Abstract
Poxviruses encode many if not all of the proteins required for viral genome replication in the cytoplasm of the host cell. In this context, we investigated the function of the vaccinia virus G5 protein because it belongs to the FEN1-like family of nucleases and is conserved in all poxviruses. A vaccinia virus G5 deletion mutant was severely impaired, as the yield of infectious virus was reduced by approximately two orders of magnitude. The mutant virions contained an apparently normal complement of proteins but appeared spherical rather than brick-shaped and contained no detectable DNA. The inability of G5 with substitutions of the predicted catalytic aspartates to complement the deletion mutant suggested that G5 functions as a nuclease during viral DNA replication. Although the amount of viral DNA produced in the absence of G5 was similar to that made by wild-type virus, the mean size was approximately one-fourth of the genome length. Experiments with transfected plasmids showed that G5 was required for double-strand break repair by homologous recombination, suggesting a similar role during vaccinia virus genome replication.
Keywords: DNA processing, poxvirus genome replication
All studied DNA replication systems include multiple nucleases that are involved in proofreading, processing of Okazaki fragments, double-strand break repair (DSBR), homologous recombination (HR), and other processes. Poxviruses are remarkable with regard to DNA synthesis because they replicate within specialized “factories” in the cytoplasm of infected animal cells (1). The poxvirus-encoded replication proteins include a DNA polymerase, primase fused to a predicted helicase, uracil DNA glycosylase, processivity factor, DNA ligase, Holliday junction resolvase, and a dedicated protein kinase (2). Two of the above proteins have nuclease activities, namely the 3′-5′ exonuclease domain of the DNA polymerase (3) and the Holliday junction endonuclease (4). In addition, a nicking-joining enzyme that can cleave at the apex of hairpin telomeres has been described (5, 6). The activity associated with the polymerase appears to be involved in both proof reading (3) and HR (7); the Holliday junction endonuclease resolves DNA concatemers into unit-length genomes (8), whereas the role of the nicking-joining enzyme is uncertain because deletion of the implicated gene has no discernible effect on replication (5).
Another putative nuclease encoded by the G5R gene of vaccinia virus (VACV), the prototype poxvirus, was predicted from computational analyses (9, 10). The G5 protein homologs, which are conserved in all poxviruses and in some related nucleocytoplasmic large DNA viruses, belong to the ubiquitous family of exo/endonucleases known as the FEN1 family after its best-characterized member, the human flap structure-specific endonuclease 1. The FEN1 family includes the 5′-3′ exo domain of the bacterial polymerase 1 catalytic subunit, archaeal and eukaryotic flap endonucleases, and a variety of other, less well-characterized nucleases (11, 12). Analysis of G5 and the orthologous proteins in other poxviruses indicates that the main structural elements of the FEN1 fold and the five aspartates that are involved in catalysis by FEN1 family nucleases are conserved, making it likely that these viral proteins possess nuclease activity (9, 10).
The G5 protein is made early after infection and is packaged in the virus core (13). Temperature-sensitive (ts) G5 mutants failed to initiate the formation of viral membranes but no defect in viral DNA synthesis was noted, and DNA concatemers were resolved into genome-length DNA at the non-permissive temperature. Based on the phenotype of the ts mutants, it was proposed that G5 was essential for an early stage in morphogenesis. However, the phenotype of the G5 mutant was enigmatic because it failed to account for the predicted nuclease activity of G5. Considering that some ts mutants of VACV have anomalous phenotypes, we decided to reevaluate the function of G5 by isolating a deletion mutant. The phenotype of the mutant was distinct from that of the previous ts mutants and consistent with a nuclease function of G5. The G5 protein was required for HR, DSBR and the production of full-length viral genomic DNA.
Results
Impaired Replication of a G5R Deletion Mutant.
Although a previous attempt to isolate a G5R deletion mutant using a neo and gus selection and screening system failed (13), we succeeded in replacing the G5R ORF with the gene encoding enhanced green fluorescent protein (GFP) under control of a VACV promoter. Small green fluorescent plaques were detected against the background of large, round, non-fluorescent parental virus plaques. The small-plaque mutant, vΔG5, was clonally purified, and deletion of the G5R gene was confirmed by DNA sequencing. The failure to isolate the deletion mutant in the previous study was probably due to partial suppression of plaque formation by the antibiotic used for selection as well as the low sensitivity of β-glucuronidase as a screening marker.
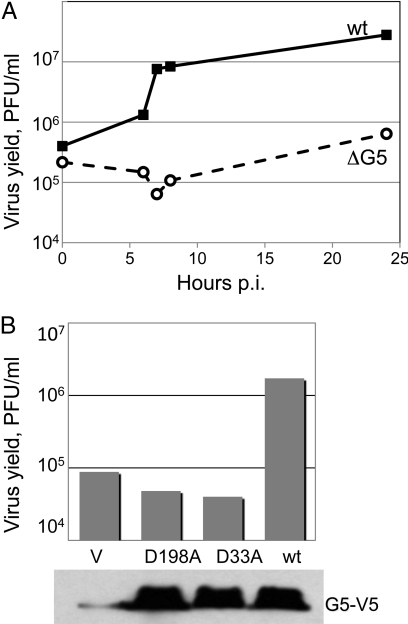
A one-step growth curve (Fig. 1A) documented the very low reproduction of vΔG5 in cell culture, which would account for the small plaque phenotype. The yield of infectious virus was estimated to range between 2–4 plaque-forming units (PFUs) per cell. The poor growth and small plaque phenotype of vΔG5 was confirmed in several other cell lines, indicating that there was a general impairment in replication rather than a restricted host range defect. To prove that an unrelated spontaneous mutation or rearrangement was not responsible for the vΔG5 phenotype, the G5R gene was restored to its original locus by HR. The revertant virus reproduced to high titer and formed plaques that were indistinguishable from wild-type (wt) VACV.
Fig. 1.
Replication of vΔG5. (A) One-step growth curve. BS-C-1 cells were infected with 10 PFU/cell of vΔG5 or wt VACV for 1 h at 37 °C, washed three times with medium, and incubated further at 37 °C. Duplicate samples were harvested at indicated times post infection (p.i.) and virus titers were determined by plaque assay on BS-C-1 cells. (B) Complementation of vΔG5 with plasmids expressing wt or mutated copies of the G5 gene. At 18 h before infection, cells were transfected with vector (V) or plasmids expressing wt G5 or G5 with amino acid substitutions D198A or D33A under control of the cytomegalovirus promoter. Western blot with antibody to the V5 tag, present on the C terminus of G5 expressed from plasmids, is shown at the bottom.
Transcomplementation of vΔG5 with wt G5 but Not with Predicted Catalytic Site Mutants.
The titer of vΔG5 increased by more than a log when the cells were transfected, before infection, with a plasmid containing the wt G5R gene regulated by the cytomegalovirus promoter (Fig. 1B), confirming that the replication defect was due to the loss of the G5R gene. Transfection of the G5R gene was routinely used to prepare high titer stocks of vΔG5 in lieu of establishing a stable complementing cell line. In contrast to the results obtained with the wt G5R, plasmids expressing copies of the G5R gene in which the predicted catalytic aspartate D33 or D198 was replaced by alanine did not complement vΔG5 (Fig. 1B). The latter result suggested that the predicted nuclease activity of G5 is required for the function of this protein in viral reproduction.
Defective Virion Morphogenesis in the Absence of G5.
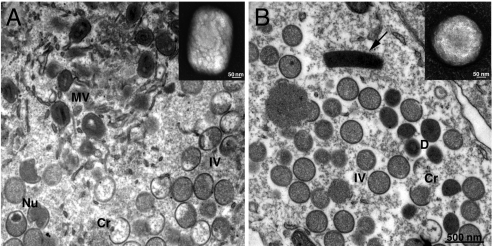
In agreement with the low yield of infectious virions per cell, transmission electron microscopy showed an almost complete absence of characteristic brick-shaped mature virions in vΔG5-infected cells. In thin sections of vΔG5-infected cells, viral structures were represented by normal looking crescents and immature virus particles, and dense, spherical rather than brick-shape particles (Fig. 2). Cytoplasmic structures resembling DNA crystalloids, seen under conditions in which mature virions do not form were also present. Occasionally, membrane wrapping of dense immature particles was observed as occurs with normal mature virions. The dense spherical particles observed in vΔG5-infected cells resembled aberrant virions lacking DNA that were previously found in cells infected with two DNA-packaging mutants (14, 15) and a mutant defective in concatemer resolution (8).
Fig. 2.
Electron microscopy of wt- and vΔG5-infected cells and purified virions. BS-C-1 cells were infected with wt VACV (A) or vΔG5 (B) for 24 h, fixed, embedded in Epon, and ultrathin sections were prepared. Abbreviations: Cr, crescent membrane; IV, immature virion, MV, mature virion; Nu, nucleoid in IV; D, dense spherical particle. Arrow points to a structure with the appearance of a DNA crystalloid. Insets show typical wt (A) and mutant (B) virions that were purified, adsorbed to grids, washed with water, and stained with 7% uranyl acetate in 50% ethanol.
The aberrant virus particles that accumulated in vΔG5-infected cells were purified in parallel with VACV mature virions from wt VACV-infected cells. Opalescent bands of similar intensity were seen in sucrose gradients containing material from cells infected with vΔG5 and wt virus. However, comparison of negatively stained particles by electron microscopy showed that the mutant virus did not have the typical brick shape of poxvirus virions but appeared more spherical, corresponding to the dense particles seen by transmission electron microscopy (Fig. 2, insets). Moreover, the specific infectivity of the mutant particles was about 100-fold lower than that of wt virions purified in parallel.
Analysis of Viral DNA in Defective Virions and Infected Cells.
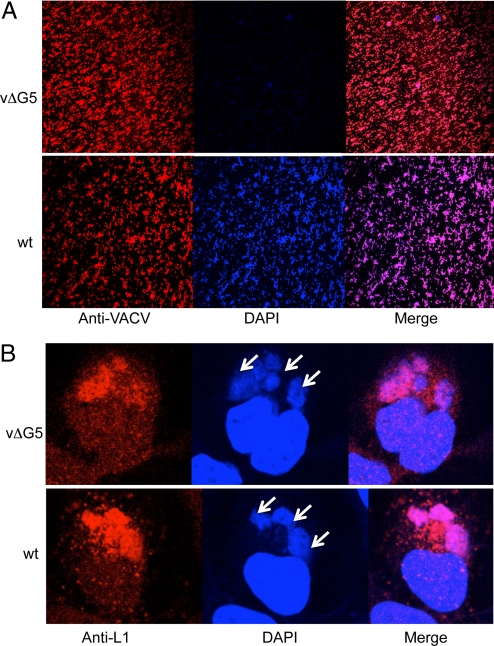
Mutant and wt particles were immobilized on cover slips and stained with 4′,6-diamidino-2-phenylindole (DAPI), which binds avidly to double-stranded DNA. Both mutant and wt virions were detected with antibody to VACV, but only the latter stained efficiently with DAPI (Fig. 3A). In contrast to the absence of DNA in mutant virions, vΔG5 DNA was present in the cytoplasmic factories, the sites of VACV DNA synthesis and virion assembly (Fig. 3B). The DAPI staining coincided with the staining by an antibody to the VACV L1 protein that was used as a marker for the factory (Fig. 3B). The normal appearance of viral factories suggested that there was no major quantitative defect in DNA synthesis in vΔG5-infected cells although the DNA was not packaged into virions.
Fig. 3.
Confocal microscopy of purified virions and cytoplasmic factories. (A) Purified vΔG5 and wt virions were adsorbed to cover slips and stained with polyclonal rabbit anti-VACV antibody R8191 followed by a secondary antibody conjugated to Alexa Fluor 594 (red) (Molecular probes, Eugene, OR) and DAPI (blue). (B) HeLa cells were infected with vΔG5 or wt VACV for 8 h and stained with DAPI (blue) or monoclonal anti-L1 antibody followed by a secondary antibody conjugated to Alexa Fluor 594 (red). Arrows point to virus factories adjacent to the larger nuclei.
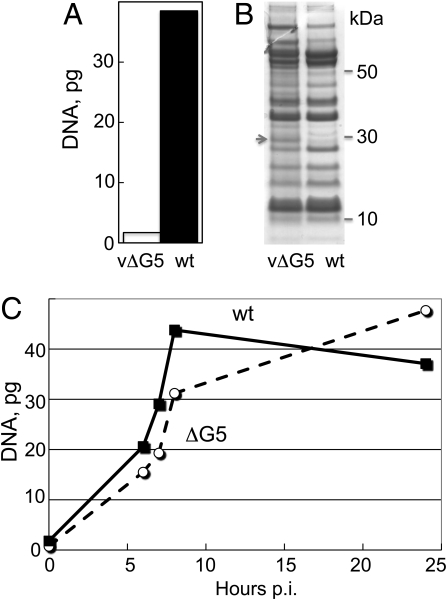
Real time PCR (qPCR) was used to quantify viral DNA in virions and infected cells. The vΔG5 virions contained at least 20-fold less DNA than wt virions as determined by analysis of the material extracted using a standard DNA isolation procedure (Fig. 4A). Nevertheless, the polypeptide compositions of the wt and mutant particles, analyzed by SDS-polyacrylamide gel electrophoresis, were nearly identical (Fig. 4B). To quantify viral DNA synthesis, total DNA was extracted from cells infected with vΔG5 or wt virus at multiple times after infection and the amount of viral DNA was determined by qPCR. Similar amounts of viral DNA accumulated over 24 h in cells infected with vΔG5 and wt virus, although there was a short delay in the former (Fig. 4C).
Fig. 4.
DNA content of virions and infected cells determined by qPCR. (A) DNA was extracted from purified virions and analyzed by qPCR with primers designed to anneal to the VACV C11L gene specific for VACV DNA. Data from a representative experiment are shown. (B) Proteins from purified vΔG5 and wt virions were analyzed by SDS-polyacrylamide gel electrophoresis. A gel stained with Coomassie blue is shown. The migration positions of marker proteins are indicated on the right. The arrow points to a 28-kDa band believed to be the precursor of the L4 protein, which was present in higher amounts in vΔG5 than wt virions. (C) DNA was extracted from infected BS-C-1 cells at indicated hours post infection (p.i.) and analyzed by qPCR as in panel A. Data from a representative experiment are shown.
Characterization of Viral DNA in Infected Cells.
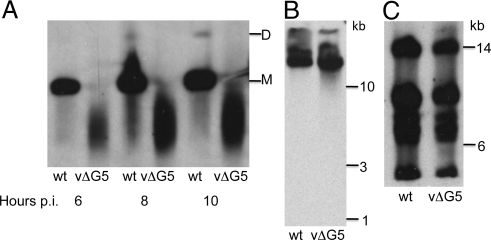
DNA from vΔG5- and wt virus-infected cells was analyzed by pulsed-field gel electrophoresis and Southern blotting. In wt virus-infected cells, genome length VACV DNA (185 kbp) along with lesser amounts of dimers and diffuse smaller fragments were detected (Fig. 5A). In contrast, most of the DNA from vΔG5-infected cells was present as a smear corresponding to genomic fragments ranging from approximately 20 to approximately 200 kb, with a mean of approximately 45 kbp (Fig. 5A). The amounts of vΔG5 and wt DNA that remained in the wells of the gel were not visibly different, suggesting that branched DNA molecules did not accumulate in the absence of G5.
Fig. 5.
Electrophoretic analysis of DNA. (A) Pulsed-field gel electrophoresis. BS-C-1 cells were infected with wt VACV or vΔG5 for the indicated times and the cell pellets were embedded in agarose plugs and lysed with SDS, EDTA and proteinase K. DNA was separated by electrophoresis, transferred to a membrane and hybridized to total VACV DNA labeled with digoxigenin. Abbreviations: M, full length VACV genome; D, VACV genome dimer. (B) Alkaline gel electrophoresis. Infected cell pellets were lysed in the wells and the DNA was denatured with 50 mM NaOH, resolved by electrophoresis in a 1% agarose gel, and detected by Southern blotting as in panel A. (C) Restriction enzyme analysis. Cells were infected as in A and DNA was extracted with the Puregene kit (Qiagen), digested with EcoRI, and analyzed by pulsed-field agarose gel electrophoresis and Southern blotting as in panel A. Only the upper part of the gel is shown.
The presence of numerous random nicks, that could cause fragmentation of the double-stranded DNA upon isolation, was ruled out by alkaline gel electrophoresis (Fig. 5B). To confirm that the entire VACV genome was represented in the DNA synthesized in cells infected with vΔG5, the DNA was digested with the restriction enzymes EcoRI and BglI and analyzed by Southern blotting using total VACV genomic DNA as the probe. As shown for EcoRI, there was no visible difference between the restriction fragments of vΔG5 and wt DNA (Fig. 5C).
Junction fragments are formed during VACV genome replication by synthesis around the hairpin termini (16). These junctions are rapidly cleaved by the Holliday junction resolvase to reform the hairpin termini of mature genomes. Unless resolution is impaired, BstE II digestion of DNA from infected cells yields predominantly 1.3-kb terminal fragments and only trace amounts of the 2.6-kb junctions (17). Precisely this situation was found when vΔG5 and wt VACV DNA was analyzed by Southern blotting of samples at 6, 8, and 24 h.
In summary, viral genomic sequences in wt and mutant virus-infected cells were replicated to approximately the same level, but the mutant DNA was fragmented so that the amount of genome-sized molecules was severely reduced. We hypothesized that this genome fragmentation was caused by accumulation of double-strand breaks (DSBs) and that the predicted nuclease activity of G5 is involved in HR mediated DSBR.
HR Is Impaired in the Absence of G5.
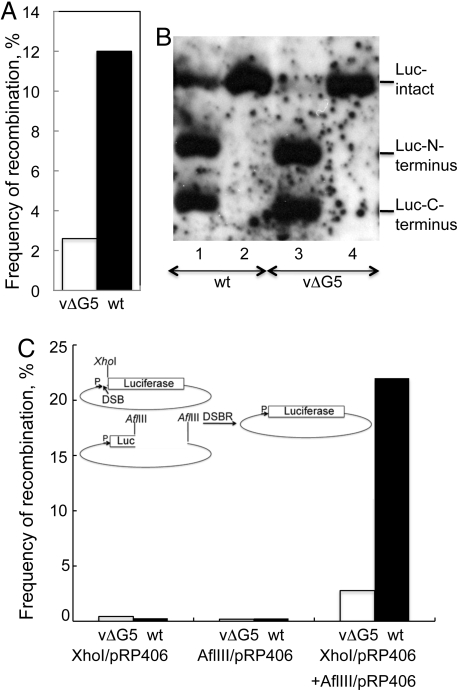
To investigate whether HR is impaired in the absence of G5, we took advantage of the ability of plasmids to replicate in cytoplasmic factories with the help of VACV-encoded proteins (18, 19). The levels of HR in wt- and vΔG5-infected cells were quantified using a firefly luciferase (LUC) assay that is dependent on the reconstitution of the LUC gene from two overlapping fragments in separate plasmids (20). The frequency of HR was 3- to 5-fold lower in the absence of G5 (Fig. 6A). In contrast, similar LUC values were obtserved when the infected cells were transfected with a plasmid containing an intact LUC gene, indicating that the reduced LUC in the vΔG5-infected cells was indeed due to a deficiency of HR rather than defects in plasmid replication or gene expression. We also assessed plasmid replication and recombination directly by Southern blotting. DNA was extracted from duplicate samples used to measure LUC expression, digested with restriction enzymes to separate the LUC gene from the rest of the plasmid and with DpnI to eliminate the input plasmid, and analyzed by Southern blotting with a LUC DNA probe. Much less full-length LUC DNA was formed when the two plasmids with overlapping LUC fragments were transfected into cells infected with vΔG5 compared to wt virus, whereas similar amounts were present when the single plasmid with an intact LUC gene was transfected (Fig. 6B).
Fig. 6.
HR and DSBR. (A) Cells infected with vΔG5 or wt virus were transfected with two plasmids containing overlapping fragments of LUC and another containing the β-gal gene. Recombination frequencies were determined in duplicate as described in Materials and Methods. Data from a representative experiment are shown. (B) DNA from duplicate samples of experiments in panel A were digested with DpnI, XhoI, and SpeI followed by agarose gel electrophoresis and Southern blotting with a LUC gene probe. Lanes: 1 and 2, wt virus; lanes 3 and 4, vΔG5. Lanes 1 and 3, transfection with two plasmids containing LUC gene fragments; lanes 2 and 4, transfection with one plasmid containing complete LUC gene. (C) Plasmid pR406 was digested with XhoI or AflIII and purified by agarose gel electrophoresis. The intact plasmid or linearized plasmids individually or together were transfected into cells infected with vΔG5 (empty bar) or wt (filled bar) virus. The frequency of recombination was determined by comparing the LUC activity in cells transfected with linearized plasmids to that of intact plasmid. Insert is a diagram of recombination needed for repair and LUC expression.
A related DSBR assay was adapted from the protocol of Gammon and Evans (21). The plasmid containing the intact LUC gene used above was either digested with XhoI to create a DSB between the promoter and the LUC ORF or with AflIII to excise a segment of the LUC gene. The plasmids were transfected individually or together into cells infected with mutant or wt virus and LUC activity was subsequently determined. As a positive control, the intact LUC plasmid was transfected. In the cells infected with wt virus and transfected with the two plasmids, DSBR resulted in 23% of the LUC activity of the intact plasmid, whereas only 3% of the activity was obtained in the transfected cells that had been infected with the mutant virus (Fig. 6C). Only low background activity was obtained when either the XhoI- or AflIII-digested plasmids were transfected individually (Fig. 6C) indicating that the DSBR was dependent on HR.
Discussion
Identification of a poxvirus-encoded primase (22) suggested that poxviruses might also encode an endonuclease that would remove RNA primers in nascent DNA. The G5 protein seemed a likely candidate as it belongs to the FEN1 nuclease family (9, 10), some members of which are flap endonucleases (11). We isolated a VACV G5R deletion mutant and found that it was defective, generating only 2–4 infectious units per cell. Isolation of a revertant virus and transcomplementation with plasmid encoding the wt G5R gene confirmed that the replication defect was specific. Moreover, mutations of predicted catalytic aspartates abrogated complementation, strongly suggesting that G5 functions as a nuclease in DNA replication. Although the level of DNA replication determined by qPCR appeared unaffected, pulsed-field gel electrophoresis showed that the viral DNA made in the absence of G5 was a mixture of fragments, with a mean of about 45 kb instead of 200-kb, full-length genomes. The DNA fragments were double-stranded and representative of the entire genome because digestion with restriction endonucleases yielded the expected pattern. Furthermore, there was an excess of mature DNA termini compared to junctions, indicating that junction resolution had occurred. The presence of numerous random nicks in the DNA or accumulation of Okazaki-size fragments was ruled out by alkaline gel electrophoresis. Therefore, it seems most likely that the subgenomic DNA fragments result from DSBs, which occur in many systems at the replication fork, and that these DSBs are not repaired in the absence of G5. Assuming that the DSBs were random (i.e., Poisson-distributed) and that the mean fragment length was 45 kb, then full-length (185 kb) genomes would be expected to comprise approximately 1% of the total viral DNA. This estimated fraction of intact genomes is compatible with the observed residual infectivity of vΔG5.
Using a quantitative plasmid recombination assay developed by the Evans laboratory (20), we found that HR was diminished by 3- to 5-fold. In a second HR DSBR assay, we found a 7- to 8-fold decrease in cells infected with the G5R deletion mutant. The deficiencies in DSBR and HR are likely to have the same mechanistic cause as the two processes are intimately related (23). In eukaryotic cells, HR-mediated DSBR involves the resection of DNA ends to produce recombinogenic 3′ single-stranded tails. This process involves multiple nucleases including Mre11, a 3′-5′ exonuclease, and Exo1, a 5′-3′ exonuclease and flap endonuclease (23). An important next step will be to characterize the putative nuclease activity of G5. Recently Gammon and Evans (21) reported that the 3′ exonuclease of VACV DNA polymerase has a role in genetic recombination. In addition, the VACV Holliday junction resolvase can cleave a variety of branched DNA structures in vitro (4, 24, 25), although it is expressed late in infection whereas G5 and the viral DNA polymerase are expressed early. Thus, two or three nucleases are potentially involved in poxvirus DSBR and HR.
Most of the DNA made in cells infected with the G5 deletion mutant was not packaged in virus particles, even though junctions were resolved to form mature termini. There might be a length requirement for packaging DNA or mature termini might be required at both ends of the genome. Still another possibility is that G5 itself is required, since it is normally present in virus cores (13). At least two other proteins are needed for packaging DNA, namely a putative ATPase encoded by the A32 gene (14) and the telomere-binding protein I6 (15). The absence of DNA probably accounted for the spherical shape of the virions because similar-shaped virions have also been found with the A32 and I6 mutants.
The herpesvirus FEN1-family member is a ribonuclease involved in host and early viral mRNA degradation (26). To determine whether G5 also possessed such an activity, we compared the steady state levels of a cellular mRNA, an early VACV mRNA and an intermediate VACV mRNA in cells infected with wt VACV and the G5 deletion mutant. No differences were found suggesting that G5 does not affect mRNA stability. This result was not surprising given that VACV and other poxviruses encode decapping enzymes, which regulate mRNA half-life (27, 28). Other nucleocytoplasmic large DNA viruses including iridoviruses, ascoviruses, mimivirus, and one of the phycodnaviruses (but not other phycodnaviruses or African Swine Fever virus) encode predicted Fen1-family nucleases. However, these proteins are no more closely related to poxvirus G5 orthologs than they are to the herpesvirus homologs, leaving the roles of these proteins in viral reproduction an open question.
Materials and Methods
Cells and Viruses.
Procedures for the preparation and maintenance of BS-C-1 cells and for the propagation, titration and purification of VACV have been described (29). Recombinant VACV were derived from the WR strain (ATCC, VR-1354) and constructed by transfection of a DNA segment with the sequence of interest and 500 bp flanks to allow HR. DNA segments were assembled by overlapping PCR. In vΔG5, the G5R gene except for the first 47 and last 46 nucleotides that overlap the promoters of adjacent genes was replaced with the GFP gene preceded by a VACV synthetic early/late promoter. A revertant, vG5-rev, was made from vΔG5 by replacing the GFP gene with the respective DNA and screening for non-fluorescent plaques.
Transcomplementation of vΔG5 with Plasmids Expressing G5.
The synthetic human codon optimized G5R gene including a coding sequence for the C-terminal V5 tag was prepared by GeneArt and cloned into the BamHI and XhoI sites of pcDNA 3.1 (+) (Invitrogen) for expression under control of cytomegalovirus promoter. Codons for aspartic acid residues D33 and D198 were replaced by codons for alanine using overlapping PCR, and mutant gene copies were cloned into the same vector. To complement vΔG5 function, plasmids with wt or mutant copies of the gene were transfected with Lipofectamine 2000 (Invitrogen) into BS-C-1 cells 18 h before infection with 0.01 PFU/cell of vΔG5. Cells were collected 72 h after infection and virus titers determined.
HR Assays.
The assays were carried out as described (20) using plasmids with the VACV P11 promoter kindly provided by D. H. Evans. Plasmids pRP406Δ and pRP403Δ, containing the N- and C-terminal parts of LUC overlapping by 366 bp, and plasmid pRP7.5lacZ containing the β-galactosidase (β-gal) gene were co-transfected into BS-C1 cells that had been infected with 5 PFU/cell of vΔG5- or wt virus and extracts were prepared 18–22 h later. Enzyme activities were measured with a luminometer using the LUC (Promega) or β-gal (Clonetech) detection kit, respectively. LUC activity was normalized for β-gal to account for possible variation in transfection efficiency. The normalized LUC activity was compared to the activity resulting from transfection of pRP406, which contains the complete LUC gene, to determine the recombination frequency. Recombination frequency was calculated using the formula: 100% × [LUC (pRP406Δ + pRP403Δ)/β-gal]/[LUC (pRP406)/β-gal] as described (21). Τhe same methods were used to measure DSBR (21).
qPCR.
DNA was extracted from purified virions or infected cells with blood kit (Qiagen) and used in qPCR reaction with SYBR Green PCR master mix (Applied Biosystems) with primers C11pF: 5′AAACACACACTGAGAAACAGCATAAA and C11pR: 5′ACTATCGGCGAATGATCTGATTATC designed to anneal to the VACV C11L gene in an Eppendorf Mastercycler with Eppendorf realplex software. Amplification conditions were 40 cycles 95°, 15 s and 60°, 60 s.
Pulsed-Field Agarose Gel Electrophoresis.
Cell pellets from single wells of a 24-well plate of infected cells were washed once with PBS and embedded in molten agarose plugs. The plugs were incubated in a solution containing SDS, EDTA and proteinase K as described in the protocol provided by Bio-Rad. Electrophoresis was performed in 1% agarose gel in CHEF-DR III apparatus (Bio-Rad) at 5.8 V with a switching time gradient of 50–90 s, 120° angle for 22 h at 14 °C.
Southern Blotting.
DNA resolved in agarose gels was fragmented with HCl, denatured with alkali, neutralized, transferred overnight to positively charged nylon membrane (Roche), and UV cross-linked to the membrane. Hybridization with digoxigenin-dUTP labeled probe was performed using DIG High Prime DNA Labeling and Detection kit (Roche).
Alkaline Agarose Gel Electrophoresis.
BS-C-1 cells (2 × 105) infected with wt or vΔG5 were lysed in the wells of 1% agarose gel in 10 mM Tris pH 8.0, 10 mM EDTA, 0.5% SDS for 15 min at room temperature; the concentration of NaOH was adjusted to 50 mM and electrophoresis was carried out in 50 mM NaOH, 1 mM EDTA at 3 V/cm for 6 h and analyzed by Southern blotting.
Transmission Electron Microscopy.
BS-C-1 cells were infected with 10 PFU vΔG5 or wt virus per cell for 1 h at 37 °C. Unadsorbed virus was removed by washing and the incubation continued for 12–20 h. Cells were fixed with 2% glutaraldehyde, embedded in Epon resin, and ultrathin sectioned samples were prepared (30).
Acknowledgments.
We thank David Evans (University of Alberta, Edmonton, Canada) for plasmids, Jerry Weir for the loan of a pulsed-field gel apparatus, Andrea Weisberg for electron microscopy, Catherine Cotter for preparation of cell cultures, and David Evans and Paula Traktman for critical comments and suggestions. The work was supported by funds from the Division of Intramural Research, National Institute of Allergy and Infectious Diseases, National Institutes of Health.
Footnotes
The authors declare no conflict of interest.
References
- 1.Moss B. In: Fields Virology. Knipe DM, Howley PM, editors. Philadelphia: Lippincott Williams & Wilkins; 2007. pp. 2905–2946. [Google Scholar]
- 2.Moss B, De Silva F. In: DNA Replication & Human Disease. DePamphilis ML, editor. Cold Spring Hrbor: Cold Spring Harbor Laboratory Press; 2006. pp. 707–727. [Google Scholar]
- 3.Challberg MD, Englund PT. Purification and properties of the deoxyribonucleic acid polymerase induced by vaccinia virus. J Biol Chem. 1979;254:7812–7819. [PubMed] [Google Scholar]
- 4.Garcia AD, Aravind L, Koonin EV, Moss B. Bacterial-type DNA Holliday junction resolvases in eukaryotic viruses. Proc Natl Acad Sci USA. 2000;97:8926–8931. doi: 10.1073/pnas.150238697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Eckert D, Williams O, Meseda CA, Merchlinsky M. Vaccinia virus nicking-joining enzyme is encoded by K4L (VACWR035) J Virol. 2005;79:15084–15090. doi: 10.1128/JVI.79.24.15084-15090.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.DeMasi J, Du S, Lennon D, Traktman P. Vaccinia virus telomeres: Interaction with the viral I1, I6, and K4 proteins. J Virol. 2001;75:10090–10105. doi: 10.1128/JVI.75.21.10090-10105.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Willer DO, Mann MJ, Zhang WD, Evans DH. Vaccinia virus DNA polymerase promotes DNA pairing and strand- transfer reactions. Virology. 1999;257:511–523. doi: 10.1006/viro.1999.9705. [DOI] [PubMed] [Google Scholar]
- 8.Garcia AD, Moss B. Repression of vaccinia virus Holliday junction resolvase inhibits processing of viral DNA into unit-length genomes. J Virol. 2001;75:6460–6471. doi: 10.1128/JVI.75.14.6460-6471.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Iyer LA, Balaji S, Koonin EV, Aravind L. Evolutionary genomics of nucleo-cytoplasmic large DNA viruses. Virus Res. 2006;117:156–184. doi: 10.1016/j.virusres.2006.01.009. [DOI] [PubMed] [Google Scholar]
- 10.Da Silva M, Shen L, Tcherepanov V, Watson C, Upton C. Predicted function of the vaccinia virus G5R protein. Bioinformatics. 2006;22:2846–2850. doi: 10.1093/bioinformatics/btl506. [DOI] [PubMed] [Google Scholar]
- 11.Liu Y, Kao HI, Bambara RA. Flap endonuclease 1: A central component of DNA metabolism. Ann Rev Biochem. 2004;73:589–615. doi: 10.1146/annurev.biochem.73.012803.092453. [DOI] [PubMed] [Google Scholar]
- 12.Allen LM, Hodskinson MR, Sayers JR. Active site substitutions delineate distinct classes of eubacterial flap endonuclease. Biochem J. 2009;418:285–292. doi: 10.1042/BJ20081637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Da Fonseca FG, Weisberg AS, Caeiro MF, Moss B. Vaccinia virus mutants with alanine sbstitutions in the conserved G5R gene fail to initiate morphogenesis at the nonpermissive temperature. J Virol. 2004;78:10238–10248. doi: 10.1128/JVI.78.19.10238-10248.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cassetti MC, Merchlinsky M, Wolffe EJ, Weisberg AS, Moss B. DNA packaging mutant: Repression of the vaccinia virus A32 gene results in noninfectious, DNA-deficient, spherical, enveloped particles. J Virol. 1998;72:5769–5780. doi: 10.1128/jvi.72.7.5769-5780.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Grubisha O, Traktman P. Genetic analysis of the vaccinia virus I6 telomere-binding protein uncovers a key role in genome encapsidation. J Virol. 2003;77:10929–10942. doi: 10.1128/JVI.77.20.10929-10942.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Baroudy BM, Venkatesan S, Moss B. Incompletely base-paired flip-flop terminal loops link the two DNA strands of the vaccinia virus genome into one uninterrupted polynucleotide chain. Cell. 1982;28:315–324. doi: 10.1016/0092-8674(82)90349-x. [DOI] [PubMed] [Google Scholar]
- 17.Baroudy BM, Venkatesan S, Moss B. Structure and replication of vaccinia virus telomeres. Cold Spring Harbor Symp Quant Biol. 1982;47:723–729. doi: 10.1101/sqb.1983.047.01.083. [DOI] [PubMed] [Google Scholar]
- 18.DeLange AM, McFadden G. Sequence-nonspecific replication of transfected plasmid DNA in poxvirus-infected cells. Proc Natl Acad Sci USA. 1986;83:614–618. doi: 10.1073/pnas.83.3.614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.De Silva FS, Moss B. Origin-independent plasmid replication occurs in vaccinia virus cytoplasmic factories and requires all five known poxvirus replication factors. Virol J. 2005;2:23. doi: 10.1186/1743-422X-2-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Parks RJ, Winchcombe-Forhan C, DeLange AM, Xing X, Evans DH. DNA ligase gene disruptions can depress viral growth and replication in poxvirus-infected cells. Virus Res. 1998;56:135–147. doi: 10.1016/s0168-1702(98)00055-0. [DOI] [PubMed] [Google Scholar]
- 21.Gammon DB, Evans DH. The 3 '-to-5 ' exonuclease activity of vaccinia virus DNA polymerase is essential and plays a role in promoting virus genetic recombination. J Virol. 2009;83:4236–4250. doi: 10.1128/JVI.02255-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.De Silva FS, Lewis W, Berglund P, Koonin EV, Moss B. Poxvirus DNA primase. Proc Natl Acad Sci USA. 2007;104:18724–18729. doi: 10.1073/pnas.0709276104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mimitou EP, Symington LS. Nucleases and helicases take center stage in homologous recombination. Trends Biochem Sci. 2009;34:264–272. doi: 10.1016/j.tibs.2009.01.010. [DOI] [PubMed] [Google Scholar]
- 24.Culyba MJ, Harrison JE, Hwang Y, Bushman FD. DNA cleavage by the A22R resolvase of vaccinia virus. Virology. 2006;352:466–476. doi: 10.1016/j.virol.2006.05.007. [DOI] [PubMed] [Google Scholar]
- 25.Culyba MJ, Minkah N, Hwang Y, Benhamou OM, Bushman FD. DNA branch nuclease activity of vaccinia A22 resolvase. J Biol Chem. 2007;282:34644–34652. doi: 10.1074/jbc.M705322200. [DOI] [PubMed] [Google Scholar]
- 26.Taddeo B, Zhang W, Roizman B. The UL41 protein of herpes simplex virus 1 degrades RNA by endonucleolytic cleavage in absence of other cellular or viral proteins. Proc Natl Acad Sci USA. 2006;103:2827–2832. doi: 10.1073/pnas.0510712103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Parrish S, Moss B. Characterization of a second vaccinia virus mRNA-decapping enzyme conserved in poxviruses. J Virol. 2007;81:12973–12978. doi: 10.1128/JVI.01668-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Parrish S, Resch W, Moss B. Vaccinia virus D10 protein has mRNA decapping activity, providing a mechanism for control of host and viral gene expression. Proc Natl Acad Sci USA. 2007;104:2139–2144. doi: 10.1073/pnas.0611685104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Earl PL, Cooper N, Wyatt LS, Moss B, Carroll MW. In: Current Protocols in Molecular Biology. Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K, editors. New York: John Wiley and Sons; 1998. pp. 16.16.11–16.16.13. [Google Scholar]
- 30.Senkevich TG, Wyatt LS, Weisberg AS, Koonin EV, Moss B. A conserved poxvirus NlpC/P60 superfamily protein contributes to vaccinia virus virulence in mice but not to replication in cell culture. Virology. 2008;374:506–514. doi: 10.1016/j.virol.2008.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]