oriC plasmid constructs were introduced into their corresponding host by PEG-mediated transformation. For each of the three mycoplasmas, tetracycline-resistant colonies appeared within 4–8 days of incubation. At that time, no spontaneous tetR colonies were observed for untransformed controls. For M.capricolum and MmmSC, transformation efficiencies with homologous plasmids were of 5.6 × 10–6 and 1.1 × 10–6 transformants/c.f.u./µg plasmid DNA, respectively (Table 1). Transformations of MmmLC were performed using the same PEG method as for the two other mycoplasmas from the mycoides cluster, but significantly higher efficiencies were observed (6.0 × 10–5 transformants/c.f.u./µg). These results suggested that the chromosomal oriC region predicted from sequence data did promote oriC plasmid replication in these mycoplasmas, as previously shown in the mollicutes M.pulmonis (18) and S.citri (24). These two latter bacteria were transformed with their respective homologous plasmids with average values of 3.1 × 10–6 and 2.0 × 10–4 transformants/c.f.u./µg, respectively. However, one has to keep in mind that M.pulmonis and S.citri have to be transformed using specific methods. Therefore, the comparison of transformation efficiencies obtained with these mollicutes and the mycoplasmas from the mycoides cluster is not meaningful.
Table 1. Transformation of mollicutes with homologous and heterologous oriC plasmids.
| Plasmid | MmmLC | MmmSC | M.capricolum | S.citri | M.pulmonis |
|---|---|---|---|---|---|
| pMYCO1 (MmmLC) | 6.0 × 10–5 | 3.0 × 10–7 | 1.2 × 10–5 | – | – |
| pMYSO1 (MmmSC) | 2.3 × 10–5 | 1.1 × 10–6 | 1.9 × 10–5 | – | – |
| pMCO3 (M.capricolum) | – | – | 5.6 × 10–6 | – | – |
| pSD4 (S.citri) | – | – | 2.7 × 10–7a | 2.0 × 10–4 | – |
| pMPO1 (M.pulmonis) | – | – | – | – | 3.1 × 10–6 |
The average transformation efficiencies from three experiments are indicated as transformants/c.f.u./µg plasmid DNA for the five species studied. The origins of the oriC plasmids are indicated in parentheses.
aCorrected value (see text).
In order to confirm that the oriC plasmids were present as free molecules in M.capricolum, MmmLC and MmmSC, genomic DNA was extracted from five independent transformants after five and 15 passages, and analysed by Southern blot hybridisation (Fig. 3). For transformations with pMCO3, pMYCO1 and pMYSO1, the EcoRI fragments of 10 and 7.5 kbp hybridising with the oriC probe correspond to wild-type chromosomal oriC and free plasmid molecules, respectively (Fig. 3D). In contrast, the presence of EcoRI fragments of 2 and >10 kbp hybridising with the same probe indicated integration of oriC plasmid into the chromosomal replication origin. Analysis of M.capricolum/pMCO3 transformants only revealed wild-type chromosomal oriC and free plasmid, at least until 15 passages (Fig. 3A). For MmmLC transformed with pMYCO1, extrachromosomal plasmid molecules were observed after five passages, but integration events at the chromosomal replication origin were also detected for four clones (Fig. 3B). After 15 passages, free plasmid molecules could not be detected in four of the five clones (Fig. 3B; clones 1, 2, 3 and 5). In the fifth clone, in which no recombination could be detected after five passages, a weak 2 kbp hybridising band was observed after 15 passages, indicating that plasmid recombination had occurred, at least in some cells. For MmmSC/pMYSO1 transformants, free plasmid was observed for the five clones at both five and 15 passages (Fig. 3C). Plasmid integration was detected only in one clone (clone 4) after 15 passages.
Figure 3.
State of integration of homologous oriC plasmids in M.capricolum, MmmLC and MmmSC. EcoRI-restricted DNAs extracted from transformants (clones 1–5) after five and 15 passages (5P, 15P) were probed with the corresponding oriC probe. Southern blot was performed for M.capricolum transformed with pMCO3 (A), MmmLC with pMYCO1 (B) and MmmSC with pMYSO1 (C). Lane P corresponds to the purified plasmid DNA, and lanes MC, LC and SC correspond to the genomic DNA from M.capricolum, MmmLC and MmmSC, respectively. Sizes are indicated in kilobase pairs. (D) Schematic representation of oriC plasmid integration by recombination at the chromosomal replication origin. pBS, plasmid pBluescript; E, EcoRI.
The presence of extrachromosomal DNA molecules in transformants shows that the oriC region predicted in silico for M.capricolum, MmmLC and MmmSC actually enables the replication of plasmids. Moreover, molecular analyses of transformants after five and 15 passages indicate a marked heterogeneity in the occurrence of homologous recombination events at the chromosomal oriC. While nearly no recombination events could be observed for M.capricolum and MmmSC, integrated plasmidic sequences were detected in MmmLC transformants after a few passages.
Host specificity of oriC plasmids
To investigate the host specificity of the oriC plasmids among mollicutes, we performed a set of heterologous transformations (Table 1). MmmLC and MmmSC were efficiently transformed with both plasmids pMYCO1 and pMYSO1. No transformants of these two species were obtained with the plasmids from M.capricolum and from the more distant species S.citri and M.pulmonis. In contrast, transformation of M.capricolum with pMYCO1 and pMYSO1 originating from MmmLC and MmmSC yielded transformants at slightly higher efficiencies (∼1.5 × 10–5 transformants/c.f.u./µg) than that obtained with the homologous plasmid pMCO3 (∼5.6 × 10–6 transformants/c.f.u./µg). In addition, M.capricolum could also be transformed with pSD4 harbouring the S.citri oriC. In this case, tetR colonies appeared only after an extended incubation time (8–10 days), and further molecular analysis of 10 clones revealed that only four of them actually contain plasmid sequences (see below). The corrected transformation efficiency was therefore only 2.7 × 10–7 transformants/c.f.u./µg. No M.capricolum transformants could be obtained with the plasmid from M.pulmonis. All attempts to transform S.citri and M.pulmonis with heterologous plasmids failed.
A molecular analysis of five transformants from each of the five successful heterologous transformations was performed (fig. S1 of the Supplementary Material available at NAR Online). For MmmSC transformed with the heterologous plasmid pMYCO1, only one clone displayed, after 15 passages, a four-band pattern indicating the presence of free plasmid and integrated molecules. This scarcity of recombinants is similar to what was observed for the MmmSC/pMYSO1 homologous transformants. In contrast, while integration events were detected in all clones of MmmLC transformed by the homologous plasmid pMYCO1, only one recombinant was evidenced among the five clones. In the case of M.capricolum transformed by pMYCO1 or pMYSO1, the Southern patterns at the 15th passage of all clones indicated the presence of free plasmids and also of some integrated molecules. This result contrasts with the lack of integration observed in the M.capricolum/pMCO3 homologous transformants. Only free plasmid molecules were detected in M.capricolum transformed with the pSD4 plasmid from S.citri.
These results as a whole indicate that the host specificity of oriC plasmids is not absolute, and that closely related mollicutes present a variable ability to replicate heterologous plasmids. Moreover, integration events of heterologous plasmids by recombination at the chromosomal oriC are observed in all three mycoplasmas from the mycoides cluster, but the occurrence of these events seems to be very different according to the mycoplasma and the plasmid.
Replication of heterologous oriC plasmids without a functional plasmid dnaA gene
To further investigate the ability of oriC plasmids to replicate in mycoplasmas, the requirement for the plasmid-encoded DnaA proteins in heterologous transformation systems was questioned.
First, with the aim of identifying putative variations within conserved domains of the DnaA proteins considered in this work, a detailed comparison of their amino acid sequences was performed (Supplementary fig. S2). Typical motifs and predicted secondary structures organisation in 15 α-helices and nine β-strands were similar to other DnaA proteins arrangements, as described by Weigel and Messer (2001, www.molgen.mpg.de/~messer). As indicated in Table 2, the DnaA primary sequence is highly conserved within the mycoides cluster (95.9–98.7% similarity) but strikingly less when compared with that of S.citri (40.0–40.9%). Compared with the sequences from the Spiroplasma group, the M.pulmonis DnaA sequence was found to be poorly conserved.
Table 2. Percentage similarity of the DnaA proteins of the studied mollicutes.
| Mollicute | MmmLC | MmmSC | M.capricolum | S.citri | M. pulmonis |
|---|---|---|---|---|---|
| MmmLC | 100.0 | 98.7 | 96.6 | 40.0 | 17.9 |
| MmmSC | – | 100.0 | 95.9 | 40.0 | 17.9 |
| M.capricolum | – | – | 100.0 | 40.9 | 18.2 |
| S.citri | – | – | – | 100.0 | 16.9 |
| M.pulmonis | – | – | – | – | 100.0 |
Within the mycoides cluster, the characteristic motifs Walker A, Walker B, RFC box, Sensor 2 and the DnaA signature motif, which have been shown to define both the affinity and specificity of DnaA box binding, are fully conserved; one single substitution was observed within the B-loop motif (also required for DNA binding). However, when extending the comparison to the five DnaA sequences studied, the motifs did not appear significantly more conserved than the whole sequence. Noticeably, comparing the DnaA sequence from S.citri with those from the mycoides cluster revealed 6/17 and 20/27 amino acid substitutions within the DnaA signature and B-loop motifs, respectively.
Considering the low level of conservation of these DnaA box-binding motifs, it was unclear whether the replication of pSD4 (from S.citri) in M.capricolum required the plasmid-encoded S.citri DnaA or the chromosomal M.capricolum DnaA. To answer this question, M.capricolum was transformed with two plasmids derived from pSD4 (Fig. 1). Plasmid pSD4m was identical to pSD4 except that the dnaA gene was disrupted as described in Materials and Methods. In pSD1, the S.citri oriC region lacks dnaA and is reduced to the DnaA box region located downstream of the dnaA gene. This plasmid was shown to replicate in S.citri (S.Duret and J.Renaudin, unpublished data). Eight days after M.capricolum transformation, tetR colonies were observed with all three plasmids. For each transformation, the genomic DNA from 10 putative transformants was extracted after five and 15 passages and analysed by Southern blot using the S.citri oriC region as a probe; due to the low level of sequence homology, this probe does not hybridise to the M.capricolum oriC. Hybridisation bands were observed for four, two and three clones transformed by plasmids pSD4, pSD4m and pSD1, respectively (Fig. 4). For all the other clones, no hybridising band could be observed, suggesting that they were spontaneously resistant to tetracycline. Only free plasmid was observed for pSD4 and pSD1 transformants (Fig. 4A and B). In contrast, in one of the two pSD4m transformants, the detection of two additional fragments of 2.1 and 10 kbp suggested that in some cells, pSD4m had integrated into the chromosome. However, hybridisation with a M.capricolum oriC probe revealed a unique 10 kbp band showing that in clone 2, plasmid recombination did not occur at the chromosomal oriC but elsewhere in the chromosome (Fig. 4D).
Figure 4.
Transformation of M.capricolum with dnaA-inactivated oriC plasmids from S.citri. Genomic DNAs from pSD4 (A) and pSD1 (B) transformants were EcoRI restricted and hybridised with an S.citri oriC probe. XbaI-restricted DNAs extracted from pSD4m transformants were hybridised using S.citri oriC (C) or M.capricolum oriC (D) as a probe. Lanes P and MC correspond to the purified plasmid DNA and the genomic DNA, respectively, from untransformed M.capricolum. Sizes are indicated in kilobase pairs.
These results show that the replication machinery from M.capricolum efficiently promotes the replication of S.citri-derived oriC plasmids.
DISCUSSION
Replicative plasmids for mycoplasmas of the mycoides cluster
In this study, we have developed oriC plasmids from three mycoplasmas belonging to the mycoides cluster. The cloned mycoplasma oriC fragment encompasses the dnaA gene and the bordering intergenic regions that contain putative DnaA boxes. As previously shown for the murine pathogen M.pulmonis (18) and the plant mollicute S.citri (17), oriC plasmids were efficiently replicated in their respective hosts, demonstrating the functionality of in silico predicted oriC regions. This functional characterisation is in agreement with previous studies of the progression of replication forks in M.capricolum (15,16). These plasmids constitute the first stable genetic vectors for three pathogenic mycoplasmas, especially for MmmSC, the aetiological agent of bovine contagious pleuropneumoniae.
Recombination of oriC plasmid at the chromosomal oriC region has been observed after a few passages in liquid medium for S.citri (32) and M.pulmonis (18). Within the mycoides cluster, various recombination patterns were observed. Homologous plasmids were mostly maintained as free molecules in most of the M.capricolum and MmmSC transformants. In contrast, integrations of the homologous plasmid were detected as early as the fifth passage for MmmLC. This noticeable difference among mollicutes of the Spiroplasma phylogenetic group may reflect a basic difference in their recombination potential. Alternatively, the presence of an additional oriC in the chromosome after recombination may be more deleterious for specific mycoplasmas.
Interestingly, we observed that, in contrast to what was observed with the homologous plasmid, the integration of the heterologous plasmids from MmmSC and MmmLC seemed to occur in M.capricolum oriC. This result indicates that homologous recombination events actually occur in this mycoplasma and suggests that gene targeting strategies based on the use of heterologous oriC plasmids could be successful with this bacterium.
Host specificity of oriC plasmids
In this work, the host specificity of oriC plasmids among mollicutes has been investigated by reciprocal transformation of M.pulmonis, S.citri and three mycoplasmas from the mycoides cluster. No inter-compatibility of oriC plasmids could be observed between M.pulmonis and mollicutes from the Spiroplasma phylogenetic group. Likewise, transformants of S.citri were exclusively obtained with homologous plasmids. In contrast, within the mycoides cluster, MmmLC and MmmSC could be transformed by reciprocal plasmids. All of these results are in agreement with the assumption that oriC plasmids are only exchangeable between closely related species, as described for several groups of bacteria (33–35). Interestingly, M.capricolum was successfully transformed not only by all oriC plasmids from the mycoides cluster but also by the S.citri oriC plasmids. The replication of these plasmids in M.capricolum is therefore one of the very few examples of oriC plasmids that are compatible between different bacterial genera (11–13). Heterologous transformations of oriC plasmids from Enterobacter aerogenes, Klebsiella pneumoniae (11), Vibrio harveyi (13) and Erwinia carotovora (12) have been successfully performed in E.coli. However, it is noteworthy that the 250 bp oriC region of these phylogenetically distant species presents similar organisations with four highly conserved DnaA boxes (R1–R4). In contrast, sequence comparison of the oriC regions of M.capricolum and S.citri has shown that the sequences and the positions of the DnaA boxes were poorly conserved. Interestingly, it has been shown that the dnaA gene is highly expressed in M.capricolum (36). It was proposed that the lack of conservation of the DnaA box sequences in the oriC of M.capricolum might be compensated by an enhanced amount of DnaA protein. This hypothesis could potentially explain the unusual ability of this mycoplasma to replicate heterologous oriC plasmids.
In this work, oriC plasmids were obtained for three mycoplasmas of the mycoides cluster, MmmSC, MmmLC and M.capricolum. These plasmids are of particular interest because: (i) they are major pathogens for ruminants (37); (ii) the complete genome sequence will soon be available for MmmSC and M.capricolum, and genetic tools are urgently required for functional studies; (iii) there is a general lack of knowledge of the virulence mechanisms for these bacteria; and (iv) the development of new vaccines against MmmSC is considered to be an international priority (38). This study may lead to the general use of oriC-based plasmids for the functional genomics of species belonging to the genus Mycoplasma since there is a real need for such genetic tools (39).
SUPPLEMENTARY MATERIAL
The molecular analyses of mycoplasma transformants by heterologous plasmids are available in the Supplementary Material available at NAR Online (Fig. S1). The alignment of the deduced DnaA proteins from MmmLC, MmmSC, M.capricolum, S.citri and M.pulmonis is also provided (Fig. S2).
Acknowledgments
ACKNOWLEDGEMENTS
We thank Dr Anja Persson for kindly providing the oriC sequence of MmmSC, Sybille Duret-Nurbel for constructing pSD1 and pSD4, and Géraldine Gourgues for technical help. This research was supported by grants from INRA, Région Aquitaine, University Victor Ségalen Bordeaux 2, and CIRAD.
DDBJ/EMBL/GenBank accession no. AY277700
REFERENCES
- 1.Messer W. (2002) The bacterial replication initiator DnaA. DnaA and oriC, the bacterial mode to initiate DNA replication. FEMSMicrobiol. Rev., 26, 355–374. [DOI] [PubMed] [Google Scholar]
- 2.Akman L., Yamashita,A., Watanabe,H., Oshima,K., Shiba,T., Hattori,M. and Aksoy,S. (2002) Genome sequence of the endocellular obligate symbiont of tsetse flies, Wigglesworthia glossinidia. Nature Genet., 32, 402–407. [DOI] [PubMed] [Google Scholar]
- 3.Messer W. and Weigel,C. (1996) Initiation of chromosome replication. In Neidhardt,F.C., Curtiss,R., Lin,E.C.C., Low,K.B., Magasanik,B., Reznikoff,W.S., Riley,M., Schaechter,M. and Umbarger,H.E. (eds), Escherichia coli and Salmonella. ASM Press, Washington, DC, pp. 1579–1601. [Google Scholar]
- 4.Yoshikawa H. and Wake,R.G. (1993) Initiation and termination of chromosome replication. In Sonenshein,A.L., Hoch,J.A. and Losick,R. (eds), Bacillus subtilis and Other Gram-positive Bacteria. ASM, Washington, DC, pp. 507–528. [Google Scholar]
- 5.Lobry J.R. (1996) Asymmetric substitution patterns in the two DNA strands of bacteria. Mol. Biol. Evol., 13, 660–665. [DOI] [PubMed] [Google Scholar]
- 6.Brewer B.J. and Fangman,W.L. (1987) The localization of replication origins on ARS plasmids in S.cerevisiae. Cell, 51, 463–471. [DOI] [PubMed] [Google Scholar]
- 7.Huberman J.A., Spotila,L.D., Nawotka,K.A., el-Assouli,S.M. and Davis,L.R. (1987) The in vivo replication origin of the yeast 2 microns plasmid. Cell, 51, 473–481. [DOI] [PubMed] [Google Scholar]
- 8.Yee T.W. and Smith,D.W. (1990) Pseudomonas chromosomal replication origins: a bacterial class distinct from Escherichia coli-type origins. Proc. Natl Acad. Sci. USA, 87, 1278–1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Skarstad K. and Boye,E. (1994) The initiator protein DnaA: evolution, properties and function. Biochim. Biophys. Acta, 1217, 111–130. [DOI] [PubMed] [Google Scholar]
- 10.Qin M.H., Madiraju,M.V. and Rajagopalan,M. (1999) Characterization of the functional replication origin of Mycobacterium tuberculosis. Gene, 233, 121–130. [DOI] [PubMed] [Google Scholar]
- 11.Harding N.E., Cleary,J.M., Smith,D.W., Michon,J.J., Brusilow,W.S. and Zyskind,J.W. (1982) Chromosomal replication origins (oriC) of Enterobacter aerogenes and Klebsiella pneumoniae are functional in Escherichia coli. J. Bacteriol., 152, 983–993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Takeda Y., Harding,N.E., Smith,D.W. and Zyskind,J.W. (1982) The chromosomal origin of replication (oriC) of Erwinia carotovora. Nucleic Acids Res., 10, 2639–2650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zyskind J.W., Cleary,J.M., Brusilow,W.S., Harding,N.E. and Smith,D.W. (1983) Chromosomal replication origin from the marine bacterium Vibrio harveyi functions in Escherichia coli: oriC consensus sequence. Proc. Natl Acad. Sci. USA, 80, 1164–1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Razin S., Yogev,D. and Naot,Y. (1998) Molecular biology and pathogenicity of mycoplasmas. Microbiol. Mol. Biol. Rev., 62, 1094–1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Miyata M., Sano,K., Okada,R. and Fukumura,T. (1993) Mapping of replication initiation site in Mycoplasma capricolum genome by two-dimensional gel-electrophoretic analysis. Nucleic Acids Res., 21, 4816–4823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Miyata M. and Fukumura,T. (1997) Asymmetrical progression of replication forks just after initiation on Mycoplasma capricolum chromosome revealed by two-dimensional gel electrophoresis. Gene, 193, 39–47. [DOI] [PubMed] [Google Scholar]
- 17.Ye F., Renaudin,J., Bove,J.M. and Laigret,F. (1994) Cloning and sequencing of the replication origin (oriC) of the Spiroplasma citri chromosome and construction of autonomously replicating artificial plasmids. Curr. Microbiol., 29, 23–29. [DOI] [PubMed] [Google Scholar]
- 18.Cordova C.M., Lartigue,C., Sirand-Pugnet,P., Renaudin,J., Cunha,R.A. and Blanchard,A. (2002) Identification of the origin of replication of the Mycoplasma pulmonis chromosome and its use in oriC replicative plasmids. J. Bacteriol., 184, 5426–5435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dybvig K. and Voelker,L.L. (1996) Molecular biology of mycoplasmas. Annu. Rev. Microbiol., 50, 25–57. [DOI] [PubMed] [Google Scholar]
- 20.Pettersson B., Leitner,T., Ronaghi,M., Bolske,G., Uhlen,M. and Johansson,K.E. (1996) Phylogeny of the Mycoplasma mycoides cluster as determined by sequence analysis of the 16S rRNA genes from the two rRNA operons. J. Bacteriol., 178, 4131–4142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chambaud I., Heilig,R., Ferris,S., Barbe,V., Samson,D., Galisson,F., Moszer,I., Dybvig,K., Wroblewski,H., Viari,A. et al. (2001) The complete genome sequence of the murine respiratory pathogen Mycoplasma pulmonis. Nucleic Acids Res., 29, 2145–2153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Freund E.A. (1983) Culture media for classic mycoplasmas. In Razin,S. and Tully,J.G. (eds), Methods in Mycoplasmology. Academic Press, San Diego, Vol. 1, p. 127. [Google Scholar]
- 23.Vigneau J.C., Bové,J.M., Saillard,C., Vogel,R., Faro,A., Venegas,L., W,S., Aoki,S., McCoy,R., Albeldawi,A.S. et al. (1980) Mise en culture de spiroplasmes à partir de matériel végétal et d’insectes provenant des pays circum-méditéranéens et du Proche-Orient. C.R. Acad. Sci. Paris, 290, 775–778. [Google Scholar]
- 24.Lartigue C., Duret,S., Garnier,M. and Renaudin,J. (2002) New plasmid vectors for specific gene targeting in Spiroplasma citri. Plasmid, 48, 149–159. [DOI] [PubMed] [Google Scholar]
- 25.Fujita M.Q., Yoshikawa,H. and Ogasawara,N. (1992) Structure of the dnaA and DnaA-box region in the Mycoplasma capricolum chromosome: conservation and variations in the course of evolution. Gene, 110, 17–23. [DOI] [PubMed] [Google Scholar]
- 26.Renaudin J., Marais,A., Verdin,E., Duret,S., Foissac,X., Laigret,F. and Bove,J.M. (1995) Integrative and free Spiroplasma citri oriC plasmids: expression of the Spiroplasma phoeniceum spiralin in Spiroplasma citri. J. Bacteriol., 177, 2870–2877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stamburski C., Renaudin,J. and Bove,J.M. (1991) First step toward a virus-derived vector for gene cloning and expression in spiroplasmas, organisms which read UGA as a tryptophan codon: synthesis of chloramphenicol acetyltransferase in Spiroplasma citri. J. Bacteriol., 173, 2225–2230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bailey T.L. and Elkan,C. (1994) Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc. Int. Conf. Intell. Syst. Mol. Biol., 2, 28–36. [PubMed] [Google Scholar]
- 29.Thompson J.D., Higgins,D.G. and Gibson,T.J. (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res., 22, 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rost B. and Sander,C. (1994) Combining evolutionary information and neural networks to predict protein secondary structure. Proteins, 19, 55–72. [DOI] [PubMed] [Google Scholar]
- 31.Geourjon C. and Deleage,G. (1995) SOPMA: significant improvements in protein secondary structure prediction by consensus prediction from multiple alignments. Comput. Appl. Biosci., 11, 681–684. [DOI] [PubMed] [Google Scholar]
- 32.Duret S., Danet,J.L., Garnier,M. and Renaudin,J. (1999) Gene disruption through homologous recombination in Spiroplasma citri: an scm1-disrupted motility mutant is pathogenic. J. Bacteriol., 181, 7449–7456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jakimowicz D., Majka,J., Messer,W., Speck,C., Fernandez,M., Martin,M.C., Sanchez,J., Schauwecker,F., Keller,U., Schrempf,H. et al. (1998) Structural elements of the Streptomyces oriC region and their interactions with the DnaA protein. Microbiology, 144, 1281–1290. [DOI] [PubMed] [Google Scholar]
- 34.Madiraju M.V., Qin,M.H., Yamamoto,K., Atkinson,M.A. and Rajagopalan,M. (1999) The dnaA gene region of Mycobacterium avium and the autonomous replication activities of its 5′ and 3′ flanking regions. Microbiology, 145, 2913–2921. [DOI] [PubMed] [Google Scholar]
- 35.Suvorov A.N. and Ferretti,J.J. (2000) Replication origin of Streptococcus pyogenes, organization and cloning in heterologous systems. FEMS Microbiol. Lett., 189, 293–297. [DOI] [PubMed] [Google Scholar]
- 36.Seto S., Murata,S. and Miyata,M. (1997) Characterization of dnaA gene expression in Mycoplasma capricolum. FEMS Microbiol. Lett., 150, 239–247. [DOI] [PubMed] [Google Scholar]
- 37.Frey J. (2002) Mycoplasmas of animals. In Razin,S. and Herrmann,R. (eds), Molecular Biology and Pathogenicity of Mycoplasmas. Kluwer Academic/Plenum Publishers, London, pp. 73–90. [Google Scholar]
- 38.Tulasne J.J., Litamoi,J.K., Morein,B., Dedieu,L., Palya,V.J., Yami,M., Abusugra,I., Sylla,D. and Bensaid,A. (1996) Contagious bovine pleuropneumonia vaccines: the current situation and the need for improvement. Rev. Sci. Tech., 15, 1373–1396. [DOI] [PubMed] [Google Scholar]
- 39.Renaudin J. (2002) Extrachromosomal elements and gene transfer. In Razin,S. and Herrmann,R. (eds.), Molecular Biology and Pathogenicity of Mycoplasmas. Kluwer Academic/Plenum Publisher, New York, NY, pp. 347–371. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.