Abstract
Members of the two expanding RNA subclasses termed C/D and H/ACA RNAs guide the 2′-O-methylations and pseudouridylations, respectively, of rRNA and spliceosomal RNAs (snRNAs). Here, we report on the identification of 13 novel human intron-encoded small RNAs (U94–U106) belonging to the two subclasses of modification guides. Seven of them are predicted to direct 2′-O-methylations in rRNA or snRNAs, while the remainder represent novel orphan RNA modification guides. From these, U100, which is exclusively detected in Cajal bodies (CBs), is predicted to direct modification of a U6 snRNA uridine, U9, which to date has not been found to be pseudouridylated. Hence, within CBs, U100 might function in the folding pathway or other aspects of U6 snRNA metabolism rather than acting as a pseudouridylation guide. U106 C/D snoRNA might also possess an RNA chaperone activity only since its two conserved antisense elements match two rRNA sequences devoid of methylated nucleotides and located remarkably close to each other within the 18S rRNA secondary structure. Finally, we have identified a retrogene for U99 snoRNA located within an intron of the Siat5 gene, supporting the notion that retro-transposition events might have played a substantial role in the mobility and diversification of snoRNA genes during evolution.
INTRODUCTION
Eukaryal, bacterial and archaeal organisms contain an unexpectedly large number of non-coding RNAs (also called non-messenger RNAs or nmRNAs). Although most of them are poorly characterised as yet, they are thought to play important roles at various steps in the control of gene expression (1–3). Experimental RNomics studies in multicellular organisms have recently begun to illustrate the complexity of the nmRNA population in several model organisms (4–9). A large fraction of nmRNAs belong to well-defined families, e.g. snRNAs, snoRNAs or miRNAs, exhibiting specific structural hallmarks, thus allowing many of them to be identified by computational searches of completely sequenced genomes. The combination of experimental and computational RNomics is now paving the way for a better understanding of the functions of this diverse class of RNA molecules (7).
In eukaryal and archaeal organisms, a substantial proportion of the nmRNAs identified so far belong to the two expanding subclasses, termed C/D and H/ACA RNAs, which guide the two prevalent types of rRNA modifications in both groups of organisms, i.e. 2′-O-methylation and pseudouridylation, respectively. For both subclasses, each guide RNA specifies the nucleotide to be modified through the formation of a canonical duplex spanning the cognate modification site (4,10–13). In eukaryotes, most of these relatively short guide RNAs (60–100 nt for C/D RNAs and 120–140 nt for H/ACA RNAs) accumulate within the nucleolus, hence they are designated as snoRNAs. In addition to targeting rRNAs, C/D and H/ACA snoRNAs also direct modifications of the Pol III transcribed spliceosomal U6 snRNA which is thought to transit through the nucleolus (14). In contrast, the C/D or H/ACA RNAs involved in the modification of the Pol II-transcribed spliceosomal snRNAs, U1, U2, U4 and U5, do not accumulate within the nucleolus but are exclusively found within the Cajal (coiled) bodies (or CBs), a subnuclear compartment in which they appear to interact with their RNA substrates (15). This subset of modification guides have been called small CB-specific RNAs or scaRNAs (16). Cellular RNAs targeted by C/D and H/ACA RNA guides also include tRNAs in Archaea as well as probably a few other RNA species in Eukarya (12,17). Intriguingly, an increasing number of ‘orphan snoRNAs’, i.e. presumptive modification guides which remain so far without an identified cellular RNA target, have been reported in mammals, some of them being expressed in a tissue-specific manner and subjected to genomic imprinting (6,17–20). The range of cellular functions mediated by both subclasses of modification guides might therefore be larger than recognised so far, particularly in complex organisms such as vertebrates or plants, stressing the need for a thorough identification of the C/D and H/ACA repertoires of such organisms.
Computational genomic searches of C/D RNAs targeting rRNA or spliceosomal snRNAs have been efficient in many organisms (21–26), mainly based on the presence of a relatively long, continuous antisense element at the target site. However, the power of this approach is considerably limited in the case of C/D specimens targeting unknown cellular RNA species. As for the H/ACA RNAs, due to the bipartite structure of their shorter antisense elements, most of them so far have remained refractory to computational searches, even for those targeting rRNAs or snRNAs, and most of them have been identified through direct experimental approaches (5,6,8).
Here, by a combination of experimental screens of rat libraries and in silico searches of the human and mouse genomes, we report on the characterisation of 10 novel C/D and three novel H/ACA small RNAs which are all intron encoded and conserved among human and rodents. Remarkably, six of these new specimens (U97, U98, U99, U100, U101 and U106) do not seem to be involved in the modification of rRNAs or snRNAs. One of them, H/ACA U100, is particularly intriguing as it is the first scaRNA predicted to target a Pol III transcript, snRNA U6, at a nucleotide position not known to be modified, suggesting that it might have a function different from a typical pseudouridylation guide.
MATERIALS AND METHODS
Unless otherwise noted, all techniques for cloning and manipulating nucleic acids were performed according to standard protocols.
Oligonucleotides
The following oligonucleotides were used: U6, 5′-CGTGTCATCCTTGCGCAGGGGCC-3′; U3, 5′-AAATGTCCCTGAAAGTATAGTCTT-3′; U94, 5′-TCCGTACCCCTGC GCCAATCATCA-3′; U97, 5′-TCATATCTCATAATCTTCGCTCATAGGACG-3′; U98, 5′-AAACAGAACTGCGACCGTCAAGGAA-3′; U99, 5′-TGTCCCGGCGTTTGAGGAT AGAACC-3′; U100, 5′-TGTATGGAGCCATCGCACAGAAAATCTGA-3′; U101, 5′-TCAGACTCTTATGTTTCACTCATAA-3′; U102, 5′-TCAGAGCCGGTGAAATGTGTT TTC-3′; U106, 5′-TCAGAACTAACTGGCAAAATATAAGACGTCA-3′; RT-18S, 5′-CCTCGTTCATGGGGAATAATTGC-3′; and U100-C6dT, 5′-GATAACTAXACAGACC CTGXCGGCAGGAACCATCTGXTTTAATGTGTGXG-3′ (amino-allyl-modified T residues are indicated by an XX).
Search for novel RNA modification guides
Human ribosomal protein genes were systematically searched for C (RUGAUGA) and D (CUGA) boxes. Pairs of correctly spaced (C box 35–140 bp upstream from D box) C and D boxes, with no more than one deviation as compared with the C and D box consensus, and flanked by complementary sequences (at least 4 bp complementarity, with no more than one G–U pair, among the five nucleotides beside each box), were then searched for murine orthologues using a UCSC blat search (http://genome.ucsc.edu/cgi-bin/hgBlat). Construction of the C/D snoRNA cDNA library has been described (19). The insert of individual clones was PCR amplified using M13 reverse and forward primers and sequenced using M13 forward primer and the BigDye terminator cycle sequencing reaction kit (PE Applied Biosystems). Sequences were analysed on an ABI Prism 377 (Perkin Elmer) sequencer using the LASERGENE sequence analysis program package.
Cell fractionation, immunoprecipitation and northern blot analysis
Subcellular fractionation of HeLa cells was performed as described (27). Immunoprecipitation of trimethylated capped RNAs with monoclonal antibody R1131 (kindly provided by Dr R. Luhrmann) was performed according to Cavaille et al. (17). RNAs were fractionated by electrophoresis in 6% acrylamide/7 M urea gels and transferred onto nylon membranes (Qiabrane Nylon Plus, Qiagen) using the Biorad semi-dry blotting apparatus (Trans-blot SD, Biorad). After immobilising RNA using the Stratagene cross-linker, nylon membranes were pre-hybridised for 30 min in 1 M sodium phosphate buffer pH 6.2, 7% SDS. Oligonucleotides complementary to the respective snoRNA species were end-labelled with [32P]ATP and T4 polynucleotide kinase; hybridisation was carried out at 58°C in 1 M sodium phosphate buffer pH 6.2, 7% SDS overnight. Blots were washed twice at room temperature in 2× SSC buffer (20 mM sodium phosphate pH 7.4, 0.3 M NaCl, 2 mM EDTA), 0.1% SDS for 15 min and exposed to Kodak MR-1 film.
In situ hybridisation
The human U100 gene was PCR amplified from HeLa cell genomic DNA and cloned into the ClaI and XhoI sites of the pCMV-globin expression vector [kindly provided by Dr T. Kiss (16)]. This pCMV-U100 vector together with pGFP-coilin (kindly provided by Dr E. Bertrand) was transfected into HeLa cells with Fugene transfection reagent according to the manufacturer’s recommendations (Roche). The U100-specific modified oligonucleotide was labelled with FluoroLink Cy3 reactive dye (Amersham), and fluorescent in situ hybridisation (FISH) was performed according to the protocol of the laboratory of Dr Singer (http://singerlab.aecom.yu.edu). Nuclear DNA was stained by 4′,6-diamidino-2-phenylindole (DAPI).
RESULTS AND DISCUSSION
Identification of novel C/D snoRNAs
By screening two C/D snoRNA-specific cDNA libraries obtained from a whole rat brain (19) and from rat testis extracts (P.Vitali and A.Hüttenhofer, unpublished data), we have identified scores of C/D RNAs. While most of them correspond to rat counterparts of previously characterised human (4) or mouse (6) C/D snoRNAs (data not shown), six novel specimens were detected among the sequenced clones. These RNAs, referred to as U94, U95, U103, U104, U105 and U106, exhibit all the C/D hallmarks, and their homologues in mouse and human have been identified by database searches (not shown).
In most cases, the small intron-encoded RNA is hosted by the same gene in human and mouse. In both mammals, two U103 gene copies are located within introns 17 and 21 of the PUM1 gene (28), while U94 and U95 are encoded within introns of the FLJ20758 and GNB2L1 genes, respectively (Table 1). In the case of U95, however, found in GNB2L1 intron 1 in both mammals, an additional gene copy is present in intron 3 of the same gene in mouse only (Fig. 1).
Table 1. Compilation and major features of the RNA modification guides described in this study.
| Name | Family | RNA target (human) | Antisense element | snoRNA host gene (human) | Accession no. |
|---|---|---|---|---|---|
| U94 | C/D | Cm62 (U6) | 13 bp/D | FLJ20758 (intron 21), conserved hypothetical protein | AY349593 |
| U95a | C/D | Am2792/Cm2801 (28S) | 13 bp/D + 10 bp/D′ | GNB2L1 (intron 1), guanine nucleotide-binding protein (G protein) | AY349594 |
| U96aa,b | C/D | Gm75 (5.8S) | 12 bp/D | GNB2L1 (intron 2) | AY349595 |
| U96b | C/D | Gm75 (5.8S) | 12 bp/D | AMMECR1 (intron 2), unknown function | AY349596 |
| U97 | C/D | ND | EIF4G2 (intron 14), translation initiation factor | AY349597 | |
| U98a | H/ACA | ND | ND | LOC85028 (intron 2), poorly characterised gene | AY349598 |
| U98b | H/ACA | ND | PPP2R5A (intron 8), protein phosphatase 2, regulatory subunit B | AY349599 | |
| U99 | H/ACA | ND | MGC2477 (intron 3), conserved hypothetical protein | AY349600 | |
| U100 | H/ACA | U9? (U6)c | (6 + 4)/3′ hairpin | FLJ20516 (intron 6), homologous to Tipin | AY349601 |
| U101 | C/D | ND | RPS12 (intron 3), ribosomal protein | AY349602 | |
| U102 | C/D | Gm4010 (28S) | 11 bp/D | RPL21 (intron 2), ribosomal protein | AY349603 |
| U103ad | C/D | Gm601 (18S) | 10 bp/D′ | PUM1 (intron 17), RNA-binding protein (Pumilio family member) | AY349604 |
| U103bd | C/D | Gm601 (18S) | 10 bp/D′ | PUM1 (intron 21) | |
| U104a | C/D | Cm1320 (28S) | 14 bp/D′ | Not annotated gene | AY349605 |
| U105 | C/D | Um799 (18S) | 15 bp/D | SSF1 (intron 3), Peter Pan homologue (contains a Brix domain) | AY349606 |
| U106 | C/D | G1536?/U1602? (18S)c | 10 bp/D + 10 bp/D′ | BC032480 (intron 1), poorly characterised gene | AY349607 |
aU95, U96 and U104 sequences have been independently submitted in the public database as: Z38, Z37 and Z12 snoRNAs, respectively.
bU96 might be the mammalian counterpart of the A.thaliana Z37a and Z37b snoRNA involved in Gm79 formation in A.thaliana 5.8S rRNA (24).
cA question mark means that no Ψ9 or Gm1536 and Um1602 have been detected in human U6 snRNA and 18S rRNA, respectively.
dU103a and U103b RNAs target the same rRNA position as MBII-251 and thus might be considered as snoRNA isoforms of the latter.
Figure 1.
Differences in the genomic organisation of the three novel snoRNAs between human and mouse. Each snoRNA sequence (small arrow) is located within an intron of the indicated genes. Exons are represented by boxes and splicing events by dotted lines. Note that AK009175, a spliced EST, and LOC85028, a poorly characterised gene, both located 15–21 kb downstream from Taf12 in mouse and human, are not related to each other. The cartoon is not drawn to scale.
Genes hosting an intron-encoded snoRNA frequently contain one (or more) additional snoRNA(s) within other intron(s). We therefore systematically searched other introns of the three above-mentioned host genes for the potential presence of additional C/D-like sequences. A likely candidate, termed U96a, was identified in intron 2 of the GNB2L1 gene. Subsequently, we noticed that the human genome contains an additional, strongly related copy of the U96 sequence, U96b, located in intron 2 of the AMMECR1 gene. This gene maps to chromosome Xq22 and is potentially involved in the pathogenesis of the AMME (Alport syndrome, mental redardation, midface hypoplasia and ellipotocytosis) contiguous gene deletion syndrome (29). Curiously, the murine Ammecr1 gene does not contain an intron-encoded, U96-like sequence (Fig. 1).
The novel C/D snoRNA candidates were searched for the presence of appropriate complementarities to rRNAs or snRNAs. U95, which contains two antisense elements, is predicted to target two neighbouring 2′-O-methylations, Cm2801 and Am2792, in 28S rRNA. As for U96, U103, U104 and U106 snoRNAs, they are predicted to direct Gm75 (5.8S RNA), Gm601 (18S rRNA), Cm1320 (28S rRNA) and Um799 (18S rRNA), respectively, through phylogenetically conserved RNA guide duplexes (Table 1).
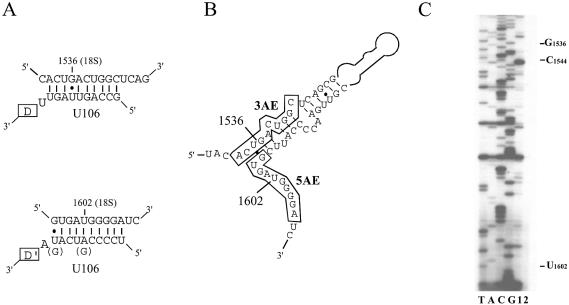
Remarkably, U106 contains two conserved, 9–10 nt long antisense elements matching two 18S rRNA segments (Fig. 2A) devoid of 2′-O-methylated nucleotides (30) and mapping very close to each other within the rRNA secondary structure (Fig. 2B). We confirmed that the two nucleotides (G1536 and U1602 in 18S rRNA) predicted to be targeted by U106 do not seem to be 2′-O-methylated as judged by the absence of reverse transcription pauses at low dNTP concentration (Fig. 2C). Many archaeal and plant C/D RNAs have been reported to carry two antisense elements interacting with neighbouring rRNA sites which might reflect their important role as RNA chaperones for rRNA folding in addition to nucleotide modification (22,23,25,26). Thus, our observation suggests that U106 might play a role distinct from directing 2′-O-methylations. Interestingly, this could also be the case for a few C/D snoRNAs, such as MBII-142, MBII-170, MBII-289, MBII-295, MBII-316 and MBII-426, which all can form canonical guide duplexes targeting apparently unmethylated rRNA sites [(6); J.P.Bachellerie, unpublished data].
Figure 2.
Interaction between C/D snoRNA U106 and 18S rRNA. (A) Predicted base pairing between U106 and 18S rRNA. The sequences shown are for human (nucleotide differences in rodents are indicated in parentheses). Note that G1536 and U1602 do not appear to be 2′-O-methylated (30). (B) Location of the sites of complementarity to snoRNA U106 within the 18S rRNA secondary structure. The 3′ and 5′ antisense elements of U106 are denoted by 3AE and 5AE, respectively, and the rRNA nucleotides potentially involved in base pairing with the snoRNA are boxed. (C) Mapping of ribose methylated nucleotides in 18S rRNA. Primer extension at low concentrations of dNTP was performed with a 5′-32P-labelled 18S rRNA-specific oligonucleotide, either at 0.04 mM (lane 1) or at 1 mM (control, lane 2). The significance of a pause at C1544 is unclear since 2′-O-methylation at this position has never been reported so far.
Finally, U94 contains an 11 nt long antisense element spanning three adjacent methylated nucleotides in the U6 snRNA, Cm60, Cm62 and Cm63, with Cm62 being paired to the fifth position upstream from the D′ box (data not shown). Although this RNA duplex contains a bulged nucleotide on the snoRNA strand (between box D′ and the targeted nucleotide), such an irregularity does not hamper efficient RNA guide activity (31) and we thus propose that U94 is the bona fide RNA guide for Cm62.
Genes hosting both subclasses of intron-encoded snoRNAs belong to the 5′ (TOP) family of vertebrate genes, including many ribosomal protein genes (32,33). In an attempt to identify additional intron-encoded C/D snoRNAs, we systematically searched all introns of the complete set of ribosomal protein genes in the human genome for the presence of C/D hallmarks. In this way, we could detect in introns of the RPS12 and RPL21 genes two novel C/D RNAs, called U101 and U102, respectively, which are strongly conserved among mammals. While U102 is predicted to direct a highly conserved 2′-O-methylation in 28S rRNA (Table 1), U101 remains without an identified target in rRNAs or snRNAs.
Finally, we re-examined all the candidate snmRNA genes previously identified by screening of a mouse brain cDNA library that could not have been assigned to a snoRNA subclass (6) essentially because they corresponded to very short, probably truncated cDNA sequences not represented in the genomic sequences in databases at that time. Following the publication of the entire human genome and large parts of the mouse genome, the 5′-terminal sequences of all these snmRNAs could be examined, leading to the definite assignment of a few of them to one of the two snoRNA subclasses. Thus, in the recently published human genome, we could identify the sequence homologous to mouse MBI-82 snmRNA. The 5′-extended human sequence, located in an intron of the EIF4-G2 gene, clearly exhibits the C/D hallmarks. However, the corresponding RNA, renamed U97, remains without a predicted target in rRNAs or snRNAs. All the novel C/D RNAs mentioned above, whether they were identified in the cDNA libraries or predicted by search of sequence databases, were experimentally verified by northern blotting analysis and found to be ubiquitously expressed (data not shown).
Identification of novel H/ACA snoRNAs
We also re-examined by reference to the updated human and mouse genomic sequences all the snmRNA candidates not yet assigned to snoRNAs in the screen of the mouse brain library mentioned above (6). We found that three of the most strongly expressed specimens in this category, MBII-367, MBII-104 and MBII-201, do in fact correspond to intronic sequences exhibiting the characteristic H/ACA hallmarks, i.e. two large hairpin domains linked by a hinge (containing the H box sequence, ANANNA) and followed by a short tail containing the ACA motif (3 nt away from the RNA 3′ end). The expression of these snmRNAs, renamed U98, U99 and U100, which are conserved in mammals, has been experimentally verified by northern blot analysis [(6); data not shown].
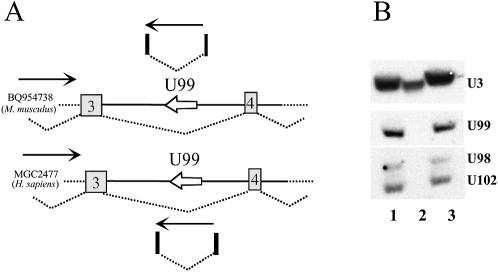
In the mouse, two identical U98 genes map very close to each other: one is encoded within the large intron 1 of the strongly conserved Taf12 gene and the other one within a poorly known gene ∼15 kb downstream from Taf12. In human, the U98 gene organisation is quite distinct, with the TAF12 gene devoid of any related U98-related sequence while a U98 gene copy is also found immediately downstream from the TAF12 gene. Moreover, another U98 gene copy, U98b, is embedded within intron 8 of the PPP2R5A gene (Fig. 1). Intriguingly, the U99 gene, which maps, in both mouse and human, to intron 3 of a gene encoding a conserved hypothetical protein (MGC2477), is transcribed in the opposite orientation to this gene (Fig. 3A). This might suggest that U99 could be independently transcribed from its own promoter in the intron, rather than being processed from the debranched intron like all known vertebrate C/D or H/ACA snoRNAs. However, lack of immunoprecipitation of U99 with the R1131 antibody (Fig. 3B) that specifically recognises the trimethyl-guanosine cap structure demarking the 5′ end of vertebrate snoRNAs transcribed from independent genes is consistent with U99 representing a processed RNA. Moreover, we noticed that several human and mouse spliced expressed sequence tags (ESTs) mapping within intron 3 of the MGC2477 gene were indicative of transcription in the direction opposite to this gene, accompanied by splicing of a U99-containing intron mapping within the boundaries of the MGC2477 intron 3, as shown in Figure 3A. While U99 is likely to be produced by intron processing of the MGC2477 antisense transcript, the biological significance of this antisense genomic organisation, never previously described for a snoRNA modification guide, remains elusive.
Figure 3.
The U99 snoRNA gene is transcribed in the opposite orientation to its host intron. (A) Schematic representation of a part of the mouse (top) and human (bottom) genes hosting U99 snoRNA. Exons 3 and 4 are represented by white boxes, while splicing events are denoted by dotted lines. The U99 snoRNA gene is schematised by a white arrow indicating transcription orientation. Thin black arrows denote the transcription orientation of the indicated genes. Several spliced ESTs (i.e. BU588934, BF219096, BU588653 and BI461867 in human, and a single one, AK011444, in the mouse) overlapping and in the same orientation as the U99 snoRNA gene are also depicted (filled boxes denote exons). Not drawn to scale. (B) U99 does not immunoprecipitate with R1131 antibody. Total RNA from HeLa cells was subjected to immunoprecipitation with R1131 antibody (specific for the trimethyl cap structure), and RNAs recovered from either the pellet or supernatant were assayed for U99 by northern blot hybridisation with a U99-specific oligonucleotide probe. 1, input RNA (1:10); 2, pellet; 3, supernatant (1:10). The 5′-trimethyl-capped snoRNA U3 and the intron-encoded U98 and U102 snoRNAs were used as positive and negative controls, respectively.
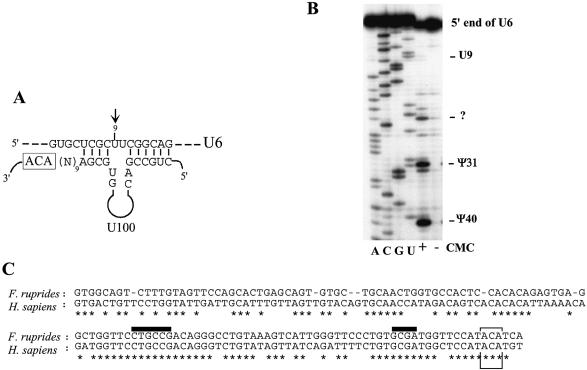
We could not identify any significant target uridine in rRNA or snRNA for U98 and U99 H/ACA RNAs. In contrast, U100 can form with snRNA U6 a canonical bipartite duplex predicted to direct pseudouridylation at position 9 within the spliceosomal RNA (Fig. 4A). A fish U100 homolog was identified by a search of the Fugu rubripes genomic sequence. Remarkably, the presumptive bipartite guide RNA duplex is conserved between Homo sapiens and F.rubripes (Fig. 4C). Intriguingly, however, position U9 in a stem–loop structure of U6 has not been reported to be pseudouridylated (34). We experimentally verified the absence of the predicted pseudouridine by a reverse transcriptase approach following chemical modification by CMC and alkaline treatment of the RNA template (Fig. 4B). Two other previously reported mouse H/ACA snoRNAs, MBI-39 and MBI-164, seem able to direct pseudouridylation on rRNA uridines which are not experimentally found to be modified (6). U100, as well as MBI-39 and MBI-164, might therefore play a role distinct from that of a pseudouridylation guide, possibly acting as RNA chaperones (see below).
Figure 4.
U100, a novel RNA guide targeting U6 snRNA. (A) Predicted base pairing of U100 with U6 snRNA. The U6 nucleotide predicted to be targeted for modification by U100 is indicated by an arrow. Note that only the the 3′ hairpin domain from U100 is shown. The sequences shown are for human. (B) Mapping of pseudouridines at the 5′ end of the spliceosomal U6 snRNA. Total RNA extracted from HeLa cells, treated (+) or not treated (–) with CMC, was subjected to primer extension analysis with a U6-specific 32P-labelled oligonucleotide. (C) U100 sequence alignment between H.sapiens and the fish F.rubripes. The fish U100 gene is located in an intron of the Huntington’s disease gene homologue (accession no. X82939). The conserved ACA motif and potential bipartite antisense element are denoted (boxed and overlined, respectively). Below the alignment, conserved nucleotides are denoted by asterisks.
U100 H/ACA RNA belongs to the scaRNA family
To date, RNA modification guides have been detected either in the nucleolus or in the nucleoplasm, according to the nature of their RNA substrates (13). Specimens located in the nucleolus include guides for the modification of rRNA as well as Pol III-transcribed snRNA U6 (14), whereas guides for the modification of Pol II-transcribed spliceosomal snRNAs, U1, U2, U4 and U5, are found within the CBs (13,15,16,35). We have investigated the intracellular localisation of the novel orphan C/D and H/ACA RNAs by isolating different subcellular fractions obtained from exponentially growing HeLa cells. All of them, except U100, are highly enriched in the nucleolar fraction (Fig. 5A). To gain further insight about the intra-nuclear location of U100 RNA, FISH with a U100-specific oligonucleotide probe has been performed in HeLa cells. As endogenous U100 signals were relatively weak (not shown), we cloned the human U100 gene into the pCMV-globin expression construct (16) and co-transfected it into HeLa cells together with a green fluorescent protein (GFP)–coilin expression plasmid which was used as a marker protein for CBs. As shown in Figure 5B, U100 signals did not distribute homogenously within the nucleoplasm but rather accumulated within distinct domains that co-localise with GFP–coilin signals. We therefore conclude that U100 is a novel member of the scaRNA family, in contrast to previously reported RNA guides for U6 modifications (as well as U94 described in this study) which have been all detected within the nucleolus [(14); Fig. 5A]. Consistent with U100 being a scaRNA, we noticed that it harbors a recently identified CB-specific localisation signal [(27); Fig. 5C]. U6 snRNA has been previously shown to transit through CBs (36). Thus, U100 is the first H/ACA RNA predicted to potentially base-pair with a Pol III transcript outside the nucleolus. Moreover, the absence of pseudouridylation at position 9 in U6 snRNA suggests an unusual role for U100 in the metabolism of U6 in CBs. Thereby, U100 could act as an RNA chaperone presumably within the CBs during U6 snRNP and/or U4/U6 snRNP assembly and trafficking within the nucleoplasm, en route to and from the nucleolus (37).
Figure 5.
U100 is a novel member of the scaRNA family. (A) Subfractionation of HeLa cells. RNA isolated either from total HeLa cells (Tot) or from cytoplasmic (Cyt), nuclei (Nu), nucleoplasmic (Np) or nucleolar (No) fractions was analysed in a 6% acrylamide/7 M urea gel and the various snoRNAs detected by northern blot analysis using specific oligonucleotide probes. Hybridisations with U3- and U6-specific probes have been used as controls of the cell fractionation procedure. (B) In situ hybridisation showing the localisation of transfected human U100. HeLa cells co-transfected with pCMV-hU100 and pGFP-coilin were hybridised with a specific U100 fluorescent oligonucleotide. The Cajal bodies are visualised by co-expressing GFP–coilin fluorescent protein. The merged picture shows that U100 co-localises with coilin. (C) Schematic representation of the terminal loops of the 5′ and 3′ hairpins of U100. The predicted Cajal body-specific localisation signals (27) are indicated in bold.
Detection of an intronic snoRNA retrogene
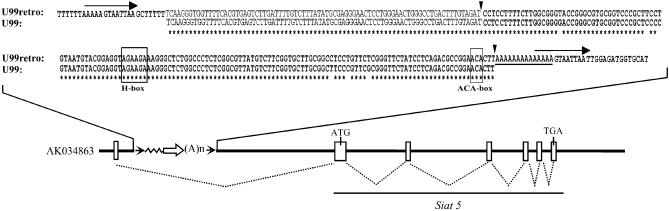
During our BLAST search of the mouse genome, in addition to the sequences encoding H/ACA RNA U99, we also detected an additional U99-like copy (Fig. 6) preceded by an 81 nt long tract identical to the intronic sequence immediately upstream from the functional U99 gene and immediately followed by a 3′ poly(A) stretch. The U99-like copy and upstream 81 nt were flanked by a pair of 13 nt long direct repeats. This probably corresponds to an H/ACA retrogene resulting from the insertion at staggered nicks in genomic DNA of a cDNA produced from an intronic RNA processing intermediate in U99 biogenesis. Intronic snoRNAs can be hosted by different genes in distant species, and retrotransposition mechanisms have been proposed to play a pivotal role in the mobility and diversification of snoRNA genes (12,38,39). In particular, retroposition into an intron may frequently result in the production of a faithfully processed snoRNA retrogene transcript, owing to the presence of internal cis-acting processing signals. Remarkably, the full-length U99 retrogene mentioned above is located in the mouse Siat5 gene (Fig. 6, bottom) within an intron of its 5′-untranslated region as reflected by the detection of many spliced ESTs. We checked by RT–PCR that the corresponding portion of the Siat5 gene intron was expressed in mouse cells (data not shown). It is noteworthy that within the U99 retrogene sequence, the tracts, which are predicted to correspond to bipartite antisense element(s) of the H/ACA RNA, are perfectly conserved by reference to the bona fide U99 gene. More generally, none of the few sequence differences exhibited by the U99 retrogene are expected to be detrimental to H/ACA snoRNA structure and function. The U99 retrogene might well encode a functional snoRNA, a possibility without precedent to our knowledge. Overall, sequences of the U99 retrogene and bona fide U99 gene diverge by only 5%, pointing to a relatively recent origin of the snoRNA retrogene in the mouse lineage. In line with this notion, the homologous intron of the human SIAT5 gene is devoid of any U99-like sequence.
Figure 6.
Identification of an intronic U99 retrogene. Sequence alignment of a U99 mouse snoRNA retrogene with its functional counterpart. Boundaries of the mature U99 sequence are denoted by vertical arrowheads. Direct repeats flanking the snoRNA retrogene are overlined (arrow), while the poly(A) stretch is underlined and the H and ACA motifs boxed. Bottom: intronic location of the U99 retrogene within the mouse Siat5 gene, with indication of a spliced EST (AK034863) connecting the U99 retrogene-containing intron to the rest of the Siat5 gene. Exons are represented by white boxes, splicing events by dotted lines, and the U99 retrogene by an open arrow (the flanking direct repeats are depicted by arrowheads). Nucleotides conserved between gene and retrogene are indicated by asterisks.
Conclusions
Among the 13 novel specimens of the C/D or H/ACA subclasses characterised in this study, six are especially intriguing. Four (C/D snoRNAs U97 and U101 and H/ACA snoRNAs U98 and U99) belong to the expanding group of orphan snoRNAs and remain without identified targets in rRNAs or snRNAs. They could either target cellular RNA species distinct from rRNA or snRNAs, or perform an entirely different function. In the case of the other two specimens, the H/ACA scaRNA U100 and the C/D snoRNA U106, our present evidence points to the second possibility, suggesting that they might be involved in an RNA chaperone function in the metabolism of U6 snRNA and 18S rRNA, respectively.
Acknowledgments
ACKNOWLEDGEMENTS
This paper is dedicated to the memory of Daniel Vitali. We thank T. Kiss for sharing unpublished data about the Cajal body-specific localisation signal, J. Brosius for critical reading of the manuscript, E. Bertrand for advice on RNA FISH, and M.-L. Bortolin and T. Rozhdestvensky for technical support. This work was supported by grants from the ‘Toulouse Génopole’, the Programme Interdisciplinaire du Centre National de la Recherche Scientifique ‘Dynamique et Réactivité des Assemblages Biologiques’ and ‘La Ligue contre le Cancer/Comité de Haute-Garonne’ (to J.C.), and by the German Human Genome Project through the BMBF (01KW9966) and an IZKF grant (Teilprojekt IKF3 G6, Münster) (to A.H.).
REFERENCES
- 1.Eddy S.R. (2001) Non-coding RNA genes and the modern RNA world. Nature Rev. Genet., 2, 919–929. [DOI] [PubMed] [Google Scholar]
- 2.Mattick J.S. and Gagen,M.J. (2001) The evolution of controlled multitasked gene networks: the role of introns and other noncoding RNAs in the development of complex organisms. Mol. Biol. Evol., 18, 1611–1630. [DOI] [PubMed] [Google Scholar]
- 3.Wassarman K.M. (2002) Small RNAs in bacteria: diverse regulators of gene expression in response to environmental changes. Cell, 109, 141–144. [DOI] [PubMed] [Google Scholar]
- 4.Kiss-Laszlo Z., Henry,Y., Bachellerie,J.P., Caizergues-Ferrer,M. and Kiss,T. (1996) Site-specific ribose methylation of preribosomal RNA: a novel function for small nucleolar RNAs. Cell, 85, 1077–1088. [DOI] [PubMed] [Google Scholar]
- 5.Ganot P., Caizergues-Ferrer,M. and Kiss,T. (1997) The family of box ACA small nucleolar RNAs is defined by an evolutionarily conserved secondary structure and ubiquitous sequence elements essential for RNA accumulation. Genes Dev., 11, 941–956. [DOI] [PubMed] [Google Scholar]
- 6.Huttenhofer A., Kiefmann,M., Meier-Ewert,S., O’Brien,J., Lehrach,H., Bachellerie,J.P. and Brosius,J. (2001) RNomics: an experimental approach that identifies 201 candidates for novel, small, non-messenger RNAs in mouse. EMBO J., 20, 2943–2953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Huttenhofer A., Brosius,J. and Bachellerie,J.P. (2002) RNomics: identification and function of small, non-messenger RNAs. Curr. Opin. Chem. Biol., 6, 835–843. [DOI] [PubMed] [Google Scholar]
- 8.Marker C., Zemann,A., Terhorst,T., Kiefmann,M., Kastenmayer,J.P., Green,P., Bachellerie,J.P., Brosius,J. and Huttenhofer,A. (2002) Experimental RNomics. Identification of 140 candidates for small non-messenger RNAs in the plant Arabidopsis thaliana. Curr. Biol., 12, 2002–2013. [DOI] [PubMed] [Google Scholar]
- 9.Yuan G., Klambt,C., Bachellerie,J.P., Brosius,J. and Huttenhofer,A. (2003) RNomics in Drosophila melanogaster: identification of 66 candidates for novel non-messenger RNAs. Nucleic Acids Res., 31, 2495–2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cavaille J., Nicoloso,M. and Bachellerie,J.P. (1996) Targeted ribose methylation of RNA in vivo directed by tailored antisense RNA guides. Nature, 383, 732–735. [DOI] [PubMed] [Google Scholar]
- 11.Ganot P., Bortolin,M.L. and Kiss,T. (1997) Site-specific pseudouridine formation in preribosomal RNA is guided by small nucleolar RNAs. Cell, 89, 799–809. [DOI] [PubMed] [Google Scholar]
- 12.Bachellerie J.P., Cavaille,J. and Huttenhofer,A. (2002) The expanding snoRNA world. Biochimie, 84, 775–790. [DOI] [PubMed] [Google Scholar]
- 13.Kiss T. (2002) Small nucleolar RNAs: an abundant group of noncoding RNAs with diverse cellular functions. Cell, 109, 145–148. [DOI] [PubMed] [Google Scholar]
- 14.Ganot P., Jady,B.E., Bortolin,M.L., Darzacq,X. and Kiss,T. (1999) Nucleolar factors direct the 2′-O-ribose methylation and pseudouridylation of U6 spliceosomal RNA. Mol. Cell. Biol., 19, 6906–6917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jady B.E., Darzacq,X., Tucker,K.E., Matera,A.G., Bertrand,E. and Kiss,T. (2003) Modification of Sm small nuclear RNAs occurs in the nucleoplasmic Cajal body following import from the cytoplasm. EMBO J., 22, 1878–1888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Darzacq X., Jady,B.E., Verheggen,C., Kiss,A.M., Bertrand,E. and Kiss,T. (2002) Cajal body-specific small nuclear RNAs: a novel class of 2′-O-methylation and pseudouridylation guide RNAs. EMBO J., 21, 2746–2756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cavaille J., Buiting,K., Kiefmann,M., Lalande,M., Brannan,C.I., Horsthemke,B., Bachellerie,J.P., Brosius,J. and Huttenhofer,A. (2000) Identification of brain-specific and imprinted small nucleolar RNA genes exhibiting an unusual genomic organization. Proc. Natl Acad. Sci. USA, 97, 14311–14316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jady B.E. and Kiss,T. (2000) Characterisation of the U83 and U84 small nucleolar RNAs: two novel 2′-O-ribose methylation guide RNAs that lack complementarities to ribosomal RNAs. Nucleic Acids Res., 28, 1348–1354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cavaille J., Vitali,P., Basyuk,E., Huttenhofer,A. and Bachellerie,J.P. (2001) A novel brain-specific box C/D small nucleolar RNA processed from tandemly repeated introns of a noncoding RNA gene in rats. J. Biol. Chem., 276, 26374–26383. [DOI] [PubMed] [Google Scholar]
- 20.Cavaille J., Seitz,H., Paulsen,M., Ferguson-Smith,A.C. and Bachellerie,J.P. (2002) Identification of tandemly-repeated C/D snoRNA genes at the imprinted human 14q32 domain reminiscent of those at the Prader–Willi/Angelman syndrome region. Hum. Mol. Genet., 11, 1527–1538. [DOI] [PubMed] [Google Scholar]
- 21.Lowe T.M. and Eddy,S.R. (1999) A computational screen for methylation guide snoRNAs in yeast. Science, 283, 1168–1171. [DOI] [PubMed] [Google Scholar]
- 22.Brown J.W., Clark,G.P., Leader,D.J., Simpson,C.G. and Lowe,T. (2001) Multiple snoRNA gene clusters from Arabidopsis. RNA, 7, 1817–1832. [PMC free article] [PubMed] [Google Scholar]
- 23.Barneche F., Gaspin,C., Guyot,R. and Echeverria,M. (2001) Identification of 66 box C/D snoRNAs in Arabidopsis thaliana: extensive gene duplications generated multiple isoforms predicting new ribosomal RNA 2′-O-methylation sites. J. Mol. Biol., 311, 57–73. [DOI] [PubMed] [Google Scholar]
- 24.Qu L.H., Meng,Q., Zhou,H., Chen,Y.Q., Liang-Hu,Q., Qing,M., Hui,Z. and Yue-Qin,C. (2001) Identification of 10 novel snoRNA gene clusters from Arabidopsis thaliana. Nucleic Acids Res., 29, 1623–1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Omer A.D., Lowe,T.M., Russell,A.G., Ebhardt,H., Eddy,S.R. and Dennis,P.P. (2000) Homologs of small nucleolar RNAs in Archaea. Science, 288, 517–522. [DOI] [PubMed] [Google Scholar]
- 26.Gaspin C., Cavaille,J., Erauso,G. and Bachellerie,J.P. (2000) Archaeal homologs of eukaryotic methylation guide small nucleolar RNAs: lessons from the Pyrococcus genomes. J. Mol. Biol., 297, 895–906. [DOI] [PubMed] [Google Scholar]
- 27.Richard P., Darzacq,X., Bertrand,E., Jady,B.E., Verheggen,C. and Kiss,T. (2003) A common sequence motif determines the Cajal body-specific localization of H/ACA scaRNAs. EMBO J., 22, 4283–4293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Spassov D.S. and Jurecic,R. (2002) Cloning and comparative sequence analysis of PUM1 and PUM2 genes, human members of the Pumilio family of RNA-binding proteins. Gene, 299, 195–204. [DOI] [PubMed] [Google Scholar]
- 29.Vitelli F., Meloni,I., Fineschi,S., Favara,F., Tiziana Storlazzi,C., Rocchi,M. and Renieri,A. (2000) Identification and characterization of mouse orthologs of the AMMECR1 and FACL4 genes deleted in AMME syndrome: orthology of Xq22.3 and MmuXF1–F3. Cytogenet. Cell Genet., 88, 259–263. [DOI] [PubMed] [Google Scholar]
- 30.Maden B.E. (1990) The numerous modified nucleotides in eukaryotic ribosomal RNA. Prog. Nucleic Acid Res. Mol. Biol., 39, 241–303. [DOI] [PubMed] [Google Scholar]
- 31.Cavaille J. and Bachellerie,J.P. (1998) SnoRNA-guided ribose methylation of rRNA: structural features of the guide RNA duplex influencing the extent of the reaction. Nucleic Acids Res., 26, 1576–1587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Smith C.M. and Steitz,J.A. (1998) Classification of gas5 as a multi-small-nucleolar-RNA (snoRNA) host gene and a member of the 5′-terminal oligopyrimidine gene family reveals common features of snoRNA host genes. Mol. Cell. Biol., 18, 6897–6909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pelczar P. and Filipowicz,W. (1998) The host gene for intronic U17 small nucleolar RNAs in mammals has no protein-coding potential and is a member of the 5′-terminal oligopyrimidine gene family. Mol. Cell. Biol., 18, 4509–4518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Massenet S., Mougin,A. and Branlant,C. (1998) Posttranscriptional Modifications in the U snRNAs. ASM Press, Washington, DC. [Google Scholar]
- 35.Kiss A.M., Jady,B.E., Darzacq,X., Verheggen,C., Bertrand,E. and Kiss,T. (2002) A Cajal body-specific pseudouridylation guide RNA is composed of two box H/ACA snoRNA-like domains. Nucleic Acids Res., 30, 4643–4649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Carmo-Fonseca M., Pepperkok,R., Carvalho,M.T. and Lamond,A.I. (1992) Transcription-dependent colocalization of the U1, U2, U4/U6 and U5 snRNPs in coiled bodies. J. Cell Biol., 117, 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stanek D., Rader,S.D., Klingauf,M. and Neugebauer,K.M. (2003) Targeting of U4/U6 small nuclear RNP assembly factor SART3/p110 to Cajal bodies. J. Cell Biol., 160, 505–516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bachellerie J.P., Nicoloso,M., Qu,L.H., Michot,B., Caizergues-Ferrer,M., Cavaille,J. and Renalier,M.H. (1995) Novel intron-encoded small nucleolar RNAs with long sequence complementarities to mature rRNAs involved in ribosome biogenesis. Biochem. Cell Biol., 73, 835–843. [DOI] [PubMed] [Google Scholar]
- 39.Maxwell E.S. and Fournier,M.J. (1995) The small nucleolar RNAs. Annu. Rev. Biochem., 64, 897–934. [DOI] [PubMed] [Google Scholar]